Abstract
Rapid identification and antimicrobial susceptibility testing remain a crucial step for early efficient therapy of bloodstream infections. Traditional methods require turnaround times of at least 2 days, while rapid procedures are often associated with extended hands-on time. The Accelerate Pheno™ System provides microbial identification results within 90 min and susceptibility data in approximately 7 h directly from positive blood cultures with only few minutes of hands-on time. The aim of this study was, therefore, to evaluate the performance of the Accelerate Pheno™ System in identification and antimicrobial susceptibility testing of both Gram-positive and Gram-negative bacteria directly from clinical blood culture samples. We analyzed 108 and 67 blood culture bottles using the Accelerate PhenoTest™ BC kit with software version v1.0 and the FDA-cleared version v1.2, respectively. Reliable identification was achieved for Enterobacteriaceae, staphylococci, and enterococci, with 76/80 (95%), 42/46 (91%), and 10/11 (91%) correct identifications. Limitations were observed in the identification of streptococci, including Streptococcus pneumoniae and Streptococcus pyogenes, and coagulase-negative staphylococci. Antimicrobial susceptibility results for Enterobacteriaceae, for amikacin, ertapenem, ciprofloxacin, gentamicin, meropenem, and piperacillin-tazobactam ranged between 86 and 100% categorical agreement. Using v1.2, results for ceftazidime showed 100% concordance with the reference method. For staphylococci, the overall performance reached 92% using v1.2. Qualitative tests for detection of methicillin or macrolide-lincosamide-streptogramin B (MLSB) resistance caused major and very major errors for isolates. Overall, the present data show that the Accelerate Pheno™ system can, in combination with Gram stain, be used as a rapid complementation to standard microbial diagnosis of bloodstream infections.
Introduction
Bloodstream infections are associated with high morbidity and mortality and remain a leading cause of death [3]. Early effective therapy improves the disease outcomes but is dependent on identification and susceptibility testing of the causative agent [2].
Automated antimicrobial susceptibility testing (AST) systems such as the Vitek 2 system (bioMeriéux) and Phoenix 100 (BD Biosciences) use simplified protocols and require shorter incubation times compared to standard methods. Also, these procedures can be combined with rapid protocols to considerably reduce the time until a resistance profile is available [14, 20, 23]. These rapid procedures are, however, associated with extended hands-on time and the need for specialized personnel and are, thus, not compatible with the implementation of automated methods.
Time-lapse microscopic imaging of bacterial colony formation in the presence of an antimicrobial agent has been suggested as a novel approach to ascertain AST directly from the positive blood culture bottles [8]. The Accelerate Pheno™ system (Accelerate Diagnostics Inc., Tucson, AZ, USA) employs this technique in a fully automated system. The system identifies 15 bacterial species/groups as well as two Candida species and determines phenotypic AST for part of the identification panel. Bacteria or yeast cells are identified by fluorescence in situ hybridization (FISH). In addition to specific probes, bacterial and eukaryotic cells are universally labeled which allows to detect the presence of off-panel microorganisms. Microorganisms are then subjected to species-specific AST. Morphokinetic cellular parameters, such as division rate and cell morphology, are recorded and a minimal inhibitory concentration (MIC) for each antibiotic is calculated using the software algorithm. Results are automatically reported after approximately 90 min for identification and 7 h for antimicrobial susceptibility.
Only a few studies that examined the clinical performance of the Accelerate PhenoTest™ BC are available. The published studies focused on a limited group of microorganisms or patients, Gram-negative bacteria or pediatric oncology patients, respectively [7, 18]. Moreover, the FDA-cleared Accelerate Pheno™ system employs a different software version than the version used in those studies. The aim of the current study was, therefore, to evaluate the performance of the Accelerate Pheno™ system, including the FDA-cleared version of the software, on a broader selection of clinical blood culture samples including Gram-positive and Gram-negative bacteria.
Materials and methods
Blood culture bottles
A total of 175 positive blood culture bottles were chosen from clinical blood culture samples sent to Karolinska University Laboratory, Stockholm, Sweden, from three tertiary care hospitals in the greater Stockholm area. Blood culture bottles BacT/Alert FA Plus, BacT/Alert FN Plus, and BacT/Alert PF Plus were incubated in the BacT/Alert 3D system. Bottles were selected per the following criteria: (i) ≤ 8 h after blood culture positivity, (ii) growth in aerobic bottles, (iii) time to positivity < 24 h, and (iv) positivity in more than one bottles from the same patient. Referring to the manufacturer’s protocol, the first point (i) was required per manufacturer’s specifications while samples that did not meet the remaining criteria were investigated only in cases where no other bottles were available such as Gram stain results and patient data, as available from the laboratory information system, were considered in cases where more than two suitable positive blood culture bottles were available. Samples suspected for contamination or off-panel organisms were avoided. Comparisons of results were made using samples taken from the same blood culture bottle.
Standard procedures for identification and susceptibility testing
Microorganisms from positive blood culture bottles were initially assessed using Gram stain. Species identification was determined by MALDI-TOF MS (Bruker Daltonik, Bremen, Germany) in nearly all cases. In the event of failure of identification by MALDI-TOF MS, Vitek 2 was used. Antimicrobial susceptibility for all isolates included in this study was tested by disk diffusion following EUCAST guidelines and breakpoints according to version 6.0. In addition, Enterobacteriaceae were tested with the Vitek 2 GN AST-N218 card. MIC values were interpreted following EUCAST breakpoints as above.
ID and AST with the Accelerate Pheno™ system
The Accelerate PhenoTest™ BC kit (Accelerate Diagnostics, Inc., Tucson, AZ, USA) was run according to manufacturer’s instructions. In brief, an aliquot of 2–5 ml blood culture broth was aseptically removed from the blood culture bottle and transferred to a sample vial and, regardless of inoculation volume, a 0.3-ml aliquot was processed and analyzed by the system. The run was then immediately started. After completing the run, the blood culture broth from the sample vial was cultured on blood agar plates and suitable selective agar plates, if indicated. All isolated bacteria were stored at − 80 °C. The first set of samples (n = 108, collected between April and September 2016) was run and evaluated by the Accelerate host software version v1.0.0.417 (v1.0); the second set of samples (n = 67, collected between January and March 2017) was run and evaluated by the Accelerate host software version v1.2.0.87 (v1.2). Quality control runs were performed each week the instrument was in use, alternating between the two modules available during the study period.
Data evaluation and statistics
AST results were analyzed for categorical agreement (S, susceptible; I, intermediate; R, resistant). Categorical errors were defined as minor (S/R for I, or I for S/R by reference method), major (R for S by reference method, false resistant), or very major (S for R by reference method, false sensitive) errors.
All data were evaluated using GraphPad Prism 6.00, GraphPad Software, La Jolla, CA, USA. Continuous data are presented with median and range, and results were compared using a Mann-Whitney test. Categorical data are given in absolute numbers and percentages, results were compared using Fisher’s exact test. Differences with P values < 0.05 were considered statistically significant.
Results
Samples and technical performance
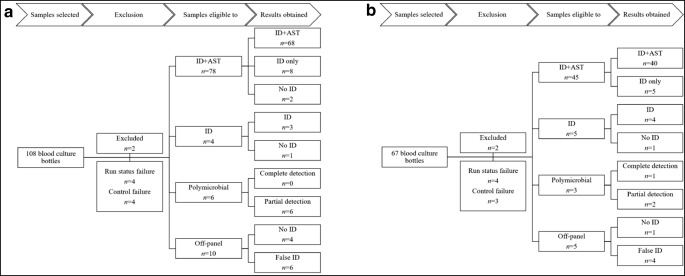
Blood culture bottles were selected based on the manufacturer’s instructions and microbiological parameters including time to detection, aerobic growth, and the number of positive bottles from the same patient. These criteria were chosen to focus on clinically relevant samples. During the first study period, a total of 108 bottles were selected and analyzed using the Accelerate host software version v1.0 (Fig. 1a). In the second study period using the new or software version, a total of 67 bottles was evaluated and software version v1.2 was applied (Fig. 1b). An overview of pre-analytical sample parameters and technical assay performance is shown in Table 1. From the initially chosen samples, ten and nine bottles from the first and second study periods, respectively, were excluded from evaluation. In the majority of cases, run status failure was related to cassette illumination failure and the run was terminated after 6–8 min. In one case, run status failure was reported at the end of the run at 8.25 h. Control failures were reported at the end of FISH analysis and bacterial identification failures were reported at 82 min. These failures gave an error message “Too few cells for analysis” in the Accelerate run report. Cultures from the blood culture broth did however not indicate low bacterial counts or impaired bacterial viability; nor was there a delay in starting the assay after blood culture positivity (3.4 h for failed samples versus 4.5 h for successfully analyzed samples, P = 0.42). In contrast, the time to detection in the blood culture system was significantly longer for samples with control failure (21 versus 14.5 h, P = 0.005). In each part of the study, two samples were excluded for reasons unrelated to the system, i.e., failures in the control culture, duplicate samples, or electricity breaks during the analysis. With these failures excluded, 15/171 (8.8%) runs could not be analyzed because of failures in the system.
Fig. 1.
Blood culture samples included in the study. Positive blood culture bottles were analyzed with the Accelerate PhenoTest™ BC using software version SW1.0 (a) or SW1.2 (b). ID, identification; AST, antimicrobial susceptibility testing
Table 1.
Sample characteristics and technical performance of the Accelerate PhenoTest™ BC kit
| v1.0 | v1.2 | P value | |
|---|---|---|---|
| Pre-analytical sample characteristics | |||
| Time to detection [h]a | 14 (5–27) | 15 (6–54) | 0.1692 |
| Time until analysis [h]a | 4.5 (0.6–15.6) | 4.1 (0.3–7.6) | 0.0436 |
| Assay performance | |||
| Run status failureb | 4/106 (3.8%) | 4/65 (6%) | 0.4807 |
| Control failureb | 4/102 (3.9%) | 3/61 (5%) | 1.000 |
| Time to ID [min]a | 82 (80–84) | 81 (80–85) | <0.0001 |
| Time to AST [h]a | 6.7 (6.5–6.9) | 6.5 (6.5–6.9) | 0.0002 |
| Completed ID + AST results (eligible samples) | 68/78 (87%) | 40/45 (89%) | 0.7807 |
aData are presented as median (range) and evaluated by Mann-Whitney test
bData are presented as n (%) and evaluated by Fisher’s exact test
Results reporting bacterial identification and AST obtained with v1.0 and v1.2 are presented separately in “Performance using version v1.0” and “Evaluation of v1.2” sections, respectively.
Performance using software version v1.0
Bacterial identification
Samples from the first study period contained 105 isolates and 94/105 (89.5%) were covered by the PhenoTest™ BC kit panel. Samples containing species that were not differentiated by the panel were counted as a single isolate, e.g., coagulase-negative staphylococci (CoNS) or α-hemolytic streptococci. As determined by Gram staining, the 105 isolates included 59 Gram-negative rods, 26 staphylococci, 11 streptococci, and 9 enterococci (Table 2). The performance of the PhenoTest™ BC kit for bacterial identification is summarized in Table 3. Identification of Gram-negative rods, staphylococci, and enterococci was highly reliable, with 84/94 (89%) of bacteria detected and correctly identified. Undetected organisms were primarily found in polymicrobial infections. In contrast, cross-reactivity was observed among the group of α-hemolytic streptococci, including three α-hemolytic streptococci reported as S. pneumoniae. Two of three off-panel β-hemolytic Streptococcus species were reported as CoNS. In addition, three Candida species and one of each Escherichia coli, Pseudomonas aeruginosa, and Streptococcus species were reported by the PhenoTest™ BC kit while not observed using standard culture. Another 13 isolates were reported as “indeterminate” (not depicted in Table 2), including Enterobacter species (n = 5, all in combination with Klebsiella or Citrobacter species), Staphylococcus species (n = 7), and Enterococcus faecium (n = 1). None of these isolates could be detected by standard methods, nor were there any other microbiological results from these patients indicating the presence of such microorganisms. Thus, in total 19 detections were regarded false positive.
Table 2.
Identification results from Accelerate PhenoTest™ BC kit in comparison to standard methods
| Gram stain | Samples (n) | Standard culture | PhenoTest™ BC kit |
|---|---|---|---|
| SW1.0 | |||
| Gram-negative rods | 26 | Escherichia coli | Escherichia coli |
| 1 | Escherichia coli, Pseudomonas aeruginosa | Escherichia coli, Candida albicans | |
| 14 | Klebsiella species | Klebsiella species | |
| 1 | Klebsiella pneumoniae | Klebsiella species, Candida albicans | |
| 1 | Klebsiella oxytoca | Klebsiella species, Streptococcusspecies | |
| 1 | Klebsiella oxytoca | Klebsiella species, Escherichia coli | |
| 1 | Klebsiella oxytoca | Unidentified organism(s) | |
| 3 | Enterobacter species | Enterobacter species | |
| 1 | Enterobacter cloacae, Escherichia colia | Enterobacter species, unidentified organism(s) | |
| 1 | Enterobacter cloacae | Unidentified organism(s) | |
| 2 | Citrobacter species | Citrobacter species | |
| 1 | Proteus mirabilis | Proteus species | |
| 1 | Haemophilus influenzae | Unidentified organism(s) | |
| Gram-negative rods, Gram-positive cocci in chains/diplococci | 1 | Enterobacter cloacae, Enterococcus faecium | Enterobacter species |
| 1 | Escherichia coli, Klebsiella pneumoniae,Enterococcus duransb | Escherichia coli | |
| Gram-positive cocci in clusters | 12 | Staphylococcus aureus | Staphylococcus aureus |
| 1 | Staphylococcus aureus | Staphylococcus aureus, Pseudomonas aeruginosa | |
| 1 | Staphylococcus aureus, Staphylococcus epidermidis | Staphylococcus aureus | |
| 8 | Coagulase-negative staphylococci | Coagulase-negative staphylococci | |
| 1 | Staphylococcus warneri | Coagulase-negative staphylococci | |
| 1 | Staphylococcus simulans | Unidentified organism(s) | |
| Gram-positive cocci in clusters, Gram-positive cocci in chains/diplococci | 1 | Enterococcus faecalis, Staphylococcus aureus | Enterococcus faecalis |
| Gram-positive cocci in chains/diplococci | 5 | Enterococcus faecalis | Enterococcus faecalis |
| 1 | Enterococcus faecium | Enterococcus faecium | |
| 1 | Streptococcus agalactiae | Streptococcus agalactiae | |
| 1 | Streptococcus pneumoniae | Streptococcus pneumoniae | |
| 1 | Streptococcus mitis-group | Unidentified organism(s) | |
| 1 | Streptococcus mitis-group | Streptococcus pneumoniae | |
| 1 | Streptococcus pyogenes | Coagulase-negative staphylococcus | |
| 1 | Streptococcus dysgalactiae | Coagulase-negative staphylococcus | |
| 2 | Streptococcus dysgalactiae | Unidentified organism(s) | |
| 2 | Streptococcus sanguinis-group | Streptococcus pneumoniae | |
| 1 | Streptococcus salivarius-group | Streptococcus species, Candida glabrata | |
| SW1.2 | |||
| Gram-negative rods | 14 | Escherichia coli | Escherichia coli |
| 5 | Klebsiella species | Klebsiella species | |
| 3 | Enterobacter species | Enterobacter species | |
| 1 | Stenotrophomonas maltophilia | Coagulase-negative staphylococci | |
| 1 | Streptococcus anginosus, Haemophilus influenzae | Streptococcus species, coagulase-negative staphylococci | |
| Gram-positive cocci in clusters | 11 | Staphylococcus aureus | Staphylococcus aureus |
| 2 | Staphylococcus aureus | Coagulase-negative staphylococci | |
| 7 | Coagulase-negative staphylococci | Coagulase-negative staphylococci | |
| 1 | Coagulase-negative staphylococci, Streptococcus salivarius-group | Coagulase-negative staphylococci | |
| Gram-positive cocci in clusters, Gram-positive cocci in chains/diplococci | 1 | Enterococcus faecalis | Enterococcus faecalis |
| Gram-positive cocci in chains/diplococci | 1 | Enterococcus faecium | Enterococcus faecium |
| 1 | Enterococcus faecium, Staphylococcus epidermidis, S. haemolyticus | Enterococcus faecium, coagulase-negative staphylococci | |
| 2 | Streptococcus mitis/oralis | Streptococcus species | |
| 1 | Streptococcus agalactiae | Streptococcus agalactiae | |
| 1 | Streptococcus pneumoniae | Streptococcus agalactiae | |
| 1 | Streptococcus pneumoniae | Four or more organisms detected. | |
| 1 | Streptococcus pyogenes | Streptococcus species | |
| 1 | Streptococcus gordonii | Streptococcus species | |
| 1 | Abiotrophia defectiva | Suspected off-panel microorganism | |
| Gram-positive rods | 1 | Arcanobacterium haemolyticum | Coagulase-negative staphylococci |
aSpecies in bold indicate non-concordant results
bSpecies underlined are not included in the Accelerate identification panel
Table 3.
Performance in species identification Accelerate PhenoTest™ BC kit in comparison to standard methods
| Identification (ID), n (%) | ||||
|---|---|---|---|---|
| Correct | Correct genusa | False | Undetected/ unidentifiedb | |
| Gram-negative rods | ||||
| v1.0 (n = 59/105, 56.2%) | 54 (92%) | 5 (8)c | ||
| SW1.2 (n = 27/61, 41%) | 23 (92%) | 2 (8) | ||
| Staphylococci | ||||
| SW1.0 (n = 26/105, 24.8%) | 23 (88) | 1 (4) | 2 (8)c | |
| SW1.2 (n = 22/61, 36%) | 20 (91) | 2 (9) | ||
| Streptococci | ||||
| SW1.0 (n = 11/105, 10.5%) | 4 (36) | 1 (9) | 5 (45) | 1 (9) |
| SW1.2 (n = 10/61, 16%) | 4 (40) | 3 (30) | 2 (20) | 1 (10) |
| Enterococci | ||||
| SW1.0 (n = 9/105, 8.6%) | 7 | 2c | ||
| SW1.2 (n = 3/61, 5%) | 3 | |||
aFor off-panel species
bFor on-panel organisms
cUndetected Enterobacteriaceae (n = 3), staphylococci (n = 2), and enterococci (n = 2) were part of a polymicrobial sample
Polymicrobial samples
The PhenoTest™ BC kit is capable of identifying several different microorganisms within one sample. Monomicrobial results may be flagged “monomicrobial,” if the specific probe signal matches the universal signal in the FISH assay. Monomicrobial results may lack this comment, suggesting that the presence of further microorganisms cannot be ruled out.
Among the six polymicrobial samples investigated, one of two isolates (five samples) or one of three isolates (one sample) was detected using the PhenoTest™ BC kit (Table 2). All polymicrobial samples were identified by the initial Gram stain where possible and morphologically similar isolates were differentiated from the first subculture. Three of the six polymicrobial samples were flagged monomicrobial in the Accelerate run report. Overall, 66/92 (72%) monomicrobial and 3/6 polymicrobial samples were flagged monomicrobial (κ = 0.078, 95% confidence interval − 0.078 to 0.233).
Antimicrobial susceptibility testing
The PhenoTest™ BC kit comprises 9–14 antibiotics for Gram-negative rods, 6–7 antibiotics and 2 qualitative resistance phenotypes for staphylococci, and 5 antibiotics for enterococci. We chose to focus our evaluation on those antimicrobial drugs which are included in the panel routinely tested in our laboratory. Disk diffusion according to EUCAST guidelines served as reference. Susceptibility data for the isolates involved in this study are presented in Table 4. For Enterobacteriaceae, the performance of the PhenoTest™ BC kit was also compared to the performance of Vitek 2 because this system is frequently used in clinical microbiology laboratories.
Table 4.
Susceptibility data for isolates using disk diffusion
| Antimicrobial agent | SW1.0 | SW1.2 | ||
|---|---|---|---|---|
| Resistant, n (%) |
Susceptible, n (%) |
Resistant, n (%) |
Susceptible, n (%) |
|
| Enterobacteriaceae | ||||
| Amikacin | 0 | 49 (100) | 0 | 19 (100) |
| Ertapenem | 0 | 49 (100) | 0 | 19 (100) |
| Ciprofloxacin | 3 (6) | 43 (94) | 3 (16) | 16 (84) |
| Ceftazidime | 5 (10) | 44 (90) | 1 (5) | 18 (95) |
| Gentamicin | 1 (2) | 48 (98) | 2 (10) | 17 (90) |
| Meropenem | 0 | 49 (100) | 0 | 19 (100) |
| Piperacillin-tazobactam | 3 (6) | 43 (94) | 1 (5) | 17 (90) |
| Staphylococcus species | ||||
| Cefoxitin | 6 (30) | 14 (70) | 6 (30) | 13 (70) |
| Erythromycin | 5 (25) | 15 (75) | 6 (30) | 13 (70) |
| Linezolid | 0 | 20 (100) | 0 | 19 (100) |
| Trimethoprim/sulfamethoxazole | 4 (21) | 15 (79) | 0 | 11 (100) |
Enterobacteriaceae
Among the 58 Gram-negative rods eligible for AST, 49/58 (89%) were correctly identified and AST results were obtained. Five isolates remained unidentified or undetected, and for four identified isolates, no AST could be provided. Results for seven antimicrobial agents were compared, including amikacin, ertapenem, ciprofloxacin, ceftazidime, gentamicin, meropenem, and piperacillin-tazobactam (Table 5). Overall, categorical agreement was reported for disk diffusion in 285/343 (83.1%) analyses, with 43/343 (12.5%) minor errors, 14/331 (4.2%) major errors, and 1/12 (8%) very major error. Ceftazidime showed poor agreement with the reference method. The majority of susceptible isolates (n = 44) were reported with intermediate (34/44, 77%) or resistant (10/44, 23%) MIC values. In addition, minor errors were recorded for 3/5 ceftazidime-resistant isolates. In comparison, Vitek 2 performed significantly better for ceftazidime (P < 0.0001). Errors, including a very major error, were also observed for piperacillin-tazobactam; however, the overall performance was comparable to Vitek 2.
Table 5.
Antimicrobial susceptibility testing results for Enterobacteriaceae when compared to the reference method, disk diffusion
| Antimicrobial agent | Categorical agreementa | Minor errors | Major errors | Very major errors | |
|---|---|---|---|---|---|
| Amikacin | |||||
| A | Vitek 2 | 49 (100) | 0 | 0 | 0 |
| SW1.0 | 49 (100) | 0 | 0 | 0 | |
| B | Vitek 2 | 19 (100) | 0 | 0 | 0 |
| SW1.2 | 19 (100) | 0 | 0 | 0 | |
| Ertapenem | |||||
| A | Vitek 2 | 49 (100) | 0 | 0 | 0 |
| SW1.0 | 49 (100) | 0 | 0 | 0 | |
| B | Vitek 2 | 19 (100) | 0 | 0 | 0 |
| SW1.2 | 19 (100) | 0 | 0 | 0 | |
| Ciprofloxacin | |||||
| A | Vitek 2 | 49 (100) | 0 | 0 | 0 |
| SW1.0 | 48 (98) | 0 | 1 (2) | 0 | |
| B | Vitek 2 | 18 (95) | 1 (5) | 0 | 0 |
| SW1.2 | 19 (100) | 0 | 0 | 0 | |
| Ceftazidime | |||||
| A | Vitek 2 | 45 (92)b | 3 (6) | 0 | 1 (2) |
| SW1.0 | 2 (4) | 37 (76) | 10 (20) | 0 | |
| B | Vitek 2 | 19 (100) | 0 | 0 | 0 |
| SW1.2 | 19 (100)c | 0 | 0 | 0 | |
| Gentamicin | |||||
| A | Vitek 2 | 49 (100) | 0 | 0 | 0 |
| SW1.0 | 47 (96) | 2 (4) | 0 | 0 | |
| B | Vitek 2 | 19 (100) | 0 | 0 | 0 |
| SW1.2 | 19 (100) | 0 | 0 | 0 | |
| Meropenem | |||||
| A | Vitek 2 | 49 (100) | 0 | 0 | 0 |
| SW1.0 | 48 (98) | 1 (2) | 0 | 0 | |
| B | Vitek 2 | 19 (100) | 0 | 0 | 0 |
| SW1.2 | 19 (100) | 0 | 0 | 0 | |
| Piperacillin-tazobactam | |||||
| A | Vitek 2 | 46 (94) | 2 (4) | 0 | 1 (2) |
| SW1.0 | 42 (86) | 3 (6) | 3 (6) | 1 (2) | |
| B | Vitek 2 | 19 (100) | 0 | 0 | 0 |
| SW1.2 | 18 (95) | 1 (5) | 0 | 0 | |
aIn comparison to reference method (disk diffusion)
bSW1.0 versus Vitek 2, Fisher’s exact test, P < 0.0001
cSW1.0 versus SW1.2, Fisher’s exact test, P < 0.0001
Staphylococci
For staphylococci, results from the cefoxitin-screen, erythromycin and the macrolide-lincosamide-streptogramin B (MLSB) phenotype, and linezolid and trimethoprim/sulfamethoxazole testing were evaluated. Results were obtained for 20/23 (87%) eligible isolates, including a Staphylococcus warneri isolate identified as CoNS. For three Staphylococcus aureus isolates, no AST results were obtained. Agreement with disk diffusion was good with 92/100 (92%) concordant results (Table 6). For trimethoprim/sulfamethoxazole, 4/4 resistant CoNS isolates were reported as intermediate. The cefoxitin-screen failed for 2/20 (10%) samples. While these samples were reported monomicrobial by the PhenoTest™ BC kit, standard culture revealed the presence of a second staphylococcal isolate resistant to cefoxitin. Methicillin-resistant S. aureus (MRSA) were not isolated during the study period.
Table 6.
Antimicrobial susceptibility testing results for staphylococci
| Categorical agreementa | Not analyzed | Minor errors | Major errors | Very major errors | |
|---|---|---|---|---|---|
| Cefoxitin-screen | |||||
| SW1.0 | 18 (90) | 2 (10) | – | 0 | 0 |
| SW1.2 | 17 (90) | 0 | – | 1 (5) | 1 (5) |
| Erythromycin | |||||
| SW1.0 | 19 (95) | 1 (5) | 0 | 0 | 0 |
| SW1.2 | 19 (100) | 0 | 0 | 0 | 0 |
| MLSB | |||||
| SW1.0 | 20 (100) | 0 | – | 0 | 0 |
| SW1.2 | 18 (95) | 0 | – | 0 | 1 (5) |
| Linezolid | |||||
| SW1.0 | 19 (95) | 1 (5) | 0 | 0 | 0 |
| SW1.2 | 19 (100) | 0 | 0 | 0 | 0 |
| Trimethoprim/sulfamethoxazole | |||||
| SW1.0 | 16 (80) | 0 | 4 (20) | 0 | 0 |
| SW1.2 | 11 (100)b | 0 | 0 | 0 | 0 |
aIn comparison to reference method (disk diffusion)
bOnly for Staphylococcus aureus
Enterococci
Enterococcus faecalis and Enterococcus faecium were tested for ampicillin, linezolid, and vancomycin resistance. Results were obtained for 5/8 isolates. Two isolates from polymicrobial samples were not detected, and for one isolate, no AST results could be obtained. Complete categorical agreement was observed for the remaining five isolates.
Evaluation of v1.2
To evaluate the performance of this commercially available version of the assay, 67 positive blood cultures were analyzed (Fig. 1b, Table 1).
Bacterial identification
Based on issues experienced with v1.0, software adjustments for microbial identification were made in v1.2. The species S. warneri and Streptococcus oralis were added to the group of detectable CoNS and Streptococcus species, respectively; S. pneumoniae was removed from the panel but instead reported on genus level. In addition, adjustments were made to minimize non-specific and indeterminate detections.
A total of 61 isolates were detected in the samples investigated during the second study period, 53/61 (87%) covered by the PhenoTest™ BC panel. By Gram stain and standard culture, 25 Gram-negative rods, 22 staphylococci, 10 streptococci, 3 enterococci as well as 1 Gram-positive rod were identified (Table 2, Table 3). The overall performance of v1.2 for bacterial identification was similar to v1.2 (Table 3). Consistent with findings using v1.0, differentiation among streptococci was poor. In addition, off-panel organisms (Stenotrophomonas maltophilia, Haemophilus influenzae, Arcanobacterium haemolyticum) were identified as CoNS, indicating poor specificity of this group-specific probe. In contrast to v1.0, false positive and “indeterminate” detections were no longer recorded (Table 2).
Polymicrobial samples
During the second study period, three polymicrobial samples with two (two samples) and three (one sample) isolates were identified by standard culture. The latter sample contained two different species of coagulase-negative staphylococci, which were counted as one isolate in this assessment. With this limitation, the sample was correctly and completely identified by the PhenoTest™ BC, however not eligible to AST because of morphological similarities of the included species, i.e., staphylococci and enterococci. In the remaining two samples, one of the isolates was missed or incorrectly identified (Table 2). Overall, 37/55 (67%) monomicrobial and 1/3 polymicrobial samples were flagged monomicrobial (κ = 0.092, 95% confidence interval − 0.082 to 0.267). In conclusion, the accuracy in detecting polymicrobial cultures was low and similar to that of SW1.0.
Antimicrobial susceptibility testing
Enterobacteriaceae
Among the 23 Gram-negative rods eligible for AST, 19/23 (83%) were correctly identified and AST results were obtained. The four remaining isolates were identified, but AST could not be provided. A major issue with SW1.0 was susceptibility testing for ceftazidime. The algorithm for this antimicrobial agent was adjusted in v1.2. Consequently, MIC values for sensitive isolates were correctly assigned (Table 5). For the remaining antimicrobial agents, the overall high degree categorical agreement with disk diffusion was confirmed.
Staphylococci
Results were obtained for 19/21 (90%) eligible isolates. Two S. aureus isolates were incorrectly identified as CoNS. For one of these isolates, AST results were presented but not considered for evaluation. Because of difficulties in detecting resistance to trimethoprim/sulfamethoxazole in CoNS using SW1.0, this antimicrobial combination was removed from the CoNS AST panel in SW1.2. With this adjustment, categorical agreement with disk diffusion was achieved for 84/87 (97%) analyses. In the cefoxitin-screen, 1/13 (8%) major and 1/6 very major error were noted for CoNS isolates. The inhibition zone diameters for these isolates were at the breakpoint, 25 and 24 mm, respectively. Therefore, they were also tested for the presence of the mecA-gene. In both cases, the molecular analysis confirmed the results obtained by disk diffusion. In addition to susceptibility to erythromycin, the PhenoTest™ BC kit includes an analysis to detect MLSB resistance. This phenotype was missed in 1/6 erythromycin-resistant isolates. This was the only isolate with an inducible resistance phenotype. Three of six isolates expressed constitutive clindamycin resistance, and 2/6 isolates displayed an efflux-mediated erythromycin resistance phenotype.
Enterococci
Results for the two enterococcal isolates eligible to AST in the second part of the study were in complete agreement with results from disk diffusion.
Discussion
The rapid emergence of resistant bacteria is an obvious challenge for choosing early appropriate antibiotic treatment for severe infections including sepsis. Escalation and de-escalation of antibiotic treatment require information on antimicrobial susceptibility. In the present study, we evaluated the performance of the Accelerate Pheno™ system on clinical blood culture samples. The PhenoTest™ BC kit provided reliable identification and AST for major pathogens causing bloodstream infection, i.e., Enterobacteriaceae and S. aureus. Limitations were experienced in the detection of polymicrobial infections.
The PhenoTest™ BC kit panel covered 147/166 (88.6%) isolates included in the study. This coverage is very similar to a prospectively performed study on samples from pediatric oncology patients [7]. Among the on-panel microorganisms, 133/147 (90.5%) isolates could be detected and correctly identified by the Accelerate Pheno™ system. In addition, 5/19 (26%) off-panel microorganisms were detected and correctly reported as unidentified organisms. Thus, identification by the Accelerate Pheno™ system was overall comparable with other rapid methods such as the FilmArray Blood Culture ID (BCID) Panel, the Verigene Blood Culture tests for Gram-negative or Gram-positive bacteria, and MALDI-TOF MS directly from blood culture broth [1, 6, 11, 12, 15, 16, 22]. Highest performance was experienced for detection and identification of Enterobacteriaceae with 76/80 (95%) correct identifications, staphylococci with 42/46 (91%) correct identifications, and for enterococci with 10/11 (91%) correct identifications. Noticeable misidentifications occurred for Streptococcus species and off-panel organisms. This included cross-reactivity with off-panel species among the α-hemolytic streptococci, but also false identifications of streptococcal species as S. pneumoniae or CoNS, reducing also the positive predictive value for these staphylococcal species. Despite the clinical significance, differentiation of streptococcal species, and especially differentiation of S. pneumoniae from α-hemolytic streptococci, remains challenging [4, 21, 25]. Results from other rapid molecular identification methods are inconsistent [1, 6, 9, 19, 24]. Consequently, the species S. pneumoniae was removed from the species panel and this pathogen is reported on genus level by the PhenoTest™ BC kit software version v1.2. In addition, five off-panel organisms, including two β-hemolytic streptococci and two S. aureus isolates, were falsely identified as CoNS. Comparable data on β-hemolytic streptococci are not included in the previously published studies [7, 18]. Interestingly, and in contrast to the experience from the present study, CoNS were repeatedly reported as S. aureus in the study performed by Brazelton de Cardenas et al. [7]. Part of these misidentifications can be resolved by Gram stain. Based on our data, this complementation appears advisable and is also recommended by FDA.
The major achievement of the Accelerate Pheno™ system in comparison to rapid molecular tests is phenotypic AST for a panel of clinically relevant antimicrobial agents. Especially for Gram-negative bacteria and in regions with low prevalence of major resistance traits, phenotypic AST is still advisable for effective treatment choices but also for de-escalation of antimicrobial therapy [5, 17]. Overall, our results were similar to the studies by Marschal et al. [18] and Brazelton de Cardenas et al. [7]. Differences may be due to different reference methods and breakpoints, as well as differences in resistance patterns and species distribution among the investigated isolates. In addition to the previously published studies, we also investigated the performance of the PhenoTest™ BC kit operated with software version v1.2, which corresponds to the version available for clinical application. Ceftazidime was the antimicrobial agent most affected by the adjustments made in v1.2, and the results presented here are not reported in the previously published studies.
Overall, AST for Enterobacteriaceae was highly reliable. After adjustments made for ceftazidime in v1.2, categorical agreement of the PhenoTest™ BC-provided results with disk diffusion improved from 83.1 to 99.2%. A limitation of our study was the low number of resistant isolates investigated (Table 4). For ceftazidime, there was only one resistant isolate analyzed with v1.2. Five ceftazidime-resistant isolates identified during the first part of the study were therefore re-analyzed from simulated blood cultures (data not shown). Using SW1.2, 3/5 isolates were identified as resistant while 2/5 isolates were identified as susceptible in test results, suggesting that resistant strains including ESBL producers might be missed by the PhenoTest™ BC.
Previous reports highlight difficulties in identifying resistance to cephalosporines and carbapenems [10, 13]. Therefore, in our laboratory and based on EUCAST recommendation, we complement Vitek 2 analyses with disk diffusion for piperacillin-tazobactam, ceftazidime and cefotaxime, as well as meropenem and ertapenem to avoid these system-related errors. The data presented here demonstrate that similar reservations might be necessary for interpretation of the PhenoTest™ BC. Only few ESBL-producing and no carbapenemase-producing Enterobacteriaceae were found among the isolates studied here. Hence, further studies are warranted to estimate the performance for these and other antibiotics to correctly identify resistant isolates.
For staphylococci, performance improved from 92% categorical agreement for SW1.0 to 97% for SW1.2. This was primarily due to the exclusion of trimethoprim/sulfamethoxazole from the CoNS AST panel. Since no trimethoprim/sulfamethoxazole-resistant S. aureus were isolated during this investigation, conclusions regarding the performance of the PhenoTest™ BC kit for trimethoprim/sulfamethoxazole are limited. Other errors occurred for qualitative analyses, i.e., cefoxitin-screen and MLSB detection. No MRSA occurred during the study period, but we tested 12 methicillin-resistant CoNS. Among these, 11 (92%) were identified correctly. Likewise, 8/8 constitutively clindamycin-resistant isolates were tested MLSB-positive; however, 1/1 inducibly resistant isolate was missed. The low frequency of these resistance phenotypes, and the lack of MRSA isolates, does not allow definitive conclusions and investigation of isolates across the entire range of MIC values is required to evaluate the performance of the PhenoTest™ BC kit.
Conclusion
Our study demonstrates that the Accelerate Pheno™ system provides overall reliable identification results for Enterobacteriaceae and S. aureus. In combination with an initial Gram stain, the assay can therefore be used to complement routine methods for rapid preliminary identification and AST of major pathogens from bloodstream infections. Conclusions from this study are limited by the low frequency of resistant isolates investigated.
The current study focused on the analytical performance of the Accelerate Pheno™ system with promising results. However, the implementation of a rapid commercial AST system in the clinical routine is complex and depends also on the size of the laboratory, the cost of the system, cost per test, and the clinical performance of the method. Studies analyzing these factors are warranted prior to the implementation of the Accelerate Pheno™ system in other clinical diagnostic settings.
Acknowledgements
No funding was received for this study. The Accelerate Pheno™ system modules and PhenoTest™ BC kits were kindly provided by Accelerate Diagnostics, Inc. The authors thank Accelerate Diagnostics, Inc. for the complementing analyses. The company had no influence on the design of the study, interpretation and presentation of the data.
Funding information
Open access funding provided by Karolinska Institute.
Compliance with ethical standards
Conflict of interest
None of the authors had conflict of interest.
Ethical approval
The authors confirm that the ethical policies of the journal, as noted on the journal’s author guidelines page, have been adhered to. No ethical approval was required as the research in this article related to microorganisms.
Informed consent
Only broth from the positive blood culture bottles were included. No informed consent was required.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Altun O, Almuhayawi M, Ullberg M, Ozenci V. Clinical evaluation of the FilmArray blood culture identification panel in identification of bacteria and yeasts from positive blood culture bottles. J Clin Microbiol. 2013;51(12):4130–4136. doi: 10.1128/JCM.01835-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altun O, Botero-Kleiven S, Carlsson S, Ullberg M, Özenci V. Rapid identification of bacteria from positive blood culture bottles by MALDI-TOF MS following short-term incubation on solid media. J Med Microbiol. 2015;64(11):1346–1352. doi: 10.1099/jmm.0.000168. [DOI] [PubMed] [Google Scholar]
- 3.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med. 2001;29(7):1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Arbique JC, Poyart C, Trieu-Cuot P, Quesne G, Carvalho Mda G, Steigerwalt AG, et al. Accuracy of phenotypic and genotypic testing for identification of Streptococcus pneumoniae and description of Streptococcus pseudopneumoniae sp. nov. J Clin Microbiol. 2004;42(10):4686–4696. doi: 10.1128/JCM.42.10.4686-4696.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee R, Teng CB, Cunningham SA, Ihde SM, Steckelberg JM, Moriarty JP, et al. Randomized trial of rapid multiplex polymerase chain reaction-based blood culture identification and susceptibility testing. Clin Infect Dis. 2015;61(7):1071–1080. doi: 10.1093/cid/civ447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatti MM, Boonlayangoor S, Beavis KG, Tesic V. Evaluation of FilmArray and Verigene systems for rapid identification of positive blood cultures. J Clin Microbiol. 2014;52(9):3433–3436. doi: 10.1128/JCM.01417-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brazelton de Cárdenas J.N., Su Y., Rodriguez A., Hewitt C., Tang L., Garner C.D., Hayden R.T. Evaluation of rapid phenotypic identification and antimicrobial susceptibility testing in a pediatric oncology center. Diagnostic Microbiology and Infectious Disease. 2017;89(1):52–57. doi: 10.1016/j.diagmicrobio.2017.06.014. [DOI] [PubMed] [Google Scholar]
- 8.Choi J, Jeong HY, Lee GY, Han S, Han S, Jin B, et al. Direct, rapid antimicrobial susceptibility test from positive blood cultures based on microscopic imaging analysis. Sci Rep. 2017;7(1):1148. doi: 10.1038/s41598-017-01278-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dodemont M, De Mendonca R, Nonhoff C, Roisin S, Denis O. Evaluation of Verigene Gram-positive blood culture assay performance for bacteremic patients. Eur J Clin Microbiol Infect Dis. 2015;34(3):473–477. doi: 10.1007/s10096-014-2250-4. [DOI] [PubMed] [Google Scholar]
- 10.Espinar M. J., Rocha R., Ribeiro M., Goncalves Rodrigues A., Pina-Vaz C. Extended-spectrum -lactamases of Escherichia coli and Klebsiella pneumoniae screened by the VITEK 2 system. Journal of Medical Microbiology. 2011;60(6):756–760. doi: 10.1099/jmm.0.024075-0. [DOI] [PubMed] [Google Scholar]
- 11.Foster AG. Rapid identification of microbes in positive blood cultures by use of the Vitek MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system. J Clin Microbiol. 2013;51(11):3717–3719. doi: 10.1128/JCM.01679-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Idelevich EA, Schule I, Grunastel B, Wullenweber J, Peters G, Becker K. Rapid identification of microorganisms from positive blood cultures by MALDI-TOF mass spectrometry subsequent to very short-term incubation on solid medium. Clin Microbiol Infect. 2014;20(10):1001–1006. doi: 10.1111/1469-0691.12640. [DOI] [PubMed] [Google Scholar]
- 13.Jang W, Park YJ, Park KG, Yu J. Evaluation of MicroScan WalkAway and Vitek 2 for determination of the susceptibility of extended-spectrum beta-lactamase-producing Escherichia coli and Klebsiella pneumoniae isolates to cefepime, cefotaxime and ceftazidime. J Antimicrob Chemother. 2013;68(10):2282–2285. doi: 10.1093/jac/dkt172. [DOI] [PubMed] [Google Scholar]
- 14.Jo SJ, Park KG, Han K, Park DJ, Park YJ. Direct identification and antimicrobial susceptibility testing of bacteria from positive blood culture bottles by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry and the Vitek 2 system. Ann Lab Med. 2016;36(2):117–123. doi: 10.3343/alm.2016.36.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ledeboer NA, Lopansri BK, Dhiman N, Cavagnolo R, Carroll KC, Granato P, et al. Identification of Gram-negative bacteria and genetic resistance determinants from positive blood culture broths by use of the Verigene Gram-negative blood culture multiplex microarray-based molecular assay. J Clin Microbiol. 2015;53(8):2460–2472. doi: 10.1128/JCM.00581-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Loonen AJ, Jansz AR, Stalpers J, Wolffs PF, van den Brule AJ. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/ALERT blood cultures by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2012;31(7):1575–1583. doi: 10.1007/s10096-011-1480-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.MacVane SH, Nolte FS. Benefits of adding a rapid PCR-based blood culture identification panel to an established antimicrobial stewardship program. J Clin Microbiol. 2016;54(10):2455–2463. doi: 10.1128/JCM.00996-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marschal Matthias, Bachmaier Johanna, Autenrieth Ingo, Oberhettinger Philipp, Willmann Matthias, Peter Silke. Evaluation of the Accelerate Pheno System for Fast Identification and Antimicrobial Susceptibility Testing from Positive Blood Cultures in Bloodstream Infections Caused by Gram-Negative Pathogens. Journal of Clinical Microbiology. 2017;55(7):2116–2126. doi: 10.1128/JCM.00181-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez RM, Bauerle ER, Fang FC, Butler-Wu SM. Evaluation of three rapid diagnostic methods for direct identification of microorganisms in positive blood cultures. J Clin Microbiol. 2014;52(7):2521–2529. doi: 10.1128/JCM.00529-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero-Gomez MP, Gomez-Gil R, Pano-Pardo JR, Mingorance J. Identification and susceptibility testing of microorganism by direct inoculation from positive blood culture bottles by combining MALDI-TOF and Vitek-2 Compact is rapid and effective. J Inf Secur. 2012;65(6):513–520. doi: 10.1016/j.jinf.2012.08.013. [DOI] [PubMed] [Google Scholar]
- 21.Simoes AS, Tavares DA, Rolo D, Ardanuy C, Goossens H, Henriques-Normark B, et al. lytA-based identification methods can misidentify Streptococcus pneumoniae. Diagn Microbiol Infect Dis. 2016;85(2):141–148. doi: 10.1016/j.diagmicrobio.2016.03.018. [DOI] [PubMed] [Google Scholar]
- 22.Ward C, Stocker K, Begum J, Wade P, Ebrahimsa U, Goldenberg SD. Performance evaluation of the Verigene® (Nanosphere) and FilmArray® (BioFire®) molecular assays for identification of causative organisms in bacterial bloodstream infections. Eur J Clin Microbiol Infect Dis. 2015;34(3):487–496. doi: 10.1007/s10096-014-2252-2. [DOI] [PubMed] [Google Scholar]
- 23.Wattal C, Oberoi JK. Microbial identification and automated antibiotic susceptibility testing directly from positive blood cultures using MALDI-TOF MS and VITEK 2. Eur J Clin Microbiol Infect Dis. 2016;35(1):75–82. doi: 10.1007/s10096-015-2510-y. [DOI] [PubMed] [Google Scholar]
- 24.Wojewoda CM, Sercia L, Navas M, Tuohy M, Wilson D, Hall GS, et al. Evaluation of the Verigene Gram-positive blood culture nucleic acid test for rapid detection of bacteria and resistance determinants. J Clin Microbiol. 2013;51(7):2072–2076. doi: 10.1128/JCM.00831-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wyllie Anne L., Pannekoek Yvonne, Bovenkerk Sandra, van Engelsdorp Gastelaars Jody, Ferwerda Bart, van de Beek Diederik, Sanders Elisabeth A. M., Trzciński Krzysztof, van der Ende Arie. Sequencing of the variable region of rpsB to discriminate between Streptococcus pneumoniae and other streptococcal species. Open Biology. 2017;7(9):170074. doi: 10.1098/rsob.170074. [DOI] [PMC free article] [PubMed] [Google Scholar]