Abstract
Chemokine-like factor (CKLF)-like MARVEL transmembrane members (CMTMs) represent a novel protein family linking the chemokine and transmembrane-4 superfamily families, which potentially play several roles in diverse physiological and pathological processes. The detailed functions and underlying molecular mechanisms of CMTMs remain elusive in breast cancer. Herein, we performed a comprehensive bioinformatic analysis to investigate the prognostic effect, potential functions, and biomolecular regulatory network of CMTMs in breast cancer. The mRNA expression level of CMTM5, in particular, was significantly downregulated in breast cancer; moreover, high mRNA expression level of CMTM5 was significantly associated with better relapse-free survival. DNA promoter hypermethylation of CMTM5 was negatively correlated with its mRNA expression levels. Furthermore, CMTM5 strongly associated with pathway in MARVEL domains, chemotaxis, cytokines, transmembrane structures, and integral component of membrane. For example, genes related to MARVEL domains, transmembrane structures, and chemokines were significantly enriched. Our findings indicate that CMTM5 can be used as a prognostic biomarker and potential therapeutic target for breast cancer.
Keywords: DNA methylation, prognosis, biomarkers, disease progression, breast neoplasms
Introduction
Breast cancer is the most common malignancy in women; approximately 2 million new cases are annually diagnosed worldwide (Ferlay et al., 2015). Various molecular markers, including estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor-2 (HER2), and Ki67, are widely used in the prognosis of breast cancer and to make treatment-related decisions (Untch et al., 2019). In recent decades, the early diagnostic techniques and clinical management of breast cancer have considerably improved (Merino Bonilla et al., 2017). However, distant metastasis remains an important risk factor related to outcomes in breast cancer, seriously affecting patient quality of life and contributing to a major socioeconomic burden. Therefore, there exists an urgent need to explore the mechanisms underlying breast cancer development and progression as well as to identify novel biomarkers to further improve its diagnosis, treatment, and prognosis.
In 2005, chemokine-like factor superfamily members 1–8 (CKLFSF1–8) were renamed CKLF-like MARVEL transmembrane domain-containing 1–8 (CMTM1–8). CKLF and CMTM1–8 represent a novel protein family linking the chemokine and transmembrane-4 superfamily (TM4SF) families, which potentially play several roles in diverse physiological and pathological processes (Han et al., 2003). CKLF and CMTM1–4 are located in a gene cluster on 16q22.1, CMTM6–8 are located in another gene cluster on chromosome 3p22, and CMTM5 is located in 14q11.2 separately. CMTM family members play a key role in the immune system, tumorigenesis, and male reproductive system (Lu et al., 2016). CMTM1–8 are known tumor suppressor genes (Bu et al., 2008; Li et al., 2011, 2015; Liu et al., 2013, 2015; Xie et al., 2014; Zhang et al., 2014, 2016; Gao et al., 2015; Xiao et al., 2015; Bei et al., 2017; Choi et al., 2018; Huang et al., 2019; Zhu et al., 2019); in addition, CMTM6 is a key regulator of PD-L1 in various human cancer cells. PD-L1 relies on CMTM6/4 to efficiently exert its inhibitory functions (Burr et al., 2017; Mezzadra et al., 2017). However, little remains known regarding the function of CMTM family members in breast cancer development and progression.
Herein, we performed a comprehensive bioinformatic analysis to investigate the prognostic effect and biomolecular regulatory network of CMTMs in breast cancer development and progression.
Materials and Methods
cBio Cancer Genomics Portal
We used the open-access cBio Cancer Genomics Portal (cBioPortal)1 to explore the relationship between CMTMs and genetic alteration frequency in 963 breast cancer samples (Cerami et al., 2012). Data from the Breast Invasive Carcinoma [The Cancer Genome Atlas (TCGA), provisional, 1108 samples] Project were used for analysis. GISTIC 2.0 was used to assess significant somatic copy-number alterations, such as deletions and amplifications (Mermel et al., 2011). The correlations among CMTM5 and related genes were also assessing with cBioPortal, and co-occurrence genes were defined as positive correlation, while genes that displayed mutual exclusivity were defined as negative correlation.
Oncomine
Oncomine2 was employed to analyze the mRNA expression levels of CMTMs in various human cancers and corresponding normal tissues (Rhodes et al., 2004). In addition, the roles of CMTM5 expression levels in response to breast cancer therapies were also investigate using Oncomine.
UALCAN
UALCAN is an open-access, interactive web portal for analyzing cancer transcriptome data from the TCGA database3 (Chandrashekar et al., 2017). We used this portal to assess CMTM mRNA expression and DNA promoter methylation levels in breast cancer tissues and corresponding normal tissues.
Kaplan–Meier Plotter
We used the Kaplan–Meier (KM) plotter to assess the potential prognostic effect of CMTMs on breast cancer4 (Lánczky et al., 2016). Survival curves (relapse-free survival, RFS) were plotted using the KM method and compared by the log-rank test.
Methylation Modification Analyses
MEXPRESS was used to assess the relationship between CMTM5 mRNA expression and DNA methylation levels in 871 invasive breast carcinoma samples (Koch et al., 2015). The relationship between CMTM5 mRNA expression and DNA promoter methylation levels in 748 invasive breast carcinoma samples was also determined using MethHC, which is a database of DNA methylation and mRNA expression profiles in human cancers (Huang et al., 2015).
Biomolecular Regulatory Network Analyses
We assessed the functional protein–protein interaction network for CMTM5 using STRING5 with a confidence score of 0.4 (Szklarczyk et al., 2015). In addition, the Database for Annotation, Visualization and Integrated Discovery (DAVID) was used for gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of CMTM5-related genes (Huang da et al., 2009a, b).
Immunohistochemistry Data
We used the open-access Human Protein Atlas6 to analyze protein expression patterns of CMTM5 in breast cancer. In general, gene expression patterns can be examined using this portal at the protein level using antibody-based protein profiling data. We used staining intensity to assess protein expression levels.
Statistical Analyses
The patients clinical characteristic parameters were extracted from TCGA database. The correlation between clinical characteristic parameters and the expression of CMTM5 in breast cancer was compared by one-way ANOVA test, and the results were expressed using mean ± standard deviation (SD). The statistical tests were performed using SPSS 22.0 (IBM Corp., Armonk, NY, United States). P values <0.05 were considered to indicate statistical significance.
Results
mRNA Expression Levels and Differences in Genetic Alteration Frequency of CMTMs in Human Cancers
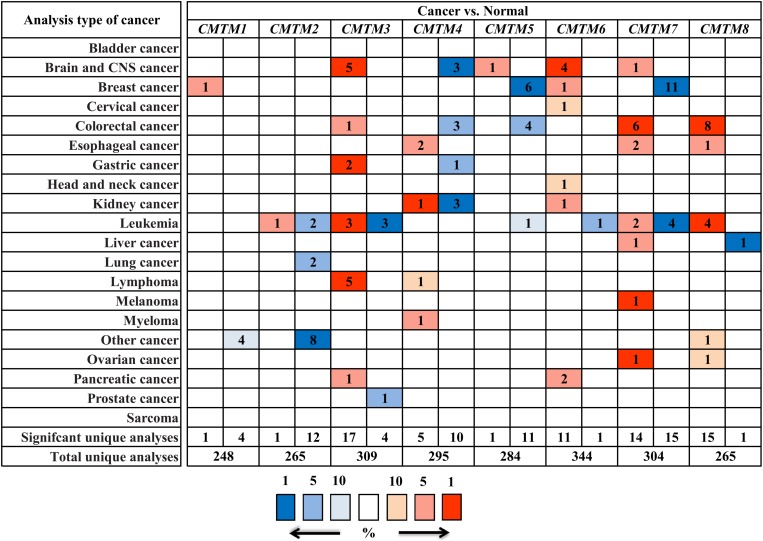
To assess the role of CMTMs in human carcinogenesis, we used Oncomine to investigate mRNA expression levels of CMTM family members in various human cancers. The mRNA expression levels of CMTM1, CMTM2, CMTM4, and CMTM5 were significantly downregulated whereas those of CMTM3, CMTM6, and CMTM8 were noticeably upregulated in human tumors than in normal tissues. In addition, the mRNA expression of CMTM7 was at a comparable level between human tumors and corresponding normal tissues (Figure 1). In breast cancer tissues, the mRNA expression levels of only CMTM1, CMTM5, CMTM6, and CMTM7 were measured. The mRNA expression levels of CMTM5 and CMTM7 were significantly downregulated while those of CMTM1 and CMTM6 were significantly upregulated in breast cancer tissues than in corresponding normal tissues (Figure 1).
FIGURE 1.
mRNA expression levels of CMTM family members in different human cancers and corresponding normal tissues from the Oncomine database.
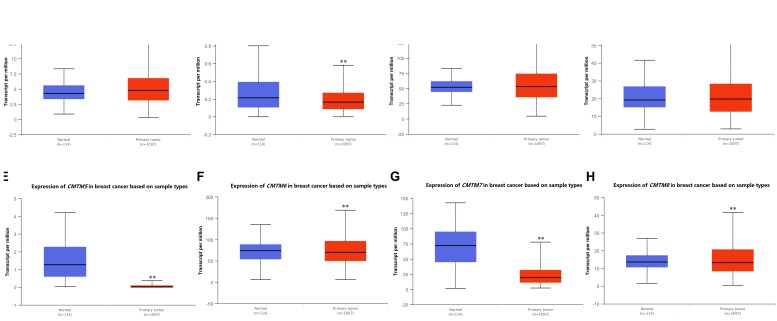
Next, we used UALCAN to confirm the mRNA expression levels of CMTM family members. We found that the expression levels of all CMTM family members were significantly dysregulated in breast cancer tissues. Among them, the mRNA expression levels of CMTM5 [normal tissues vs. breast cancer tissues, 45.36-fold (P = 1.44e−08)] and CMTM7 [normal tissues vs. breast cancer tissues, 3.72-fold (P < 1e−12)] were significantly downregulated, whereas those of CMTM1 (1.11-fold, P = 8.41e−12) and CMTM3 (1.02-fold, P = 3.43e−04) were only slightly upregulated and of CMTM2 (1.28-fold, P = 1.15e−02), CMTM6 (1.07-fold, P = 1.29e−02), and CMTM8 (1.03-fold, P = 1.90e−05) were only slightly downregulated in breast cancer tissues than in corresponding normal tissues (Figure 2).
FIGURE 2.
mRNA expression levels of CMTM family members in invasive breast carcinoma samples and corresponding normal tissues from the UALCAN database (∗∗P < 0.01). (A) CMTM1, (B) CMTM2, (C) CMTM3, (D) CMTM4, (E) CMTM5, (F) CMTM6, (G) CMTM7, and (H) CMTM8.
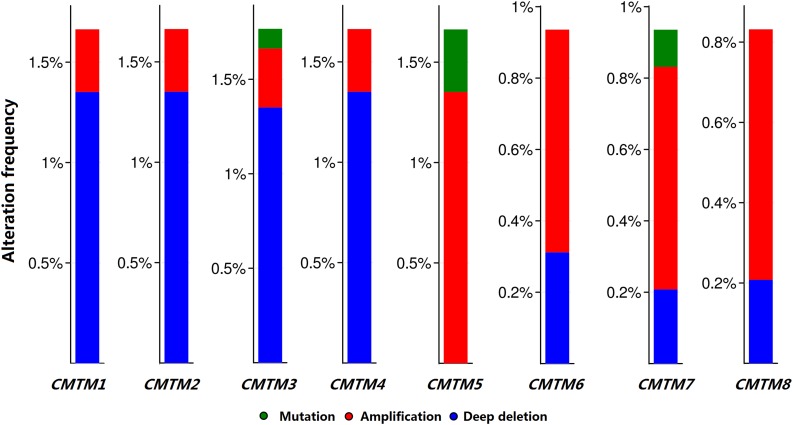
Moreover, we analyzed the genetic alteration frequency data pertaining to CMTM family members in 963 breast cancer samples using cBioPortal. Alterations and amplifications were more likely to observe in case of CMTM5, and CMTM1–4 were more likely to have a deep deletion (Figure 3). However, there were no significant differences between disease-free survival (P = 0.723) and overall survival (P = 0.904) in breast cancer patients with or without CMTM5 alterations.
FIGURE 3.
Genetic alteration frequency data of CMTM family members in breast cancer using cBioPortal.
Prognostic Effects of CMTMs on Breast Cancer
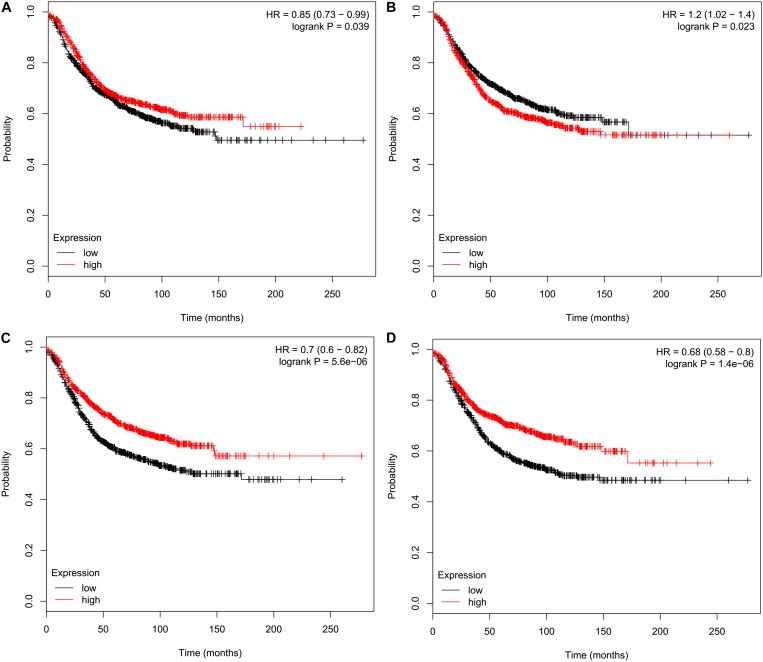
We performed survival analyses to assess prognostic effects of CMTMs on breast cancer using the KM plotter. The obtained results indicated that high mRNA levels of CMTM3 [hazard ratio (HR) 0.85, 95% confidence interval (CI) 0.73–0.99, P = 0.039] (Figure 4A), CMTM5 (HR 0.70, 95% CI 0.60–0.82, P = 5.60e−06) (Figure 4C), and CMTM7 (HR 0.68, 95% CI 0.58–0.80, P = 1.40e−06) (Figure 4D) were significantly related to better RFS; however, high mRNA levels of CMTM4 (HR 1.2, 95% CI 1.02–1.40, P = 0.023) were significantly related to poor RFS (Figure 4B). In addition, mRNA levels of CMTM1 (P = 0.510), CMTM2 (P = 0.051), CMTM6 (P = 0.970), and CMTM8 (P = 0.910) were not correlated at all with RFS. These findings suggest that CMTM5 has a higher prognostic effect on breast cancer than other CMTM family members; therefore, we selected CMTM5 for further analyses.
FIGURE 4.
Relapse-free survival analysis of CMTM family members in breast cancer using the Kaplan–Meier plotter. (A) CMTM3, (B) CMTM4, (C) CMTM5, and (D) CMTM7.
CMTM5 mRNA Expression Levels and Prognostic Effect According to Clinicopathological Characteristics
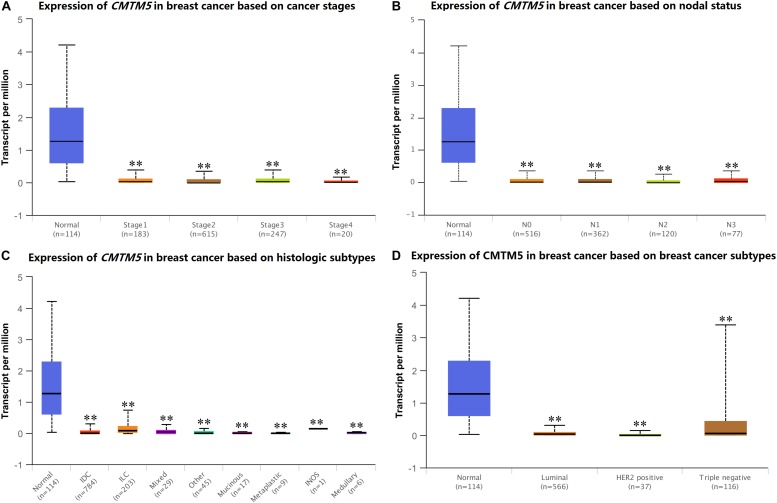
We used UALCAN to assess CMTM5 mRNA expression levels in breast cancer according to clinicopathological characteristics. We found that the mRNA expression levels of CMTM5 in breast cancer tissues were still significantly lower than in normal tissues after stratification by tumor stage, nodal stage, histologic subtypes, and breast cancer subtypes (Figures 5A–D).
FIGURE 5.
mRNA expression levels of CMTM5 in breast cancer according to different clinicopathological characteristics from the UALCAN database (∗∗P < 0.01). (A) Tumor stage, (B) nodal stage, (C) histologic subtypes, and (D) breast cancer subtypes.
After assessing the expression and prognosis of CMTM5 in breast cancer, we further analyzed the correlation between patients clinical characteristics and the CMTM5 expression levels. The patient clinical characteristic parameters have listed in Table 1. The levels of CMTM5 expression were significantly associated with age (P = 0.016), ER status (P < 0.001), and PR status (P < 0.001). Patients with older age, and ER positive and PR positive disease had lower levels of CMTM5 expression.
TABLE 1.
The correlation between clinical characteristic parameters and the expression of CMTM5 in breast cancer.
| Variables | Number | Mean ± SD | P |
| Age (years) | |||
| <50 | 295 | 0.26 ± 1.34 | 0.016 |
| ≥50 | 1090 | 0.11 ± 0.71 | |
| Gender | |||
| Male | 12 | 0.01 ± 0.01 | 0.585 |
| Female | 1078 | 0.15 ± 0.93 | |
| Histology (n = 1089) | |||
| Invasive ductal carcinoma | 779 | 0.14 ± 0.83 | 0.286 |
| Invasive lobular carcinoma | 203 | 0.09 ± 0.19 | |
| Other | 107 | 0.30 ± 1.90 | |
| Tumor stage (n = 1087) | |||
| T1 | 279 | 0.18 ± 1.13 | 0.870 |
| T2 | 631 | 0.14 ± 0.94 | |
| T3 | 137 | 0.12 ± 0.31 | |
| T4 | 40 | 0.11 ± 0.50 | |
| Nodal stage (n = 1070) | |||
| N0 | 514 | 0.19 ± 0.99 | 0.658 |
| N1 | 360 | 0.14 ± 1.07 | |
| N2 | 120 | 0.08 ± 0.40 | |
| N3 | 76 | 0.11 ± 0.37 | |
| Distant metastatic status (n = 923) | |||
| No | 901 | 0.17 ± 1.01 | 0.750 |
| Yes | 22 | 0.10 ± 0.25 | |
| ER status (n = 1040) | |||
| Negative | 237 | 0.47 ± 1.74 | <0.001 |
| Positive | 803 | 0.06 ± 0.48 | |
| PR status (n = 1037) | |||
| Negative | 343 | 0.34 ± 1.40 | <0.001 |
| Positive | 694 | 0.06 ± 0.60 | |
| HER2 status (n = 913) | |||
| Negative | 702 | 0.17 ± 0.98 | 0.808 |
| Equivocal or Indeterminate | 26 | 0.10 ± 0.23 | |
| Positive | 185 | 0.13 ± 1.16 | – |
ER, estrogen receptor; HER2, human epidermal growth factor receptor-2; N, node; PR, progesterone receptor; SD, standard deviation; T, tumor.
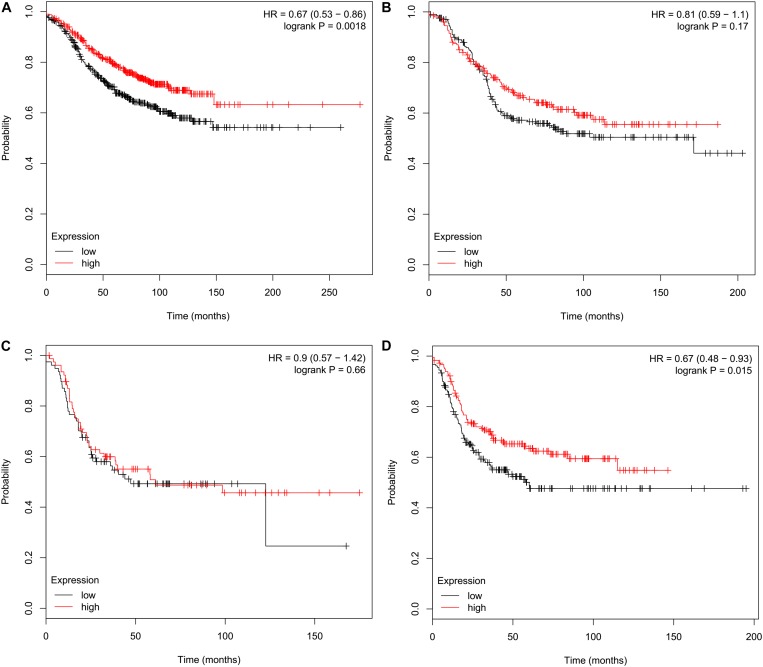
Relapse-free survival analyses using the KM plotter indicated that high CMTM5 mRNA expression levels were related to better RFS in luminal A (HR 0.67, 95% CI 0.53–0.86, P = 0.0018) and basal-like (HR 0.67, 95% CI 0.48–0.93, P = 0.015) subtypes (Figures 6A,D); however, the mRNA expression levels of CMTM5 were not associated with RFS at all in luminal B (P = 0.170) (Figure 6B) and HER2+ (P = 0.660) (Figure 6C) subtypes. Moreover, CMTM5 mRNA expression levels did not correlate with RFS after stratification by tumor grade, nodal status, and TP53 status.
FIGURE 6.
Effect of CMTM5 mRNA expression levels on relapse-free survival in different breast cancer subtypes using the Kaplan–Meier plotter. (A) Luminal A, (B) luminal B, (C) HER2+, and (D) basal-like subtypes.
With regard to the immunohistochemical staining of CMTM5, among the 11 breast cancer patients with pertinent data in the Human Protein Atlas, only one (9.1%) showed low expression of CMTM5, whereas no expression of CMTM5 in tumor tissues was observed in the other 10 (90.9%) patients (Figures 7A,B).
FIGURE 7.

Representative immunohistochemical staining of CMTM5 in breast cancer tissue samples from the Human Protein Atlas database (original magnification 50×). (A) Invasive ductal carcinoma (staining: not detected, intensity: negative, and quantity: none). (B) Invasive lobular carcinoma (staining: not detected, intensity: negative, and quantity: none).
The Roles of CMTM5 in Breast Cancer Therapies
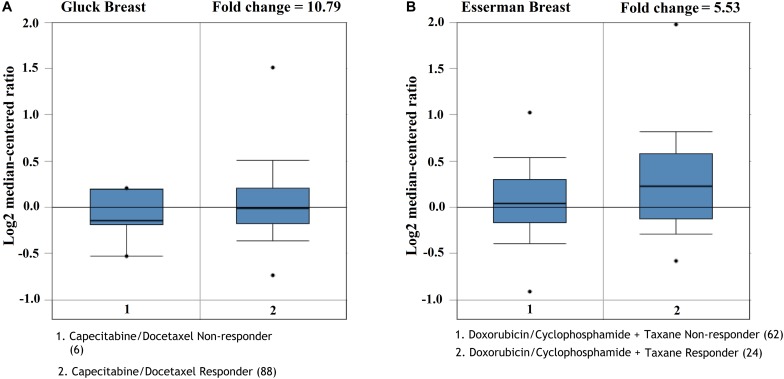
We future analyzed the roles of CMTM5 expression levels in response to breast cancer therapies using Oncomine. The results showed that lower expression of CMTM5 was associated with lower response to systemic therapy compared to those with higher expression of CMTM5 including the regimens of capecitabine/docetaxel (Gluck Breast database) and doxorubicin/cyclophosphamide + taxane (Esserman Breast database) (Figures 8A,B). These findings showed that lower expression of CMTM5 might be related to inferior response of breast cancers to chemotherapeutics.
FIGURE 8.
The influence of the expression of CMTM5 on the response to chemotherapeutics in breast cancer (A, Gluck Breast database; B, Esserman Breast database).
Promoter Methylation Levels of CMTM5
DNA promoter methylation plays an important role in the epigenetic regulation of chromatin structure and gene transcription, and DNA promoter hypermethylation can induce stable epigenetic inhibition of gene expression. Herein we used UALCAN, MethHC, and MEXPRESS to investigate whether the dysregulated expression of CMTM5 is related to DNA promoter methylation status. As per the analyses involving UALCAN, a significantly higher promoter methylation level of CMTM5 was observed in breast cancer tissues than in normal tissues (1.17-fold, P < 1e−12) (Figure 9A), which was also confirmed upon using MethHC (P < 0.01) (Figures 9B,C). Moreover, MEXPRESS showed that the CMTM5 mRNA expression level was negatively associated with DNA promoter methylation status (Figure 9D). The analyses involving MEXPRESS also indicated that the CMTM5 mRNA expression level was significantly lower in breast cancer tissues than in normal tissues and correlated with ER (P = 5.76e−5), PR (P = 3.84e−6), and menopause status (r = −0.0954) (Figure 9D). These results suggested that DNA promoter hypermethylation of CMTM5 negatively correlates with the mRNA expression level of CMTM5 in breast cancer.
FIGURE 9.
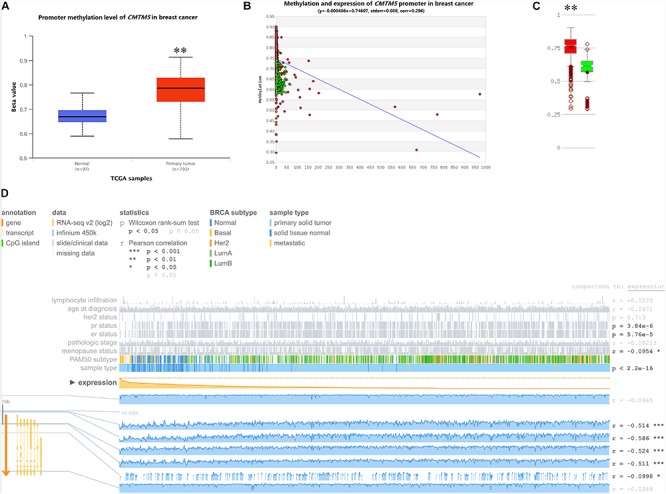
Impact of epigenetic alterations on CMTM5 mRNA expression levels in breast cancer (∗∗P < 0.01). (A) Promoter methylation levels of CMTM5 as per the analyses involving UALCAN. (B,C) Promoter methylation levels of CMTM5 as per the analyses involving MethHC. (D) CMTM5 expression levels in breast cancer related to DNA promoter methylation as per the analyses involving MEXPRESS.
CMTM5-Regulated Biomolecular Network
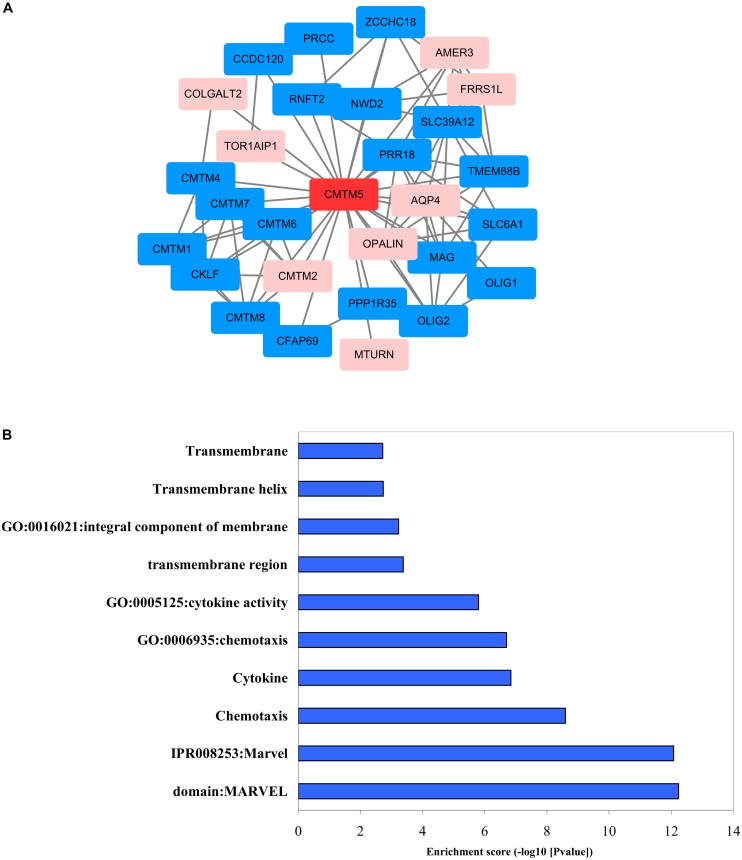
We used STRING and Cytoscape networks to assess the effects of regulatory genes on the carcinogenesis and progression of breast cancer. Functional interaction network analysis indicated that CMTM5 is closely correlated with 28 genes including 20 genes were positive correlation with CMTM5 and 8 genes were negative correlation with CMTM5 (Figure 10A). We then performed GO enrichment and KEGG pathway analyses using DAVID; the obtained results showed that most significantly enriched gene sets related to MARVEL domains, chemotaxis, cytokines, transmembrane structures, and integral component of membrane (Figure 10B) were significantly enriched.
FIGURE 10.
Biomolecular regulatory network of CMTM5. (A) Protein–protein interaction network complex and modular analysis using STRING (blue nodes indicate positive correlation and pink nodes show negative correlation). (B) Top 10 clusters identified via GO enrichment and KEGG pathway analysis using DAVID.
Discussion
Herein we used Oncomine to analyze the expression of CMTM family members in various human cancers. We focused on breast cancer and CMTM5, and we report the results of a comprehensive bioinformatic analysis on the survival outcomes and potential molecular mechanisms using mRNA and protein expression levels, prognostic effects, genetic and epigenetic alterations, and corresponding biomolecular regulatory networks.
CMTM family members play a crucial role in the immune, male reproductive, and hematopoiesis systems (Lu et al., 2016); moreover, they serve as tumor suppressor genes and are associated with tumor progression in various human cancers (Bu et al., 2008; Li et al., 2011, 2015; Liu et al., 2013, 2015; Xie et al., 2014; Zhang et al., 2014, 2016; Gao et al., 2015; Xiao et al., 2015; Bei et al., 2017; Choi et al., 2018; Huang et al., 2019; Zhu et al., 2019). In fact a previous study even indicated that higher CMTM1 and CMTM3 expression levels are significantly associated with shorter overall survival in glioblastoma (Delic et al., 2015). In this study, we report that the mRNA expression levels of CMTM1, CMTM2, CMTM4, and CMTM5 are significantly downregulated whereas those of CMTM3, CMTM6, and CMTM8 are significantly upregulated in human cancers; therefore, our results indicate that the mRNA expression levels of CMTM family members can be highly divergent.
The dysfunction of CMTMs is closely correlated with the progression of numerous cancer types, and many of them may be strongly involved in the prognosis of patients with cancer; however, at present, little is known about the potential underlying mechanisms or their prognostic value in breast cancer. In addition to determining the mRNA expression levels of several CMTM family members, we herein used UALCAN and found that CMTM5 was significantly dysregulated in breast cancer relative to other CMTM family members (45.36-fold vs. 1.02–3.72-fold). In addition, 11 breast cancer patients with available immunohistochemical staining of CMTM5 in Human Protein Atlas, only 1 patient had low expression and 10 reflected no expression of CMTM5. Therefore, we chose CMTM5 for further analyses.
Unlike other CMTM family members, CMTM5 is localized separately on 14q11.2, a locus frequently altered in many types of cancers. CMTM5 is widely expressed in normal tissues and its expression levels are either downregulated or silenced in liver (Guan et al., 2018), kidney (Cai et al., 2017), and prostate (Xiao et al., 2015) cancers. Shao et al. (2007) indicated that CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. They assessed the mRNA expression levels of CMTM5 in breast cancer cell lines and reported undetectable or downregulated expression levels in all the cell lines. Moreover, CMTM5 promoter methylation was observed in all downregulated or silenced breast cancer cell lines; a methylation-mediated mechanism was predicted to be involved in the development of breast cancer (Shao et al., 2007). To the best of our knowledge, this is the first study to confirm the downregulation of CMTM5 mRNA and protein expression levels in breast cancer. Furthermore, we believe that promoter hypermethylation is a key reason for the inhibition of CMTM5 mRNA expression and transcription.
In this study, we also found that lower expression of CMTM5 was associated with inferior response to chemotherapeutics of breast cancers. CMTM5 is located in 14q11.2, a chromosome region which have showed gains significantly more frequently in the chemoresistant disease subgroup compared to the chemosensitive disease subgroup in serous ovarian cancer (Kim et al., 2007). In addition, hypermethylator cancer cells also contribute to chemotherapy resistance of breast cancer (Sandhu et al., 2012). To the best of our knowledge, there were no studies assessing the role of CMTM5 in chemotherapy sensitivity in human cancers. According to our study, CMTM5 may be available as potential target for development of new treatment strategies of breast cancer.
Tumor heterogeneity refers to the difference in genes and phenotypes between cells of different tumors, it has been considered to be the biggest obstacle to the clinical management of breast cancer. In addition, heterogeneity is a fundamental and key element in cancer initiation, progression, and distant metastasis (Turashvili and Brogi, 2017). In our study, we found that patients with ER and PR positive diseases had lower levels of CMTM5 expression. However, low mRNA levels of CMTM5 were associated with lower RFS in our study, while ER and PR positive diseases were associated with better survival outcomes of breast cancer (Untch et al., 2019). Moreover, our study found that lower levels of CMTM5 expression were related to lower RFS in luminal A and basal-like subtypes, but not in luminal B and HER2+ subtype, which indicated that this was also present with tumor heterogeneity in the CMTM5 expression. The magnitude of the influence of tumor heterogeneity on CMTM5 expression and its clinical significance still need further exploration.
By browsing biomolecular regulatory networks for genes of interest and via functional enrichment analyses, we found that CMTM5 is closely correlated with 28 genes that are involved in pathways that potentially play a crucial role in breast cancer development and progression. For example, as anticipated, genes related to MARVEL domains, transmembrane structures, and chemokines were significantly enriched. The product encoded by CMTM5 is closer to TM4SF (Han et al., 2003). TM4SF includes several types of proteins possessing the four transmembrane-helix structure, such as the classical TM4SF (tetraspanin) and MARVEL-domain containing proteins, and such proteins have been linked to breast cancer development and progression (Bouchagier et al., 2014; Jiang et al., 2019; Lara-Lemus, 2019). CMTM5 also encodes a protein that potentially has a four transmembrane-helix architecture, which provides a theoretical basis for the new tumor suppressor gene of CMTM5.
Chemotaxis has a key role in cancer progression (Roussos et al., 2011). Some recent studies have reported that chemotaxis occurs during metastasis in many types of cancers, including breast cancers (Wang et al., 2004; Friedl and Alexander, 2011; Roussos et al., 2011). Several upregulated chemokines in human cancers are reportedly prominent tumor promoters (El-Haibi et al., 2013; Singh et al., 2016). Chemokines can generate a tumor immunosuppressive microenvironment and support cancer cells in their metastatic journey toward secondary tumors (Singh et al., 2011). In breast cancer, chemokines are mainly involved in the generation of a protumoral microenvironment and organ-directed metastasis; moreover, they mediate disease progression and contribute to the growth and proliferation of tumor cells (Palacios-Arreola et al., 2014).
Another important pathway that was identified to be associated with CMTM5 was the negative regulation of cytokines. Cytokines are low molecular weight proteins that are involved in cell-to-cell communication by autocrine or paracrine signaling. Cancer cells communicate with the host mainly through cytokines and use this system for shaping the tumor microenvironment, promote tumor dissemination, and for epithelial–mesenchymal transition, motility, and invasion (Yamaguchi and Condeelis, 2007). In addition, inflammatory cytokines play a role in the invasiveness and migration of breast cancer cells (Jorcyk et al., 2006; Bolin et al., 2012), and they are also implicated in the differentiation, apoptosis, angiogenesis, and survival of breast cancer cells (Scheller et al., 2006, 2011). Moreover, within the sites of metastasis, distinct molecules, such as cytokines and chemokines, are involved in attracting tumor cells that express the corresponding cell surface receptors (Ben-Baruch, 2003).
Tawara et al. (2019) reported that a high expression of cytokine or cytokine receptors in breast cancer was associated with lower survival. Moreover, patients with high expression of cytokines showed lower survival than those with low expression of cytokines in HER2−, but not in HER2+, disease (Tawara et al., 2019). Our study also confirmed that high mRNA expression levels of CMTM5 were associated with better RFS in luminal A and basal-like subtypes (HER2− disease) and not associated to RFS at all in luminal B and HER2+ subtypes (HER2+ disease). An earlier study proposed that interleukin-6 family cytokines can be used as profiling biomarkers for HER2− breast cancer tissues, particularly in cases of triple-negative breast cancer, but they had no significant association with HER2+ cancer tissues (Niu et al., 2002). Therefore, these results suggest the presence of redundancy between CMTM5- and HER2-induced signaling.
One limitation of this study is that most analyses involved determining mRNA expression levels; further studies should thus be conducted to validate our results at a protein level by performing, for example, Western blotting. Although our study highlights the prognostic value of CMTM5 in breast cancer and predicts several potential pathways implicated in its function, more studies are warranted so as to elucidate the specific roles of CMTM5 and underlying mechanisms that affect breast cancer development and progression.
Conclusion
In conclusion, using comprehensive bioinformatic analysis, we report that CMTM5 is a putative tumor suppressor gene that can serve as a potential negative prognostic biomarker in breast cancer. We believe that our findings should be helpful for investigating the detailed function and potential carcinogenic mechanisms of CMTM5 in breast cancer, which might promote the development of a novel target treatment for breast cancer.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: www.cbioportal.org, http://www.oncomine.org, http://ualcan.path.uab.edu, http://kmplot.com/analysis/index.php?p=service&cancer=breast, http://string-db.org, and https://www.proteinatlas.org.
Author Contributions
JZ, JW, S-GW, and Z-YH are lead authors who participated in data collection, manuscript drafting, table and figures creation, and manuscript revision. C-LL, LH, and JL aided in data collection. JZ, JL, and JW are senior authors who aided in drafting the manuscript and manuscript revision. Z-YH and S-GW are the corresponding authors who initially developed the concept and drafted and revised the manuscript. All authors read and approved the final manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This work was partly supported by the National Natural Science Foundation of China (Nos. 81802600, 81872459, 81570818, and 81803050), the Commission Young and Middle-Aged Talents Training Project of Fujian Health Commission (No. 2019-ZQNB-25), and the Science and Technology Planning Projects of Xiamen Science & Technology Bureau (No. 3502Z20174070).
References
- Bei C., Zhang Y., Wei R., Zhu X., Wang Z., Zeng W., et al. (2017). Clinical significance of CMTM4 expression in hepatocellular carcinoma. Onco Targets Ther. 10 5439–5443. 10.2147/OTT.S149786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Baruch A. (2003). Host microenvironment in breast cancer development: inflammatory cells, cytokines and chemokines in breast cancer progression: reciprocal tumor-microenvironment interactions. Breast Cancer Res. 5 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolin C., Tawara K., Sutherland C., Redshaw J., Aranda P., Moselhy J., et al. (2012). Oncostatin m promotes mammary tumor metastasis to bone and osteolytic bone degradation. Genes Cancer 3 117–130. 10.1177/1947601912458284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchagier K. A., Assimakopoulos S. F., Karavias D. D., Maroulis I., Tzelepi V., Kalofonos H., et al. (2014). Expression of claudins-1, -4, -5, -7 and occludin in hepatocellular carcinoma and their relation with classic clinicopathological features and patients’ survival. In Vivo 28 315–326. [PubMed] [Google Scholar]
- Bu L., Jiang G., Wang J. (2008). Expression of cklf1,cmtm1, cmtm2 and cmtm4 in non-small cell lung cancer [in Chinese]. Chin. J. Exp. Surg. 25 1126–1127. [Google Scholar]
- Burr M. L., Sparbier C. E., Chan Y. C., Williamson J. C., Woods K., Beavis P. A., et al. (2017). CMTM6 maintains the expression of PD-L1 and regulates anti-tumour immunity. Nature 549 101–105. 10.1038/nature23643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai B., Xiao Y., Li Y., Zheng S. (2017). CMTM5 inhibits renal cancer cell growth through inducing cell-cycle arrest and apoptosis. Oncol. Lett. 14 1536–1542. 10.3892/ol.2017.6350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., et al. (2012). The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2 401–404. 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar D. S., Bashel B., Balasubramanya S. A. H., Creighton C. J., Ponce-Rodriguez I., Chakravarthi B. V. S. K., et al. (2017). UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 19 649–658. 10.1016/j.neo.2017.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. H., Kim Y. B., Ahn J. M., Kim M. J., Bae W. J., Han S. U., et al. (2018). Identification of genomic aberrations associated with lymph node metastasis in diffuse-type gastric cancer. Exp. Mol. Med. 50:6. 10.1038/s12276-017-0009-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delic S., Thuy A., Schulze M., Proescholdt M. A., Dietrich P., Bosserhoff A. K., et al. (2015). Systematic investigation of CMTM family genes suggests relevance to glioblastoma pathogenesis and CMTM1 and CMTM3 as priority targets. Genes Chromosomes Cancer 54 433–443. 10.1002/gcc.22255 [DOI] [PubMed] [Google Scholar]
- El-Haibi C. P., Sharma P., Singh R., Gupta P., Taub D. D., Singh S., et al. (2013). Differential G protein subunit expression by prostate cancer cells and their interaction with CXCR5. Mol. Cancer 12:64. 10.1186/1476-4598-12-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., et al. (2015). Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer 136 E359–E386. 10.1002/ijc.29210 [DOI] [PubMed] [Google Scholar]
- Friedl P., Alexander S. (2011). Cancer invasion and the microenvironment: plasticity and reciprocity. Cell 147 992–1009. 10.1016/j.cell.2011.11.016 [DOI] [PubMed] [Google Scholar]
- Gao D., Hu H., Wang Y., Yu W., Zhou J., Wang X., et al. (2015). CMTM8 inhibits the carcinogenesis and progression of bladder cancer. Oncol. Rep. 34 2853–2863. 10.3892/or.2015.4310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan L., Ji D., Liang N., Li S., Sun B. (2018). Up-regulation of miR-10b-3p promotes the progression of hepatocellular carcinoma cells via targeting CMTM5. J. Cell Mol. Med. 22 3434–3441. 10.1111/jcmm.13620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han W., Ding P., Xu M., Wang L., Rui M., Shi S., et al. (2003). Identification of eight genes encoding chemokine-like factor superfamily members 1-8 (CKLFSF1-8) by in silico cloning and experimental validation. Genomics 81 609–617. 10.1016/s0888-7543(03)00095-8 [DOI] [PubMed] [Google Scholar]
- Huang W. Y., Hsu S. D., Huang H. Y., Sun Y. M., Chou C. H., Weng S. L., et al. (2015). MethHC: a database of DNA methylation and gene expression in human cancer. Nucleic Acids Res. 43 D856–D861. 10.1093/nar/gku1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z. M., Li P. L., Yang P., Hou X. D., Yang Y. L., Xu X., et al. (2019). Overexpression of CMTM7 inhibits cell growth and migration in liver cancer. Kaohsiung J. Med. Sci. 35 332–340. 10.1002/kjm2.12058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009a). Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37 1–13. 10.1093/nar/gkn923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A. (2009b). Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jiang L., Zhang X., Geradts J., Wei Q., Hochwald S., Xu H., et al. (2019). Expression of tetraspanins NET-6 and CD151 in breast cancer as a potential tumor biomarker. Clin. Exp. Med. 19 377–384. 10.1007/s10238-019-00554-x [DOI] [PubMed] [Google Scholar]
- Jorcyk C. L., Holzer R. G., Ryan R. E. (2006). Oncostatin M induces cell detachment and enhances the metastatic capacity of T-47D human breast carcinoma cells. Cytokine 33 323–336. 10.1016/j.cyto.2006.03.004 [DOI] [PubMed] [Google Scholar]
- Kim S. W., Kim J. W., Kim Y. T., Kim J. H., Kim S., Yoon B. S., et al. (2007). Analysis of chromosomal changes in serous ovarian carcinoma using high-resolution array comparative genomic hybridization: potential predictive markers of chemoresistant disease. Genes Chromosomes Cancer 46 1–9. 10.1002/gcc.20384 [DOI] [PubMed] [Google Scholar]
- Koch A., De Meyer T., Jeschke J., Van Criekinge W. (2015). MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics 16:636. 10.1186/s12864-015-1847-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lánczky A., Nagy Á., Bottai G., Munkácsy G., Szabó A., Santarpia L., et al. (2016). miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res. Treat. 160 439–446. 10.1007/s10549-016-4013-7 [DOI] [PubMed] [Google Scholar]
- Lara-Lemus R. (2019). On the role of myelin and lymphocyte protein (MAL) in cancer: a puzzle with two faces. J. Cancer 10 2312–2318. 10.7150/jca.30376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Liu K., Li L., Yang M., Gao W., Feng J., et al. (2011). Reduced CMTM5 expression correlates with carcinogenesis in human epithelial ovarian cancer. Int. J. Gynecol. Cancer 21 1248–1255. 10.1097/IGC.0b013e3182259c31 [DOI] [PubMed] [Google Scholar]
- Li T., Cheng Y., Wang P., Wang W., Hu F., Mo X., et al. (2015). CMTM4 is frequently downregulated and functions as a tumour suppressor in clear cell renal cellcarcinoma. J. Exp. Clin. Cancer Res. 34:122. 10.1186/s13046-015-0236-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Su Y., Li T., Yuan W., Mo X., Li H., et al. (2015). CMTM7 knockdown increases tumorigenicity of human non-small cell lung cancer cells and EGFR-AKT signaling by reducing Rab5 activation. Oncotarget 6 41092–41107. 10.18632/oncotarget.5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q., Su Y., Jiang G. C., Zhou Z. L., Liu B. C., Bu L., et al. (2013). Change of CMTM7 expression, a potential tumor suppressor, is associated with poor clinical outcome in human non-small cell lung cancer. Chin. Med. J. 126 3006–3012. [PubMed] [Google Scholar]
- Lu J., Wu Q. Q., Zhou Y. B., Zhang K. H., Pang X. B., Li L., et al. (2016). Cancer research advances regarding the CKLF-like MARVEL transmembrane domain containing family. Asian Pac. J. Cancer Prevent. 17 2741–2744. 10.7150/ijbs.33733 [DOI] [PubMed] [Google Scholar]
- Merino Bonilla J. A., Torres Tabanera M., Ros Mendoza L. H. (2017). Breast cancer in the 21st century: from early detection to new therapies. [Article in English. Spanish]. Radiologia 59 368–379. 10.1016/j.rx.2017.06.003 [DOI] [PubMed] [Google Scholar]
- Mermel C. H., Schumacher S. E., Hill B., Meyerson M. L., Beroukhim R., Getz G. (2011). GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 12:R41. 10.1186/gb-2011-12-4-r41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mezzadra R., Sun C., Jae L. T., Gomez-Eerland R., de Vries E., Wu W., et al. (2017). Identification of CMTM6 and CMTM4 as PD-L1 protein regulators. Nature 549 106–110. 10.1038/nature23669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu G., Wright K. L., Huang M., Song L., Haura E., Turkson J., et al. (2002). Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene 21 2000–2008. 10.1038/sj.onc.1205260 [DOI] [PubMed] [Google Scholar]
- Palacios-Arreola M. I., Nava-Castro K. E., Castro J. I., García-Zepeda E., Carrero J. C., Morales-Montor J. (2014). The role of chemokines in breast cancer pathology and its possible use as therapeutic targets. J. Immunol. Res. 2014 849720. 10.1155/2014/849720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D. R., Yu J., Shanker K., Deshpande N., Varambally R., Ghosh D., et al. (2004). ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia 6 1–6. 10.1016/s1476-5586(04)80047-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussos E. T., Condeelis J. S., Patsialou A. (2011). Chemotaxis in cancer. Nat. Rev. Cancer 11 573–587. 10.1038/nrc3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu R., Rivenbark A. G., Coleman W. B. (2012). Enhancement of chemotherapeutic efficacy in hypermethylator breast cancer cells through targeted and pharmacologic inhibition of DNMT3b. Breast Cancer Res. Treat. 131 385–399. 10.1007/s10549-011-1409-2 [DOI] [PubMed] [Google Scholar]
- Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. (2011). The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 1813 878–888. 10.1016/j.bbamcr.2011.01.034 [DOI] [PubMed] [Google Scholar]
- Scheller J., Grötzinger J., Rose-John S. (2006). Updating interleukin-6 classic and trans-signaling. Signal. Transduct. 6 240–259. 10.1002/sita.200600086 [DOI] [Google Scholar]
- Shao L., Cui Y., Li H., Liu Y., Zhao H., Wang Y., et al. (2007). CMTM5 exhibits tumor suppressor activities and is frequently silenced by methylation in carcinoma cell lines. Clin. Cancer Res. 13 5756–5762. 10.1158/1078-0432.ccr-06-3082 [DOI] [PubMed] [Google Scholar]
- Singh R., Kapur N., Mir H., Singh N., Lillard J. W., Jr., Singh S. (2016). CXCR6-CXCL16 axis promotes prostate cancer by mediating cytoskeleton rearrangement via Ezrin activation and αvβ3 integrin clustering. Oncotarget 7 7343–7353. 10.18632/oncotarget.6944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R., Lillard J. W., Jr., Singh S. (2011). Chemokines: key players in cancer progression and metastasis. Front. Biosci. 3:1569–1582. 10.2741/s246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D., Franceschini A., Wyder S., Forslund K., Heller D., Huerta-Cepas J., et al. (2015). STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43 D447–D452. 10.1093/nar/gku1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawara K., Scott H., Emathinger J., Ide A., Fox R., Greiner D., et al. (2019). Co-expression of VEGF and IL-6 family cytokines is associated with decreased survival in HER2 negative breast cancer patients: subtype-specific IL-6 family cytokine-mediated VEGF secretion. Transl. Oncol. 12 245–255. 10.1016/j.tranon.2018.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turashvili G., Brogi E. (2017). Tumor heterogeneity in breast cancer. Front. Med. 4:227. 10.3389/fmed.2017.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untch M., Thomssen C., Bauerfeind I., Braun M., Brucker S. Y., Felberbaum R., et al. (2019). Primary therapy of early breast cancer: evidence, controversies, consensus: spectrum of opinion of German specialists on the 16th St. Gallen international breast cancer conference (Vienna 2019). Geburtshilfe Frauenheilkd 79 591–604. 10.1055/a-0897-6457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. J., Saadi W., Lin F., Nguyen C. M. C., Jeon N. L. (2004). Differential effects of EGF gradient profiles on MDAMB- 231 breast cancer cell chemotaxis. Exp. Cell Res. 300 180–189. 10.1016/j.yexcr.2004.06.030 [DOI] [PubMed] [Google Scholar]
- Xiao Y., Yuan Y., Zhang Y., Li J., Liu Z., Zhang X., et al. (2015). CMTM5 is reduced in prostate cancer and inhibits cancer cell growth in vitro and in vivo. Clin. Transl. Oncol. 17 431–437. 10.1007/s12094-014-1253-z [DOI] [PubMed] [Google Scholar]
- Xie J., Yuan Y., Liu Z., Xiao Y., Zhang X., Qin C., et al. (2014). CMTM3 is frequently reduced in clear cell renal cell carcinoma and exhibits tumor suppressor activities. Clin. Transl. Oncol. 16 402–409. 10.1007/s12094-013-1092-3 [DOI] [PubMed] [Google Scholar]
- Yamaguchi H., Condeelis J. (2007). Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 1773 642–652. 10.1016/j.bbamcr.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Nan X., Li X., Chen Y., Zhang J., Sun L., et al. (2014). CMTM5 exhibits tumor suppressor activity through promoter methylation in oral squamous cell carcinoma. Biochem. Biophys. Res. Commun. 447 304–310. 10.1016/j.bbrc.2014.03.158 [DOI] [PubMed] [Google Scholar]
- Zhang S., Pei X., Hu H., Zhang W., Mo X., Song Q., et al. (2016). Functional characterization of the tumor suppressor CMTM8 and its association with prognosis in bladder cancer. Tumour Biol. 37 6217–6225. 10.1007/s13277-015-4508-6 [DOI] [PubMed] [Google Scholar]
- Zhu X., Qi G., Li C., Bei C., Tan C., Zhang Y., et al. (2019). Expression and clinical significance of CMTM6 in hepatocellular carcinoma. DNA Cell Biol. 38 193–197. 10.1089/dna.2018.4513 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: www.cbioportal.org, http://www.oncomine.org, http://ualcan.path.uab.edu, http://kmplot.com/analysis/index.php?p=service&cancer=breast, http://string-db.org, and https://www.proteinatlas.org.