Abstract
The study reports 147 full-length WRKY genes based on the transcriptome analysis of Glycyrrhiza genus (G. glabra and G. uralensis). Additional motifs in G. glabra included DivIVA (GgWRKY20) and SerS Superfamily (GgWRKY21) at the C-terminal, and Coat family motifs (GgWRKY55) at the N-terminal of the proteins, while Exo70 exo cyst complex subunit of 338 amino acid (GuWRKY9) was present at the N-terminal of G. uralensis only. Plant Zn cluster super-family domain (17 WRKYs) and bZIP domain (2 WRKYs) were common between the two species. Based on the number of WRKY domains, sequence alignment and phylogenesis, the study identified GuWRKY27 comprising of 3 WRKY domains in G. uralensis and a new subgroup-IIf (10 members), having novel zinc finger pattern (C-X4-C-X22-HXH) in G. glabra. Multiple WRKY binding domains (1–11) were identified in the promoter regions of the GgWRKY genes indicating strong interacting network between the WRKY proteins. Tissue-specific expression of 25 GgWRKYs, under normal and treated conditions, revealed 11 of the 18 induction factor triggered response corroborating to response observed in AtWRKYs. The study identified auxin-responsive GgWRKY 55 & GgWRKY38; GA3 responsive GgWRKYs15&59 in roots and GgWRKYs8, 20, 38, 57 &58 in the shoots of the treated plant. GgWRKYs induced under various stresses included GgWRKY33 (cold), GgWRKY4 (senescence), GgWRKYs2, 28 & 33 (salinity) and GgWRKY40 (wounding). Overall, 23 GgWRKYs responded to abiotic stress, and 17 WRKYs were induced by hormonal signals. Of them 13 WRKYs responded to both suggesting inter-connection between hormone signalling and stress response. The present study will help in understanding the transcriptional reprogramming, protein-protein interaction and cross-regulation during stress and other physiological processes in the plant.
Subject terms: Plant molecular biology, Abiotic
Introduction
The complexity in plant cell organization can be directly related to intricate inter-connections between genes and regulatory network inside the cell. This observation is further substantiated by studies on vast genomic and transcriptomic sequence information available in the public domain1. The inter-cellular biological circuit in higher plants is governed at several discreet levels, one of them is regulated by a specific group of DNA binding proteins, the WRKYs2 are among the ten largest families of transcription factors (TFs) in higher plants. The literature cites several papers after the first report on Ipomea batata (SPF1) in 19943, from dicots4, monocots5, orchids6 to unicellular eukaryote (Giardia lamblia) and the slime mold (Dictyostelium discoideum), revealing their evolutionary significance and complex organization7,8. The 60 amino-acid characteristic conserved sequence of WRKY transcription factor (TFs) are most commonly identified by specific hepta-nucleotide signature sequence (WRKYGQK), the W-Box, which binds to the promoter sequence of target gene(s) modulating its activity9. The large WRKY super-family is phylogenetically classified into three groups (I, II &III) based on the number of WRKY domains and type of zinc finger sequences at the C-terminal. WRKY proteins classified in group I is characterized by two WRKY domains and zinc-finger motifs (C2H2), while group II and III WRKY proteins constitute single WRKY domain. Zinc-finger motif in group II & III comprise of C2H2 and C2HC zinc-finger pattern, respectively10. Studies have shown WRKY binding motifs (W-boxes) are present in multiple numbers in WRKY responsive gene promoters11. The promoters of 83% genes of the 72 WRKYs in Arabidopsis, contain at least two perfect W-boxes (TTGACC/T), and 58% had four or more core element sequence (TTGAC)11. Some WRKYs had 11 to 12 (AtWRKY66, AtWRKY17) core elements in the promoter fragment as analysed by Dong et al.12. Interestingly, studies confirm the presence of W-boxes also in the promoter region of WRKY genes, suggesting a potentially strong transcriptional networking between WRKY proteins11. Studies using co-transfection assays have revealed role of WRKY proteins on the promoters of their own genes and on other WRKY genes thereby modulating reporter gene13. Also in-vitro DNA-protein binding assays have highlighted single WRKY binding to several target gene promoters as elucidated in WRKY53 binding to three different WRKY genes, confirming complex interactive regulatory network. Microarray experiments using Arabidopsis genome illustrated more than 70% (45 out of 61) of the WRKY genes are co-regulated with other WRKYs14 and transcription factors12. Biological role of WRKYs are being studied in several plants15. They have been found to regulate several target genes in response to stress16 including metal stress17, development18 and secondary metabolite biosynthesis1. WRKYs have shown regulatory role in pathogen-induced response12 resulting in concerted activation of variety of genes. WRKY TFs have been found to rapidly and transiently regulate gene induction in response to signalling molecule19, wounding, stress, physiological processes like flowering20, seed germination and development21 and senescence4. Expressed Sequence Tags (ESTs) and other plant database have revealed presence of several hundred WRKYs in various tissues under different physiology, stress18, cold22, stomatal movement23 and defense24,25 implying their predominant role in varied biological functions. However, under normal growth conditions also, WRKY proteins have demonstrated broad-spectrum regulatory role as reported in morphogenesis and development of trichomes26 embryo development18, senescence13, dormancy27, plant growth28, immunity29, systematic acquired resistant and metabolic pathways30.
Two decades of studies on WRKY TFs has resulted in more than 14500 WRKY genes from 165 plant species31 with most of the species from eudicots (100 species) followed by monocots (38 species) and chlorophytae (16 species)31. Legumes with 12 species contributed to 1094 WRKY genes32. No report on WRKY transcription factors has been published from Glycyrrhiza species, though transcriptome, genome and EST databases are available in public domain from G. uralensis.
Glycyrrhiza belongs to Fabaceae sub-family of Leguminoseae family. The underground roots (Licorice) of the genus (G. uralensis, G. glabra and G. echinata) are commercially valued for its pharmaceutical, flavour enhancer natural sweetener, and cosmaceutical properties33. Roots of the plant are rich in bioactive flavonoids and tri-terpenoid saponins including glycyrrhizin34. Glycyrrhizin molecule is pharmaceutically sought molecule for its multitude of bioactivities33. The global demand of the roots of Glycyrrhiza is evident by a market report, as per Transparency Market report (ALBANY, New York, April 4, 2017 /PRNewswire). Where projected compound annual growth rate was estimated to be 5.7% during 2017–2025 equivalent to USD 2,393.9 million by 2025.
Present research underlines the transcriptome-wide identification and characterization of 147 WRKY TFs from Glycyrrhiza genus. Here, we analysed 87 WRKY genes from G. glabra and 60 from G. uralensis, categorized them into different structural groups based on conserved motif composition. We also predicted functions based on STRING prediction algorithm in G. glabra WRKY members. Subsequently their expression profiles were investigated under various stress conditions in the aerial tissues of the in vitro cultured plant. We also characterized 31 promoters (between 0.5 kb to 4.1 kb) of the 87 GgWRKY genes (from the transcriptomic data) to get an insight into its functioning and regulation of secondary metabolites.
Results and Discussion
Transcriptome-wide analysis and characterization of Glycyrrhiza WRKY TF
We have done the transcriptomics of G. glabra plant and mined the data for the WRKY transcription factor. Among the 125 sequences that matched WRKY genes on BLAST and PF03106 HMM profile searches, 87 GgWRKYs had complete CDS, and 38 gene sequences were partial (Table 1). All of these were revalidated using Uniprot (https://www.uniprot.org/) resulting in 78 sequences with best hits, while 47 sequences were found unique. Out of these, 55 (UniProt hits) and 32 (unique) sequences were full length, and 23 (UniProt hits) and 15 (unique) were partial sequences (Table 1). Further, we used the publicly available G. uralensis transcriptome data as a reference source (http://ngs-data-archive.psc.riken.jp/Gur-genome/download.pl.) to retrieve the WRKY transcription factor using BLAST and PF03106 HMM profile searches, we could identify 60 WRKY genes from G. uralensis. Subsequently, all the full-length protein sequences (147) were re-examined for the presence of WRKY domains using conserved domain database (https://www.ncbi.nlm.nih.gov/cdd/) and through HMMScan (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). The GgWRKY sequences were submitted to NCBI, and their accession numbers are given in (Supplementary File 1). The identified GuWRKY protein sequences were included in the sequence alignment and phylogenetic studies only (Supplementary File 2). The detailed GgWRKY protein sequence features are listed in Table 2. The deduced GgWRKY proteins had amino acid residues between 112 (GgWRKY67) to 760 (GgWRKY12). The coding sequences of 87 full-length GgWRKYs ranged from 339 bp (GgWRKY67) to 2283 bp (GgWRKY12), and their molecular weight (MW) varied between 13291.91 Da (GgWRKY67) to 82181.16 Da (GgWRKY12) (Table 3). The isoelectric point (pI) of 44 GgWRKYs were acidic, one (GgWRKY55) was neutral with pI value equal to 7.0, and the remaining 42 were basic proteins. According to the instability index proteins with index value higher than 40.0 is unstable35. In the present study most of the GgWRKYs were found to be unstable, having maximum instability index of 68.68 (GgWRKY34) with the exception of ten GgWRKYs namely, GgWRKY10 (30.20), GgWRKY16 (38.82), GgWRKY48 (39.86), GgWRKY50 (33.92), GgWRKY60 (39.40), GgWRKY73 (33.70), GgWRKY80 (39.40), GgWRKY83 (37.16), GgWRKY84 (32), GgWRKY86 (35.94) (Table 3). Additionally, the WoLFPSORT prediction showed that 81 GgWRKY proteins were localised in nucleus, suggesting that they play regulatory role predominantly in cell nucleus, while 4 GgWRKYs (-23, 32, 75, 84) had chloroplast orientation. GgWRKY73 had mitochondrial and GgWRKY 86 had cytoplasmic subcellular localization (Table 3). Further, five GgWRKY members (GgWRKYs 10,-33,-67,-68,-87) had WRKYGKK domain instead of the common WRKYGQK (Table 2). Earlier studies have also reported replacement of Q by K as common variant. Rice WRKYs have shown 19 variants, where the characteristic WRKY is substitution by WRRY,WSKY,WKKY, WVKY or WKKY motifs5.
Table 1.
Sequence information of WRKY genes in G. glabra.
| Sequence type | Uniprot matches | Unique | Total |
|---|---|---|---|
| Full length sequences | 55 | 32 | 87 |
| Partial sequences | 23 | 15 | 38 |
| Total | 78 | 47 | 125 |
Table 2.
Sequence features of WRKY genes in G. glabra.
| GgWRKYs | Gene ID | CDS (bp) | ORF (aa) | group | Conserved motif | Domain pattern | Zinc finger |
|---|---|---|---|---|---|---|---|
| GgWRKY1 | MK511239 | 1563 | 520 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY2 | MK511240 | 1155 | 384 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY3 | MK511241 | 804 | 267 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY4 | MK511242 | 840 | 279 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY5 | MK511243 | 762 | 253 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY6 | MK511244 | 603 | 200 | 2f | WRKYGQK | C-X4-C-X22-HNH | C2H2 |
| GgWRKY7 | MK511245 | 1767 | 588 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY8 | MK511246 | 1128 | 375 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY9 | MK511247 | 1440 | 479 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY10 | MK511248 | 507 | 168 | 2c | WRKYGKK | C-X4-C-X23-HXH | C2H2 |
| GgWRKY11 | MK511249 | 1104 | 367 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY12 | MK511250 | 2283 | 760 | 1 | 2(WRKYGQK) | C-X4-C-X23-HNH | C2H2 |
| GgWRKY13 | MK511251 | 1104 | 367 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY14 | MK511252 | 1266 | 421 | 2f | WRKYGQK | C-X4-C-X22-HXH | C2H2 |
| GgWRKY15 | MK511253 | 1305 | 434 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY16 | MK511254 | 1206 | 401 | 1 | 2(WRKYGQK) | C-X4-C-X23-HNH | C2H2 |
| GgWRKY17 | MK511255 | 669 | 222 | 2f | WRKYGQK | C-X4-C-X22-HXH | C2H2 |
| GgWRKY18 | MK511256 | 1608 | 535 | 1 | 2(WRKYGQK) | C-X4-C-X22-HNH | C2H2 |
| GgWRKY19 | MK511257 | 1527 | 507 | 1 | 2(WRKYGQK) | C-X4-C-X23-HXH | C2H2 |
| GgWRKY20 | MK511258 | 1974 | 657 | 2b | WRKYGQK | C-X5-C-X23-HXH | C2H2 |
| GgWRKY21 | MK511259 | 1743 | 580 | 2b | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY22 | MK511260 | 1035 | 344 | 1 | 2(WRKYGQK) | C-X4-C-X22-HXH | C2H2 |
| GgWRKY23 | MK511261 | 723 | 240 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| GgWRKY24 | MK511262 | 777 | 258 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY25 | MK511263 | 624 | 207 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY26 | MK511264 | 624 | 207 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY27 | MK511265 | 1617 | 538 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY28 | MK511266 | 612 | 203 | 2f | WRKYGQK | C-X4-C-X22-HXH | C2H2 |
| GgWRKY29 | MK511267 | 1143 | 380 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY30 | MK511268 | 1068 | 355 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| GgWRKY31 | MK511269 | 1437 | 478 | 1 | 2(WRKYGQK) | C-X4-C-X22-HXH | C2H2 |
| GgWRKY32 | MK511270 | 495 | 165 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| GgWRKY33 | MK511271 | 498 | 165 | 2c | WRKYGKK | C-X4-C-X23-HXH | C2H2 |
| GgWRKY34 | MK511272 | 966 | 321 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| GgWRKY35 | MK511273 | 1029 | 342 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY36 | MK511274 | 579 | 192 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY37 | MK511275 | 717 | 238 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY38 | MK511276 | 1158 | 385 | 2b | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY39 | MK511277 | 1881 | 626 | 2b | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY40 | MK511278 | 789 | 262 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY41 | MK511279 | 1248 | 415 | 1 | 2(WRKYGQK) | C-X4-C-X22-HXH | C2H2 |
| GgWRKY42 | MK511280 | 1029 | 342 | 2f | WRKYGQK | C-X4-C-X22-HXH | C2H2 |
| GgWRKY43 | MK511281 | 1149 | 382 | 1 | 2(WRKYGQK) | C-X4-C-X22-HXH | C2H2 |
| GgWRKY44 | MK511282 | 1296 | 431 | 2b | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY45 | MK511283 | 786 | 261 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY46 | MK511284 | 1356 | 451 | 1 | 2(WRKYGQK) | C-X4-C-X22-HXH | C2H2 |
| GgWRKY47 | MK511285 | 1530 | 509 | 1 | 2(WRKYGQK) | C-X4-C-X22-HXH | C2H2 |
| GgWRKY48 | MK511286 | 594 | 197 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY49 | MK511287 | 789 | 262 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY50 | MK511288 | 468 | 155 | 2f | WRKYGQK | C-X4-C-X22-HXH | C2H2 |
| GgWRKY51 | MK511289 | 894 | 297 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| GgWRKY52 | MK511290 | 732 | 243 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY53 | MK511291 | 945 | 314 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY54 | MK511292 | 867 | 288 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY55 | MK511293 | 882 | 293 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY56 | MK511294 | 750 | 248 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY57 | MK511295 | 783 | 260 | 2f | WRKYGQK | C-X4-C-X22-HNH | C2H2 |
| GgWRKY58 | MK511296 | 936 | 311 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| GgWRKY59 | MK511297 | 1014 | 337 | 2f | WRKYGQK | C-X4-C-X22-HXH | C2H2 |
| GgWRKY60 | MK511298 | 651 | 216 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY61 | MK511299 | 732 | 243 | 2a | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY62 | MK511300 | 429 | 142 | 2a | WRKYGQK | C-X5-C-X13-HN | C2H2 |
| GgWRKY63 | MK511301 | 1092 | 363 | 2d | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY64 | MK511302 | 1050 | 349 | 2d | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY65 | MK511303 | 1065 | 354 | 2e | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY66 | MK511304 | 1047 | 348 | 2d | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY67 | MK511305 | 339 | 112 | 2c | WRKYGKK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY68 | MK511306 | 588 | 195 | 2c | WRKYGKK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY69 | MK511307 | 933 | 310 | 2d | WRKYGQK | Zinc cluster | |
| GgWRKY70 | MK511308 | 981 | 326 | 2d | WRKYGQK | Zinc cluster | |
| GgWRKY71 | MK511309 | 1071 | 356 | 2d | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY72 | MK511310 | 957 | 318 | 2d | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY73 | MK511311 | 387 | 128 | 2d | WRKYGQK | C-X5-C-X23-HNH | C2H2 |
| GgWRKY74 | MK511312 | 1089 | 362 | 2f | WRKYGQK | C-X4-C-X22-HNH | C2H2 |
| GgWRKY75 | MK511313 | 690 | 229 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY76 | MK511314 | 1005 | 334 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY77 | MK511315 | 1068 | 355 | 3 | WRKYGQK | C-X7-C-X23-HXC | C2HC |
| GgWRKY78 | MK511316 | 723 | 240 | 2f | WRKYGQK | C-X4-C-X22-HNH | C2H2 |
| GgWRKY79 | MK511317 | 690 | 229 | 2c | WRKYGQK | C-X4-C-X23-HXH | C2H2 |
| GgWRKY80 | MK511318 | 651 | 216 | 2c | WRKYGQK | C-X4-C-X23-HNH | C2H2 |
| GgWRKY81 | MN625734 | 912 | 303 | 2d | WRKYGQK | Zinc cluster | |
| GgWRKY82 | MN625735 | 780 | 259 | 2d | WRKYGQK | Zinc cluster | |
| GgWRKY83 | MK511319 | 591 | 196 | NG | WRKYGQK | ||
| GgWRKY84 | MK511320 | 531 | 176 | NG | WRKYGQK | ||
| GgWRKY85 | MN625736 | 654 | 217 | NG | WRKYGQK | ||
| GgWRKY86 | MN625737 | 552 | 183 | NG | WRKYGQK | ||
| GgWRKY87 | MN625738 | 426 | 141 | NG | WRKYGKK |
Table 3.
Physical parameters of GgWRKY genes.
| GgWRKYs | pI | Mw(Da) | Instability Index | Aliphatic index | GRAVY | Subcellular localization |
|---|---|---|---|---|---|---|
| GgWRKY1 | 6.64 | 56446.57 | 61.13 | 58.15 | −0.760 | Nucleus |
| GgWRKY2 | 6.61 | 42812.25 | 57.19 | 57.60 | −1.054 | Nucleus |
| GgWRKY3 | 4.87 | 30524.69 | 67.49 | 59.14 | −1.006 | Nucleus |
| GgWRKY4 | 6.55 | 31717.61 | 41.31 | 63.19 | −0.743 | Nucleus |
| GgWRKY5 | 6.51 | 28190.94 | 53.76 | 48.50 | −1.096 | Nucleus |
| GgWRKY6 | 6.17 | 22124.23 | 61.94 | 44.85 | −1.109 | Nucleus |
| GgWRKY7 | 7.18 | 64879.58 | 59.87 | 41.12 | −0.988 | Nucleus |
| GgWRKY8 | 8.89 | 41166.98 | 53.56 | 46.53 | −0.984 | Nucleus |
| GgWRKY9 | 8.63 | 52606.42 | 52.87 | 52.55 | −0.903 | Nucleus |
| GgWRKY10 | 5.30 | 19325.09 | 30.20 | 43.99 | −1.340 | Nucleus |
| GgWRKY11 | 5.91 | 39455.37 | 50.96 | 49.92 | −0.843 | Nucleus |
| GgWRKY12 | 5.74 | 82181.16 | 54.43 | 52.75 | −0.836 | Nucleus |
| GgWRKY13 | 5.91 | 39455.37 | 50.96 | 49.92 | −0.843 | Nucleus |
| GgWRKY14 | 5.63 | 45531.95 | 51.96 | 55.39 | −0.773 | Nucleus |
| GgWRKY15 | 6.12 | 48273.38 | 66.44 | 38.82 | −1.086 | Nucleus |
| GgWRKY16 | 7.70 | 44055.51 | 38.82 | 55.39 | −0.859 | Nucleus |
| GgWRKY17 | 5.52 | 24792.96 | 45.33 | 50.05 | −1.027 | Nucleus |
| GgWRKY18 | 5.81 | 58714.96 | 51.32 | 61.23 | −0.758 | Nucleus |
| GgWRKY19 | 6.34 | 56060.64 | 52.16 | 59.04 | −1.002 | Nucleus |
| GgWRKY20 | 6.45 | 70757.15 | 58.09 | 61.11 | −0.700 | Nucleus |
| GgWRKY21 | 6.86 | 63675.28 | 53.72 | 64.60 | −0.622 | Nucleus |
| GgWRKY22 | 8.10 | 38457.09 | 52.91 | 70.20 | −0.784 | Nucleus |
| GgWRKY23 | 5.81 | 26383.71 | 47.29 | 48.25 | −1.033 | Chloroplast |
| GgWRKY24 | 5.51 | 28633.96 | 61.55 | 58.22 | −0.728 | Nucleus |
| GgWRKY25 | 5.73 | 23136.95 | 55.89 | 59.32 | −0.679 | Nucleus |
| GgWRKY26 | 5.72 | 23236.34 | 54.16 | 65.89 | −0.515 | Nucleus |
| GgWRKY27 | 6.01 | 59133.88 | 60.52 | 54.94 | −0.827 | Nucleus |
| GgWRKY28 | 6.86 | 22645.0 | 61.66 | 65.27 | −1.052 | Nucleus |
| GgWRKY29 | 8.22 | 41362.95 | 60.27 | 59.34 | −0.720 | Nucleus |
| GgWRKY30 | 5.32 | 40108.97 | 49.44 | 63.18 | −0.690 | Nucleus |
| GgWRKY31 | 6.88 | 52267.98 | 57.49 | 55.46 | −0.992 | Nucleus |
| GgWRKY32 | 5.96 | 18879.89 | 55.37 | 53.15 | −1.062 | Chloroplast |
| GgWRKY33 | 6.30 | 18917.71 | 60.53 | 48.97 | −1.024 | Nucleus |
| GgWRKY34 | 5.44 | 36176.01 | 68.68 | 69.84 | −0.771 | Nucleus |
| GgWRKY35 | 6.23 | 38723.22 | 62.50 | 48.22 | −1.141 | Nucleus |
| GgWRKY36 | 9.60 | 22200.15 | 58.51 | 60.94 | −0.886 | Nucleus |
| GgWRKY37 | 6.25 | 26645.01 | 52.90 | 74.20 | −0.690 | Nucleus |
| GgWRKY38 | 8.72 | 40884.31 | 52.12 | 61.53 | −0.570 | Nucleus |
| GgWRKY39 | 5.56 | 67756.87 | 46.23 | 59.11 | −0.720 | Nucleus |
| GgWRKY40 | 6.32 | 29579.37 | 51.55 | 75.57 | −0.747 | Nucleus |
| GgWRKY41 | 7.63 | 45591.88 | 53.87 | 62.46 | −0.896 | Nucleus |
| GgWRKY42 | 6.48 | 37329.15 | 62.93 | 56.14 | −1.051 | Nucleus |
| GgWRKY43 | 9.09 | 41765.72 | 55.64 | 55.60 | −0.958 | Nucleus |
| GgWRKY44 | 7.99 | 47206.16 | 55.82 | 75.66 | −0.515 | Nucleus |
| GgWRKY45 | 5.42 | 28438.55 | 60.89 | 58.35 | −0.670 | Nucleus |
| GgWRKY46 | 7.22 | 49789.82 | 50.10 | 66.08 | −0.735 | Nucleus |
| GgWRKY47 | 7.28 | 55366.60 | 54.68 | 60.33 | −0.935 | Nucleus |
| GgWRKY48 | 8.96 | 21525.73 | 39.86 | 56.85 | −0.812 | Nucleus |
| GgWRKY49 | 6.75 | 28771.61 | 45.23 | 57.98 | −0.842 | Nucleus |
| GgWRKY50 | 7.77 | 17309.33 | 33.92 | 59.74 | −0.684 | Nucleus |
| GgWRKY51 | 7.09 | 32735.09 | 63.33 | 46.09 | −0.864 | Nucleus |
| GgWRKY52 | 9.23 | 27088.46 | 43.69 | 63.74 | −0.833 | Nucleus |
| GgWRKY53 | 8.70 | 34991.34 | 51.44 | 61.72 | −0.825 | Nucleus |
| GgWRKY54 | 8.98 | 31973.96 | 50.45 | 59.86 | −0.840 | Nucleus |
| GgWRKY55 | 7.00 | 32897.26 | 52.03 | 76.45 | −0.568 | Nucleus |
| GgWRKY56 | 8.88 | 27837.52 | 52.35 | 71.20 | −0.610 | Nucleus |
| GgWRKY57 | 5.69 | 29064.88 | 64.72 | 53.27 | −0.994 | Nucleus |
| GgWRKY58 | 6.06 | 34087.41 | 45.82 | 54.15 | −0.973 | Nucleus |
| GgWRKY59 | 7.16 | 35981.76 | 53.20 | 57.63 | −0.706 | Nucleus |
| GgWRKY60 | 9.13 | 23611.08 | 39.40 | 57.27 | −0.838 | Nucleus |
| GgWRKY61 | 9.23 | 27088.46 | 43.69 | 63.74 | −0.833 | Nucleus |
| GgWRKY62 | 9.65 | 16437.91 | 45.33 | 74.15 | −0.896 | Nucleus |
| GgWRKY63 | 9.70 | 40591.79 | 54.87 | 66.61 | −0.769 | Nucleus |
| GgWRKY64 | 9.68 | 38091.79 | 59.72 | 63.52 | −0.724 | Nucleus |
| GgWRKY65 | 5.58 | 38415.26 | 54.79 | 53.79 | −0.706 | Nucleus |
| GgWRKY66 | 9.73 | 39237.38 | 51.34 | 61.93 | −0.809 | Nucleus |
| GgWRKY67 | 9.61 | 13291.91 | 58.98 | 41.70 | −1.143 | Nucleus |
| GgWRKY68 | 6.23 | 22180.23 | 60.65 | 45.95 | −1.086 | Nucleus |
| GgWRKY69 | 9.86 | 34964.59 | 52.37 | 67.94 | −0.742 | Nucleus |
| GgWRKY70 | 9.89 | 36444.10 | 55.54 | 68.19 | −0.728 | Nucleus |
| GgWRKY71 | 9.24 | 38874.59 | 47.53 | 66.29 | −0.631 | Nucleus |
| GgWRKY72 | 9.91 | 34646.20 | 51.39 | 65.06 | −0.586 | Nucleus |
| GgWRKY73 | 9.96 | 14291.46 | 33.70 | 58.67 | −0.802 | Mitochondria |
| GgWRKY74 | 8.36 | 39382.65 | 55.22 | 53.40 | 0.858 | Nucleus |
| GgWRKY75 | 7.60 | 25909.53 | 42.02 | 51.05 | −0.895 | Chloroplast |
| GgWRKY76 | 6.27 | 37066.81 | 56.89 | 53.95 | −0.845 | Nucleus |
| GgWRKY77 | 5.45 | 40568.91 | 49.05 | 57.13 | −0.830 | Nucleus |
| GgWRKY78 | 4.93 | 26513.78 | 63.03 | 54.08 | −1.010 | Nucleus |
| GgWRKY79 | 8.31 | 26029.34 | 43.79 | 62.14 | −0.738 | Nucleus |
| GgWRKY80 | 9.13 | 23611.08 | 39.40 | 57.27 | −0.838 | Nucleus |
| GgWRKY81 | 9.96 | 33185.38 | 63.25 | 62.54 | −0.746 | Nucleus |
| GgWRKY82 | 10.05 | 28317.13 | 59.62 | 64.40 | −0.639 | Nucleus |
| GgWRKY83 | 9.11 | 22167.36 | 37.16 | 66.68 | −0.463 | Nucleus |
| GgWRKY84 | 7.73 | 19597.61 | 32 | 57.05 | −0.807 | Chloroplast |
| GgWRKY85 | 5.58 | 23670.73 | 64.14 | 37.83 | −1.053 | Nucleus |
| GgWRKY86 | 9.05 | 20003.87 | 35.94 | 76.72 | −0.551 | Cytoplasm |
| GgWRKY87 | 6.28 | 16102.53 | 55.86 | 43.55 | −1.207 | Nucleus |
Conserved domain in Glycyrrhiza WRKY members
Generally, similar domains in a protein impart similar function. Transcription factors gene families have a common conserved domain involved in DNA binding. All the 147 WRKYs (G. glabra & G. uralensis) had a distinctive hepta-peptide DNA binding sequence (WRKYG[Q/K]K), the identifying character of the WRKY family. In the present study, 28 WRKYs showed the presence of additional motif besides the WRKY domain (Figs. 1 and S1). GgWRKY55 possessed Coat family motif (30 amino acid residues) at the N-terminal while GgWRKY20 and GgWRKY21 had DivIVA super-family (63 amino acid residues) and SerS Superfamily motif (61 amino acid residues), respectively at the C-terminal. G. uralensis, on the other hand, had GuWRKY9 having Exo70 exo cyst complex subunit (338 amino acid) and Flac-arch super (GuWRKY26) at the N-terminal and PAT1 (GuWRKY23) and SGNH_hydrolase (GuWRKY60) at the C-terminal. However two motifs were common in both the species-Plant Zinc Cluster (26–40 amino acid) in 16 WRKYs and bZIP domain (42amino acid) in 2 WRKYs was present in both the species. Plant Zn cluster super-family domain was present in nine GgWRKYs (GgWRKYs63,64, 66, 69,70,71,72,73 and 86) and seven GuWRKYs (GuWRKYs1,13,18,29,35,49 and 56). All the domains simultaneously reported in the present study in both the species of Glycyrrhiza were reported individually in different plants in earlier studies. We have not come across any report mentioning DivIVA, SerS Superfamily, PAT1, Exo70 exo cyst complex subunit SGNH_hydrolase, Flac-arch super & Coat family protein in plant WRKY proteins. However, bZIP & plant zinc cluster have been reported from A. thaliana earlier36. The analysis of sequence motifs using MEME platform (http://meme.nbcr.net/meme/cgi-bin/meme.cgi)37 displaying common and unique motifs within the GgWRKY sequences are shown in Fig. 2.
Figure 1.
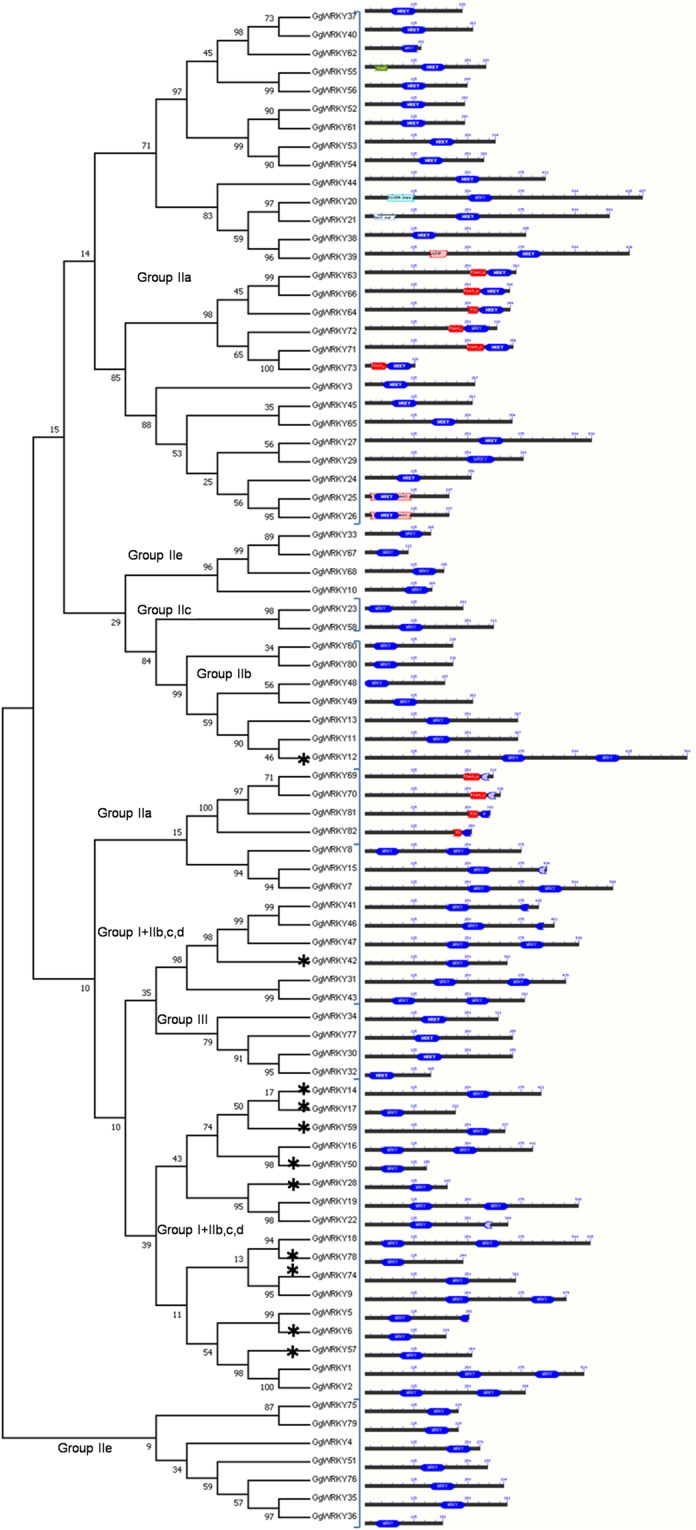
Classification of full-length GgWRKY amino acid sequences with different conserved domains (DivIVA, SerS, bZIP, Coat & Plant Zn cluster, WRKY). The conserved domains were investigated by CDD; * are exceptions in the classified groups and sub-groups in the phylogeny.
Figure 2.
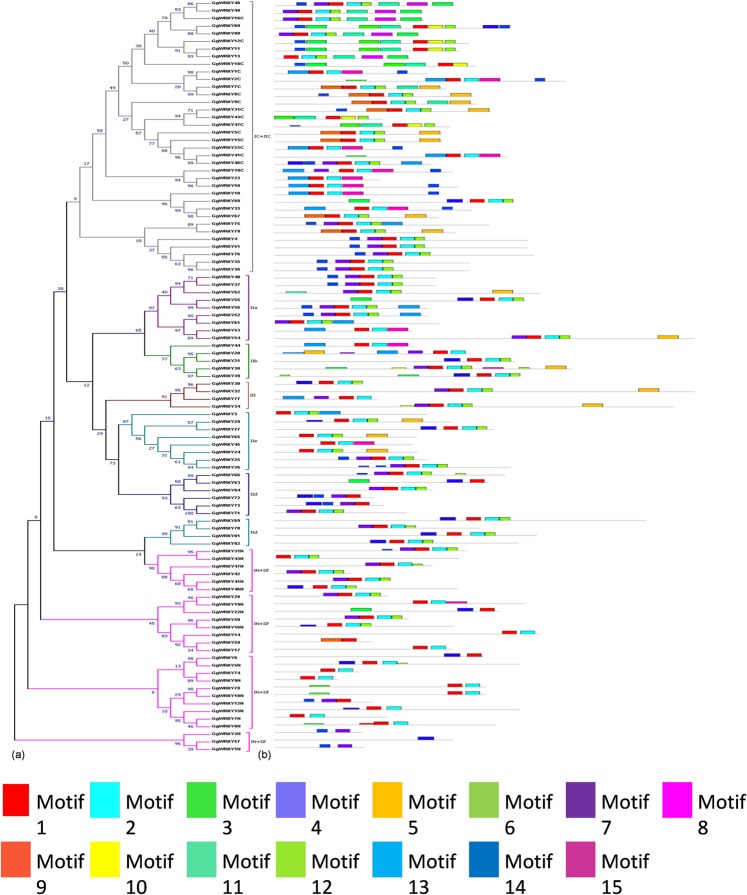
(a) Visualization of classification of 82 GgWRKY proteins. Conserved regions of GgWRKYs were used to construct the NJ phylogenetic tree with 1000 bootstrap value. (b) Architecture of 15 conserved protein motifs in GgWRKYs. Each motif is represented in different color (Motif 1–15). The conserved motifs were predicted by MEME program.
Phylogeny
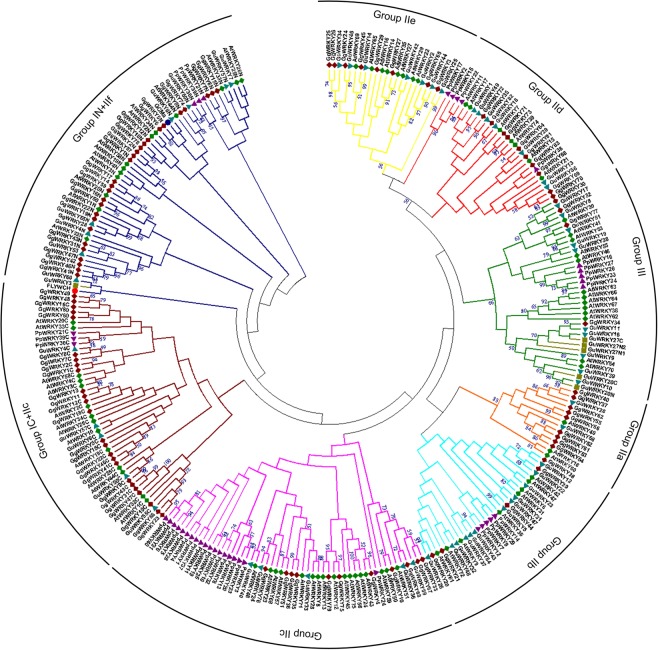
The relatedness among 136 Glycyrrhiza WRKY proteins with the 109WRKYs identified from Arabidopsis thaliana, Psychometrella patens, Human FLYWCH CRAa and GCMa were investigated (Fig. 3) and tabulated in Table 4. The phylogeny of 136 WRKY proteins from the genus Glycyrrhiza displayed 22WRKYs (17GgWRKYs & 5GuWRKYs) belonging to group-I, 98 WRKYs (61 GgWRKYS & 37 GuWRKYs) clustering in group-II and 16 WRKY members comprising of group-III (4GgWRKYS & 12GuWRKYs). Group-II was further sub-divided into five sub-groups, IIa (11), IIb (17), IIc (16 + 8), IId (17), IIe (15) and an additional novel sub-group IIf (14) based on WRKY transcription factor rules adopted in Arabidopsis9. The present paper reports few exceptions observed in the WRKY members identified in the genus Glycyrrhiza. The GuWRKY27 possessed three WRKY domains (N1, N2 &C). Few recent publications have also reported more than 2 WRKY domains in Gossypium raimondii38, Linum usitatissimum Lupinus angustifolius, Aquilegia coerulea and Setaria italic32. Phylogenetic analysis of the indicated proteins, however clubbed them into different subgroups. For example, in G. raimondii (WRKY108) the three domains (WRKY108N1, WRKY108N2 &WRKY108C) were clustered into IIc, III & IId sub-groups, respectively. In the present study, however, all the three WRKY domains (N1, N2 &C) of GuWRKY27 were found to be clustered into Group-III having Zn finger pattern similar to groupIII. This implies that the GuWRKY27 protein sequences are highly homologous to the group III WRKY member proteins, unlike the earlier published reports. Another exception was seen in GuWRKY20, where the protein was classified into group I based on the number of WRKY domains (2). However, it was clustered into group-III in the phylogenetic classification. MSA revealed that both the WRKY domains had Zn finger pattern similar to Group-III (C-X7-C-X23-HXC). The third exception was observed in GuWRKY3 whose Zn finger pattern was unlike any of the existing subgroups of group II. It could be the starting point for the evolution of a new subgroup in group II.
Figure 3.
Neighbour-Joining JTT model of phylogenetic tree comprising of 82 Glycyrrhiza glabra (maroon), 54 Glycyrrhiza uralensis (cyan blue), 70 Arabidopsis thaliana (dark green), 37 Psycometrella patens (violet), with GCMa (blue) and FLYWCH CRAa (red) WRKY domains. Suffix ‘N’ and ‘C’ indicates the N-terminal and the C-terminal of 60 amino acids WRKY domains of Group I.
Table 4.
Phylogenetic classification of WRKY domains identified from G. glabra, A. thaliana, P. patens and G. uralensis WRKY proteins.
| Group | Sub group | Gene number | GgWRKYs | AtWRKYs | PpWRKYs | GuWRKYs | |||
|---|---|---|---|---|---|---|---|---|---|
| GgWRKYs | AtWRKYs | PpWRKYs | GuWRKYs | ||||||
| I | IN | 17 | 13 | 3 | 5 |
41,46,47, 31,43,1,2,15,9,16, 19,22,18, 12,7,8,5 |
58,20,1, 32,3,4,19,44,2,34, 33,25,26 |
30,39,21 | 4,57,6,50,55 |
| IC | 17 | 13 | 3 | 5 |
19,41,46, 22,15,5, 47,31,43, 12,18,9,1,2,7,8,16 |
58,4,3,33,20,2,26, 34,19,25, 44,32,1 |
39,30,21 | 4,57,6,50,55 | |
| II | IIa | 9 | 3 | 0 | 2 |
53,54,61, 52,55,56, 62,37,40 |
40,60,18 | -------- | 58,28 |
| IIb | 5 | 8 | 5 | 12 |
20,21,44, 38,39 |
36,6,31, 42,47,61, 9,72 |
7,9,12, 14,29 |
54,43,5,12, 22,23,47,31,37,48,32,26 |
|
| IIc | 11 | 17 | 19 | 5 |
36,35,76, 51,79,75, 4,10,68, 67,33 |
45,75,43, 24,56,48, 57,23,68, 71,8,28, 13,12,50, 51,59 |
37,11,25, 10,3,23, 20,8,13, 32,1,4,19,31,40,28, 5,6,24 |
40,24,33,36,59 | |
| IId | 10 | 7 | 5 | 7 |
70,69,81, 82,66,63, 64,72,71, 73 |
11,17,15, 21,39,74, 7 |
15,22,17, 2,38 |
1,49,35,13, 56,29,18 |
|
| IIe | 8 | 8 | 0 | 7 |
27,29,3, 65,25,26, 24,45 |
65,69,29, 27,22,16, 14,35 |
-------- |
46,34,14,2, 44,25,42 |
|
| IC + IIc | 8 | 1 | 0 | 0 |
58,23,60, 80,48,49, 11,13 |
10 | --------- | --------- | |
| IN + IIf | 10 | 0 | 0 | 4 |
78,28,50, 14,59,17, 74,6,57,42 |
--------- | ------------ | 53,7,60,3 | |
| III | 4 | 13 | 5 | 12 |
34,30,32, 77 |
63,64,66, 67,38,62, 54,70,55, 46,30,41, 53 |
16,27,26, 33,34 |
9,16,11,39, 10,19,38,8, 17,51,20N, 20C,27N1, 27N2,27C |
|
| Total | 82 | 70 | 37 | 54 | |||||
It was further observed among the 82 GgWRKY proteins in the phylogenetic tree, Group IN (17 members) clubbed with ten GgWRKYs (GgWRKYs 59,-14,-17,-28,-50,-42,-6,-57,-74,-78) belonging to Group-II with unique Zn finger pattern (C-X4-C-X22-HXH) which was not reported earlier in this group. We propose a new subgroup-IIf based on the present findings which could be the initiation of divergence into a new sub-group maintaining the characteristic WRKY domain.
The phylogenetic analysis of the 60 amino acid region of the Glycyrrhiza WRKY proteins indicated their diverse origin. The N-terminal and the C-terminal of Group I of the WRKY proteins clustered them into different clades indicating their dissimilar background. Further, the majority of the subgroup IIc proteins (8 proteins) were found to assemble with group IC indicating their common origin with respective clusters. Contrary to our results, Zhu et al.39 found that subgroup IIc WRKY domain in Triticum aestivum, originated from the N-terminal WRKY domain of group I. However, recent study on legumes have revealed that IIc sub-groups have multiple origins32. The present study also showed gathering of sub-groups IIa & IIb, while sub-group IId + IIe were clustered with group III, signifying close relationship with members with respective groups. Previously Zhang & Wang8 proposed a phylogenetic tree based evolutionary relationships which classified the WRKY gene family into four clades including groups I + IIc, groups IIa + IIb, group IId, and group IIe. But according to Rinerson et al.40 hypothesis the WRKY protein evolution may have followed two alternative paths, “Group I Hypothesis” which proposed that all WRKY proteins evolved from the C-terminal WRKY domains of group I proteins, and the “ IIa + b Separate Hypothesis” which suggested that groups IIa and IIb have evolved directly from a single domain algal gene separated from a group I-derived lineage. It is hard to explain the origin of the WRKY gene family on the basis of any one hypothesis, as mounting number of studies have demonstrated their multiple origins. Based on our phylogenetic analyses, we found that a phylogenetic cluster was a mix of WRKY genes from at least two different groups or sub-group indicating their dynamic nature.
Further, the present study could identify eleven WRKY proteins (GgWRKY-83 to-87 & GuWRKY-15, -21, -30, -41, -45 &-52), not included in the phylogenetic analyses, that possessed WRKY domain but had truncated characteristic zinc finger motif. Earlier studies on Vitis venifera41 and rice42 had also shown loss of Zn finger motifs in WRKY proteins. The phylogenetic clustering was further examined at sequence level by multiple sequence alignment (MSA).
Multiple sequence alignment of the identified WRKY proteins
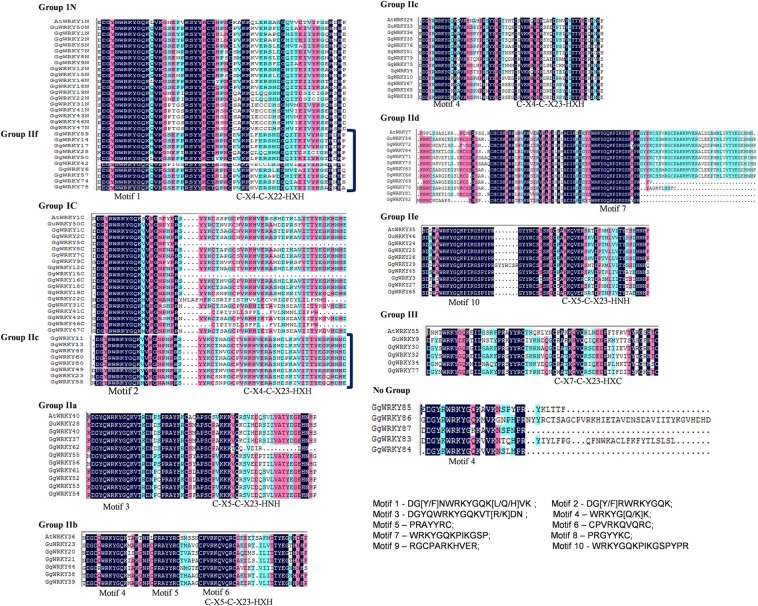
The multiple sequence alignment of 60 amino acids conserved region of all the 87 GgWRKY proteins were clustered in 9 different groups and sub-groups with very high homology (>70%) as shown in (Fig. 4). Group IN displayed conserved motif 1 (DG[Y/F]NWRKYGQK[L/Q/H]VK) and zinc finger pattern of C-X4-C-X22-HXH showing conservancy with 27 GgWRKY proteins, 17 of them belonged to Group IN and 10 GgWRKYs (59,-14,-17,-28,-50,-42,-6,-57,-74,-78) belonging to new sub-group IIf, having Zn finger motif (C-X4-C-X22-HXH) which was similar to Zn finger domain of group IN unlike group II members. While Group IC had 17 GgWRKYs belonging to group IC and 8 GgWRKYs from group IIc (GgWRKYs 11,-13,-48,-60,-80,-49,-23,-58). All the 25 GgWRKY members in Group IC displayed conserved motif 2 (DG[Y/F]RWRKYGQK), zinc finger pattern of C-X4-C-X23-HXH and high identity (70.4%) as shown in Fig. 4. The third group IIa had nine GgWRKY proteins (GgWRKYs40,-37,-62,-55,-56,-61,-52,-53,-54) displaying motif 3 (DGYQWRKYGQKVT[R/K] DN) and a zinc finger motif pattern of C-X5-C-X23-HNH having 86.2% identity, except GgWRKY62 which had C-X5-C-X13-HN Zn finger pattern. Group IIb had five GgWRKYs (GgWRKYs 20,-21,-44,-38,-39) with three conserved motifs, motif 4 (WRKYG[Q/K]K), motif 5 (PRAYYRC) and motif 6 (CPVRKQVQRC) with 85.8% identity, while Group IIc comprised of 11 sequences (GgWRKYs35,-36,-76,-51,-79,-75,-4,-10,-67,-68,-33) with conserved sequence motif 4 (WRKYG[Q/K]K) and a zinc finger motif pattern of C-X4-C-X23-HXH (77.6% identity). The 60 amino acid signature sequence of Group IId proteins comprising of 10 GgWRKYs, when aligned together showed only 32% identity (Fig. 5). Six of the ten members had zinc finger, while four (GgWRKYs 69,-70,-81,82) had no zinc finger present in them; instead they had 50 amino acid zinc cluster domain at the N terminal. Based on the conservancy, when this group was divided into two sub-groups IId1 (GgWRKYs 69,-70,-81,-82) & IId2 (GgWRKYs 72, -64,-71,-73,-63,-66,) the identity was significantly increased from 32% to 54.9% and 83.7%, respectively. The members clustered in group IId1 showed conserved motif 7(WRKYGQKPIKGSP) and no zinc finger at the C-terminal end, while subgroup IId2 displayed conservancy of three motifs- 7(WRKYGQKPIKGSP), 8(PRGYYKC) & 9(RGCPARKHVER) along with a common zinc finger pattern C-X5-C-X23-HNH (Fig. 5). Further, when conserved domain sequence of 60amino acid of all the 10 GgWRKYs of sub-group IId, was increased to 110 amino acids the two sub-groups (IId1&IId2) combined into a single group (IId) displaying 70.68% identity among all the members. We also confirmed the conservancy of each group and subgroup with the WRKY members belonging to A. thaliana and G. uralensis (Figs. 4 and 5). The MSA further proves the dynamic nature of GgWRKYs.
Figure 4.
Multiple sequence alignment (MSA) of conserved GgWRKY domain. The alignment was performed using Clustal W program and displayed using DNAMAN software. Conserved motifs (1–10) and type of zinc-finger pattern are indicated within groups or sub-groups. Blue color represents 100% sequence identity. Pink color is for more than 75% while cyan color is for less than 75% sequence identity.
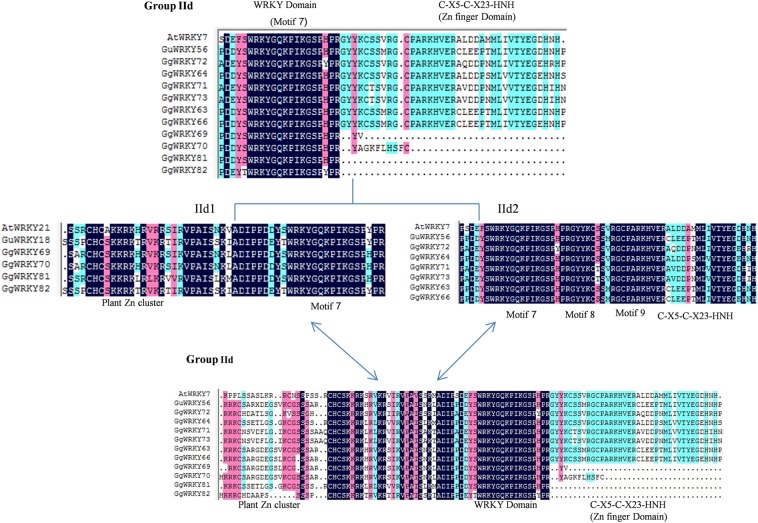
Figure 5.
Multiple sequence alignment (MSA) profile of group IId (10 sequences). Initially conserved 60 amino acids region is used to build alignment that showed low sequence identity (32%). When it was separated in two groups (IId1&IId2), identity increased significantly (54.9& 83.7%). Sub-group IId1 (4 sequences) with sequences upstream to WRKY domain having Plant Zinc cluster with motif 7and no zinc finger; subgroup IId2 (6 sequences) with 7, 8, 9 motifs and Zn finger. When four sequences of sub group IId1 were extended 50 amino acids towards N’terminal (total110 amino acid), sequence identity of sub groupIId increased to 70.68%.
Promoter analysis
The upstream region of 31 GgWRKY genes was examined for the presence of Cis-regulatory elements. Several stress-responsive elements like UV, salinity, ABA, GA signalling, etiolation, water stress, auxin and sulphur responsive elements were identified (Fig. 6). Also, several copies of WRKY binding motifs were identified in the promoter region of GgWRKY genes. The DNA binding WRKY motifs in the promoter region ranged from 1 (GgWRKY20, 23) to 11 (GgWRKYs 18 &62). Overall, twenty-seven GgWRKYs had three or more W-boxes in their promoter region. Observation revealed presence of multiple W-box elements mostly in the stress-related genes, which is following the earlier studies6,12. Additionally promoters of several glycyrrhizin biosynthesis genes (CYP88D6, CYP72A154 & squalene epoxidase) contained W boxes (unpublished data) suggesting regulatory role of WRKY in glycyrrhizin biosynthesis, thereby providing a platform to understand its regulation.
Figure 6.
Analysis of cis-regulatory elements (CREs) in GgWRKY promoter region. Total ten stress responsive elements were mapped on sense and anti-sense strand using RSAT tool.
Protein-protein interaction
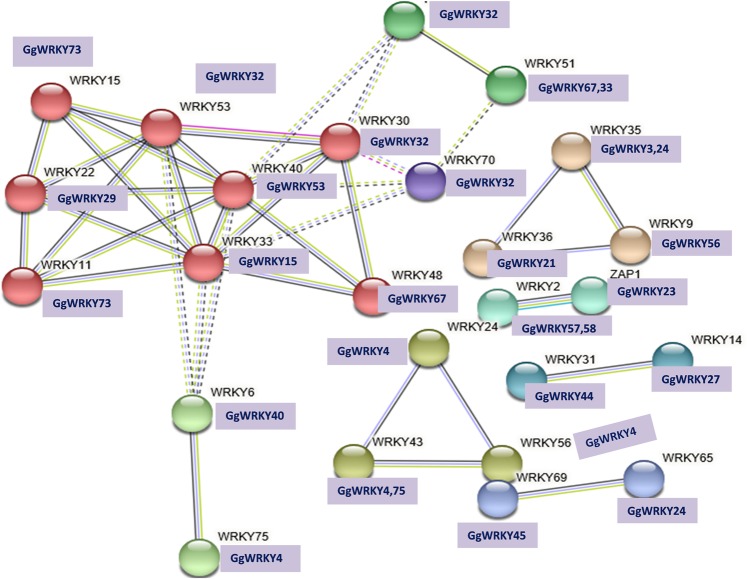
The protein-protein interaction of GgWRKYs was predicted by STRING43 with A. thaliana (taxonomic ID 3702) as a model using Markov clustering (MCL) having inflation factor of 8.5. The STRING software is a prediction pipeline for deducing protein-protein associations from co-expression data and interaction conservation (Fig. S2; Supplementary File 3). It predicts interaction between orthologs in taxonomically different organism. The corresponding GgWRKY orthologs selected had more than 60% protein sequence homology having one WRKY domain (PF03106) as predicted by Pfam and three domains (IPR003657, IPR003657 and IPR017412) as analysed by INTERPRO. The analysis revealed 74, 08 and 03 GO term significantly enriched in biological processes, molecular function and cellular components, respectively (Supplementary File 4). The MCL clustering displayed 8 distinct groups, largest being associated with 8 WRKY proteins (red) showing strong interaction (AtWRKYs 15,22,11,33,40,53,30 &48) corresponding to predicted orthologs GgWRKYs 73, 29, 73, 15, 53, 32, 32 & 67, respectively (Fig. 7; Supplementary File 5). These specific associations indicated that these proteins jointly contributed to a shared function of cis or trans in nature as inferred from curated databases or experimentally determined data available in public domains43. The AtWRKYs and corresponding GgWRKYs were shown to be involved in various biological processes including ROS induced modulation, plant growth and osmotic stress (AtWRKY15/GgWRKY73), development (AtWRKY22/GgWRKY29), Jasmonic acid-induced response (AtWRKY11/GgWRKY73), wound-induced response and positive regulator of stress (AtWRKY33/GgWRKY15), senescence (AtWRKY40/GgWRKY53), leaf development and senescence (AtWRKY53/GgWRKY32), abiotic stress and senescence (AtWRKY30/GgWRKY32), and hormonal signal response and defense (AtWRKY48/GgWRKY67). Strong association between 8 AtWRKYs and corresponding ortholog GgWRKYs indicated co-regulation of several biological processes related to senescence, Jasmonate response, hormonal signaling and wound induced response (Supplementary File 5).
Figure 7.
Protein-Protein interaction of GgWRKYs transcription factor based on AtWRKYs orthologs as predicted by STRING search tool.
The associated proteins need not physically connect in a protein-protein interaction of a specific step instead, they may form functional protein linkages especially in transcriptional or post-transcriptional regulation of a process. Also, it has been observed that evolutionarily related proteins usually maintain their three-dimensional structure, even when they have diverged43,44. This interaction between orthologs is expected to display high degree of interaction conservation more so in indirect or transient types of protein-protein associations. Based on protein conservancy of GgWRKYs with AtWRKYs, we assessed the putative functions of GgWRKYs and verified the expression profile of few of the predicted functions of GgWRKYs experimentally in Lab (Table 5) under abiotic stress.
Table 5.
AtWRKYs, their induction factor and experimentally verified responses in GgWRKYs.
| S.N | GgWRKYs | Orthologs AtWRKYs |
Orthologs (% Identity) |
Reported Induction factor (AtWRKYs) | Experimentally verified Induction factor (GgWRKYs) |
|---|---|---|---|---|---|
| 1 | GgWRKY2 | AtWRKY4 | 53% | P. syringae, Salicyclic acid (SA), Jasmonic acid (JA), sucrose, senescence, cold, salinity | Senescence, Carbon starvation, NAA, GA3 |
| 2 | GgWRKY4 | AtWRKY24 | 74% | unknown | salinity, Carbon starvation |
| 3 | GgWRKY5 | AtWRKY4 | 58% | P. syringae, SA, JA, sucrose, senescence, cold, salinity | Senescence, Salinity, dark, GA3 |
| 4 | GgWRKY8 | AtWRKY33 | 61% |
Salinity, mannitol, cold, heat, H2O2, ozone, UV, chitin, B. cinerea, P. syringae, A. brassiciola |
Senescence, Salinity, Carbon starvation, NAA, GA3 |
| 5 | GgWRKY14 | AtWRKY20 | 53% | unknown | Heat, cold, Salinity. UV, GA3 |
| 6 | GgWRKY15 | AtWRKY33 | 62% |
Salinity, mannitol, cold, heat, H2O2, ozone, UV, chitin, B. cinerea, P. syringae, A. brassiciola |
Senescence, cold, Carbon starvation, NAA, GA3 |
| 7 | GgWRKY20 | AtWRKY61 | 51% | unknown | GA3 |
| 8 | GgWRKY24 | AtWRKY65 | 52% | Fe starvation | Dark, heat, UV, Salinity |
| 9 | GgWRKY29 | AtWRKY22 | 64% | H2O2, dark, chitin, flagellin | Senescence, cold, Nwrky |
| 10 | GgWRKY36 | AtWRKY28 | 77% | Salinity, mannitol, H2O2 | Salinity, heat, UV, dark, Carbon starvation |
| 11 | GgWRKY38 | AtWRKY61 | 77% | unknown | Senescence, dark, NAA, GA3 |
| 12 | GgWRKY40 | AtWRKY6 | 60% |
H2O2, methyl viologen, Pi and B starvation |
Cold, dark |
| 13 | GgWRKY44 | AtWRKY31 | 67% | unknown | GA3 |
| 14 | GgWRKY45 | AtWRKY69 | 64% | unknown | Senescence, dark, Carbon starvation, GA3 |
| 15 | GgWRKY51 | AtWRKY71 | 75% | unknown | Dark, Carbon starvation, GA3 |
| 16 | GgWRKY53 | AtWRKY40 | 55% | ABA signaling, SA, chitin, wounding | Wounding, cold, dark |
| 17 | GgWRKY54 | AtWRKY40 | 58% | ABA signaling, SA, chitin, wounding | Senescence, wounding, cold, dark, NAA, GA3 |
| 18 | GgWRKY55 | AtWRKY40 | 43% | ABA signaling, SA, chitin, wounding | Senescence, UV, cold, NAA, GA3 |
| 19 | GgWRKY56 | AtWRKY9 | 51% | unknown | Cold, dark, NAA |
| 20 | GgWRKY57 | AtWRKY2 | 78% | Salinity,manitol,ABA | Salinity, cold, dark, GA3 |
| 21 | GgWRKY58 | AtWRKY2 | 64% | Salinity, manitol, ABA | Cold, Carbon starvation, GA3 |
| 22 | GgWRKY59 | AtWRKY20 | 54% | unknown | Cold, GA3 |
| 23 | GgWRKY62 | AtWRKY18 | 69% | ABA, SA | Salinity, heat, cold |
| 24 | GgWRKY69 | AtWRKY 21 | 65% | unknown | Cold, dark |
| 25 | GgWRKY70 | AtWRKY 21 | 63% | unknown | Salinity, cold, dark |
Real-time expression analysis
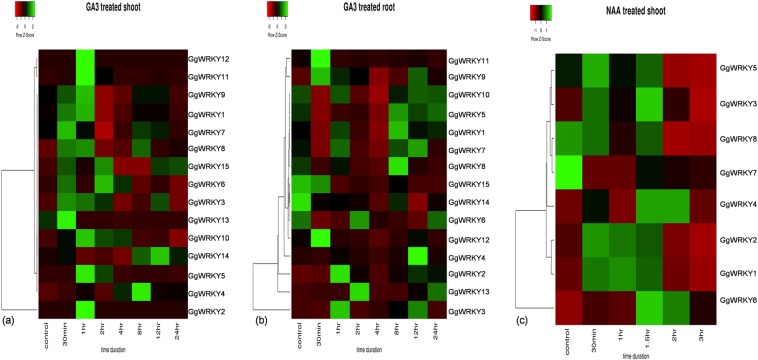
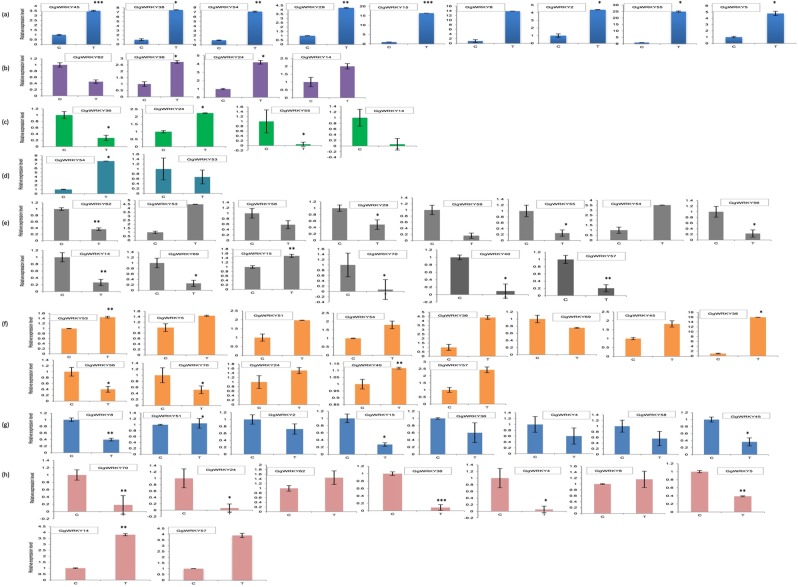
The A. thaliana based protein conservancy for the functional prediction of putative orthologs in G. glabra was experimentally performed. The expression profile of twenty-five GgWRKY genes was investigated post-hormonal treatments (NAA & GA3) and under eight abiotic stress treatments including carbon starvation, salinity, heat, cold, dark, UV, senescence and wounding administered to the aerial tissues of the in-vitro cultured G. glabra plant. Out of the 25 GgWRKYs examined, eight GgWRKYs responded to the NAA treatment (Fig. 8). As can be seen from the heat map, transcripts of GgWRKYs 8, 15 & 29 accumulated maximum (4.1, 3.3 &1.6 folds, respectively) between 0.5 to 1.00 hrs, GgWRKY55 took longer (1.30 hrs) to display its maxima (17.5 folds). GgWRKYs 54 & 56 were mostly up-regulated all the time, while GgWRKYs 4 & 38 were down-regulated in the specified time of study. It seems GgWRKY56 & GgWRKY 38 had definite positive (3.3 folds) & negative regulatory (0.001folds) effects, respectively on the aerial tissues of the plant treated with auxin. GA3 treatment, on the other hand (Fig. 8), revealed GgWRKY 58 was highly up-regulated (257.3 folds) in the aerial tissues of the plant, while GgWRKY15 was up-regulated (43.6 fold) in the underground tissues of the plant as compared to the control. Most of the GgWRKYs responded within 1.5 hrs of GA3 treatment, except GgWRKY 20 which took longer to show their maxima (2.0 hrs) in the root tissues. The results inferred from the present study were compared with the earlier published reports on the functions of AtWRKYs45 which are presented in Table 5. Of the 25 GgWRKYs assessed for abiotic stress treatment, maximum showed response to post-cold treatment (17), followed by dark (13). Nine GgWRKYs responded to senescence and salinity, while eight triggered a response on carbon starvation. Maximum number of GgWRKYs were up-regulated (10) after dark treatment followed by senescence (9). Darkness induced up-regulation of GgWRKYs 5, 24, 36, 38, 40, 45, 51, 53, 54 and 57, while GgWRKYs 56, 69 & 70 were down-regulated. Nine GgWRKYs 2, 5, 8, 15, 29, 38, 45, 54 & 55 were up-regulated during senescence, GgWRKYs 5, 14, 24 & 54 were down-regulated under saline conditions. The transcript levels of GgWRKYs 14, 24 & 36 were more under heat stress, while GgWRKY 24 was up-regulated on UV treatment. The injured plant showed up-regulated transcripts of GgWRKY 54, while GgWRKY53 was found to be down-regulated. Cold treated samples showed higher transcript levels of GgWRKYs 15, 53 & 54, while GgWRKYs14, 40, 55, 56, 57, 58, 59, 62, 69 &70 were down-regulated. The Carbon starved plants showed up-regulation of only GgWRKY51 while GgWRKYs 2, 4, 8, 15, 36, 45 & 58 were found to be down-regulated (Fig. 9). Significantly up-regulated (P ≤ 0.001) GgWRKYs were observed only in senescence (GgWRKYs 45&15), while in salinity, GgWRKY36 was significantly down-regulated. Out of the ten different treatments performed to assess the role of GgWRKYs in abiotic stress, predicted by STRING based on AtWRKY protein conservancy, the response of 11 GgWRKYs corroborated very well with 15 AtWRKYs whose functions were reported in literature (Table 5). Among the 25 GgWRKYs examined, 23 responded to abiotic stress, 17 were induced by hormone while 13 were common to both, suggesting role of hormone under stress conditions. Further study on these functionally assigned GgWRKYs will throw light on their role in underlying molecular mechanism. On comparing the experimental data with the STRING predicted data, it was found that our results corroborated well with the earlier reports on the induction of AtWRKY4 on senescence, AtWRKY40 on wounding, AtWRKYs 2, 28 &33 during salinity and AtWRKY33 under cold treatments. Few AtWRKYs whose functions were not assigned, like AtWRKYs 21, 24, 31, 38, 61, 69 and 71, were also designated putative function based on identity percentage.
Figure 8.
GgWRKY genes are represented as rows and treatment time duration as columns in the matrix. Expression analysis of selected GgWRKY genes displaying differential expression pattern in shoot and roots under various hormonal stress. Heat map showing- Cluster analysis of GgWRKY genes according to their expression profiles in (a) shoots and (b) roots after GA3 treatment for 0.5, 1, 2, 4, 8, 12 and 24 h time interval; (c) Cluster analysis of GgWRKY genes according to their expression profiles in shoots after NAA treatment for 0.5, 1, 1.5, 2 and 3 h time interval.
Figure 9.
Expression profiles of selected GgWRKY genes under eight different stresses. The Y-axis indicates relative expression level and X-axis indicates control shoot tissues (C) and treated shoot tissues (T). (a) expression patterns under etiolated conditions; (b–h) expression profiles under heat, UV, wounding, cold, dark, carbon starvation and salinity, respectively. Actin was used as internal reference. Three biological replicates were used to calculate error bars using standard deviation. Asterisks indicate that the corresponding gene was significantly up- or down regulated in a given treatment (*P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001).
In any biological process, understanding the role of transcription factor provides an insight into its regulatory mechanism. WRKY transcriptional factors have been extensively studied in a plant for plant growth, development, and response to biotic and abiotic stresses. However, WRKY genes present in Glycyrrhiza species have not been elucidated. In conclusion, we identified and characterized 147 full length putative WRKY genes in the genus Glycyrrhiza. These putative genes were grouped based on the number of WRKY domains & zinc finger pattern and further analysed for various properties like molecular weight, iso-electric point, instability index, sub-cellular location. The phylogenetic analysis categorised more than one group/sub-group together, indicating their multiple origins. The present paper highlights several findings not reported earlier, like the novel Zn finger motif of C-X4-CX22-HXH type (sub-group IIf). Also these group-II members shared homology with group IN WRKY members, unlike the other members of group II. This paper also reports several additional domains (DivIVA, SerS, Coat, Exo70 exo cyst complex subunit, Flac-arch super, PAT1and SGNH_hydrolase) apart from the conserved WRKY domain in the WRKY proteins. MSA based 60 amino acid signature sequence of group IId showed very low sequence identity (32%), however when its length was increased to 110 amino acid the identity increased to 70.7%. A closer look at the subgroup IId showed presence of 50 amino acid plant Zn cluster domain upstream to the WRKY domain in four members. However, characteristic Zn finger motif was absent in these members.
Additionally, putative functions were assigned to the identified GgWRKYs, based on STRING database which comprised of both theoretically reported and experimentally verified data. Verification of the data in the Lab displayed 11 out of 15 functions as assigned. The study provides significant evidence to further investigate and validate the role of WRKYs in Glycyrrhiza species in growth, under stress condition and in secondary metabolite biosynthesis.
Materials and Methods
Identification and sequence annotation of WRKY genes
Transcriptome-wide identification of WRKY genes in G. glabra and G. uralensis transcriptome data was done by local similarity (tblastn) search and HMM profile methods. Initially, seventy AtWRKY proteins were downloaded from Arabidopsis Information Resource (TAIR; http://www.Arabidopsis.org/), and HMM profile of WRKY family with accession number PF03106 was retrieved from the Pfam protein family database (https://pfam.xfam.org/). The A. thaliana (AtWRKYs) and PF03106 profile were used as a query sequence to search against the transcriptome data of G. glabra and G. uralensis. An e-value cut off of 1e−50 was applied for the homologue recognition. Parsing the BLAST data from G. glabra, a total of 125 contig hits were found. All these contigs were further analysed in ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) to get the full-length CDS of 87 GgWRKY sequences. Publicly available transcriptome database of G. uralensis (http://ngs-data-archive.psc.riken.jp/Gur-genome/download.pl.) was used to get 60 GuWRKYs. The retrieved coding sequences (CDSs) were then translated by ExPASy translate (https://web.expasy.org/translate/) tool and validated using the Uniprot protein database (https://www.uniprot.org/), conserved domain database (https://www.ncbi.nlm.nih.gov/cdd/) and HMMScan (https://www.ebi.ac.uk/Tools/hmmer/search/hmmscan). The molecular weight (MW), Theoretical isoelectric point (pI), instability index, aliphatic index, Grand average of hydropathicity (GRAVY) of GgWRKY proteins were predicted via the ProtParam (http://web.expasy.org/protparam/). Additionally, subcellular localisation was also predicted by an advanced protein subcellular localisation prediction tool WoLFPSORT (https://wolfpsort.hgc.jp/).
Multiple sequence alignment, phylogenetic analysis and classification
The multiple sequence alignment (MSA) of 245 WRKY proteins was performed using 82 WRKY proteins of G. glabra (GgWRKY), 54 WRKY proteins from G. uralensis (GuWRKY), 70 from A. thaliana (AtWRKYs), 37 from P. patens (PpWRKYs) and one each from Human FLYWCH CRAa and GCMa. The protein sequences of Arabidopsis were downloaded from TAIR (http://www.Arabidopsis.org/), GuWRKYs from (http://ngs-data-archive.psc.riken.jp/Gur-genome/download.pl.), PpWRKYs were obtained from P. patens v3.3 (https://phytozome.jgi.doe.gov/pz/portal.html#!info?alias = Org_Ppatens), Human FLYWCH CRAa (EAW85440) and GCMa (BAA13651) were retrieved from NCBI (http://www.ncbi.nlm.nih.gov/protein/). The conserved regions of 60 amino acids for the WRKY proteins were searched using HMMScan and aligned using CLUSTALW for the construction of the phylogenetic tree. For the GgWRKY based phylogenetic tree, complete protein sequences were used. The tree was constructed using MEGA 7.0 with neighbor-joining method using JTT substitution model and pair-wise deletion method with 1000 bootstrap value. The 60 amino acid conserved region of MSA of G. glabra was visualised using DNAMAN. The MSA included conserved region of WRKY members representing each group and subgroup from A. thaliana and G. uralensis as reference.
Protein-protein interaction analysis and motif detection
The conserved motifs of GgWRKY proteins were analyzed using Multiple Expectation Maximization for Motif Elicitation (MEME: http://meme-suite.org/tools/meme) with the following parameters: minimum and maximum motif widths 6 and 50, respectively and the maximum number of motifs 15. Protein-protein interactions were predicted by STRING43 with A. thaliana as model using Markov clustering with inflation factor of 8.5.
Analysis of cis-regulatory elements in GgWRKYs promoter regions
Promoter sequences of 31 GgWRKYs of up to 2.5 kb (kilobase) upstream to the transcription start site were retrieved manually (Supplementary File 6). These promoter sequences were used as queries to scan the presence of various Cis-regulatory elements in Plant Cis-acting Regulatory DNA Elements (PLACE, http://www.dna.affrc.go.jp/PLACE/)46. The position of identified CREs (biotic and abiotic stress-responsive elements) was mapped on both sense and anti-sense strand using RSAT47 (http://rsat.sb-roscoff.fr/feature-map_form.cgi) drawing tool.
Plant material and treatments
Five months old in-vitro cultured plants grown in SPB medium48 under controlled conditions of 25 °C (±1.5) temperature and a 16 h light/8 h dark cycle (light intensity of 200mmol m–2 s–1), were exposed to various treatments including hormone, temperature, salinity, senescence and wounding. The plantlets, grown in liquid SPB medium were individually subjected to 50 µM auxin (NAA) for 0.5, 1.0, 1.5, 2.0 & 3.0 hrs, and 10 µM of gibberellin (GA3) treatments for 0.5, 1.0, 2.0, 4.0, 8.0, 12 & 24 hrs. Controls were sprayed with water. Different sets of plants were independently subjected to different abiotic treatments like NaCl (500 mM) for 72 hrs, dark, cold (4 °C) and heat (55 °C) treatments for 48, 24 and 8 hrs, respectively. For the Ultra-violet treatment, plants were kept under UV-C for 30 minutes. Mechanically injured plants were examined after 8 hrs of injury. Yellow aerial tissues of plants were used for senescence study, and Carbon starvation was given to the plant for 48 hrs in SPB medium having no sugar. All the respective controls were kept under culture conditions. The control and treated plants were harvested at the appropriate times as indicated, frozen in liquid nitrogen and stored at −80 °C for RNA extraction. Each treatment was used in triplicate and was repeated at least twice.
RNA extraction and quantitative real-time reverse transcription PCR (qRT-PCR)
Total RNA of control and treated shoots and root tissues were extracted using the Pure Link RNA Mini Kit (Invitrogen, US). RNA integrity was analysed on a 1.5% agarose gel and quantity was determined using a NanoDrop 2000C spectrophotometer (Thermo Scientific, USA). cDNA synthesis was carried out using SuperScript™ VILO™ cDNA Synthesis Kit (Thermo Scientific, USA). qRT-PCRs were performed using the SYBR Green PCR Master Mix (Takara, Japan) and carried out in triplicate for each tissue sample. Gene-specific RT primers were designed manually (Supplementary File 7). The amplicons size ranged between 200 to 250 bp. Actin gene was selected as an internal reference gene. The amplification was done in a ten μl reaction volume, which contained 5.0 μl of SYBR Green PCR Master Mix, 0.2 μl of each primer (10 pc), 0.2 μl of ROX, 1.0 μl cDNA template (150 ng/μl), and 3.4 μl ddH2O. PCRs with no-template controls were also performed for each primer pair. The real-time PCRs were performed employing7500 Fast Real-Time PCR System and software (Applied Biosystems, USA). All the PCRs were performed under following conditions: 30 sec at 95 °C, 3 sec at 95 °C, respective optimized Tm for 1 min (40 cycles) followed by 95 °C (15 seconds), 60 °C (30 sec) and 95 °C (15 sec) in MicroAmp fast reaction tubes (Applied Biosystems, USA). The specificity of amplicons was verified by melting curve analysis (55 to 95 °C) after 40 cycles.
Supplementary information
Acknowledgements
P.G. sincerely acknowledges University Grant Commission (UGC), India for the award of JRF fellowship (327955). M.M. acknowledge Council of Scientific & Industrial Research (CSIR), India for SRF fellowship. Authors also acknowledge the Science and Engineering Research Board (SERB), India for the project grant (SERB/SB/SO/PS/90/2013). CSIR-IIIM-communication No.CSIR-IIIM/IPR/00103.
Author contributions
P.G. has done the complete work experimentally; M.M., D.S. has done the associated work; S.G., R.A.V. & M.K.D. has contributed in conceptualisation and writing of Ms.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-57232-x.
References
- 1.Schluttenhofer C, Yuan L. Regulation of Specialized Metabolism by WRKY Transcription Factors. Plant Physiology. 2015;167:295–306. doi: 10.1104/pp.114.251769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chi Y, et al. Protein-Protein Interactions in the Regulation of WRKY Transcription Factors. Molecular Plant. 2013;6:287–300. doi: 10.1093/mp/sst026. [DOI] [PubMed] [Google Scholar]
- 3.Ishiguro S, Nakamura K. Characterization of a cDNA encoding a novel DNA-binding protein, SPF1, that recognizes SP8 sequences in the 5′ upstream regions of genes coding for sporamin and β-amylase from sweet potato. Molecular and General Genetics MGG. 1994;244:563–571. doi: 10.1007/bf00282746. [DOI] [PubMed] [Google Scholar]
- 4.Chen L, Xiang S, Chen Y, Li D, Yu D. Arabidopsis WRKY45 interacts with the DELLA protein RGL1 to positively regulate age-triggered leaf senescence. Mol Plant. 2017;10:1174–1189. doi: 10.1016/j.molp.2017.07.008. [DOI] [PubMed] [Google Scholar]
- 5.Jiang C, et al. Transcriptome analysis of WRKY gene family in Oryza officinalis Wall ex Watt and WRKY genes involved in responses to Xanthomonas oryzae pv. oryzae stress. PloS one. 2017;12:e0188742. doi: 10.1371/journal.pone.0188742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He C, et al. A Genome-Wide Identification of the WRKY Family Genes and a Survey of Potential WRKY Target Genes in Dendrobium officinale. Scientific Reports. 2017;7:9200. doi: 10.1038/s41598-017-07872-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohanta TK, Park Y-H, Bae H. Novel Genomic and Evolutionary Insight of WRKY Transcription Factors in Plant Lineage. Scientific Reports. 2016;6:37309. doi: 10.1038/srep37309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang Y, Wang L. The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC evolutionary biology. 2005;5:1–1. doi: 10.1186/1471-2148-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulgem T, Rushton P, Robatzek S, Somssich I. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000;5:199–206. doi: 10.1016/S1360-1385(00)01600-9. [DOI] [PubMed] [Google Scholar]
- 10.Rushton PJ, Somssich IE, Ringler P, Shen QJ. WRKY transcription factors. Trends in Plant Science. 2010;15:247–258. doi: 10.1016/j.tplants.2010.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Llorca, C. M., Potschin, M. & Zentgraf, U. bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Frontiers in plant science5, 10.3389/fpls.2014.00169 (2014). [DOI] [PMC free article] [PubMed]
- 12.Dong J, Chen C, Chen Z. Expression profiles of the Arabidopsis WRKY gene superfamily during plant defense response. Plant Molecular Biology. 2003;51:21–37. doi: 10.1023/a:1020780022549. [DOI] [PubMed] [Google Scholar]
- 13.Potschin M, Schlienger S, Bieker S, Zentgraf U. Senescence Networking: WRKY18 is an Upstream Regulator, a Downstream Target Gene, and a Protein Interaction Partner of WRKY53. Journal of Plant Growth Regulation. 2013;33:106–118. doi: 10.1007/s00344-013-9380-2. [DOI] [Google Scholar]
- 14.Berri S, et al. Characterization of WRKYco-regulatory networks in rice and Arabidopsis. BMC plant biology. 2009;9:120. doi: 10.1186/1471-2229-9-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ãœlker B, Somssich IE. WRKY transcription factors: from DNA binding towards biological function. Current Opinion in Plant Biology. 2004;7:491–498. doi: 10.1016/j.pbi.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 16.Chen L, et al. The role of WRKY transcription factors in plant abiotic stresses. Biochimica et Biophysica Acta (BBA) - Gene Regulatory Mechanisms. 2012;1819:120–128. doi: 10.1016/j.bbagrm.2011.09.002. [DOI] [PubMed] [Google Scholar]
- 17.Aprile A, et al. Activation of a gene network in durum wheat roots exposed to cadmium. BMC plant biology. 2018;18:238. doi: 10.1186/s12870-018-1473-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakshi M, Oelmuller R. WRKY transcription factors: Jack of many trades in plants. Plant signaling & behavior. 2014;9:e27700–e27700. doi: 10.4161/psb.27700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Banerjee A, Roychoudhury A. WRKY proteins: signaling and regulation of expression during abiotic stress responses. TheScientificWorldJournal. 2015;2015:807560–807560. doi: 10.1155/2015/807560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen L, Zhang L, Yu D. Transcription factor WRKY75 interacts with DELLA proteins to affect flowering. Plant Physiology. 2018;176:790–803. doi: 10.1104/pp.17.00657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luo M, Dennis E, Berger F, Peacock JW, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proc Natl Acad Sci USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao T-T, et al. Expression and Functional Analysis of WRKY Transcription Factors in Chinese Wild Hazel, Corylus heterophylla Fisch. PloS one. 2015;10:e0135315–e0135315. doi: 10.1371/journal.pone.0135315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phukan UJ, Jeena GS, Shukla RK. WRKY Transcription Factors: Molecular Regulation and Stress Responses in. Plants. Frontiers in plant science. 2016;7:760–760. doi: 10.3389/fpls.2016.00760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eulgem T, Somssich IE. Networks of WRKY transcription factors in defense signaling. Current Opinion in Plant Biology. 2007;10:366–371. doi: 10.1016/j.pbi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 25.He Y, et al. Genome-Wide Identification and Expression Analysis of WRKY Transcription Factors under Multiple Stresses in Brassica napus. PloS one. 2016;11:e0157558–e0157558. doi: 10.1371/journal.pone.0157558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishida T, et al. Arabidopsis TRANSPARENT TESTA GLABRA2 is directly regulated by R2R3 MYB transcription factors and is involved in regulation of GLABRA2 transcription in epidermal differentiation. The Plant Cell. 2007;19:2531–2543. doi: 10.1105/tpc.107.052274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pnueli Lilach, Hallak-Herr Elza, Rozenberg Mira, Cohen Mira, Goloubinoff Pierre, Kaplan Aaron, Mittler Ron. Molecular and biochemical mechanisms associated with dormancy and drought tolerance in the desert legume Retama raetam. The Plant Journal. 2002;31(3):319–330. doi: 10.1046/j.1365-313X.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen Chunhong, Chen Zhixiang. Potentiation of Developmentally Regulated Plant Defense Response by AtWRKY18, a Pathogen-Induced Arabidopsis Transcription Factor. Plant Physiology. 2002;129(2):706–716. doi: 10.1104/pp.001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishihama N, Yoshioka H. Post-translational regulation of WRKY transcription factors in plant immunity. Current Opinion in Plant Biology. 2012;15:431–437. doi: 10.1016/j.pbi.2012.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Sun Chuanxin, Palmqvist Sara, Olsson Helena, Borén Mats, Ahlandsberg Staffan, Jansson Christer. A Novel WRKY Transcription Factor, SUSIBA2, Participates in Sugar Signaling in Barley by Binding to the Sugar-Responsive Elements of the iso1 Promoter. The Plant Cell. 2003;15(9):2076–2092. doi: 10.1105/tpc.014597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin J, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Research. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song H, Sun W, Yang G, Sun J. WRKY transcription factors in legumes. BMC plant biology. 2017;18:243–243. doi: 10.1186/s12870-018-1467-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Asl MN, Hosseinzadeh H. Review of pharmacological effects of Glycyrrhiza sp. and its bioactive compounds. Phytother. Res. 2008;22:709–724. doi: 10.1002/ptr.2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Al-Hazimi HMA, Al-Jaber NA. Phenolic compounds from Glycyrrhiza plants (Leguminosae) review article I. J. Saudi Chem. Soc. 2005;9:347–380. [Google Scholar]
- 35.Yang X, et al. Identification and expression analyses of WRKY genes reveal their involvement in growth and abiotic stress response in watermelon (Citrullus lanatus) PloS one. 2018;13:e0191308–e0191308. doi: 10.1371/journal.pone.0191308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jakoby M, et al. bZIP transcription factors in Arabidopsis. Trends in Plant Science. 2002;7:106–111. doi: 10.1016/S1360-1385(01)02223-3. [DOI] [PubMed] [Google Scholar]
- 37.Bailey TL, et al. MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cai C, et al. Genome-wide analysis of the WRKY transcription factor gene family in Gossypium raimondii and the expression of orthologs in cultivated tetraploid cotton. The Crop. Journal. 2014;2:87–101. doi: 10.1016/j.cj.2014.03.001. [DOI] [Google Scholar]
- 39.Zhu Xinlei, Liu Shuwei, Meng Chen, Qin Lumin, Kong Lina, Xia Guangmin. WRKY Transcription Factors in Wheat and Their Induction by Biotic and Abiotic Stress. Plant Molecular Biology Reporter. 2013;31(5):1053–1067. doi: 10.1007/s11105-013-0565-4. [DOI] [Google Scholar]
- 40.Rinerson CI, Rabara RC, Tripathi P, Shen QJ, Rushton PJ. The evolution of WRKY transcription factors. BMC plant biology. 2015;15:66. doi: 10.1186/s12870-015-0456-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang M, et al. Genome and transcriptome analysis of the grapevine (Vitis vinifera L.) WRKY gene family. Horticulture Research. 2014;1:14016. doi: 10.1038/hortres.2014.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie Zhen, Zhang Zhong-Lin, Zou Xiaolu, Huang Jie, Ruas Paul, Thompson Daniel, Shen Qingxi J. Annotations and Functional Analyses of the Rice WRKY Gene Superfamily Reveal Positive and Negative Regulators of Abscisic Acid Signaling in Aleurone Cells. Plant Physiology. 2004;137(1):176–189. doi: 10.1104/pp.104.054312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szklarczyk D, et al. STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thornton JM, Orengo CA, Todd AE, Pearl FMG. Protein folds, functions and evolution. Journal of Molecular Biology. 1999;293:333–342. doi: 10.1006/jmbi.1999.3054. [DOI] [PubMed] [Google Scholar]
- 45.Grzechowiak M. The role of WRKY transcription factors in plants. BioTechnologia. 2014;95:215–233. doi: 10.5114/bta.2014.54797. [DOI] [Google Scholar]
- 46.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database. Nucleic Acids Research 27. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Medina-Rivera A, et al. RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 2015;43:W50–W56. doi: 10.1093/nar/gkv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gupta S, et al. Direct rhizogenesis, in vitro stolon proliferation and high-throughput regeneration of plantlets in Glycyrrhiza glabra. Acta Physiologiae Plantarum. 2013;35:2699–2705. doi: 10.1007/s11738-013-1302-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.