Abstract
Cyanobacteria are attractive microbial hosts for production of chemicals using light and CO2. However, their low productivity of chemicals is a major challenge for commercial applications. This is mostly due to their relatively slow growth rate and carbon partitioning toward biomass rather than products. Many cyanobacterial strains synthesize sucrose as an osmoprotectant to cope with salt stress environments. In this study, we harnessed the photosynthetic machinery of the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 to produce sucrose under salt stress conditions and investigated if the high efficiency of photosynthesis can enhance the productivity of sucrose. By expressing the sucrose transporter CscB, Synechococcus 2973 produced 8 g L−1 of sucrose with a highest productivity of 1.9 g L−1 day−1 under salt stress conditions. The salt stress activated the sucrose biosynthetic pathway mostly via upregulating the sps gene, which encodes the rate-limiting sucrose-phosphate synthase enzyme. To alleviate the demand on high concentrations of salt for sucrose production, we further overexpressed the sucrose synthesis genes in Synechococcus 2973. The engineered strain produced sucrose with a productivity of 1.1 g L−1 day−1 without the need of salt induction. The engineered Synechococcus 2973 in this study demonstrated the highest productivity of sucrose in cyanobacteria.
Subject terms: Applied microbiology, Bacterial genetics
Introduction
Microbial production of fuels and commodity chemicals provides alternative solutions to reduce the reliance on fossil fuel. However, the requirement of sugar feedstock is one of the challenges for sustainable bioproduction. Cyanobacteria are photosynthetic prokaryotes that use light, CO2, and trace amounts of minerals for growth. Compared to terrestrial plants, cyanobacteria have higher efficiencies to utilize solar energy1. In recent years, many synthetic biology tools have been developed for cyanobacteria2. These tools have enabled metabolic engineering of cyanobacteria to produce various chemicals, including fuels3, petrochemicals4, sugars5, fragrances6, and biopolymers7. Although cyanobacteria demonstrate the potential of converting CO2 into desired products, most of the reported titers and productivities are still too low for commercial applications8,9. A more efficient photosynthetic chassis is needed to improve CO2 utilization and carbon partitioning toward products.
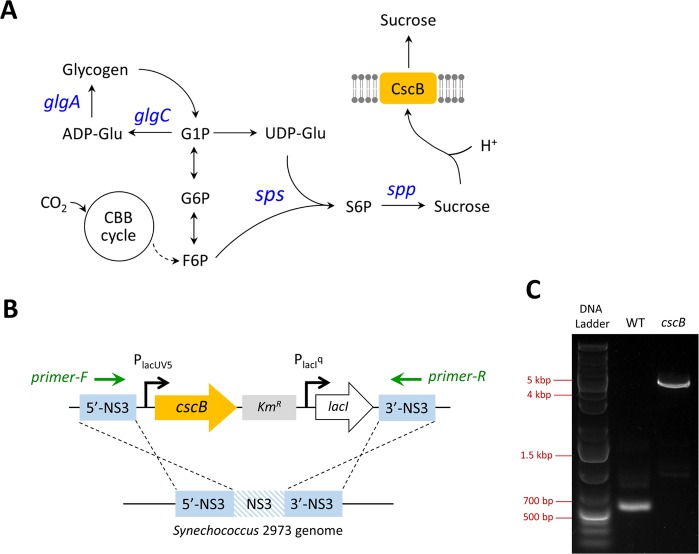
Sucrose is an important feedstock in food industry and bioethanol production. Cyanobacteria synthesize sucrose as a compatible solute to tolerate high salt environments. By synthesizing sucrose, the osmotic pressure can be maintained to avoid desiccation in salt stress conditions. Studies of various cyanobacterial strains showed that more than 60 strains accumulate sucrose under high salt conditions10. In cyanobacterial cells, sucrose is synthesized from uridine diphosphate glucose (UDP-Glu) and fructose 6-phosphate (F6P) by sucrose-phosphate synthase (SPS) and sucrose-phosphate phosphatase (SPP) (Fig. 1A).
Figure 1.
Engineering of Synechococcus elongatus UTEX 2973 for production of sucrose. (A) Metabolic pathways of carbohydrate biosynthesis in Synechococcus 2973. The sucrose permease CscB was expressed for sucrose excretion. Genes highlighted in blue encode enzymes for glycogen and sucrose production. In Synechococcus 2973, the SPS enzyme has both sucrose-phosphate synthase and sucrose-phosphate phosphatase activities. (B) Introduction of the cscB gene into the neutral site 3 (NS3). The primers used for PCR genotyping are highlighted in green. (C) PCR genotyping to confirm complete segregation of cscB. The wild type (WT) Synechococcus 2973 was used as a control. The PCR product of the WT gene is 514 base pairs (bp). No WT copy was present in the cscB-expressing strain. ADP-Glu, adenosine diphosphate glucose; CBB cycle, Calvin-Benson-Bassham cycle; CscB, sucrose permease; F6P, fructose 6-phosphate; G1P, glucose 1-phosphate; G6P, glucose 6-phosphate; S6P, sucrose 6-phosphate; UDP-Glu, uridine diphosphate glucose; glgA, glycogen synthase; glgC, ADP-glucose pyrophosphorylase; spp, sucrose-phosphate phosphatase; sps, sucrose-phosphate synthase.
CscB is a sucrose/H+ symporter which belongs to the oligosaccharide: H+ symporter family of the Major Facilitator Superfamily11. Bacteria with CscB are able to take up sucrose by utilizing the H+ gradient across plasma membranes12. Ducat et al. reported the first study of using the sucrose permease CscB as a sucrose exporter in cyanobacteria13. Upon expression of the E. coli CscB in Synechococcus elongatus PCC 7942, the higher pH value in the extracellular medium creates a reversed proton gradient, which enables the engineered strain to export sucrose13. The cscB-expressing Synechococcus 7942 exported up to 2.6 g L−1 of sucrose with a rate of 0.9 g L−1 day−1 in high salt medium13.
Additionally, a sucrose biosynthetic pathway was also engineered in Synechocystis sp. PCC 6803 by overexpressing the sps and spp genes, leading to a 2-fold enhancement in intracellular sucrose concentration14. Importantly, this study identified that the SPS-catalyzed reaction is the rate-limiting step of sucrose biosynthesis in cyanobacteria14. Recently, the fast-growing cyanobacterium Synechococcus elongatus UTEX 2973 was engineered for sucrose production by expressing the cscB gene5. However, the fast-growing property of Synechococcus 2973 did not lead to a higher sucrose titer or productivity compared to those from its closely-related strain Synechococcus 79425. Compared to Synechococcus 7942, Synechococcus 2973 has a higher carbon flux in the sugar phosphate pathway15 and accumulates a greater amount of glycogen5,16. Hence, sucrose productivity in Synechococcus 2973 is expected to be higher than that in Synechococcus 7942. Therefore, these results demanded further research to uncover the potential of sugar production in this fast-growing strain.
Inspired by these facts, we explored the capability of sucrose production in engineered Synechococcus 2973 and investigated whether the rapid growth rate and strong carbon fluxes in sugar metabolism can lead to a higher production titer of sucrose. In this work, we reported a high sucrose productivity to 1.9 g L−1 day−1 by the engineered Synechococcus 2973. Moreover, we discussed the relationship between glycogen synthesis and sucrose production, providing insights on carbon partitioning of cyanobacteria in salt stress conditions. Finally, we further engineered Synechococcus 2973 to produce sucrose without induction by salt stress. We determined that SPS and SPP are both important enzymes in sucrose biosynthesis, and the expression level of SPP is critical for enhanced sucrose production.
Results
Engineering of Synechococcus 2973 for sucrose production
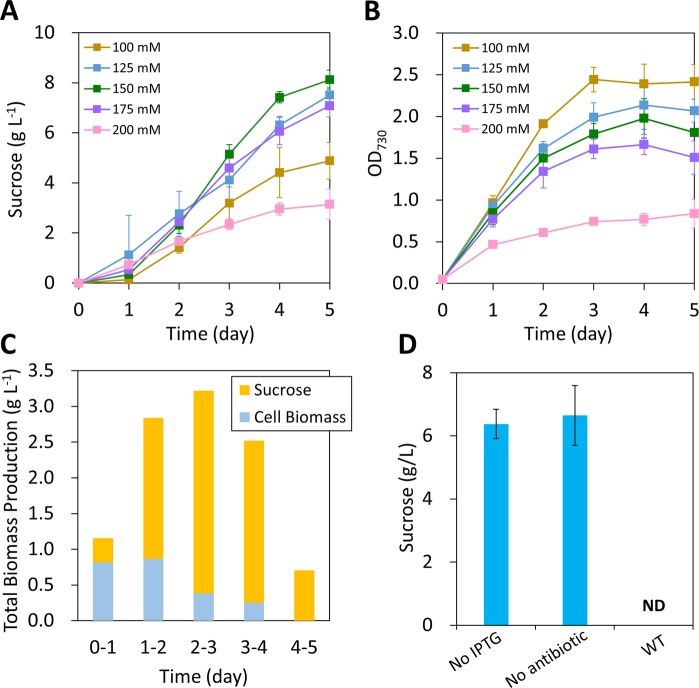
To engineer a sucrose-exporting Synechococcus 2973 strain, the sucrose permease cscB gene was cloned into a neutral site 3 (NS3)-targeting plasmid17, which was then introduced into Synechococcus 2973 via bacterial conjugation (Fig. 1B). A fully segregated strain was obtained by re-patching the plates multiple times, and the corrected genome modification was confirmed by PCR genotyping (Fig. 1C). To test sucrose production in Synechococcus 2973, the engineered 2973-cscB strain (Table 1) was grown in BG11 with 100–200 mM NaCl (Fig. 2A). IPTG (1 mM) was added to induce cscB gene expression. The supernatants were collected for sucrose analysis. The highest sucrose titer, 8 g L−1 in 5 days, was observed in BG11 with 150 mM NaCl. The highest volumetric productivity was 1.9 g L−1 day−1 in 4 days, which corresponds to a sucrose yield of 3.1 g.g−1 cell biomass. This productivity is over 2-fold higher than that from the closely-related strain Synechococcus 7942 (Supplementary Fig. S1). The pH of the medium was lower under sucrose-producing conditions because protons are exported with sucrose by the CscB sucrose/H+ symporter (Supplementary Table S1). Additionally, we observed that salt stress resulted in impaired growth of Synechococcus 2973 (Fig. 2B), leading to decreased OD730 values as NaCl concentrations increased.
Table 1.
Plasmids and strains used in this study.
| Name | Description | Reference |
|---|---|---|
| Plasmids | ||
| pRL443 | conjugal plasmid for bacterial conjugation | 29 |
| pRL623 | helper plasmid for bacterial conjugation | 29 |
| pSL2985 | PlacUV5-cscB (E. coli), lacI, NS3-targeting [CmR] | 13 |
| pSL3005 | PlacUV5-cscB (E. coli), lacI, NS3-targeting [KanR] | This study |
| pSL3037 | PlacUV5-cscB (E. coli), lacI, bom site, NS3-targeting [KanR] | This study |
| pSL3360 | Ptrc1O-sps (Synechocystis 6803), RSF1010 plasmid [GmR] | This study |
| pSL3361 | Ptrc1O-sps-spp (Synechocystis 6803), RSF1010 plasmid [GmR] | This study |
| Synechococcus 2973 strains | ||
| 2973-cscB | NS3::PlacUV5-cscB (created using pSL3037) | This study |
| 2973-cscB-spp | 2973-cscB strain with plasmid pSL3360 | This study |
| 2973-cscB-sps-spp | 2973-cscB strain with plasmid pSL3361 | This study |
Figure 2.
Sucrose production in cscB-expressing Synechococcus 2973. (A) Time-course of sucrose production in the cscB-expressing strain in BG11 medium with various concentrations of NaCl. (B) OD730 of the cscB-expressing strain. (C) Carbon partitioning between sucrose and cell biomass (BG11 with 150 mM NaCl). (D) Sucrose production in different culture conditions with 150 mM NaCl (5 days). The WT Synechococcus 2973 was included as a control. The data represents mean ± sd from three biological replicates. ND, not detected.
To investigate how much photosynthetically fixed carbon is directed to sucrose production under salt stress conditions, we analyzed carbon partitioning between sucrose and cell biomass (Fig. 2C). During the first 24 hours of growth, 71% of the fixed carbon (0.8 g L−1) was used for cell biomass synthesis, and only 0.3 g L−1 of sucrose was produced in this period. From day 1 to day 4, the sucrose titer increased dramatically. During each of these days, the sucrose productivity was greater than 2 g L−1 day−1. The highest amount of sucrose was produced between day 3–4, during which sucrose productivity reached 2.8 g L−1. After day 3, over 88% of the fixed carbon was directed to sucrose. The titer decreased during day 4–5, in which only 0.7 g L−1 sucrose was produced.
Furthermore, we tested the capacity of sucrose production without the addition of IPTG or the antibiotic kanamycin. Without IPTG induction, the 2973-cscB strain produced 6.4 g L−1 sucrose (Fig. 2D), which is 21% lower than that under IPTG-induced conditions (Fig. 2A). A recent study reported that IPTG inducible promoters (trc, lac, and LlacO-1) are leaky in Synechococcus 297318. Our data also indicate that the lacUV5 promoter is leaky in Synechococcus 2973. Similarly, the 2973-cscB strain produced 6.6 g L−1 sucrose without addition of kanamycin (Fig. 2D). Although the cscB gene was inserted into the genome, this result show that kanamycin is required to maintain high sucrose productivity in Synechococcus 2973.
Investigation of carbohydrate metabolism in sucrose-producing Synechococcus 2973
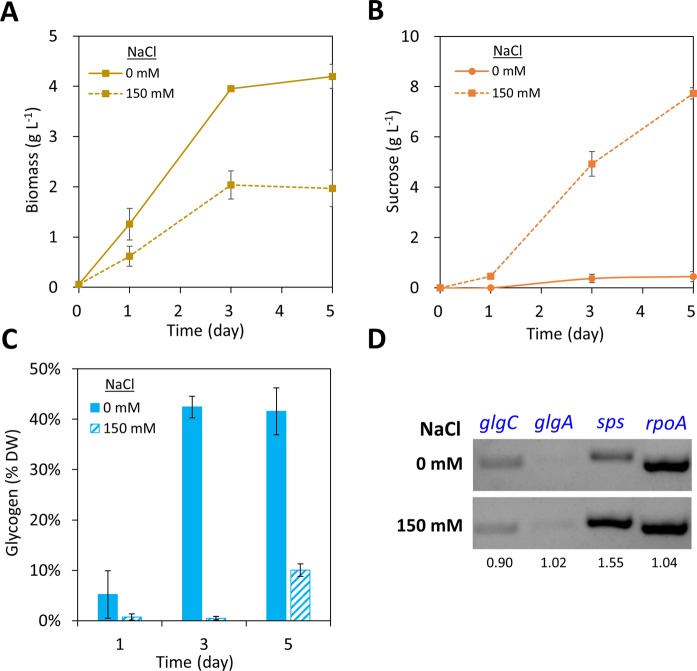
After its exponential growth phase, Synechococcus 2973 accumulates a significant amount of glycogen to reserve excess fixed carbon16. To further understand the carbon allocation between glycogen and sucrose, we analyzed the glycogen content in the sucrose-producing Synechococcus 2973 strain. The 2973-cscB cells were grown in BG11 with or without 150 mM NaCl, and the amounts of glycogen and sucrose were analyzed. The cell biomass was 2-fold higher in BG11 medium without addition of NaCl (Fig. 3A). In the absence of NaCl, Synechococcus 2973 produced only 0.4 g L−1 of sucrose in 5 days, which is 19-fold lower than that in the presence of the salt (Fig. 3B). In contrast, the glycogen content significantly decreased under sucrose-producing conditions. In the presence of 150 mM NaCl, the 2973-cscB strain accumulated glycogen less than 1% of cell dry weight (DW) during day 1 and day 3 (Fig. 3C). On day 5, the glycogen content increased to 10% of DW, corresponding to 0.2 g L−1 of glycogen (40-fold lower than sucrose). Without addition of NaCl, cells accumulated glycogen to more than 40% of DW after 3 days (Fig. 3C). These observations further confirmed that salt stress redirected carbon fluxes from glycogen accumulation to sucrose production in this engineered strain.
Figure 3.
Analysis of carbohydrate metabolism in Synechococcus 2973 cscB-expressing strain. Cells were grown in BG11 medium with or without addition of 150 mM NaCl. (A) Biomass accumulation. (B) Sucrose production. (C) Glycogen content. (D) Semi-quantitative RT-PCR for monitoring gene expression in glycogen and sucrose syntheses. The housekeeping gene rpoA was used as a control. The numbers below the gel represent the relative band intensities from samples growing with 150 mM NaCl versus 0 mM NaCl. The full-length gels are presented in Supplementary Fig. S2. The data in Panels A–C represents mean ± sd from three biological replicates. DW, cell dry weight; rpoA, RNA polymerase subunit α.
We next conducted a semi-quantitative RT-PCR to examine transcription of genes involved in glycogen and sucrose biosynthesis, including glgC, glgA, and sps (Fig. 1A). GlgC (ADP-glucose pyrophosphorylase) and GlgA (glycogen synthase) are the enzymes responsible for glycogen biosynthesis. In Synechococcus elongatus, the SPS enzyme has both sucrose-phosphate synthase and sucrose-phosphate phosphatase activities19. Therefore, expression of the sps gene was monitored. The housekeeping gene rpoA (encoding RNA polymerase subunit α) was used as a control for this RT-PCR experiment20. Transcript levels of glgC and glgA were not significantly different with or without addition of NaCl (Fig. 3D). In contrast, the sucrose synthesis gene sps was highly expressed under salt stress condition (Fig. 3D), suggesting that increased sucrose production in the salt medium is due to upregulation of the sucrose biosynthetic pathway.
Engineering of the sucrose biosynthetic pathway
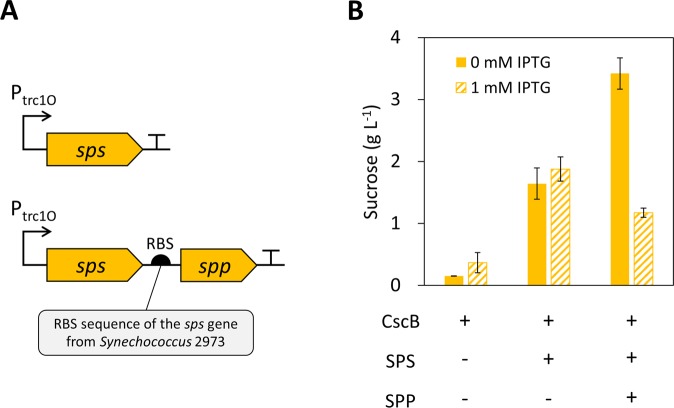
In cyanobacteria, salt stress is required to induce sucrose production. The sucrose biosynthesis gene sps is up-regulated in salt BG11 medium (Fig. 3D). To expand the applications of sucrose production in cyanobacteria, we engineered Synechococcus 2973 to produce sucrose in BG11 medium without added NaCl. The sucrose biosynthetic pathway from Synechocystis sp. PCC 6803 was expressed in the 2973-cscB strain (Fig. 4A). It is known that overexpression of the sps and spp genes from Synechocystis 6803 leads to a 2-fold improvement in intracellular sucrose production under salt stress conditions14. To express the Synechocystis 6803 sps and spp genes in Synechococcus 2973, we cloned them into the self-replicating RSF1010 plasmid21 under the control of an IPTG-inducible trc1O promoter. The plasmids expressing either sps alone or sps-spp genes were then introduced into the 2973-cscB strain (Table 1). Both sps and sps-spp overexpression strains were able to produce sucrose in BG11 medium without additional NaCl (Fig. 4B). In the absence of IPTG, the sps and sps-spp strains produced 1.6 g L−1 and 3.4 g L−1 of sucrose in 3 days, respectively, which were 11-fold and 23-fold higher than the control 2973-cscB strain (Fig. 4B). For the sps strain, the sucrose titers had no significant difference with or without addition of 1 mM IPTG. Interestingly, the sucrose titer decreased significantly upon addition of 1 mM IPTG in the sps-spp strain (Fig. 4B), indicating that full induction of sps and spp expression may be detrimental to sucrose production.
Figure 4.
Engineering of the sucrose biosynthetic pathway in Synechococcus 2973. (A) Schematics of expressing the Synechocystis 6803 sps and spp genes in the 2973-cscB strain. (B) Sucrose production in sps and sps-spp expression strains. Cells were cultured in BG11 medium for 3 days without addition of NaCl. The “+” symbol indicates that the strain contains the indicated overexpressed enzyme(s). The data represents mean ± sd of three biological replicates. RBS, ribosome binding site.
Discussion
During this study, we engineered the fast-growing cyanobacterium Synechococcus UTEX 2973 to produce sucrose. By overexpressing the sucrose permease CscB, the engineered strain produced over 8 g L−1 of sucrose with the highest productivity of 1.9 g L−1 day−1 in 4 days (Fig. 2A). Remarkably, a significant amount of sucrose was produced at the stationary growth phase. From day 2 to day 5, 5.8 g L−1 of sucrose was produced, while the cell biomass increased only 0.7 g L−1 (Fig. 2C). Nearly 90% of the photosynthetic carbon was directed to sucrose synthesis. Surprisingly, the self-shading of light in dense cell cultures had no significant impact on sucrose production. We reasoned that the high productivity of sucrose is a result of push-and-pull of carbon fluxes toward sucrose synthesis. In Synechococcus 2973, the high photosynthetic efficiency16 and strong sugar phosphates pathways15 provide a strong pushing force for sucrose production. Meanwhile, expression of the sucrose permease CscB generates a carbon sink by exporting sucrose out of the cells, thus resulting in continuous sucrose production to avoid desiccation in salt stress conditions.
Compared to all other chemicals produced from cyanobacteria, sucrose production in this study has the highest productivity. Moreover, this is the first study demonstrating cyanobacteria can produce a chemical with a rate over 1 g L−1 day−1 using light and CO2. A recent study reported 12.6 g L−1 of 2,3-butanediol production with a rate of 1.1 g L−1 day−1 from Synechococcus 794222. However, the strain was cultured under photomixotrophic conditions with 15 g L−1 of glucose added as an additional carbon source. The sucrose production in Synechococcus 2973 in the current study used CO2 as the sole carbon source.
Previously engineered Synechococcus 29735 produced sucrose at 0.9 g L−1 day−1, which is 2.1-fold lower than that from our 2973-cscB strain (Supplementary Fig. S1). The discrepancy between the two studies is presumably due to differences in strain design and experimental conditions. In their study, the cscB was controlled by an E. coli trp/lac promoter17, whereas the 2973-cscB strain in our study used the lacUV5 promoter to express cscB13. Moreover, the inoculum preparation and experimental conditions are different. Their experiments were initiated by salt induction of late exponential phase cultures (cell biomass = 1.9 g L−1 dry weight)5. The cultures were grown in 3% CO2 and 250 μmol photons m−2 s−1 light in a column bioreactor5. In our study, the 2973-cscB strain was first acclimated in BG11 medium with 150 mM NaCl for 24 hours, and the experiments were initiated at an OD730 of 0.05 (equivalent to 0.06 g L−1 dry weight) with IPTG induction. The 2973-cscB strain was cultured in 0.5–0.6% CO2 and 250 μmol photons m−2 s−1 light in shaker flasks. We reasoned that the acclimation of cells to salt stress medium enabled the cscB-expressing strain to produce a higher amount of sucrose. Since the salt stress inhibits the activity of photosystems23 and impairs the growth of Synechococcus 2973 (Fig. 2B), the previously reported salt induction strategy may not be able to unlock the potential of sucrose production in Synechococcus 2973. The promoter choice for cscB expression and salt acclimation strategy in our study have led to a higher sucrose productivity by Synechococcus 2973.
The levels of glycogen and sucrose are opposite when the 2973-cscB strain were grown with or without salt stress (Fig. 3B,C). This suggests that glycogen and sucrose syntheses compete for carbon substrates, and the competition can be affected by culture media conditions. The semi-quantitative RT-PCR results showed that the sps gene was significantly upregulated by salt induction (Fig. 3D), which led to significant increase in sucrose production (Fig. 3B). However, expression of the glycogen synthesis genes (glgC and glgA) remained similar upon salt induction (Fig. 3D), indicating that the decreased glycogen content was not due to downregulation of glycogen synthesis. This also suggests that the sucrose biosynthetic pathway is more efficient than the glycogen synthesis pathway in utilizing the carbon substrates in the 2973-cscB strain. In fact, our results corroborate the finding that deletion of the glycogen synthesis pathway has led to a limited improvement on sucrose production in cyanobacteria13. Since a negligible amount of glycogen is produced under salt stress conditions (Fig. 3C), the sucrose titer is unlikely to be further improved by deletion of the glycogen pathway. In fact, a recent study reported that glycogen synthesis may serve as a carbon pool for sucrose production in cyanobacteria24. By using a theophylline-induced riboswitch system, reduced expression of glgC led to decreased sucrose titer in Synechococcus 794224.
By overexpressing the sucrose biosynthesis genes, sucrose production of 2973-cscB cells increased significantly in BG11 medium (Fig. 4B). This is the first study to engineer cyanobacteria for sucrose production without induction of salt stress. The sucrose-producing cyanobacteria can be used as carbon feedstock for heterotrophs to produce useful chemicals25. However, the salt stress media may not be compatible with the culture media for heterotrophs. Engineering a sucrose-producing strain without the need of salt induction could expand the application of using cyanobacteria for co-culture systems. Expression of sps and spp improved the sucrose titer by 23-fold with a productivity of 1.1 g L−1 day−1 (Fig. 4B). With or without addition of 1 mM IPTG, the sps strain produced similar amounts of sucrose (1.6–1.8 g L−1). Notably, sucrose production of the sps-spp coexpression strains were 1.2 g L−1 and 3.4 g L−1 with or without addition of IPTG, respectively (Fig. 4B). The productivity increased 2.9-fold in the absence of IPTG, suggesting that the expression level of spp greatly affects the sucrose titer. The cscB and sps-spp genes were both controlled by IPTG-inducible promoters, while only one copy of the lacI repressor gene was expressed in the strain (Figs. 1B and 4A). The amount of LacI repressor may be insufficient to control gene expression of all 3 genes. Hence, leaky expression of the genes (cscB, sps and spp) led to lower gene expression but significantly improved sucrose production when compared to full induction of the spp gene in the present of 1 mM IPTG (Fig. 4B). We also observed impaired growth of the sps-spp strain with 1 mM IPTG induction (Supplementary Table S2), suggesting that overexpression of the spp gene may cause cell toxicity and reduce sucrose productivity. It is known that SPS is the rate-limiting enzyme of sucrose biosynthesis in cyanobacteria14. Our results suggest that SPP also plays an important role in sucrose production, in that its expression level is critical for enhancing sucrose production.
Conclusions
We have demonstrated the significant potential of engineered Synechococcus UTEX 2973 for photosynthetic sugar production. Expression of the sucrose transporter CscB led to continuous sucrose production with a rate of 1.9 g L−1 day−1 in salt stress conditions. Similar concepts can be applied to harness the strong sugars fluxes in Synechococcus 2973 to produce other valuable sugar compounds26. The salt stress activates the sucrose biosynthetic pathway, which redirects a significant portion of the fixed CO2 toward sucrose production rather than biomass and glycogen accumulation. By engineering the sucrose synthesis pathway in Synechococcus 2973, the mutant strains produced sucrose without salt stimulation, which could expand the versatility of using cyanobacteria for sugar production. Furthermore, we identified SPS and SPP as important enzymes for sucrose synthesis in Synechococcus 2973, while the expression level of SPP is particularly critical for enhancing sucrose production.
Methods
Growth conditions
Synechococcus elongatus UTEX 2973 was grown in BG11 liquid medium at 38 °C, 250 μmol photons m−2 s−1 light, and 0.5–0.6% CO2 in an AlgaeTron growth chamber (Photon Systems Instruments, Czech Republic). For the mutant strains, appropriate antibiotics (10 μg/mL kanamycin and 4 μg/mL gentamicin) were applied in BG11 agar plates or liquid medium. To test sucrose production, strains were first grown in BG11 medium for 24 hours, and then diluted 40-fold to BG11 with 150 mM NaCl to grow for another 24 hours. Afterwards, the salt-acclimated cultures were diluted to an OD730 of 0.05 (0.06 g L−1 cell dry weight) in 10 mL of BG11 medium in 50-mL flasks with desired concentrations of NaCl to initiate the experiments. Water was added daily to compensate for evaporation of BG11 medium. The evaporation rate is 5–7% per day.
Strain construction
All cloning was conducted using the Gibson isothermal DNA assembly method27. The sucrose permease cscB gene from E. coli (ATCC 700927) was introduced into the neutral site 3 (NS3) of Synechococcus UTEX 297317. The cscB was controlled by the lacUV5 promoter. A lac repressor gene (lacI) was included in the plasmid to control cscB expression. The cscB NS3-targeting plasmid was a.pngt from Prof. Daniel Ducat at Michigan State University13. The kanamycin resistance cassette was cloned into the plasmid to replace the chloramphenicol resistance gene. Further, the basis of mobility (bom) sequence derived from plasmid pBR322 was cloned into the plasmid. The bom site is required for gene transfer via bacterial conjugation28. The workflow of plasmid construction is described in Supplementary Fig. S3. The sucrose biosynthesis pathway was expressed in the RSF1010 self-replicating plasmid21. The sps (sll0045) and spp (slr0953) genes were amplified from the genomic DNA of Synechocystis 6803. The ribosome binding site (RBS) sequence of the sps gene in Synechococcus 2973 (20 base pairs proceeding the coding sequence) was used to control translation of spp (Fig. 4A). The trc1O promoter was used to control expression of the genes. The plasmids and mutant strains used in this study are listed in Table 1.
Bacterial conjugation was used to introduce the cscB NS3-targeting or self-replicating plasmids (Table 1) into Synechococcus 2973. E. coli HB101 with plasmid pRL443 was the conjugal strain, whereas E. coli HB101 with plasmid pRL623 and the plasmid carrying the genes of interest was the helper strain. Both E. coli strains and Synechococcus 2973 were grown in LB and BG11 liquid cultures overnight, respectively. 100 μL of E. coli cultures and 400 μl of Synechococcus 2973 culture (OD730 = 0.5) were washed twice with sterilized water and BG11 medium, respectively. Centrifugation was performed at 4,000 × g for 5 mins. The E. coli cells were gently stirred during the wash step. The washed E. coli and Synechococcus 2973 strains were mixed together with 200 μL of BG11 medium and transferred onto a nitrocellulose membrane (0.45 μm pore size, Millipore Sigma, USA) on a BG11 + 5% LB (v/v) agar plate. The plates were incubated at 38 °C, 100 μmol photons m−2 s−1 light, and 0.5% CO2 overnight. The membrane was transferred to new BG11 agar plates with appropriate antibiotics (50 μg/mL kanamycin and 4 μg/mL gentamicin). Colonies appeared on the membranes after 3–5 days of incubation.
Semi-quantitative RT-PCR
RNA was extracted from the 2973-cscB cultures using RNAwiz kit (Ambion/Thermo Fisher Scientific). The RNA samples were treated with DNase. Afterwards, cDNA was synthesized using a Superscript II Reverse Transcriptase kit (Thermo Fischer Scientific). PCR was performed at 24 cycles for semi-quantitative analysis of mRNA transcripts. The band intensities were quantified using ImageJ software. The primers used for PCR are listed in Supplementary Table S3.
Measurements of sucrose, glycogen and biomass
Sucrose in the supernatant was measured using the sucrose/D-glucose assay kit (Megazyme). The glycogen assay was based on the method described in our previous study16. The correlation of OD730 with cell dry weight was DW (g L−1) = 1.3244 × OD730–0.2459 (Supplementary Fig. S4).
Supplementary information
Acknowledgements
Funding to support this work was provided by the Office of Science (BER), U. S. Department of Energy to H.B.P. P.C.L. was partially supported by a fellowship from the McDonnell International Scholars Academy at Washington University. We thank all members of the Pakrasi research group for collegial discussions. The cscB gene was a kind gift from Prof. Daniel Ducat at Michigan State University.
Author contributions
P.C.L. performed experiments and analyzed data. All authors designed research, wrote the manuscript, and read and approved the final manuscript.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
is available for this paper at 10.1038/s41598-019-57319-5.
References
- 1.Dismukes GC, Carrieri D, Bennette N, Ananyev GM, Posewitz MC. Aquatic phototrophs: efficient alternatives to land-based crops for biofuels. Curr. Opin. Biotechnol. 2008;19:235–240. doi: 10.1016/j.copbio.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 2.Sengupta, A., Pakrasi, H. B. & Wangikar, P. P. Recent advances in synthetic biology of cyanobacteria. Applied microbiology and biotechnology, 1–15 (2018). [DOI] [PubMed]
- 3.Gao Z, Zhao H, Li Z, Tan X, Lu X. Photosynthetic production of ethanol from carbon dioxide in genetically engineered cyanobacteria. Energy & Environ. Sci. 2012;5:9857–9865. doi: 10.1039/C2EE22675H. [DOI] [Google Scholar]
- 4.Ungerer J, et al. Sustained photosynthetic conversion of CO2 to ethylene in recombinant cyanobacterium Synechocystis 6803. Energy & Environ. Sci. 2012;5:8998–9006. doi: 10.1039/c2ee22555g. [DOI] [Google Scholar]
- 5.Song K, Tan X, Liang Y, Lu X. The potential of Synechococcus elongatus UTEX 2973 for sugar feedstock production. Appl. microbiology Biotechnol. 2016;100:7865–7875. doi: 10.1007/s00253-016-7510-z. [DOI] [PubMed] [Google Scholar]
- 6.Lin P-C, Saha R, Zhang F, Pakrasi HB. Metabolic engineering of the pentose phosphate pathway for enhanced limonene production in the cyanobacterium Synechocysti s sp. PCC 6803. Sci. Rep. 2017;7:17503. doi: 10.1038/s41598-017-17831-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner J, et al. Co-production of bio-oil and propylene through the hydrothermal liquefaction of polyhydroxybutyrate producing cyanobacteria. Bioresour. Technol. 2016;207:166–174. doi: 10.1016/j.biortech.2016.01.114. [DOI] [PubMed] [Google Scholar]
- 8.Lin PC, Pakrasi HB. Engineering cyanobacteria for production of terpenoids. Planta. 2019;249:145–154. doi: 10.1007/s00425-018-3047-y. [DOI] [PubMed] [Google Scholar]
- 9.Knoot CJ, Ungerer J, Wangikar PP, Pakrasi HB. Cyanobacteria: promising biocatalysts for sustainable chemical production. J. Biol. Chem. 2018;293:5044–5052. doi: 10.1074/jbc.R117.815886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hagemann M. Molecular biology of cyanobacterial salt acclimation. FEMS microbiology Rev. 2011;35:87–123. doi: 10.1111/j.1574-6976.2010.00234.x. [DOI] [PubMed] [Google Scholar]
- 11.Vadyvaloo V, Smirnova IN, Kasho VN, Kaback HR. Conservation of residues involved in sugar/H(+) symport by the sucrose permease of Escherichia coli relative to lactose permease. J. Mol. Biol. 2006;358:1051–1059. doi: 10.1016/j.jmb.2006.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sahin-Toth M, Frillingos S, Lengeler JW, Kaback HR. Active transport by the CscB permease in Escherichia coli K-12. Biochemical biophysical Res. Commun. 1995;208:1116–1123. doi: 10.1006/bbrc.1995.1449. [DOI] [PubMed] [Google Scholar]
- 13.Ducat DC, Avelar-Rivas JA, Way JC, Silver PA. Rerouting carbon flux to enhance photosynthetic productivity. Appl. Environ. microbiology. 2012;78:2660–2668. doi: 10.1128/AEM.07901-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du W, Liang F, Duan Y, Tan X, Lu X. Exploring the photosynthetic production capacity of sucrose by cyanobacteria. Metab. Eng. 2013;19:17–25. doi: 10.1016/j.ymben.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 15.Abernathy MH, et al. Deciphering cyanobacterial phenotypes for fast photoautotrophic growth via isotopically nonstationary metabolic flux analysis. Biotechnol. biofuels. 2017;10:273. doi: 10.1186/s13068-017-0958-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ungerer, J., Lin, P. C., Chen, H. Y. & Pakrasi, H. B. Adjustments to Photosystem Stoichiometry and Electron Transfer Proteins Are Key to the Remarkably Fast Growth of the Cyanobacterium Synechococcus elongatus UTEX 2973. mBio9, 10.1128/mBio.02327-17 (2018). [DOI] [PMC free article] [PubMed]
- 17.Niederholtmeyer H, Wolfstadter BT, Savage DF, Silver PA, Way JC. Engineering cyanobacteria to synthesize and export hydrophilic products. Appl. Environ. microbiology. 2010;76:3462–3466. doi: 10.1128/AEM.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li S, Sun T, Xu C, Chen L, Zhang W. Development and optimization of genetic toolboxes for a fast-growing cyanobacterium Synechococcus elongatus UTEX 2973. Metab. Eng. 2018;48:163–174. doi: 10.1016/j.ymben.2018.06.002. [DOI] [PubMed] [Google Scholar]
- 19.Martinez-Noel GM, Cumino AC, Kolman Mde L, Salerno GL. First evidence of sucrose biosynthesis by single cyanobacterial bimodular proteins. FEBS Lett. 2013;587:1669–1674. doi: 10.1016/j.febslet.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 20.Kim YS, et al. Expression of OsTPX Gene Improves Cellular Redox Homeostasis and Photosynthesis Efficiency in Synechococcus elongatus PCC 7942. Front. plant. Sci. 2018;9:1848. doi: 10.3389/fpls.2018.01848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang HH, Camsund D, Lindblad P, Heidorn T. Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res. 2010;38:2577–2593. doi: 10.1093/nar/gkq164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno M, Carroll AL, Atsumi S. Global metabolic rewiring for improved CO2 fixation and chemical production in cyanobacteria. Nat. Commun. 2017;8:14724. doi: 10.1038/ncomms14724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allakhverdiev SI, Murata N. Salt stress inhibits photosystems II and I in cyanobacteria. Photosynthesis Res. 2008;98:529–539. doi: 10.1007/s11120-008-9334-x. [DOI] [PubMed] [Google Scholar]
- 24.Qiao, C. et al. Effects of Reduced and Enhanced Glycogen Pools on Salt-Induced Sucrose Production in a Sucrose-Secreting Strain of Synechococcus elongatus PCC 7942. Applied and environmental microbiology84, 10.1128/AEM.02023-17 (2018). [DOI] [PMC free article] [PubMed]
- 25.Weiss TL, Young EJ, Ducat DC. A synthetic, light-driven consortium of cyanobacteria and heterotrophic bacteria enables stable polyhydroxybutyrate production. Metab. Eng. 2017;44:236–245. doi: 10.1016/j.ymben.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 26.Frigaard, N.-U. In Genetically EngineeredFoods 31–47 (Elsevier, 2018).
- 27.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 28.Finnegan J, Sherratt D. Plasmid ColE1 conjugal mobility: the nature of bom, a region required in cis for transfer. Mol. Gen. Genet. MGG. 1982;185:344–351. doi: 10.1007/BF00330810. [DOI] [PubMed] [Google Scholar]
- 29.Elhai J, Vepritskiy A, Muro-Pastor AM, Flores E, Wolk CP. Reduction of conjugal transfer efficiency by three restriction activities of Anabaena sp. strain PCC 7120. J. Bacteriol. 1997;179:1998–2005. doi: 10.1128/JB.179.6.1998-2005.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.