Abstract
Precise gene editing such as CRISPR/Cas9-mediated homology directed repair (HDR) can increase our understanding of gene function and improve traits of importance for aquaculture. This fine-tuned technology has not been developed for farmed fish including Atlantic salmon. We performed knock-in (KI) of a FLAG element in the slc45a2 gene in salmon using sense (S), anti-sense (AS) and double-stranded (ds) oligodeoxynucleotide (ODN) templates with short (24/48/84 bp) homology arms. We show in vivo ODN integration in almost all the gene edited animals, and demonstrate perfect HDR rates up to 27% in individual F0 embryos, much higher than reported previously in any fish. HDR efficiency was dependent on template concentration, but not homology arm length. Analysis of imperfect HDR variants suggest that repair occurs by synthesis-dependent strand annealing (SDSA), as we show for the first time in any species that indel location is dependent on template polarity. Correct ODN polarity can be used to avoid 5′-indels interrupting the reading frame of an inserted sequence and be of importance for HDR template design in general.
Subject terms: Mutagenesis, Genetic engineering
Introduction
Aquaculture continues to grow faster than any other major food production sector and is quickly becoming the main source of seafood in human diets. In this context, Norway is the largest producer of farmed Atlantic salmon (Salmo salar) worldwide. In later years, the production of salmon in Norway has ceased to grow due to sustainability challenges linked to open sea-cage rearing. Genetic introgression of farmed salmon into wild stocks and the marine parasite, salmon louse, are recognized as the two major concerns1. The high prevalence of salmon lice in most Norwegian fjords, due to open sea-cage farming, cause high lethality in wild salmonids and is hindering expansion of sea-cage farming. The consequences of genetic introgression caused by escapees remain uncertain, but existing knowledge indicates that it may lead to changes in life‐history traits, with potential ecological impacts2–5. Sequencing of the salmon genome6 has permitted more detailed studies on the link between genes and key traits, and we and others have shown that single nucleotide polymorphisms (SNPs) to a certain degree can explain the time of maturity1 and disease resistance7,8. In this context, New Breeding Technologies (NBTs) by gene editing may offer a solution to some of the problems in salmon farming, with a possible production of salmon displaying traits such as disease resistance and sterility9–12.
We have previously demonstrated the feasibility of double allelic KO in F0 salmon using CRISPR/Cas9, by targeting genes essential for pigmentation9, elongation of polyunsaturated fatty acids13 and reproduction10. At the same time, CRISPR/Cas9 KO-mutations targeting various phenotypes have been shown by others in several farmed fish species such as tilapia14–21, sea bream22, sterlet23, channel catfish24,25, southern catfish26, common carp27, sturgeon28 and rainbow trout29. CRISPR/Cas9 KOs are produced by a Cas9-induced double-stranded DNA break (DSB) followed by activation of the endogenous error-prone non-homologous end joining (NHEJ) pathway, which introduce indels at the repair junction. In fish with a long generation time such as salmon (3–4 years), it is a necessity to study the KO in the F0 generation, however the mosaicism caused by NHEJ may include partly functional in-frame indels affecting the result. As such, controlled insertions utilizing the homology directed repair (HDR) mechanism instead of NHEJ, may for example be used to effectively insert a stop codon which can increase the homogeny of the KO already in F030.
DSB-repair by HDR can occur by several pathways31 such as synthesis-dependent strand annealing (SDSA), or by the formation of Holliday junctions that can be resolved with or without crossing-over32. HDR occurs naturally during meiosis but can also be activated by supplying a DNA repair template homologous to the CRISPR/Cas9 target sequence, as shown from studies in cells and model organisms33. This approach allows incorporation of the desired genetic changes into the repair template. A functional protocol for CRISPR/Cas9-mediated HDR in salmon offers the potential to insert genes, SNPs or regulatory elements without introducing transgenes and is therefore also interesting in the context of NBT, where such technology can be useful to target for example gene expression levels or to insert stop codons. With the exception of very low efficiency KI of a gene encoding red fluorescent protein in rohu carp34, such advanced and fine-tuned genome editing has not been developed for farmed fish, and it may be useful to learn from protocols already established in model fish species. While only a few studies have reported HDR in medaka35,36, several KI-strategies have been reported in zebrafish using either donor plasmids37–39 or single-stranded oligodeoxynucleotides (ssODNs)40–45, or both46. Knowledge from other fish studies are somewhat inconclusive when it comes to deciding the strategy for applying HDR in salmon, as there is a lack of consensus regarding the impact of different repair templates, homology arm length and strand complementarity in the above-mentioned studies.
Here, we aimed to establish an efficient method for controlled KI of a FLAG element in F0 salmon, targeting the pigmentation gene solute carrier family 45 member 2 (slc45a2) (Fig. 1). Using next generation sequencing (NGS), we could characterize the efficiency, accuracy and types of integrations formed, which was dependent on template concentration, but not on homology arm length. Interestingly, we obtained a high level of perfect integration, up to 27%. Also, we observed that the rate of in-frame integration was higher using anti-sense (AS) as compared to sense (S) and double-stranded (ds) ODNs.
Figure 1.
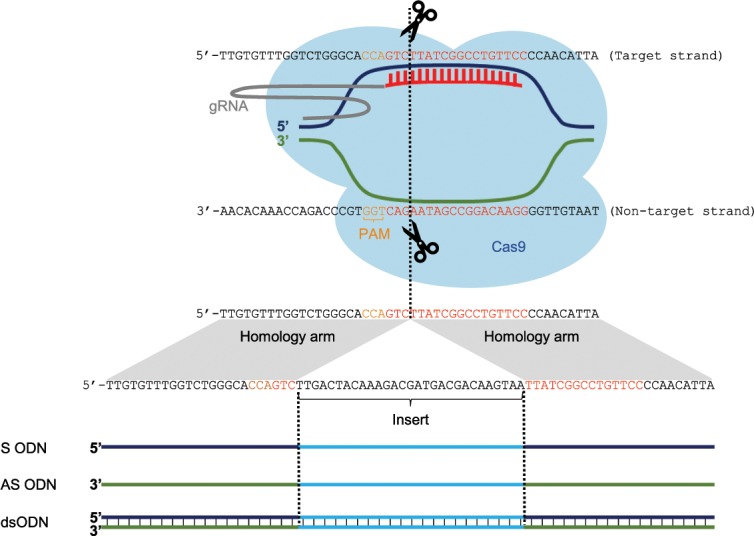
CRISPR target sequence and the donor DNA templates for the Atlantic salmon slc45a2 gene. The ODNs were designed by copying 24/48/84 nucleotides on each side of the CRISPR cut site (indicated with a dotted line), and an insert consisting of FLAG followed by a STOP codon (TAA). In order to keep the open reading frame of FLAG, two nucleotides (TT) were added in the 5′end of the FLAG sequence. The ODN S is sense relative to the Atlantic salmon slc45a2 gene.
Results
Targeting the pigmentation gene slc45a2, we have performed KI of a FLAG element in F0 salmon using CRISPR/Cas9 and symmetrical DNA repair templates (Fig. 1). Wierson et al.39 have shown that short homology arms from 24–48 bp is enough to direct precise and efficient knock-in in zebrafish. Based on this we have tested 24, 48 and 84 bp homology arms in this study. We also explored the effect of different templates, concentrations and polarity on the HDR efficiency.
Identification of FLAG-positive slc45a2-mutants
In order to identify fish containing the FLAG insert, positive slc45a2-mutants were selected by visual inspection (Supplementary Fig. 1), followed by DNA extraction from fin clips and PCR (Table 1 and Supplementary Fig. 2). We were able to detect FLAG in as many as 94–100% of the mutants injected with the highest ODN concentration (1.5 µM), and in 69% of the mutants injected with the lowest concentration (0.15 µM). In contrast, FLAG was only detected in 10–12% of the mutants injected with the two different concentrations (2.5 and 10 ng/µl) of the plasmid (see Methods for description of the plasmid). Moreover, the PCR-screening did not show any difference in efficiency between the different homology arm lengths (24, 48 and 84 bp) tested in this study (Table 1).
Table 1.
Comparison of integrate efficiency among different repair templates and concentrations, analyzed by PCR screening.
| Repair template | Concentration | # samples | # FLAG positive | FLAG positive (%) |
|---|---|---|---|---|
| Plasmid 24 | 2.5 ng/µl (0.001 µM) | 49 | 5 | 10 |
| 10 ng/µl (0.004 µM) | 50 | 6 | 12 | |
| S 24 ODN | 1.5 µM | 36 | 36 | 100 |
| AS 24 ODN | 0.15 µM | 13 | 9 | 69 |
| 1.5 µM | 34 | 32 | 94 | |
| ds 24 ODN | 1.5 µM | 34 | 34 | 100 |
| S 48 ODN | 1.5 µM | 48 | 48 | 100 |
| S 84 ODN | 1.5 µM | 48 | 48 | 100 |
NGS analysis of CRISPR-mutants
Next, we deep sequenced 76 FLAG-positive fish covering the different templates and concentrations applied in this study. The samples were selected based on the initial PCR-screening (Table 1), and the results are shown in Fig. 2, Supplementary Fig. 3 and Supplementary Table 1.
Figure 2.
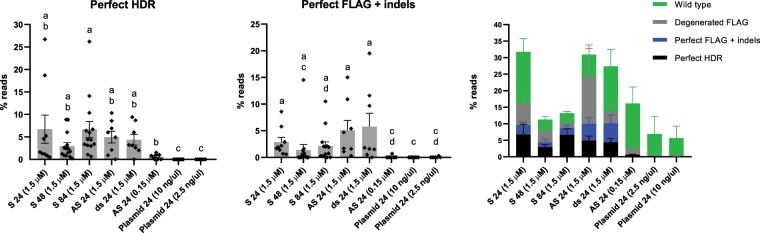
NGS results. A fragment covering the entire CRISPR target site was amplified (76 fish) prior to Illumina MiSeq sequencing. When reporting read counts with the inserted sequence, we distinguished between the following groups; (a) Perfect HDR (reads with a perfect match to the entire target sequence), and (b) Perfect FLAG + indels (reads with a correct insert sequence but mismatches/indels in the homology arms). We also reported reads with mismatches in the insert sequence (referred to as degenerated FLAG) and wild type reads (Supplementary Fig. 3). All the data are summarized in (c). Read counts for each group are given in % of the total number of reads with at least 100 identical reads, for each sample. Individual samples are represented by black diamonds, and grouped for each of the different repair templates, at different concentrations (represented by grey bars). The error bars indicate the SEM of the mean for each group. Non-parametric statistics (Kruskall-Wallis) were performed to analyze the differences in HDR efficiencies between the different repair templates. Different lower-case letters indicate significant differences (P < 0.05).
Perfect repair
We aimed to analyze whether any of the repair templates performed better with respect to both accurate repair and integration efficiency. We did not see a significant difference between the ODNs when used at the highest concentration (1.5 µM), although the individual variation was large. Average perfect reads were detected as follows: 6.7% (std = 9.4), 3.0% (std = 2.9), 6.7% (std = 6.7), 5.0% (std = 3.7) and 4.4% (std = 3.3) for S 24, S 48, S 84, AS 24 and ds 24 ODNs, respectively (Fig. 2a). Two fish from each of the S 24 and S 84 groups showed very high rate of perfect integration, displaying 18.7, 26.7, 14.1 and 26.2% perfect HDR, respectively. An example of an alignment (sample 14, S 24 ODN) is shown in Supplementary Fig. 4.
The lowest ODN concentration (0.15 µM, tested for AS 24) resulted in only 0.6% average perfect HDR. This was substantially lower than the results obtained for the highest ODN concentration (1.5 µM), with average perfect reads ranging from 3.0–6.7% (Fig. 2a). Likewise, the plasmid template displayed very low integration efficiency and no reads containing FLAG could be detected following the standard read sequence filtering (Methods and Supplementary Fig. 5). However, when we analyzed the raw material (prior to filtering), perfect FLAG sequences were detected, but only in <0.01% of the total reads.
Erroneous repair
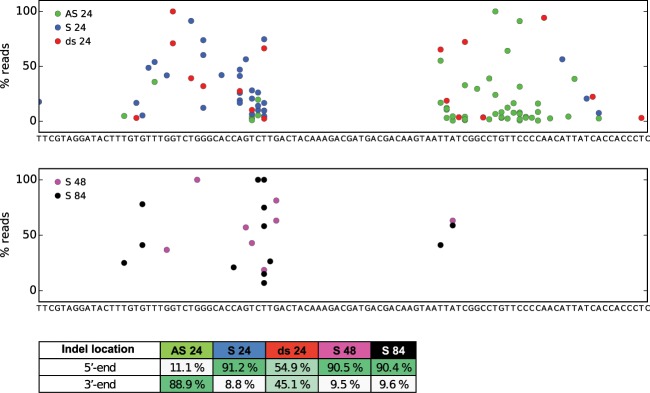
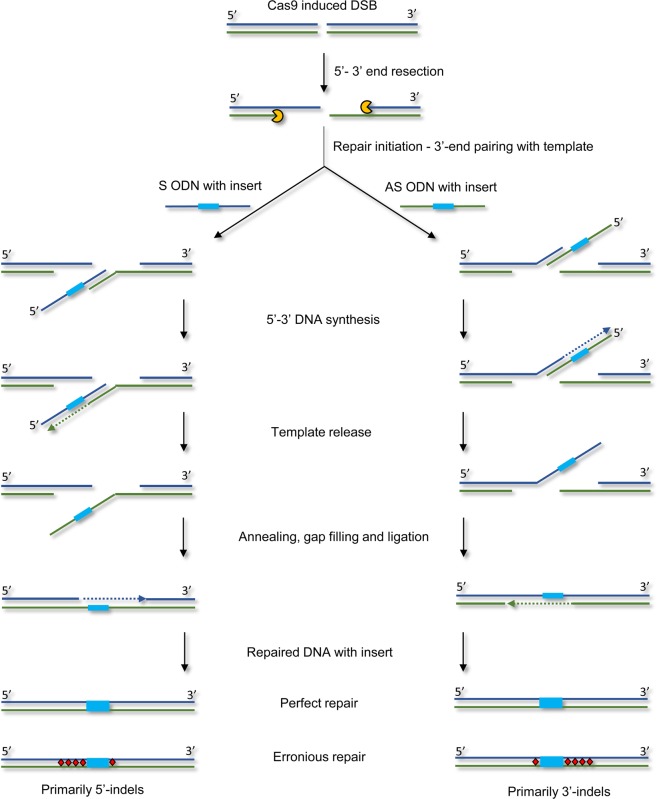
In addition to perfect HDR, we detected several imperfect HDR variants with a correct FLAG insert, but various indels within the homology arms. These reads are referred to as “Perfect FLAG + indels” (Fig. 2b). Interestingly, the type of ODN (S, AS or ds) strongly determined the location of these indels (Fig. 3). When using the AS 24 ODN, 88.9% of the indels were located on the 3′-side of the insert. Similar, when using the S 24, 48 and 84 ODNs > 90% of the indels ended up on the 5′-side of the insert. Using the dsODN the indels were equally distributed on the 5′- and 3′-sides of the insert. Based on these results, we suggest that ODN-mediated HDR initiates the SDSA repair pathway. SDSA repair proceeds via distinct steps, starting with resection to yield 3′ overhangs on both sides of the DSB. The 3′ overhangs pair with the ODN template and are extended by DNA synthesis copying template sequences. Bridging of the DSB is completed when the newly synthesized strands withdraw from the donor and anneal back at the locus47. An outline showing the steps in this pathway and ODN-determined location of the indels is shown in Fig. 4.
Figure 3.
Analysis of indel locations. All the sequence variants were extracted from the group called “Perfect FLAG + indels”. The sequences were aligned to the reference gene containing the inserted sequence. The read count for each indel-containing variant was then converted to percentage of the total read count of variants from the “Perfect FLAG + indels” for all individuals. The percentages were plotted on the reference sequence as colored dots (a) AS 24 ODN (green), S 24 ODN (blue) and ds 24 ODN (red). (b) S 48 ODN (pink) and S 84 ODN (black). In order to analyze the difference in indel positions between the different groups, the percentages of indels located either at the 5′- or 3′-end of the inserted sequence was calculated for each group and is shown in (c).
Figure 4.
Steps in SDSA and ODN-determined location of the indels. SDSA repair proceeds via distinct steps, starting with resection to yield 3′ overhangs on both sides of the Cas9 induced DSB. The 3′ overhangs pair with the ODN templates and are extended by DNA synthesis copying template sequences. Bridging of the DSB is completed when the newly synthesized strands withdraw from the donor and anneal back at the locus. This results in both perfect and erroneous repair, with indels primarily on the 5′ or 3′-end, depending on the polarity of the ODN. The FLAG insert is shown in light blue, and indels are shown as red diamonds.
Discussion
Here, we aimed to establish an efficient method for controlled KI of a FLAG element in F0 salmon using CRISPR/Cas9 and a symmetrical DNA repair template (Fig. 1). To explore the method we tested different templates, concentrations, homology arm lengths and strand complementary (summarized in Table 1). The initial PCR screening revealed a striking difference between the ODNs and the plasmid template, and the ODNs were by far the most efficient. In order to obtain in-depth information about the level of mosaicism and the nature of the integration, we deep-sequenced 76 FLAG-positive fish. We detected average perfect reads within the range of 3.0–6.7%, but the individual variation was large, and some fish displayed perfect repair rates up to 27%. To our knowledge, none have reported this level of perfect HDR in F0 fish. An important aspect hindering a direct comparison with several previous studies is the different methods used to evaluate the HDR efficiency, such as PCR- or restriction-assays, sequencing of a limited number of clones, genotyping based on high-resolution melting analysis, fluorescence etc.37–40,42,44,46. Only three studies in zebrafish have used NGS, showing lower perfect repair rates of 1–4%41, 1.7–3.5%45 and <1%43.
Comparing the different repair templates with respect to perfect repair and integration efficiency, our results showed no significant difference between 24, 48 and 84 bp homology using the S ODN. This is in contrast to Boel et al.41 who reported homology arm length to be the most influential factor and 60 bp homology arms to be the optimal length for symmetrical templates. Moreover, no apparent difference was detected between, ss- and dsODNs (when used at 1.5 µM), which is in contrast to previous findings in Drosophila48. Our results indicate that ss- vs. dsDNA template is not the main reason for the observed difference in efficiency between the plasmid and ODNs. We hypothesized that the result could be explained by the fact that the plasmid was injected in a substantially lower molar concentration (10 ng/µl is equivalent to 0.004 µM) than the ODNs (1.5 µM). Taken together, these results suggest a concentration dependent mechanism for ODN-mediated HDR in salmon embryos. Likewise, it has been reported that a 0.7 kb insert generated 75% edits when injected at 0.5pmol/l, but only 9% edits when injected at 0.1 pmol/l, in C. elegans49. Ideally, we would have liked to test a range of different concentrations for all the templates in our study, but this has not been feasible. Although salmon experiments face several challenges in terms of availability in material and slow development, we still believe it is crucial to test out HDR in salmon, as the outcome of the method seems to vary in different species. We hypothesize that the cold rearing temperature and slow development of Atlantic salmon may be an advantage in the context of HDR, allowing a longer timeframe for the integration to occur. The early ontogeny of Atlantic salmon has been described in detail by Gorodilov50, who showed how the duration of the developmental stages from fertilization is dependent on temperature. For example, if the eggs are kept at 6 °C, it takes about three months until hatching, in stark contrast to two days for zebrafish. Interestingly, it has been reported that cold shock-treatment increases the frequency of HDR gene editing in induced pluripotent stem cells51.
In addition to perfect reads, we detected several reads showing erroneous repair. These reads contained the FLAG insert, but also indels within the homology arms. Most interestingly, we found that the location of these indels were strongly dependent on the polarity of the ODNs. Using the AS ODN 89% of the indels were located on the 3′-side and using the S ODN 90% of the indels were located on the 5′-side. Similarly, when using the dsODN the indels were equally distributed on the 5′- and 3′-sides of the insert, indicating that the repair machinery has no preference regarding the template polarity (S vs. AS ODN). This, in combination with the similarity of the inserts with the ODN template sequence, also strongly indicates that DSB repair using ODNs initiates the SDSA pathway, as previously suggested for C. elegans49 and zebrafish41. Our findings suggest that the 3′-end pairing with the template and initial DNA synthesis occur with high fidelity, while the steps involving annealing, gap filling, and ligation are more prone to errors. The cause of these errors is unclear, but various mechanisms of template switching have been suggested41. Our data supports the template switching theory, as the origin of the inserts predominantly have high similarity with the ODN template sequence (Supplementary Fig. 6). To our knowledge, the ODN (S vs. AS) dependent location of indels has not been reported by others. We suggest taking this information into account when designing ODN repair templates for HDR. To obtain a high rate of in frame integration 5′- end indels must be avoided, making AS ODNs the preferred template.
We have in this study observed that ODNs (S, AS and ds) with 24, 48 and 84 bp homology arms integrates perfectly at a relatively high rate (up to 27%) into salmon embryos. These results are obtained from sequencing of DNA from fin clips, which might not perfectly reflect germline transmission efficiency. However, considering the high fecundity of salmon females (8000–10000 eggs), a potential quick integration into broodstock is possible by crossing F0s. For example, if parental F0 fish have 15% perfect integration, crosses will produce ~180–225 F1 offspring with double allelic KI. To increase the efficiency further studies could focus on the concentration of ODN template, as this clearly affects the efficiency of integration (Fig. 2). However, focus could also be aimed at Cas9. Currently we are using Cas9 mRNA, this probably results in more variants compared to Cas9 protein as observed previously52. Unfortunately, although we have performed multiple trials with Cas9 protein, we have not yet been able to successfully use it in salmon embryos. It is also possible to explore other nucleases53 to improve efficiency and accuracy of the CRISPR KI protocol. Another possibility would be to use short-life Cas9 variants, which have been reported to reduce toxicity and off-target editing54,55.
A challenge with the ODN technology is the possibility to make these ODNs long enough for, for example full gene integration. While synthesis of ODNs were previously restricted to a maximum length of <200 nucleotides, recent technologies now allow generation of longer sequences56,57, and simple ssDNA synthesis over 10 kb using asymmetric PCR has been demonstrated58. Commercial manufacturers also offer synthesis of long ssDNA, although at a relatively high cost. Nevertheless, this enables the insertion of longer sequences such as reporters, gene tags, regulatory elements or even genes. However, editing efficiency is sensitive to insert size, elegantly shown by Paix and colleagues by taking advantage of the split-GFP system47.
We have compared various DNA repair templates for HDR in salmon, and our results show that ODN templates induce highly efficient HDR integration at the target site, much higher than previously observed in any fish species. Our results also indicate that the integration occurs via the SDSA repair pathway and is dependent on template concentration. Interestingly our data also gives further clues to how the SDSA repair pathway may work, as we for the first time in any species show in detail that the distribution of indels is dependent on ODN polarity.
Methods
Ethics statement
This experiment was approved by the Norwegian Animal Research Authority (NARA, permit number 5741) and the use of these experimental animals was in accordance with the Norwegian Animal Welfare Act.
Preparation of Cas9 RNA and gRNA
The slc45a2 CRISPR target sequence is described in Edvardsen et al.9. The target sequence was blasted against the reference genome of salmon and show no other hits than to the gene in question. Preparation of gRNA and cas9 mRNA was performed as previously described9 with the following exceptions: for in vitro transcription of gRNA we used the HighScribe T7 Quick High Yield RNA Synthesis Kit (NEB) according to the protocol for short transcripts. The RNeasy MiniKit spin column (Qiagen) was used to purify the gRNA.
Design and preparation of donor DNA templates for slc45a2
S- and AS ODNs were ordered from Integrated DNA Technologies (Leuven, Belgium). They were designed by copying 24/48/84 nucleotides on each side of the CRISPR cut-site, with a 29 bp insert comprised of TT-FLAG-TAA. TT was included to keep the open reading frame of FLAG, and the STOP codon (TAA) was added to ensure an albino phenotype for slc45a2 CRISPR mutants, regardless of a successful KI-event. Aiming to compare ss- vs. dsDNA, we prepared a dsODN (with 24 bp homology arms) by annealing S and AS. The design is illustrated in Fig. 1. Another pair of S and AS ODNs (24 bp homology arms) were designed with the purpose of cloning into a plasmid (pCRTM4-TOPO vector). The design is identical to the one described above, with the addition of gRNA target sequences on each side for in vivo release of the template and A-overhangs in the 3′ends. The S and AS ODNs were annealed, and cloning performed according to the TOPO® TA Cloning® Kit for Sequencing. The different repair templates are described in Table 2.
Table 2.
Description of the different repair templates used. *All repair templates were symmetrical, with both left and right homology arms of the same length. **The polarity of the ssODNs are relative to slc45a2.
| Repair template name | Repair template characteristics | |||
|---|---|---|---|---|
| Template | Homology arm length (bp)* | ss/ds DNA | Polarity** | |
| Plasmid 24 | Plasmid | 24 | ds | |
| S 24 ODN | ODN | 24 | ss | S |
| AS 24 ODN | ODN | 24 | ss | AS |
| ds 24 ODN | ODN | 24 | ds | |
| S 48 ODN | ODN | 48 | ss | S |
| S 84 ODN | ODN | 84 | ss | S |
Microinjection
Salmon eggs and sperm were delivered by Aquagen (Trondheim, Norway). Fertilization and microinjections were carried out as described previously9 using 50 ng/µl gRNA and 150 ng/µl cas9 mRNA in nuclease free water and a FemtoJet®4i (Eppendorf) microinjector. The ODNs (S, AS or ds) were added to the injection mix with a final concentration of 1.5 or 0.15 µM, and the plasmid with a final concentration of 2.5 or 10 ng/µl (corresponding to 0.001 and 0.004 µM, respectively).
Analysis of mutants
When kept at 6–8 °C, the salmon eggs will hatch approximately three months post fertilization. The slc45a2 CRISPR mutants are easily recognized in newly hatched embryos and in juveniles, due to the lack of pigment, and these individuals (albinos) were selected for further DNA analyses. DNA was extracted from caudal fins using DNeasy Blood & Tissue kit (Qiagen). To ensure complete homogenization, the tissue was incubated overnight at 56 °C using a thermomixer. DNA was eluted in 30 µl nuclease free water. To identify FLAG-positive mutants PCR was performed on genomic DNA, with the forward primer targeting the FLAG-sequence (5′-CTACAAAGACGATGACGAC) and the reverse primer targeting slc45a2 (5′-CGCAACGACTACACATTAT). The PCR-products were evaluated on 1% agarose gels. In order to verify insertion of FLAG and to assess the level of mosaicism, a fragment covering the entire target site was amplified in selected samples (n = 76) with a two-step fusion PCR to prepare for sequencing by Illumina MiSeq, as described in30. The following primer sequences were used in the first PCR-step; 5′-tctttccctacacgacgctcttccgatctCAGATGTCCAGAGGCTGCTGCT and 5′-tggagttcagacgtgtgctcttccgatctTGCCACAGCCTCAGAATGTACA (gene specific sequence indicated in capital letters).
Analysis of MiSeq data
Fastq files were filtered and trimmed with Cutadapt59, and variants were called using a custom script (Supplementary Fig. 5). Finally, read counts were reported for the variants containing the inserted sequence, separating those with a perfect match to the entire target sequence (referred to as perfect HDR), and those with a correct insert sequence but various mismatches in the rest of the target sequence (referred to as perfect FLAG + indels) (Fig. 2). In addition, read counts were reported for variants containing degenerated insert sequences (≥50% intact insert sequence, referred to as degenerated FLAG), and wild type sequences (Supplementary Fig. 3).
Analysis of indel locations in the “Perfect FLAG + indels” group
All the sequence variants were extracted (after filtration in the previous variant analysis) from the group called “Perfect FLAG + indels”. Using Geneious, the sequences were aligned to the reference gene containing the inserted sequence using the “Highest Sensitivity” option. The alignment was used to extract the information about indel positions, and for each deletion the location of the 5′ end of the deleted sequence was chosen to represent the position. The read count for each indel-containing variant was converted to percentage of the total read count of variants from the category “Perfect FLAG” for all individuals. The percentages were plotted on the reference sequence with colors showing AS 24 (green), S 24 (blue), ds 24 (red), S 48 (pink) and S 84 (black) (Fig. 3). In order to analyze the variation in indel positions between the different templates, the percentages of indels located either at the 5′- or 3′-side of the inserted sequence was calculated for each group.
Statistical analyses
D’Agostino Person normality test (column statistics) were used to asses normal distribution of the data. Non-parametric statistical analyses were performed using a Kruskall-Wallis test, followed by Dunn’s multiple comparison test. The tests were carried out using GraphPad Prism 8.0.1.
Supplementary information
Acknowledgements
Our study was funded by the EU-COFASP project: “AQUACRISPR: Optimization of the CRISPR/Cas9 knock-in technology and application in salmon and trout”. We would like to thank Lise Dyrhovden and Ivar Helge Matre for their assistance in rearing. We are also grateful for the assistance of Laura Minsaas for running the MiSeq and Aquagen (Trondheim, Norway) for providing eggs and sperm.
Author contributions
R.B.E. and A.W. designed the project. A.H.S., E.K.S., K.O.S., H.G., L.K., A.W. and R.B.E. performed the microinjections. AHS and RBE designed the ODN templates and collected the tissue samples. A.H.S. made the plasmid template, gRNA and Cas9 RNA, purified the DNA and ran the PCR screening. K.O.S. prepared the Illumina sequencing libraries. E.K.S. performed the bioinformatic analysis. A.H.S., A.W. and R.B.E. analyzed the results and wrote the paper. All authors read and approved the final manuscript.
Data availability
Data generated or analyzed during this study are included in this article (and its Supplementary Information File).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-57295-w.
References
- 1.Ayllon F, et al. The vgll3 Locus Controls Age at Maturity in Wild and Domesticated Atlantic Salmon (Salmo salar L.) Males. PLoS Genet. 2015;11:e1005628. doi: 10.1371/journal.pgen.1005628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glover KA, et al. Half a century of genetic interaction between farmed and wild Atlantic salmon: Status of knowledge and unanswered questions. Fish. Fish. 2017;18:890–927. doi: 10.1111/faf.12214. [DOI] [Google Scholar]
- 3.Bolstad, G. H. et al. Gene flow from domesticated escapes alters the life history of wild Atlantic salmon. Nat Ecol Evol1, doi:UNSP 012410.1038/s41559-017-0124 (2017). [DOI] [PubMed]
- 4.Glover, K. A., Solberg, M. F., Besnier, F. & Skaala, O. Cryptic introgression: evidence that selection and plasticity mask the full phenotypic potential of domesticated Atlantic salmon in the wild. Sci Rep-Uk8, doi:ARTN 1396610.1038/s41598-018-32467-2 (2018). [DOI] [PMC free article] [PubMed]
- 5.Wringe BF, et al. Extensive hybridization following a large escape of domesticated Atlantic salmon in the Northwest Atlantic. Commun. Biol. 2018;1:108. doi: 10.1038/s42003-018-0112-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lien S, et al. The Atlantic salmon genome provides insights into rediploidization. Nature. 2016;533:200–205. doi: 10.1038/nature17164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kjaerner-Semb E, et al. Atlantic salmon populations reveal adaptive divergence of immune related genes - a duplicated genome under selection. BMC Genomics. 2016;17:610. doi: 10.1186/s12864-016-2867-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moen T, et al. Epithelial Cadherin Determines Resistance to Infectious Pancreatic Necrosis Virus in Atlantic Salmon. Genetics. 2015;200:1313–1326. doi: 10.1534/genetics.115.175406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edvardsen RB, Leininger S, Kleppe L, Skaftnesmo KO, Wargelius A. Targeted mutagenesis in Atlantic salmon (Salmo salar L.) using the CRISPR/Cas9 system induces complete knockout individuals in the F0 generation. PLoS One. 2014;9:e108622. doi: 10.1371/journal.pone.0108622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wargelius A, et al. Dnd knockout ablates germ cells and demonstrates germ cell independent sex differentiation in Atlantic salmon. Sci. Rep. 2016;6:21284. doi: 10.1038/srep21284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wargelius A. Application of genome editing in aquatic farm animals: Atlantic salmon. Transgenic Res. 2019;28:101–105. doi: 10.1007/s11248-019-00163-0. [DOI] [PubMed] [Google Scholar]
- 12.Gratacap RL, Wargelius A, Edvardsen RB, Houston RD. Potential of Genome Editing to Improve Aquaculture Breeding and Production. Trends Genet. 2019;35:672–684. doi: 10.1016/j.tig.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Alex, K. et al. CRISPR/Cas9-mediated ablation of elovl2 in Atlantic salmon (Salmo salar L.) inhibits elongation of polyunsaturated fatty acids and induces Srebp-1 and target genes. Sci Rep-Uk (in press) (2019). [DOI] [PMC free article] [PubMed]
- 14.Jiang DN, et al. CRISPR/Cas9-induced disruption of wt1a and wt1b reveals their different roles in kidney and gonad development in Nile tilapia. Dev. Biol. 2017;428:63–73. doi: 10.1016/j.ydbio.2017.05.017. [DOI] [PubMed] [Google Scholar]
- 15.Li M, et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics. 2014;197:591–599. doi: 10.1534/genetics.114.163667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xie QP, et al. Haploinsufficiency of SF-1 Causes Female to Male Sex Reversal in Nile Tilapia, Oreochromis niloticus. Endocrinology. 2016;157:2500–2514. doi: 10.1210/en.2015-2049. [DOI] [PubMed] [Google Scholar]
- 17.Li MH, Liu XY, Dai SF, Xiao HS, Wang DS. High Efficiency Targeting of Non-coding Sequences Using CRISPR/Cas9 System in Tilapia. G3-Genes Genom. Genet. 2019;9:287–295. doi: 10.1534/g3.118.200883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang, X. B. et al. Isolation of Doublesex- and Mab-3-Related Transcription Factor 6 and Its Involvement in Spermatogenesis in Tilapia. Biol Reprod91, doi:ARTN 13610.1095/biolreprod.114.121418 (2014). [DOI] [PubMed]
- 19.Feng, R. J. et al. Retinoic acid homeostasis through aldh1a2 and cyp26a1 mediates meiotic entry in Nile tilapia (Oreochromis niloticus). Sci Rep-Uk5, doi:ARTN 1013110.1038/srep10131 (2015). [DOI] [PMC free article] [PubMed]
- 20.Li M, et al. A Tandem Duplicate of Anti-Mullerian Hormone with a Missense SNP on the Y Chromosome Is Essential for Male Sex Determination in Nile Tilapia, Oreochromis niloticus. PLoS Genet. 2015;11:e1005678. doi: 10.1371/journal.pgen.1005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen J, et al. Heterozygous mutation of eEF1A1b resulted in spermatogenesis arrest and infertility in male tilapia, Oreochromis niloticus. Sci. Rep. 2017;7:43733. doi: 10.1038/srep43733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kishimoto K, et al. Production of a breed of red sea bream Pagrus major with an increase of skeletal muscle mass and reduced body length by genome editing with CRISPR/Cas9. Aquaculture. 2018;495:415–427. doi: 10.1016/j.aquaculture.2018.05.055. [DOI] [Google Scholar]
- 23.Chen J, et al. Efficient Gene Transfer and Gene Editing in Sterlet (Acipenser ruthenus) Front. Genet. 2018;9:117. doi: 10.3389/fgene.2018.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Elaswad, A. et al. Effects of CRISPR/Cas9 dosage on TICAM1 and RBL gene mutation rate, embryonic development, hatchability and fry survival in channel catfish. Sci Rep-Uk8, doi:ARTN 1649910.1038/s41598-018-34738-4 (2018). [DOI] [PMC free article] [PubMed]
- 25.Khalil, K. et al. Generation of Myostatin Gene-Edited Channel Catfish (Ictalurus punctatus) via Zygote Injection of CRISPR/Cas9 System. Sci Rep-Uk7, doi:ARTN 730110.1038/s41598-017-07223-7 (2017). [DOI] [PMC free article] [PubMed]
- 26.Li MH, et al. Retinoic acid triggers meiosis initiation via stra8-dependent pathway in Southern catfish, Silurus meridionalis. Gen. Comp. Endocr. 2016;232:191–198. doi: 10.1016/j.ygcen.2016.01.003. [DOI] [PubMed] [Google Scholar]
- 27.Zhong, Z. M. et al. Targeted disruption of sp7 and myostatin with CRISPR-Cas9 results in severe bone defects and more muscular cells in common carp. Sci Rep-Uk6, doi:ARTN 2295310.1038/srep22953 (2016). [DOI] [PMC free article] [PubMed]
- 28.Baloch Abdul Rasheed, Franěk Roman, Tichopád Tomáš, Fučíková Michaela, Rodina Marek, Pšenička Martin. Dnd1 Knockout in Sturgeons By CRISPR/Cas9 Generates Germ Cell Free Host for Surrogate Production. Animals. 2019;9(4):174. doi: 10.3390/ani9040174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cleveland BM, Yamaguchi G, Radler LM, Shimizu M. Editing the duplicated insulin-like growth factor binding protein-2b gene in rainbow trout (Oncorhynchus mykiss) Sci. Rep. 2018;8:16054. doi: 10.1038/s41598-018-34326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gagnon JA, et al. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS One. 2014;9:e98186. doi: 10.1371/journal.pone.0098186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jasin M, Rothstein R. Repair of strand breaks by homologous recombination. Cold Spring Harb. Perspect. Biol. 2013;5:a012740. doi: 10.1101/cshperspect.a012740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jasin M, Haber JE. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair. (Amst.) 2016;44:6–16. doi: 10.1016/j.dnarep.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salsman J, Dellaire G. Precision genome editing in the CRISPR era. Biochem. Cell Biol. 2017;95:187–201. doi: 10.1139/bcb-2016-0137. [DOI] [PubMed] [Google Scholar]
- 34.Chakrapani V, et al. Establishing targeted carp TLR22 gene disruption via homologous recombination using CRISPR/Cas9. Dev. Comp. Immunol. 2016;61:242–247. doi: 10.1016/j.dci.2016.04.009. [DOI] [PubMed] [Google Scholar]
- 35.Murakami Y, Ansai S, Yonemura A, Kinoshita M. An efficient system for homology-dependent targeted gene integration in medaka (Oryzias latipes) Zool. Lett. 2017;3:10. doi: 10.1186/s40851-017-0071-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gutierrez-Triana, J. A. et al. Efficient single-copy HDR by 5′ modified long dsDNA donors. Elife7, 10.7554/eLife.39468 (2018). [DOI] [PMC free article] [PubMed]
- 37.Irion U, Krauss J, Nusslein-Volhard C. Precise and efficient genome editing in zebrafish using the CRISPR/Cas9 system. Development. 2014;141:4827–4830. doi: 10.1242/dev.115584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kimura Y, Hisano Y, Kawahara A, Higashijima S. Efficient generation of knock-in transgenic zebrafish carrying reporter/driver genes by CRISPR/Cas9-mediated genome engineering. Sci. Rep. 2014;4:6545. doi: 10.1038/srep06545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wierson, W. A. et al. GeneWeld: a method for efficient targeted integration directed by short homology (2019). [DOI] [PMC free article] [PubMed]
- 40.Armstrong GA, et al. Homology Directed Knockin of Point Mutations in the Zebrafish tardbp and fus Genes in ALS Using the CRISPR/Cas9 System. PLoS One. 2016;11:e0150188. doi: 10.1371/journal.pone.0150188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boel Annekatrien, De Saffel Hanna, Steyaert Wouter, Callewaert Bert, De Paepe Anne, Coucke Paul J., Willaert Andy. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Disease Models & Mechanisms. 2018;11(10):dmm035352. doi: 10.1242/dmm.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Farr Gist H., Imani Kimia, Pouv Darren, Maves Lisa. Functional testing of a human PBX3 variant in zebrafish reveals a potential modifier role in congenital heart defects. Disease Models & Mechanisms. 2018;11(10):dmm035972. doi: 10.1242/dmm.035972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prykhozhij SV, et al. Optimized knock-in of point mutations in zebrafish using CRISPR/Cas9. Nucleic Acids Res. 2018;46:9252. doi: 10.1093/nar/gky674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tessadori Federico, Roessler Helen I., Savelberg Sanne M. C., Chocron Sonja, Kamel Sarah M., Duran Karen J., van Haelst Mieke M., van Haaften Gijs, Bakkers Jeroen. Effective CRISPR/Cas9-based nucleotide editing in zebrafish to model human genetic cardiovascular disorders. Disease Models & Mechanisms. 2018;11(10):dmm035469. doi: 10.1242/dmm.035469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hruscha A, et al. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development. 2013;140:4982–4987. doi: 10.1242/dev.099085. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y, Zhang Z, Ge W. An efficient platform for generating somatic point mutations with germline transmission in the zebrafish by CRISPR/Cas9-mediated gene editing. J. Biol. Chem. 2018;293:6611–6622. doi: 10.1074/jbc.RA117.001080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Paix A, et al. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl Acad. Sci. USA. 2017;114:E10745–E10754. doi: 10.1073/pnas.1711979114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Beumer KJ, Trautman JK, Mukherjee K, Carroll D. Donor DNA Utilization During Gene Targeting with Zinc-Finger Nucleases. G3 (Bethesda) 2013;3:657–664. doi: 10.1534/g3.112.005439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paix A, Schmidt H, Seydoux G. Cas9-assisted recombineering in C. elegans: genome editing using in vivo assembly of linear DNAs. Nucleic Acids Res. 2016;44:e128. doi: 10.1093/nar/gkw502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gorodilov YN. Description of the early ontogeny of the Atlantic salmon, Salmo salar, with a novel system of interval (state) identification. Env. Biol. Fish. 1996;47:109–127. doi: 10.1007/Bf00005034. [DOI] [Google Scholar]
- 51.Guo Q, et al. ‘Cold shock’ increases the frequency of homology directed repair gene editing in induced pluripotent stem cells. Sci. Rep. 2018;8:2080. doi: 10.1038/s41598-018-20358-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burger A, et al. Maximizing mutagenesis with solubilized CRISPR-Cas9 ribonucleoprotein complexes. Development. 2016;143:2025–2037. doi: 10.1242/dev.134809. [DOI] [PubMed] [Google Scholar]
- 53.Anna Wargelius, K. O. S. Gene editing can solve major sustainability issues in Atlantic salmon farming. TRANSGENIC RESEARCH27 (2019).
- 54.Yang Su, Li Shihua, Li Xiao-Jiang. Shortening the Half-Life of Cas9 Maintains Its Gene Editing Ability and Reduces Neuronal Toxicity. Cell Reports. 2018;25(10):2653-2659.e3. doi: 10.1016/j.celrep.2018.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kelkar Anju, Zhu Yuqi, Groth Theodore, Stolfa Gino, Stablewski Aimee B., Singhi Naina, Nemeth Michael, Neelamegham Sriram. Doxycycline-Dependent Self-Inactivation of CRISPR-Cas9 to Temporally Regulate On- and Off-Target Editing. Molecular Therapy. 2020;28(1):29–41. doi: 10.1016/j.ymthe.2019.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miura H, Quadros RM, Gurumurthy CB, Ohtsuka M. Easi-CRISPR for creating knock-in and conditional knockout mouse models using long ssDNA donors. Nat. Protoc. 2018;13:195–215. doi: 10.1038/nprot.2017.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoshimi, K. et al. ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nat Commun7, doi:ARTN 1043110.1038/ncomms10431 (2016). [DOI] [PMC free article] [PubMed]
- 58.Veneziano R, et al. In vitro synthesis of gene-length single-stranded DNA. Sci. Rep. 2018;8:6548. doi: 10.1038/s41598-018-24677-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin Marcel. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;17(1):10. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated or analyzed during this study are included in this article (and its Supplementary Information File).