Abstract
Podocyte function is tightly linked to the complex organization of its cytoskeleton and adhesion to the underlying glomerular basement membrane. Adhesion of cultured podocytes to a variety of substrates is reported to correlate with podocyte health. To identify novel genes that are important for podocyte function, we designed an in vitro genetic screen based on podocyte adhesion to plates coated with either fibronectin or soluble Fms-like tyrosine kinase-1 (sFLT1)/Fc. A genome-scale pooled RNA interference screen on immortalized human podocytes identified 77 genes that increased adhesion to fibronectin, 101 genes that increased adhesion to sFLT1/Fc, and 44 genes that increased adhesion to both substrates when knocked down. Multiple shRNAs against diphthamide biosynthesis protein 1−4 (DPH1−DPH4) were top hits for increased adhesion. Immortalized human podocyte cells stably expressing these hairpins displayed increased adhesion to both substrates. We then used CRISPR-Cas9 to generate podocyte knockout cells for DPH1, DPH2, or DPH3, which also displayed increased adhesion to both fibronectin and sFLT1/Fc, as well as a spreading defect. Finally, we showed that Drosophila nephrocyte-specific knockdown of Dph1, Dph2, and Dph4 resulted in altered nephrocyte function. In summary, we report here a novel high-throughput method to identify genes important for podocyte function. Given the central role of podocyte adhesion as a marker of podocyte health, these data are a rich source of candidate regulators of glomerular disease.
Keywords: adhesion, diphthamide, genetic screen, glomerulus, podocyte
INTRODUCTION
The most important function attributed to the kidney is the “filtration” of blood and excretion of nitrogenous waste. Filtration is a process by which blood components above a certain molecular weight and possessing a particular charge are retained in the blood while smaller components, such as low-molecular-weight proteins and solutes, are freely filtered. This “perm-selective” process takes place in the renal glomerulus, which houses the glomerular filtration barrier (GFB). The integrity of the GFB results from a complex set of interactions in three-dimensional space between two cell types, podocytes and endothelial cells, and their intervening basement membrane. Podocytes are highly arborized with primary major processes that branch as they extend around the underlying glomerular capillary loops and basement membrane until they form terminal foot processes. Foot processes of neighboring podocytes interdigitate and are connected to one another by a specialized intercellular junction known as the slit diaphragm. Proper slit diaphragm assembly and foot process adhesion regulate podocyte morphology and are therefore crucial to the integrity of the GFB. The interplay between podocyte adhesion and actin cytoskeletal morphology occurs through the interaction of GBM components with cell surface extracellular matrix receptors or charged entities on the podocyte cell surface, which, in turn, transduce signals to downstream intracellular effector proteins or cytoskeletal linker proteins (27).
The canonical podocyte adhesion pathway occurs via the α3β1-integrin heterodimer, which binds laminin 5-2-1 in the basement membrane of mature glomeruli (27). Additionally, podocytes express αvβ3-integrin, which binds to fibronectin (4). There is ample in vivo functional genetic evidence in both humans and mice that integrin-mediated adhesion is crucial to the maintenance of the GFB (10, 12, 25, 26, 41). In vitro, immortalized human podocytes rapidly adhere to laminin, fibronectin, and collagen (34a), a process that is sensitive to integrin-blocking antibodies and divalent cation (Ca2+ and Mg2+) chelation (17).
Although the integrin-mediated mode of adherence is well documented in podocytes, integrin-independent adhesion pathways have also been identified, at least in vitro. Cultured human podocytes rapidly adhere to plates coated with soluble Fms-like tyrosine kinase-1 (sFLT1) but not to other soluble tyrosine kinase receptors. Furthermore, podocyte-specific deletion of Flt1 results in proteinuria and podocyte foot process effacement in mice, which is rescued by a single Flt1 kinase-dead allele, suggesting that the soluble form of the receptor regulates podocyte function (17). In contrast to integrin-mediated adhesion, adhesion of podocytes triggered by sFLT1 is resistant to integrin-blocking antibodies but abolished by the addition of heparin or heparinases, consistent with a charge-based interaction (17). These findings strongly suggest that there are additional genetic pathways that regulate podocyte adhesion that remain to be elucidated.
Advances in RNA interference (RNAi)-based genetic screens have demonstrated the power of this approach to identify novel signaling pathways in a variety of cell types (7). The development of such a high-throughput screen in podocytes has been hampered by the complexity of podocyte morphology and their interactions with other components of the GFB. Given the potential of this technology and the fact that in vitro adhesive phenotypes to fibronectin and sFLT1 correlate with dramatic in vivo functional effects in mice, we hypothesized that podocyte adhesion may be used as a surrogate for podocyte function. In the present study, we report on a high-throughput genome-wide screen designed to identify candidate regulators of podocyte function, specifically with respect to integrin- or sFLT1-mediated adhesion pathways. In addition to the identification of known podocyte-expressed genes that affect adhesion, we also identified novel genes that serve to advance our understanding of podocyte biology. Furthermore, the platform and analysis described may be adapted to efficiently study other pathways of interest in podocytes.
METHODS
Pooled genome-wide shRNA screen.
Optimal puromycin selection conditions, multiplicity of infection (MOI), and adhesion time course were determined in immortalized human podocytes before the screen was performed. The 90k RNAi Consortium library was infected at 500-fold representation in immortalized human podocytes with a target MOI of 0.3 to minimize spurious loss of hairpins in the course of screening and cells harboring multiple integration events (14). Given that the viral titer of each preparation of the 90,000-hairpin library is different, the MOI was determined empirically using a dose response of viral infection followed by 48 h of puromycin selection. Briefly, 15-cm plates of human podocytes were plated with a density of 3 × 106 cells/plate. Increasing doses of virus were administered to two plates for each dose (0, 0.5, 1, 2, and 5 mL). Puromycin was administered at a dose that resulted in 100% lethality in 48 h to one of the plates for each viral dose, whereas the other plate was used as a control. After 48 h of selection, dead cells were washed away and the surviving cells were counted. Percent lethality was calculated by comparing the selected plate with the control plate to determine the appropriate MOI. Twenty-four hours after infection, cells were selected with puromycin for 48 h and then expanded for 7 days. For screening, nonenzymatic cell dissociation was performed to create a dissociated pool of podocytes. Briefly, cells were serum starved overnight in RPMI-1640 with antibiotics. PBS-based enzyme-free cell dissociation solution (Millipore, Darmstadt, Germany), serum-free RPMI 1640, and PBS without Ca2+ and Mg2+ (PBS−/−) were preheated to 33oC. Cells were washed with 10 mL of cell dissociation solution and then incubated for 15 min at 33oC in 10 mL of the same solution. Manual pipetting was then used to dislodge the cells, which were then pelleted and resuspended in 30 mL of serum-free RPMI-1640. The pool was separated into thirds; one-third was frozen to assess baseline hairpin representation, and each of the other two-thirds were plated onto fibronectin- or sFLT1/Fc-coated plates and allowed to adhere for 1 h. Two technical replicates were harvested for the adherent fractions, whereas the floating fractions were pooled to ensure sufficient hairpin counts. After adhesion, the adherent and floating fractions were separated for each substrate, pelleted, and frozen. Genomic DNA was extracted from each cell pellet using the QIAamp Blood Maxi kit (Qiagen, Hilden, Germany). Hairpin sequences were then PCR-amplified using primers that contain adaptor sequences for Illumina sequencing. Each pool was then deconvoluted using an Illumina HiSeq 2500, targeting a lower limit of 200 reads/hairpin (Illumina, San Diego, CA).
Screen data analysis.
Hairpin counts were normalized to the total read counts, and the average baseline representation for each hairpin was logarithmically transformed and plotted as a density distribution. Hairpins that fell outside the normal distribution were discarded. Data analysis was performed in R (R Foundation, Vienna, Austria) separately for hairpins that increased adhesion and hairpins that decreased adhesion to each substrate. To increase the specificity of our data analysis pipeline, we harnessed a unique feature of the screen in that hairpins enriched in adhesion should be correspondingly depleted in the floating fraction and vice versa. We built this biological constraint into the data analysis methods by creating a composite z score for each gene that takes this into account. A composite z score of >3 was considered a hit. Figure 1D shows the data analysis workflow for “increased adhesion.”
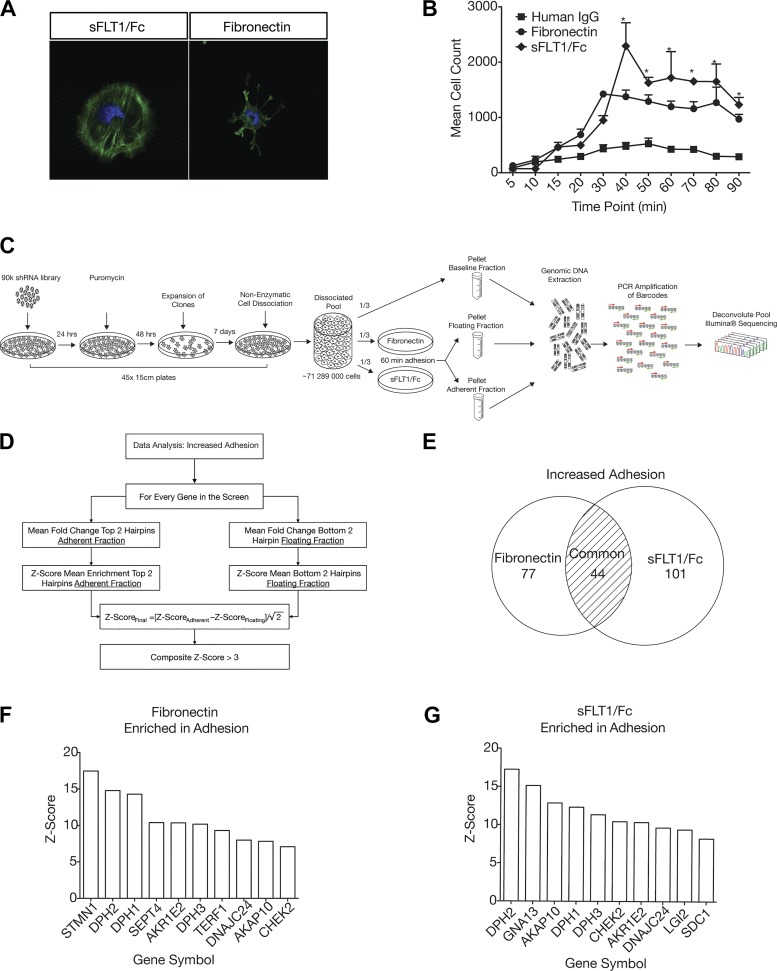
Fig. 1.
Outline of the genome-wide screening protocol to identify genes that regulate podocyte adhesion. A: podocytes plated on soluble Fms-like tyrosine kinase-1 (sFLT1)/Fc and fibronectin have a distinct actin cytoskeletal morphology. Actin is green (phalloidin-Alexa 488); nuclei are blue (Hoechst 33342). B: adhesion kinetics of podocytes to either human IgG, fibronectin, or sFLT1/Fc. Mean cell count and SEs are pictured for each time point and substrate. *Time points at which podocyte adhesion to sFLT1/Fc and fibronectin were statistically significantly higher than human IgG by two-way ANOVA and a post hoc Bonferroni test (P < 0.001). C: protocol for genome-wide adhesion assay screen in podocytes. Plates (45 × 15 cm) of immortalized human podocytes were infected with the TRC 90k pooled genome-wide shRNA library and selected with puromycin for 48 h. Surviving clones were then expanded for 7 days. After expansion of clones, 45 × 15-cm plates were dissociated by nonenzymatic cell dissociation, and the pool was divided into the following three groups: one group was pelleted and frozen to check baseline hairpin representation and the other two groupos were plated onto 5 × 15-cm plates coated with either fibronectin or sFLT1/Fc. After 1 h, adherent and nonadherent cells were collected separately, pooled, spun down, and frozen. Once all assays were complete, genomic DNA was extracted from each pool and sent for Illumina sequencing. D: data analysis of the remaining hairpins was done separately for increased and decreased adhesion based on the “top two” hairpin method. The example of increased adhesion is shown in the flow chart. E: Venn diagram showing that the knockdown of 77 genes increased adhesion to fibronectin, 101 genes increased adhesion to sFLT1/Fc, and 44 genes increased adhesion to both substrates. F: top 10 gene hits for increased adhesion to fibronectin ranked by composite z score. G: top 10 gene hits for increased adhesion to sFLT1/Fc ranked by composite z score. STMN1, stathmin; DPH1−DPH3, diphthamide biosynthesis protein 1−3; SEPT4, septin-4; AKR1E2, aldo-keto reductase family 1 member E2; TERF1, telomeric repeat binding factor 1; DNAJC24, diphthamide biosynthesis protein 4 (DPH4); AKAP10, A-kinase anchoring protein 10; CHEK2, checkpoint kinase 2; GNA13, G protein subunit-α13; SDC1, syndecan 1.
Lentiviral production.
Individual hairpin clones were arrayed from The RNAi Consortium library and expanded overnight using Stbl3 chemically competent bacteria (Life Technologies, Carlsbad, CA). Plasmid DNA was purified using a Qiagen Miniprep kit according to the manufacturer’s instructions. Packaging plasmid (pCMV-dR8.74psPAX2) and envelop plasmid (pMD2.G) were similarly prepared. Human embryonic kidney-293T cells were used for lentiviral packaging by transfecting the corresponding plasmids using X-tremeGENE 9 transfection reagent per the manufacturer’s instructions (Roche, Basel, Switzerland). Virus was harvested at 48 and 72 h posttransfection and then pooled.
Quantitative PCR.
Quantitative PCR was used to determine the degree of mRNA transcript knockdown in shRNA-bearing podocyte cell lines. Briefly, mRNA was extracted using TRIzol (Life Technologies) according to the manufacturer’s instructions. Extracted mRNA was converted to cDNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions. Quantitative PCR primers for diphthamide biosynthesis protein 1−4 (DPH1−DPH4) were designed using the National Center for Biotechnology Information Primer-BLAST tool (see Supplemental Table S2; all Supplemental Data are available online at https://osf.io/8tkpv/?view_only=973d0374e0134d56a9c45c9cec141f65). To further ensure the accuracy of expression data, primers were tested against serial dilutions of pooled cDNA (1:1, 1:10, 1:100, 1:1,000, and 1:10,000) to check their linearity, with the exception of hypoxanthine-guanine phosphoribosyltransferase (HPRT), which had been previously validated. Quantitative PCR was performed using SYBR green master mix (Bio-Rad Laboratories) as per the manufacturer’s instructions on a Bio-Rad CFX384 Real-Time PCR detection system. Data analysis was performed using the ΔΔCt (where Ct is threshold cycle) method.
Podocyte single guide RNA-Cas9 system.
Cas9-expressing podocyte cell lines were generated using lentiviral expression of a modified pXPR_001 plasmid (Addgene, Cambridge, MA). The lentiviral vector pLKOTRC005 was modified to accommodate a BfuAI Golden Gate cloning site (pLCKO) and used as the lentivector for expressing single guide RNA (sgRNA). Individual sgRNA sequences (Supplemental Table S3) were ordered as forward and reverse oligonucleotide sequences with BspMI/BfuAI overhangs. Complementary oligos were phosphorylated and annealed and then ligated into precut pLCKO vector. Ligated vector was then amplified using the method described above. Successful ligation of the appropriate sgRNA sequence was then confirmed by Sanger sequencing using a U6 sequencing primer.
sgRNA lethality assay.
Cas9 editing efficiency in the Pod-Cas9-3 cell line was assessed by expressing sgRNAs against essential genes and quantifying lethality as follows. Pod-Cas9-3 cells were infected with lentivirus-expressing previously validated sgRNAs against luciferase (1 guide), proteasome subunit-β2 (PSMB2; 2 guides), proteasome 26S subunit, non-ATPase 1 (PSMD1; 2 guides), or eukaryotic initiation factor 3 (EIF3; 1 guide) at a target MOI of 0.3–0.5 (Supplemental Table S3). After puromycin selection, cell lines were replated in triplicate and allowed to grow for 8 days. Cells were then counted, and the essential sgRNA-expressing wells (PSMB2, PSMD1, and EIF3) were compared with luciferase sgRNA wells to assess percent lethality.
Multiple cell line adhesion assay.
To validate individual hits, we developed a protocol for assessing the adhesion of multiple podocyte knockdown or knockout lines in the same assay. Cell lines of interest were dissociated using nonenzymatic cell dissociation, counted, and then plated onto a CellCarrier 96-well plate coated with either fibronectin or sFLT1/Fc. Each adhesion experiment used four biological replicates per cell line per condition. After 45 min of adhesion, cells were fixed and stained with Hoechst, and nuclei were then counted using an InCell Analyzer 2000 epifluorescent high-throughput microscope (GE Healthcare, Little Chalfont, UK) and CellProfiler (5) high-content imaging software.
Western blot analysis.
Cells were lysed in buffer F [10 mM Tris (pH 7.05), 50 mM NaCl, 30 mM Na pyrophosphate, 50 mM NaF, 10% glycerol, and 0.5% Triton X-100] and then centrifuged for 10 min. The supernatant was collected, and protein concentration was determined using the Bradford method (Bio-Rad Laboratories). Protein (25 μg) was resolved on 10% bis-Tris gels (Life Technologies) and transferred to Immobilon-P nitrocellulose membranes (Millipore, Burlington, MA). Subsequently, proteins were detected using the following antibodies: rabbit anti-DPH2 (1:500, catalog no. ab54785, Abcam), mouse anti-β-actin (1:10,000, catalog no. 8226, Abcam), and anti-FLAG M2 (1:1,000, catalog no. ab49763, Abcam) for Cas9 detection. The appropriate horseradish peroxidase-conjugated secondary antibody was then used to visualize protein bands on X-ray film using Super Signal chemiluminescence reagent (ThermoFisher Scientific, Waltham, MA) according to the manufacturer’s instructions.
Immunofluorescence.
Glass coverslips (Electron Microscopy Sciences, Hatfield, PA) were put into 12-well tissue culture plates. Cells were either plated at low density on the coverslips directly or adhesion assays were performed as described above. When ready, cells were fixed with 4% paraformaldehyde in PBS for 30 min followed by permeabilization with 0.01% Triton X-100 in PBS. Coverslips were blocked at room temperature with 5% skim milk and PBS−/− with 0.05% Tween 20 (PBS-T) and incubated overnight at 4oC with rabbit monoclonal anti-DPH3 antibody (1:50, catalog no. ab122265, Abcam) or rabbit anti-DPH2 (1:500, catalog no. ab54785, Abcam) in 5% milk and PBS-T. Coverslips were then washed in PBS-T and incubated at room temperature for 1 h with phalloidin Alexa-488 (1:1,000, catalog no. A12379, Molecular Probes), Hoechst 33342 (1:10,000, catalog no. H3570, Molecular Probes), and goat anti-rabbit Alexa-568 (1:1,000, catalog no. A-11077, Molecular Probes) in 5% milk and PBS-T. Coverslips were then washed in PBS-T and mounted onto double-frosted microscope slides (ThermoFisher Scientific) using Dako fluorescent mounting medium (Agilent Dako, Santa Clara, CA). Imaging was performed on a Leica DMI 6000B with a Yokagawa CSU10 confocal scanner and a Hamamatsu EM-CCD camera.
Drosophila studies.
Drosophila homologs for the diphthamide biosynthesis genes were identified using the online Drosophila RNAi Screening Center integrative ortholog prediction tool (http://www.flyrnai.org/diopt) (15). Nephrocyte expression for diphthamide biosynthesis genes was determined using publicly available RNA sequencing fragments per kilobase of exon model per million reads mapped data from www.flybase.org (11). Flies were reared on standard fly food at room temperature or at 29oC for experiments involving Gal4. The flies used in these experiments carried green fluorescent protein (GFP) driven by a Hand promoter (Hand-GFP), yeast transcriptional activation protein Gal4 driven by a Dot promoter (Dot-Gal4), and rat atrial natriuretic factor fused to red fluorescent protein driven by myosin heavy chain (MHC-ANF-RFP) (42). Upstream activation sequence (UAS)-RNAi gene silencing lines for Dph1, Dph2, Dph3, and Dph4 were obtained from the Bloomington Drosophila stock center. Drosophila lines expressing Hand-GFP, Dot-Gal4, MHC-ANF-RFP, and the corresponding UAS-RNAi were generated using standard breeding techniques (see Fig. 5B). Flies carrying the appropriate transgenes were allowed to lay eggs at 25°C. One day after the eggs were laid, they were transferred to 29°C. RFP uptake by pericardial nephrocytes was assessed in adult flies on the first day of life by dissecting cardiac tissue into Drosophila Schneider’s medium (ThermoFisher Scientific) and studying them by fluorescence microscopy. Twenty nephrocytes were analyzed from each fly. For microscopy, Drosophila tissues were fixed in 4% paraformaldehyde in PBS. Confocal imaging was performed with a Zeiss ApoTome.2 microscope using a 20 Plan-Apochromat 0.8 numerical aperture air objective. ImageJ software (version 1.49) was used for image processing.
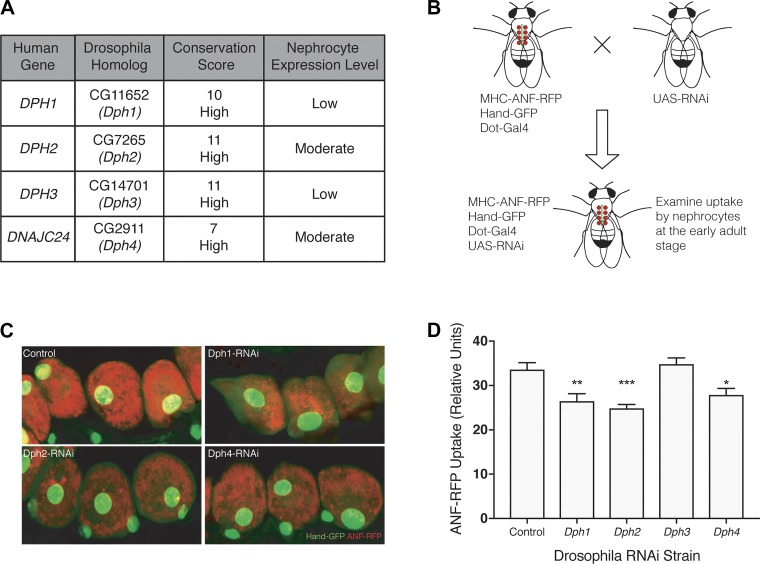
Fig. 5.
Knockdown of diphthamide biosynthesis genes in Drosophila nephrocytes results in altered function. A: diphthamide biosynthesis protein 1−4 (DPH1, DPH2, DPH3, and DPH4) were shown to be well conserved between humans and Drosophila using the DIOPT scoring system. CG11652 (Dph1), CG7265 (Dph2), and CG2911 (Dph4) show moderate expression in nephrocytes, whereas CG14701 (Dph3) has low expression levels. The nephrocyte-specific expression level was determined using publicly available RNA sequencing and microarray data. B: transgenic flies expressing myosin heavy chain (MHC)-atrial natriuretic factor (ANF)-red fluorescent protein (RFP), Hand-green fluorescent protein (GFP), and Dot-Gal4 were crossed with transgenic flies carrying upstream activation sequence (UAS)-RNA interface (RNAi) against Dph1, Dph2, Dph3, or Dph4 to produce flies with an in vivo functional readout for nephrocytes when a specific Dph gene is silenced specifically in nephrocytes. C: Drosophila nephrocytes expressing Hand-GFP (green) took up secreted RFP from the hemolymph. Nephrocyte-specific knockdown of Dph1, Dph2, and Dph4, but not Dph3, resulted in decreased ANF-RFP uptake compared with controls. D: quantification of relative RFP uptake in Drosophila nephrocytes confirmed the statistically significant decreased uptake in Drosophila-expressing nephrocyte-specific RNAi targeting Dph1 (**P = 0.0036), Dph2 (***P = 0.0001), and Dph4 (*P = 0.0162), but not Dph3.
RESULTS
Genetic screen identifies candidate regulators of podocyte adhesion.
Podocytes plated on fibronectin or sFLT1/Fc exhibit distinct actin cytoskeletal morphologies but similar adhesion kinetics (Fig. 1, A and B). Maximal adhesion is achieved on either substrate 40 min after plating. These adhesion properties were used to design a lentivirus-based genome-wide pooled RNAi (18) genetic screen and subsequent analytic approach to identify gene products that regulate podocyte adhesion to either substrate (Fig. 1, C and D). Using this method, we identified 77 genes that increased adhesion to fibronectin, 101 genes that increased adhesion to sFLT1/Fc, and 44 genes that increased adhesion to both substrates when knocked down (Fig. 1E and Supplemental Table S1). Furthermore, we identified 66 genes that decreased adhesion to fibronectin and 106 genes that decreased adhesion to sFLT1 when knocked down (Supplemental Table S1). Interestingly, there was no overlap between these two gene lists. A complete and annotated list of identified genes is available in Supplemental Table S1.
We cross-referenced our hits to a curated set of integrin-related genes derived from publicly available mass spectrometry-derived integrin adhesome complex (IAC) data (13, 38). This analysis showed considerable overlap between the two gene sets for all experimental conditions (Supplemental Table S1). Although this analysis showed the presence of integrin-associated genes upon adhesion to both substrates, integrin-related hits were more common in the fibronectin groups, with 21 IAC genes returning as hits for decreased adhesion to fibronectin and 19 genes as hits for increased adhesion to fibronectin versus 10 and 17 genes, respectively, for sFLT1/Fc. Moreover, the screen was able to identify three integrin-related genes that have previously been described as important to podocyte health: integrin-linked kinase (ILK) (8), parvin-α (PARVA) (40), and PDZ and LIM domain 2 (PDLIM2) (31).
Furthermore, a manual cross-referencing of the screen hits to previously described genes involved in podocyte function, either in human disease or animal models, returned 10 podocyte-related genes with various functions, including adhesion and actin regulation [syndecan 1 (SDC1) (17), podocalyxin like (PODXL) (2), collagen type IV-α3 (COL4A3) (35), ILK (8), PARVA (40), and PDLIM2 (31)] and metabolism [decaprenyl diphosphate synthase subunit 1 (PDSS1) (28), serum and glucocorticoid-regulated kinase 1 (SGK1) (24), tuberous sclerosis protein 1 (TSC1) (16), and coenzyme Q7 (COQ7) (1)] (Supplemental Table S1).
Among the top 10 hits for increased adhesion to either fibronectin or sFLT1, 6 genes were present in both lists [DPH1, DPH2, DPH3, DNAJC24 (DPH4), aldo-keto reductase family 1 member E2 (AKR1E2), and A-kinase anchoring protein 10 (AKAP10); Fig. 1, F and G]. Therein was the most striking observation from the screen: that multiple genes involved in the first step of diphthamide biosynthesis emerged as regulators of adhesion to both substrates. To visualize this effect better, enrichment data for all hairpins from the RNAi library against DPH1 (10 shRNAs), DPH2 (5 shRNAs), DPH3 (5 shRNAs), and DNAJC24 (DPH4; 5 shRNAs), regardless of their level of significance, were extracted from the screen data (Fig. 2, A and B). As a comparison, hairpins against DPH5 (5 shRNAs) and DPH7 (10 shRNAs), which play a role in subsequent steps of diphthamide biosynthesis, were not enriched in either fraction on either substrate (Fig. 2, A–C).
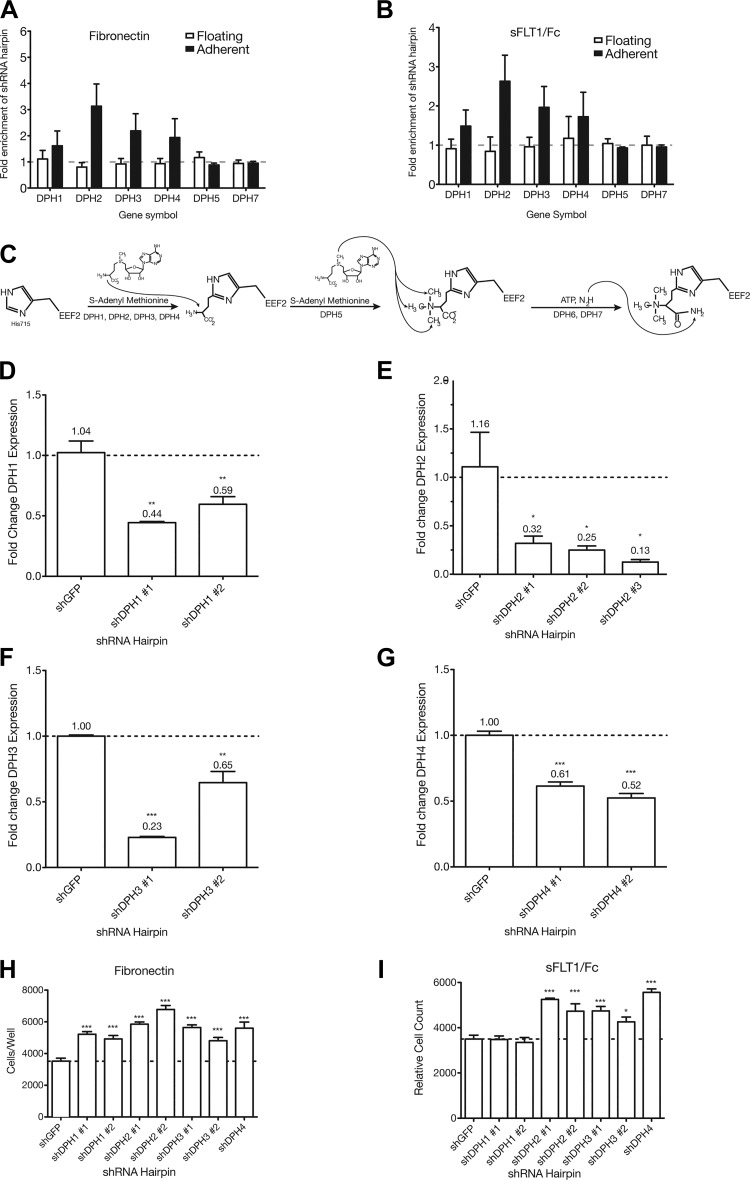
Fig. 2.
shRNA knockdown of diphthamide biosynthesis protein (DPH) genes increases podocyte adhesion to fibronectin and soluble Fms-like tyrosine kinase-1 (sFLT1)/Fc. A and B: mean fold enrichment of all hairpins (significantly enriched and depleted) present in the 90K library against each of DPH1, DPH2, DPH3, and DPH4 in the floating and adherent fraction on either fibronectin or sFLT1/Fc. Other members of the biosynthetic pathway, DPH5 and DPH7, which were not enriched in either fraction on either substrate, are also shown for comparison. C: the diphthamide biosynthesis pathway. DPH1, DPH2, DPH3, and DPH4 all play a role in the first step of diphthamide biosynthesis. DPH5 and DPH7 are involved in subsequent steps. D–G: mean fold change in mRNA expression of the target gene for a given shRNA relative to small hairpin green fluorescent protein (shGFP) in stable podocyte cell lines. SEs and exact values are shown above the bars. D: DPH1 gene expression was diminished to 44% and 59% of shGFP levels in shDPH1 no. 1- and no. 2-expressing cells, respectively. E: DPH2 gene expression was diminished to 32%, 25%, and 13% of shGFP levels in shDPH2 no. 1-, no. 2-, and no. 3-expressing cells, respectively. F: DPH3 gene expression was diminished to 23% and 65% of shGFP levels in shDPH3 no. 1- and no. 2-expressing cells, respectively. G: DPH4 gene expression was diminished to 61% and 52% of shGFP levels in shDPH4 no. 1- and no. 2-expressing cells, respectively. H and I: two hairpins for DPH1, DPH2, and DPH3 and one hairpin for DPH4 were selected to make stable cell lines and were simultaneously assessed for increased adhesion to fibronectin and sFLT1/Fc relative to shGFP-expressing cells. ***P < 0.001; **P < 0.01; *P < 0.05.
Diphthamide biosynthesis genes regulate adhesion in podocytes and other cell types.
Since shRNAs against the diphthamide biosynthesis genes were highly enriched in the adherent fraction on both substrates and are known to function in the same pathway (Fig. 2C), we hypothesized that these genes were good candidates with potential for biological relevance to validate our screen. Cell lines stably expressing shRNA hairpins against DPH1, DPH2, DPH3, and DPH4 showed a statistically significant decrease in mRNA expression of the target gene to between 61% and 13% of control (Fig. 2, D–G). Using these knockdown lines, a high-throughput adhesion assay to confirm their adhesive phenotype on fibronectin and sFLT1/Fc was performed. All stable shRNA-expressing cell lines showed increased adhesion to fibronectin compared with small hairpin green fluorescent protein-expressing cell lines (P < 0.001; Fig. 2H). shDPH1 no. 1- and shDPH1 no. 2-expressing cell lines did not show increased adhesion to sFLT1/Fc compared with small hairpin green fluorescent protein-expressing cell lines, but all other cell lines did (P < 0.001 except for shDPH3 no. 2, where P < 0.05; Fig. 2I). These results not only validated the shRNA “hits” identified in our screen but also supported the previously undescribed diphthamide biosynthesis genes as being involved in regulating podocyte adhesion.
To further validate that the diphthamide biosynthesis genes play a critical role in podocyte adhesion using an orthogonal technology, we used a sgRNA-Cas9 system in podocytes. Stable immortalized human podocyte cell lines were created expressing different levels of FLAG-tagged Cas9 (Fig. 3A). Editing efficiency was tested in the Pod-Cas9-3 cell line (hereafter referred to as Pod-Cas9) with a previously validated lethality assay for “screenability” using sgRNAs against the essential genes PSMB2, PSMD1, and EIF3 (see Supplemental Data). In Pod-Cas9 cells, transduction with sgPSMB2 caused an 80% (P < 0.001) reduction in cell viability, sgPSMD1 caused a 90% (P < 0.001) reduction in cell viability, and sgEIF3 caused a 60% (P < 0.001) reduction in cell viability relative to control sgRNA targeting the luciferase gene (Fig. 3B). These results indicate that the editing efficiency in Pod-Cas9 cells is sufficiently high to detect large changes in cell viability (>50%) after introduction of sgRNAs targeting essential genes. Editing efficiency was further confirmed in this cell line using three different sgRNAs against DPH2 (Fig. 3C) followed by Western blot analysis, which showed almost complete loss of protein expression (Fig. 3D).
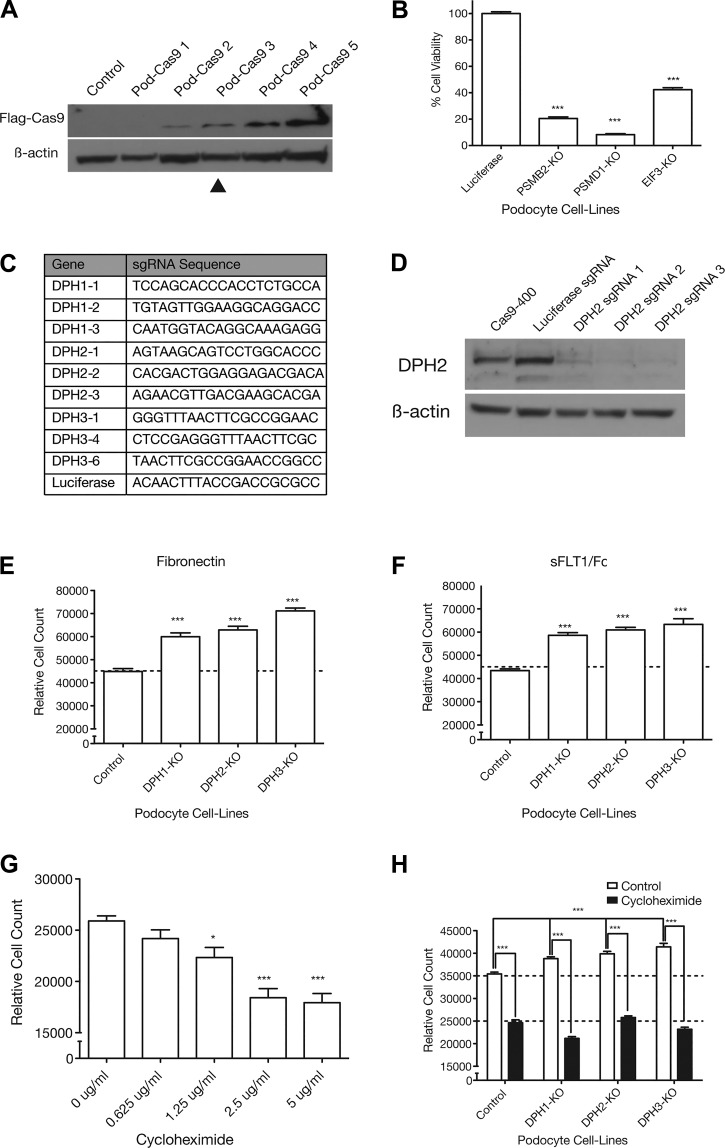
Fig. 3.
The increased adhesion phenotype in podocytes can be recapitulated using lentiCRISPR knockout (KO) of diphthamide biosynthesis genes and requires protein translation. A: Western blot showing increasing Cas9 expression levels in different Cas9-expressing podocyte cell lines. β-Actin was used as a loading control. B: previously validated single guide RNAs (sgRNAs) targeting essential genes [proteasome subunit-β2 (PSMB2), proteasome 26S subunit, non-ATPase 1 (PSMD1), and eukaryotic initiation factor 3 (EIF3)] were used to quantify editing efficiency in the Pod-Cas9-3 cell line by lethality. ***P < 0.001. C: guide sequences for three sgRNAs against each of diphthamide biosynthesis protein 1, 2, and 3 (DPH1, DPH2, and DPH3) and a luciferase control. D: DPH2 editing efficiency was tested in Pod-Cas9-3 cells by infecting three different sgRNAs against DPH2 at low multiplicity of infection. Western blot for DPH2 showed almost complete knockout of DPH2 protein with all three sgRNAs. E and F: three sgRNAs targeting each of DPH1, DPH2, and DPH3 were expressed in Pod-Cas9-3 cells to validate the adhesive phenotypes observed in the screen. Separate guide data were consolidated into one data point for easier visualization. (Individual guide data are available in Supplemental Fig. S1, A and B.) All DPH KO cell lines showed a statistically significant increase in adhesion to fibronectin and soluble Fms-like tyrosine kinase-1 (sFLT1)/Fc compared with control cells. ***P < 0.001. G: blockade of translation elongation by incubating podocytes overnight in cycloheximide caused a dose-dependent decrease in adhesion to fibronectin. H: blockade of translation by overnight incubation with 5 μg/mL cycloheximide in DPH1, DPH2, and DPH3 KO podocytes abolished the increased adhesive phenotype. Deconvoluted sgRNA data are provided in Supplemental Fig. S1F. ***P < 0.001; *P < 0.05.
Once the Pod-Cas9 human podocyte cell line was validated, three sgRNAs were designed against each of DPH1, DPH2, and DPH3 (Fig. 3C) and introduced into Pod-Cas9 cells to create knockout populations. Using the above multicell adhesion protocol, all cell lines expressing sgRNAs against DPH1, DPH2, or DPH3 showed a statistically significant increased adhesiveness to fibronectin (Fig. 3E) and sFLT1/Fc (Fig. 3F) compared with luciferase sgRNA. For clarity, relative cell count is plotted as the mean pooled across sgRNAs. Individual sgRNA data for all experiments are provided in Supplemental Fig. S1.
The diphthamide biosynthesis family of genes is broadly expressed in mammalian tissues. We were therefore interested in investigating if the increased adhesion phenotype might be generalizable to other human cell types. We stably expressed sgRNAs against DPH1, DPH2, DPH3, and luciferase (Supplemental Fig. S2) in previously validated, clonally selected Cas9-expressing retinal pigment epithelial cells (RPE1), colon cancer cells (HCT116), and melanoma cells (A375). Knockout of all three diphthamide biosynthesis genes led to significantly increased adhesion of all cell types to fibronectin compared with luciferase controls (Supplemental Fig. S2).
Inhibition of translation decreases podocyte adhesion.
Previous studies have shown that knockout of DPH1 results in decreased translational efficiency in mouse embryonic fibroblasts because of a loss of the diphthamide modification on eukaryotic elongation factor 2 (EEF2) (21). Given that podocyte diphthamide biosynthesis gene knockout lines should also lack the diphthamide modification, we sought to assess the effect of translation on podocyte adhesiveness using cycloheximide, a well-known bacterial antibiotic that inhibits translation by blocking translational elongation. Incubation of podocytes with increasing doses of cycloheximide caused a dose-dependent decrease in adhesion to fibronectin (1.25 μg/mL, P < 0.05, and 2.5 and 5 μg/mL, P < 0.001; Fig. 3G). Additionally, Pod-Cas9 cells expressing sgRNAs against DPH1, DPH2, DPH3, or luciferase control were treated with either vehicle or 5 μg/mL cycloheximide and then assessed for adhesion to fibronectin. Vehicle-treated cells expressing sgRNAs against either DPH1, DPH2, or DPH3 all showed a statistically significant increase in adhesion to fibronectin compared with luciferase (P < 0.001; Fig. 3H), which was abolished by pretreatment with cycloheximide (P < 0.05; Fig. 3H).
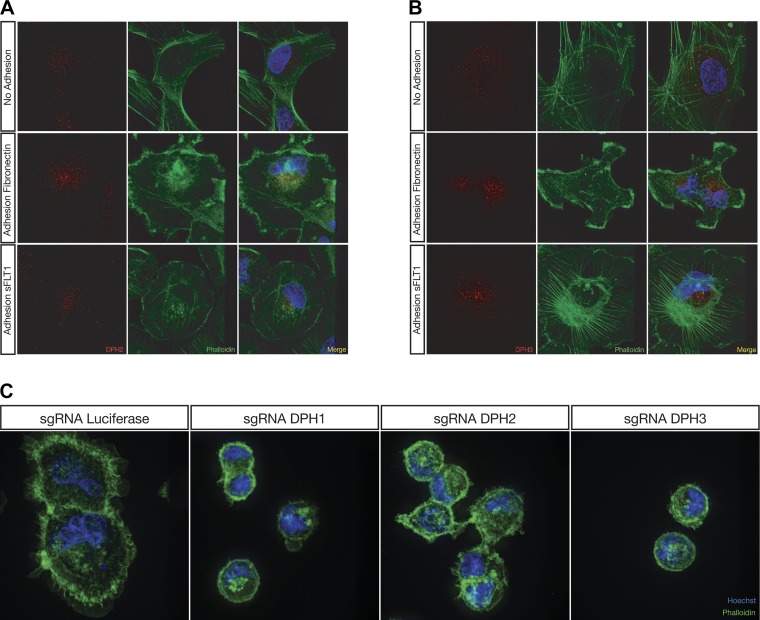
Cellular localization of DPH2 and DPH3.
To explore the localization of DPH2 and DPH3 and gain insight into their role in podocyte function, immunofluorescence was performed on human podocytes stained with antibodies recognizing DPH2 or DPH3 after adhesion to fibronectin or sFLT1/Fc. Under quiescent serum-starved conditions, DPH2 and DPH3 showed a diffuse cytoplasmic distribution (Fig. 4, A and B), whereas upon adhesion to both fibronectin and sFLT1/Fc, the staining patterns for DPH2 (Fig. 4A) and DPH3 (Fig. 4B) localized to a perinuclear compartment, suggesting that a specific spatiotemporal localization is needed for the function of these gene products during cell adhesion.
Fig. 4.
Diphthamide biosynthesis genes localize to a distinct subcellular compartment during adhesion, whereas lentiCRISPR-mediated knockout of diphthamide biosynthesis genes in human podocytes results in a morphological phenotype when adhering to fibronectin. The red channel represents either diphthamide biosynthesis protein (DPH)2 (A) or DPH3 (B), the green channel represents actin staining with phalloidin, and the blue channel is nuclear staining with Hoechst 33342. Podocytes grown on glass coverslips and serum starved overnight exhibited a diffuse cytoplasmic localization of DPH2 (A) and DPH3 (B) protein. Upon adhesion to either fibronectin or soluble Fms-like tyrosine kinase-1 (sFLT1)/Fc, DPH2 (A) and DPH3 (B) protein localized to a perinuclear compartment. Pod-Cas9 cells expressing single guide RNAs (sgRNAs) against DPH1, DPH2, and DPH3 exhibited a spreading defect when plated on fibronectin compared with luciferase sgRNA (C).
Diphthamide biosynthesis genes regulate both cell adhesion and morphology.
We also assessed actin cytoskeletal morphology in the diphthamide biosynthesis knockout podocyte cell lines upon adhesion to fibronectin using phalloidin (Fig. 4C). Compared with control cells, podocyte cells expressing sgDPH1, sgDPH2, or sgDPH3 exhibited a pronounced spreading defect. The spreading defect is characterized by diminished surface area and a lack of typical filipodia and membrane ruffles. These data indicate that the diphthamide biosynthesis genes are important for podocyte spreading.
Diphthamide biosynthesis genes are required for Drosophila nephrocyte function.
Finally, we sought to further validate the role of the diphthamide biosynthesis genes in an in vivo model. The Drosophila nephrocyte has both structural and functional similarities to the human podocyte and offers a wide range of genetic tools that allow for high-throughput validation of genomic data. As such, it is an ideal in vivo model for further validating and triaging our screen hits. First, CG11652 (Dph1), CG7265 (Dph2), CG14701 (Dph3), and CG2911 (Dph4) were identified as Drosophila homologs of the human diphthamide biosynthesis genes using the DIOPT resource and showed high conservation between Drosophila and humans (Fig. 5A). Nephrocyte-specific expression of Dph2 and Dph4 is moderate, whereas nephrocyte-specific expression of Dph1 and Dph3 is low. Drosophila lines expressing MHC-ANF-RFP, Hand-GFP, Dot-Gal4, and the corresponding UAS-RNAi were generated and showed GFP-labeled nephrocytes taking up soluble RFP from the endolymph (Fig. 5, B and C). Nephrocytes expressing RNAi targeting Dph1, Dph2, or Dph4 show visibly diminished ANF-RFP uptake compared with control RNAi (Fig. 5C), whereas Dph3 RNAi-expressing nephrocytes showed no difference (data not shown). Quantification of the fluorescent signal confirmed decreased uptake of ANF-RFP in Dph1 (P = 0.0036), Dph2 (P = 0.0001), and Dph4 (P = 0.0162) but not Dph3 RNAi lines compared with control RNAi (Fig. 5D). These results confirmed that the hits identified from our in vitro genome-wide shRNA screen have direct relevance to in vivo nephrocyte function.
DISCUSSION
In vivo podocyte structure and function is the result of a complex interplay between podocyte actin cytoskeletal dynamics, cell-cell attachments at foot processes, and cell adhesion to the basement membrane. These processes are all linked at focal adhesions (29). Although it is difficult to consider each of these cellular processes in isolation in podocytes, there is strong evidence that several glomerular disease states are a result of decreased podocyte adhesion and podocyte loss (19) or increased podocyte adhesion through activation of integrin signaling (37). In these settings, increased or decreased integrin-mediated podocyte attachment in vitro correlates with in vivo pathology. Our group has also shown that there is an sFLT1-mediated, integrin-independent, heparinase-sensitive pathway regulating podocyte function in vivo, which correlates with podocyte adhesion in vitro (17). Taken together, this would suggest that, although in vitro adhesion, both increased and decreased adhesion, does not completely capture the complexity of podocyte cytoskeletal dynamics, it does correlate with in vivo pathology and can be used to interrogate very focused questions related to podocyte function.
With this in mind, we performed the first shRNA-based genome-wide screen in podocytes to identify gene products that enhance human podocyte adhesion to plates coated with fibronectin or sFLT1/Fc. We hypothesized that podocyte adhesion in vitro may be used as a high-throughput and efficient surrogate assay for podocyte health in vivo. To test this hypothesis, we developed a pooled genome-wide screening and data analysis methodology to identify gene products that regulate podocyte adhesion to fibronectin and/or sFLT1/Fc, which we then validated in vitro and in a Drosophila model. Similar to other work in the field, we have shown by using this approach that, in fact, podocyte adhesion to the basement is likely to be a carefully titrated phenomenon in which too much or too little can cause pathology.
Given the amount of data generated by genome-wide screens, we took several steps to increase the specificity of our screen. First, we used two adhesion substrates with biologically distinct molecular mechanisms. Fibronectin is a well-known substrate for adhesion that binds integrins on the cell surface of podocytes and other cell types. In contrast, sFLT1-mediated adhesion occurs via a membrane complex that contains the glycosphingolipid GM3 and is abolished by the addition of heparin. We posit that “hits” obtained in our screen would identify pathways of convergence and divergence between these two substrates. The second feature of our screen that provides increased specificity is the data analysis algorithm. Many genome-wide screens rely on statistical methods alone to identify hits (14). Although these statistical methods can be powerful, they are not often anchored in biology. To address this, we harnessed a unique feature of our screen, namely that hairpins enriched in adhesion should be correspondingly depleted in the floating fraction and vice versa. This biological constraint was built into the data analysis methods by creating a composite z score for each gene depending on the adhesion feature being explored.
Although this is a novel method for evaluating both podocyte and adhesion biology, the results of this forward genetic screen are provided “face validity” by the presence of hits that align with previously described genes involved in both integrin-mediated adhesion and podocyte health. By cross-referencing our list of hits against a curated set of mass spectrometry-derived IAC components, we showed that integrin-related hits were more common in the fibronectin screening conditions than in the sFLT1/Fc conditions. We also identified, in an unbiased fashion, three integrin-related genes previously described as playing a role in podocyte health: ILK (8), PARVA (40), and PDLIM2 (31). Furthermore, a manual interrogation of each hit from the screen identified 10 other previously described podocyte-related genes involved in adhesion, actin cytoskeletal organization, and metabolism.
One of the most surprising findings from the screen was the considerable overlap of genes that increased adhesion to both substrates upon knockdown, whereas there was no overlap between genes that diminished adhesion to each substrate when knocked down (Fig. 1E). In two adhesion pathways that are expected to be biologically distinct, one might expect little overlap. The findings in the increased adhesion arm suggest that there are nonspecific cell autonomous processes that govern the adhesiveness of podocytes regardless of the substrate in question but that adhesion to a given substrate requires a specific interaction between specific membrane proteins and their downstream effectors. In this article, we have chosen to focus on genes that increase adhesion, as adhesion has been suggested to promote “podocyte health” (27), and these targets are of potential therapeutic interest.
Most notably, four related genes [DPH1, DPH2, DPH3, and DPH4 (DNAJC24)] were among the top 10 hits for increased adhesion to both fibronectin and sFLT1/Fc. We used both shRNA (Fig. 2, H and I) and a CRISPR-Cas9 system (Fig. 3, E and F) to individually confirm that DPH1, DPH2, DPH3, and DPH4 knockdown increase podocyte adhesion. Independent confirmation of these hits with two targeting methods, as well as their coordinate biological role, suggests that these hits were not spurious and provides the first report that this pathway is indeed involved in the regulation of adhesion.
Although genome-wide screens, such as the one carried out here, when applied to study surrogate in vitro phenotypes, such as adhesion, can provide a high-throughput means of elucidating genes that may play a role in podocyte function, the gold standard remains rodent podocyte-specific knockout models or similar studies. However, pursuing such studies can be time consuming and impractical for validating the large number of gene hits from genome-wide screens without extensive triaging. Drosophila melanogaster is a model organism for which high-throughput genetic tools have been developed to facilitate validation of large genomic data sets. Furthermore, the Drosophila pericardial nephrocyte bears a number of morphologic and functional similarities to the renal podocyte. These similarities extend to the gene products that regulate their function; a recent study showed that 34 of the 40 known genes that cause steroid-resistant nephrotic syndrome in humans also result in a phenotype when their homologs are knocked down in nephrocytes (9). With this view, we saw Drosophila as an excellent model organism for further validating hits from our screen.
The diphthamide biosynthesis genes are a well-conserved set of genes among eukaryotes and have been shown to affect the diphthamide modification of EEF2 in organisms from yeast to higher mammals (34). We therefore used the diphthamide biosynthesis genes as a proof of principle and showed that knockdown of three of four diphthamide biosynthesis genes (Dph1, Dph2, and Dph4) resulted in a nephrocyte phenotype (Fig. 5). Although it is unclear why Dph3 knockdown did not produce a phenotype, it is possible that low gene expression level and/or poor efficiency of the RNAi reagents for this gene resulted in suboptimal gene targeting. Interestingly, in this model, the in vitro phenotype of increased adhesion corresponded with a loss of function in vivo phenotype. Although this may be attributable to the limitations of pericardial nephrocytes in accurately modeling the more complex podocytes (23), it may also speak to the role of adhesion itself in podocytes: namely, that podocytes require a delicate balance of adhesion to the basement membrane to maintain their structure and function.
In addition to being well conserved between species, the diphthamide biosynthesis genes are also widely expressed in different types of mammalian cells. We were therefore interested in assessing whether the diphthamide biosynthesis genes play a role in adhesion in other human cell types. Retinal pigment epithelial cells are specialized perivascular epithelial cells that exhibit similar adhesion properties as podocytes on both fibronectin and sFLT1. We therefore sought to determine if this phenotype was reproducible in RPE1 cells and the unrelated HCT116 colon cancer cell line. Interestingly, the phenotype of increased adhesion was conserved in both of these cell types. Another group has previously reported that disruption of Dph3 in the B16F10 murine melanoma cell line resulted in decreased metastases and cell adhesion (36). To corroborate these findings, we used the A375 human melanoma cell line to investigate whether the phenotype of increased adhesion to fibronectin upon diphthamide biosynthesis gene loss was context and cell type dependent. Contrary to the findings of Wang et al. (36), we observed that knockout out of DPH1, DPH2, or DPH3 increased adhesion in A375 cells, suggesting that there may be a broader role for these genes in adhesion in many cell types.
The most-studied function for the DPH1, DPH2, DPH3, and DPH4 gene products is the catalysis of the first step of diphthamide biosynthesis (Fig. 2C). Diphthamide is a uniquely modified histidine residue found only on EEF2 (21). It is so named because it was first described as an ADP-ribosylation site catalyzed by diphtheria toxin using nicotinamide adenine dinucleotide as an ADP-ribosyl donor (21). EEF2 is a GTPase that catalyzes the translocation of peptidyl-tRNAs from the ribosomal A site to the P site where the amino acid residue is added to the nascent polypeptide chain and the tRNA is released. Upon ADP-ribosylation at the diphthamide residue, the function of EEF2 is blocked and translation is inhibited.
One expected feature of the DPH1, DPH2, DPH3, and DPH4 knockdown and knockout podocytes is the lack of diphthamide biosynthesis, raising the possibility that this unique modification of EEF2, beyond being the target of diphtheria toxin, might be playing a role in inhibiting adhesion. To study the biological role of diphthamide modification in the absence of diphtheria toxin, one group generated a Dph1−/− mouse line, which exhibited completely penetrant perinatal lethality (6, 20). Mouse embryonic fibroblasts from the Dph1−/− mouse exhibit a 20–30% impairment in translational efficiency. In light of these findings, we would expect that our DPH1, DPH2, DPH3, or DPH4 knockout podocytes would also exhibit diminished translation, which raises the question of whether globally diminished translation enhances adhesion. The likelihood of this hypothesis is low based on two key results. First, blockade of translation in wild-type podocytes using cycloheximide shows a dose-dependent decrease in adhesion (Fig. 3G). Second, it would appear that translation is necessary for the increased adhesive phenotype of DPH1, DPH2, and DPH3 knockout podocyte cell lines on fibronectin, as blocking translation with cycloheximide abolishes the phenotype (Fig. 3H).
Given that global translational suppression is unlikely to explain the increased adhesive phenotype observed in our DPH knockout cells, it is likely that there are novel roles for these genes in the regulation of adhesion, either related to their function in translation or to other less well-delineated cellular functions.
With respect to their role in translation, one recent study (22) has identified the diphthamide modification on EEF2 as being essential to the translation of selenoproteins, a family of proteins containing the amino acid selenocysteine, which play a role in the maintenance of redox homeostasis. Cells deficient in selenoproteins show increased basal oxidative stress and NF-κB activity (22). Furthermore, overexpression of selenoproteins appears to inhibit adhesion of both breast and lung cancer cells to endothelial monolayers in a NF-κB-dependent manner (39). Mice lacking selenoproteins in podocytes through the podocyte-specific deletion of the Trsp gene, a tRNA required for selenocysteine incorporation, display no podocyte phenotype at baseline or when challenged with diabetes (3). Although it is difficult to interpret these findings in the context of our results, it does suggest that decreased selenoprotein levels, as one would expect to find in diphthamide-deficient cells, are not harmful to podocytes in vivo and may in fact be consistent with increased adhesion.
One other well-delineated function is the role of DPH3 in regulating the exocyst complex as a guanine nucleotide exchange factor-interacting protein for SERGEF (DelGEF) (33). SERGEF is a guanine nucleotide exchange factor that binds to the exocyst component EXOC2 (also known as Sec5) in a Mg2+- and GTP-dependent fashion (32). DPH3 also stimulates this interaction. Knockdown of either SERGEF or DPH3 leads to increased proteoglycan release, and combined knockdown enhances the secretory phenotype further. Studies of the exocyst in podocytes have been limited. In cultured podocytes, the exocyst has been shown to localize to podocyte processes (30). Interestingly, our DPH1, DPH2, and DPH3 knockout podocytes exhibit a cell spreading defect during adhesion, which might point to a failure of process formation at the leading edge (Fig. 4C). Taken together, these data point to a model in which the DPH genes may regulate the exocyst, which, in turn, plays a role not only in adhesion but also in actin cytoskeletal dynamics.
Despite the success of current methods for dissecting the genetics of kidney disease, the underlying cause of many glomerular diseases remain poorly understood, underscoring the need for the development of novel approaches that lead to a better understanding of podocyte biology. In the present study, we have demonstrated the feasibility of leveraging high-throughput shRNA screening technology to study functional genetics in podocytes. We have also shown that gene hits from the screen stand up to rigorous in vitro validation followed by in vivo validation in Drosophila. Further studies can use this validated high-throughput pipeline to continue the systematic interrogation of gene hits from this screen to yield a short list of genes to be further explored in higher-order animal models.
GRANTS
This work was supported by National Institutes of Health Grants R01-HL-124120, P30-DK-114857, and R01-EY-025799 (to S. E. Quaggin) and R01-DK-098410 (to Z. Han), Canadian Institutes of Health Research Grants M0P62931 and M0P77756 (to S. E. Quaggin), and Terry Fox Foundation Grant 105268 (to S. E. Quaggin). S. E. Quaggin holds the Charles Mayo Chair of Medicine at the Feinberg School of Medicine.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
AUTHOR CONTRIBUTIONS
D.P.C., T.K., Y.F., Z.H., M.S., J.M., and S.E.Q. conceived and designed research; D.P.C., T.K., K.R.B., M.C., P.M., C.L., T.O., and Y.F. performed experiments; D.P.C., T.K., K.R.B., P.M., Y.F., Z.H., J.M., and S.E.Q. analyzed data; D.P.C., T.K., K.R.B., M.C., T.O., Y.F., Z.H., J.M., and S.E.Q. interpreted results of experiments; D.P.C., P.M., and Y.F. prepared figures; D.P.C. drafted manuscript; D.P.C., K.R.B., M.C., P.M., T.O., Y.F., Z.H., J.M., and S.E.Q. edited and revised manuscript; K.R.B., M.C., P.M., Z.H., M.S., J.M., and S.E.Q. approved final version of manuscript.
ACKNOWLEDGMENTS
The authors thank Dr. Laurence Pelletier and Dr. Gagan Gupta for initial guidance with methodological approaches to genetic screening. The authors also thank Sonali Weerawardane and Jasmyne Carnevale for assistance in executing the genetic screen and Dr. Traver Hart for consulting on data analysis.
REFERENCES
- 1.Ashraf S, Gee HY, Woerner S, Xie LX, Vega-Warner V, Lovric S, Fang H, Song X, Cattran DC, Avila-Casado C, Paterson AD, Nitschké P, Bole-Feysot C, Cochat P, Esteve-Rudd J, Haberberger B, Allen SJ, Zhou W, Airik R, Otto EA, Barua M, Al-Hamed MH, Kari JA, Evans J, Bierzynska A, Saleem MA, Böckenhauer D, Kleta R, El Desoky S, Hacihamdioglu DO, Gok F, Washburn J, Wiggins RC, Choi M, Lifton RP, Levy S, Han Z, Salviati L, Prokisch H, Williams DS, Pollak M, Clarke CF, Pei Y, Antignac C, Hildebrandt F. ADCK4 mutations promote steroid-resistant nephrotic syndrome through CoQ10 biosynthesis disruption. J Clin Invest 123: 5179–5189, 2013. doi: 10.1172/JCI69000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barua M, Shieh E, Schlondorff J, Genovese G, Kaplan BS, Pollak MR. Exome sequencing and in vitro studies identified podocalyxin as a candidate gene for focal and segmental glomerulosclerosis. Kidney Int 85: 124–133, 2014. doi: 10.1038/ki.2013.354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blauwkamp MN, Yu J, Schin MA, Burke KA, Berry MJ, Carlson BA, Brosius FC III, Koenig RJ. Podocyte specific knock out of selenoproteins does not enhance nephropathy in streptozotocin diabetic C57BL/6 mice. BMC Nephrol 9: 7, 2008. doi: 10.1186/1471-2369-9-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borza CM, Borza DB, Pedchenko V, Saleem MA, Mathieson PW, Sado Y, Hudson HM, Pozzi A, Saus J, Abrahamson DR, Zent R, Hudson BG. Human podocytes adhere to the KRGDS motif of the alpha3alpha4alpha5 collagen IV network. J Am Soc Nephrol 19: 677–684, 2008. doi: 10.1681/ASN.2007070793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, Golland P, Sabatini DM. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100, 2006. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CM, Behringer RR. Ovca1 regulates cell proliferation, embryonic development, and tumorigenesis. Genes Dev 18: 320–332, 2004. doi: 10.1101/gad.1162204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genet 7: 373–384, 2006. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 8.El-Aouni C, Herbach N, Blattner SM, Henger A, Rastaldi MP, Jarad G, Miner JH, Moeller MJ, St-Arnaud R, Dedhar S, Holzman LB, Wanke R, Kretzler M. Podocyte-specific deletion of integrin-linked kinase results in severe glomerular basement membrane alterations and progressive glomerulosclerosis. J Am Soc Nephrol 17: 1334–1344, 2006. doi: 10.1681/ASN.2005090921. [DOI] [PubMed] [Google Scholar]
- 9.Fu Y, Zhu J-Y, Richman A, Zhao Z, Zhang F, Ray PE, Han Z. A Drosophila model system to assess the function of human monogenic podocyte mutations that cause nephrotic syndrome. Hum Mol Genet 26: 768–780, 2017. doi: 10.1093/hmg/ddw428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldberg S, Adair-Kirk TL, Senior RM, Miner JH. Maintenance of glomerular filtration barrier integrity requires laminin alpha5. J Am Soc Nephrol 21: 579–586, 2010. doi: 10.1681/ASN.2009091004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gramates LS, Marygold SJ, Santos GD, Urbano J-M, Antonazzo G, Matthews BB, Rey AJ, Tabone CJ, Crosby MA, Emmert DB, Falls K, Goodman JL, Hu Y, Ponting L, Schroeder AJ, Strelets VB, Thurmond J, Zhou P; the FlyBase Consortium . FlyBase at 25: looking to the future. Nucleic Acids Res 45: D663–D671, 2017. doi: 10.1093/nar/gkw1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Has C, Spartà G, Kiritsi D, Weibel L, Moeller A, Vega-Warner V, Waters A, He Y, Anikster Y, Esser P, Straub BK, Hausser I, Bockenhauer D, Dekel B, Hildebrandt F, Bruckner-Tuderman L, Laube GF. Integrin α3 mutations with kidney, lung, and skin disease. N Engl J Med 366: 1508–1514, 2012. doi: 10.1056/NEJMoa1110813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horton ER, Byron A, Askari JA, Ng DHJ, Millon-Frémillon A, Robertson J, Koper EJ, Paul NR, Warwood S, Knight D, Humphries JD, Humphries MJ. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat Cell Biol 17: 1577–1587, 2015. doi: 10.1038/ncb3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu G, Luo J. A primer on using pooled shRNA libraries for functional genomic screens. Acta Biochim Biophys Sin (Shanghai) 44: 103–112, 2012. doi: 10.1093/abbs/gmr116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics 12: 357, 2011. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inoki K, Mori H, Wang J, Suzuki T, Hong S, Yoshida S, Blattner SM, Ikenoue T, Rüegg MA, Hall MN, Kwiatkowski DJ, Rastaldi MP, Huber TB, Kretzler M, Holzman LB, Wiggins RC, Guan KL. mTORC1 activation in podocytes is a critical step in the development of diabetic nephropathy in mice. J Clin Invest 121: 2181–2196, 2011. doi: 10.1172/JCI44771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin J, Sison K, Li C, Tian R, Wnuk M, Sung HK, Jeansson M, Zhang C, Tucholska M, Jones N, Kerjaschki D, Shibuya M, Fantus IG, Nagy A, Gerber HP, Ferrara N, Pawson T, Quaggin SE. Soluble FLT1 binds lipid microdomains in podocytes to control cell morphology and glomerular barrier function. Cell 151: 384–399, 2012. doi: 10.1016/j.cell.2012.08.037. [DOI] [PubMed] [Google Scholar]
- 18.Ketela T, Heisler LE, Brown KR, Ammar R, Kasimer D, Surendra A, Ericson E, Blakely K, Karamboulas D, Smith AM, Durbic T, Arnoldo A, Cheung-Ong K, Koh JLY, Gopal S, Cowley GS, Yang X, Grenier JK, Giaever G, Root DE, Moffat J, Nislow C. A comprehensive platform for highly multiplexed mammalian functional genetic screens. BMC Genomics 12: 213, 2011. doi: 10.1186/1471-2164-12-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lennon R, Randles MJ, Humphries MJ. The importance of podocyte adhesion for a healthy glomerulus. Front Endocrinol (Lausanne) 5: 160, 2014. doi: 10.3389/fendo.2014.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liang M, Ayanga B, Du S, Godwin AK, Hartsock JK, Evans SC. Ovca1, a candidate gene of the genetic modifier of Tp53, Mop2, affects mouse embryonic lethality. Genes Chromosomes Cancer 47: 315–325, 2008. doi: 10.1002/gcc.20535. [DOI] [PubMed] [Google Scholar]
- 21.Liu S, Bachran C, Gupta P, Miller-Randolph S, Wang H, Crown D, Zhang Y, Wein AN, Singh R, Fattah R, Leppla SH. Diphthamide modification on eukaryotic elongation factor 2 is needed to assure fidelity of mRNA translation and mouse development. Proc Natl Acad Sci USA 109: 13817–13822, 2012. doi: 10.1073/pnas.1206933109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mayer K, Mundigl O, Kettenberger H, Birzele F, Stahl S, Pastan I, Brinkmann U. Diphthamide affects selenoprotein expression: diphthamide deficiency reduces selenocysteine incorporation, decreases selenite sensitivity and pre-disposes to oxidative stress. Redox Biol 20: 146–156, 2019. doi: 10.1016/j.redox.2018.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Na J, Cagan R. The Drosophila nephrocyte: back on stage. J Am Soc Nephrol 24: 161–163, 2013. doi: 10.1681/ASN.2012121227. [DOI] [PubMed] [Google Scholar]
- 24.Nagase M, Yoshida S, Shibata S, Nagase T, Gotoda T, Ando K, Fujita T. Enhanced aldosterone signaling in the early nephropathy of rats with metabolic syndrome: possible contribution of fat-derived factors. J Am Soc Nephrol 17: 3438–3446, 2006. doi: 10.1681/ASN.2006080944. [DOI] [PubMed] [Google Scholar]
- 25.Noakes PG, Miner JH, Gautam M, Cunningham JM, Sanes JR, Merlie JP. The renal glomerulus of mice lacking s-laminin/laminin beta 2: nephrosis despite molecular compensation by laminin beta 1. Nat Genet 10: 400–406, 1995. doi: 10.1038/ng0895-400. [DOI] [PubMed] [Google Scholar]
- 26.Pozzi A, Jarad G, Moeckel GW, Coffa S, Zhang X, Gewin L, Eremina V, Hudson BG, Borza DB, Harris RC, Holzman LB, Phillips CL, Fassler R, Quaggin SE, Miner JH, Zent R. Beta1 integrin expression by podocytes is required to maintain glomerular structural integrity. Dev Biol 316: 288–301, 2008. doi: 10.1016/j.ydbio.2008.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sachs N, Sonnenberg A. Cell-matrix adhesion of podocytes in physiology and disease. Nat Rev Nephrol 9: 200–210, 2013. doi: 10.1038/nrneph.2012.291. [DOI] [PubMed] [Google Scholar]
- 28.Saiki R, Lunceford AL, Shi Y, Marbois B, King R, Pachuski J, Kawamukai M, Gasser DL, Clarke CF. Coenzyme Q10 supplementation rescues renal disease in Pdss2kd/kd mice with mutations in prenyl diphosphate synthase subunit 2. Am J Physiol Renal Physiol 295: F1535–F1544, 2008. doi: 10.1152/ajprenal.90445.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schell C, Huber TB. The evolving complexity of the podocyte cytoskeleton. J Am Soc Nephrol 28: 3166–3174, 2017. doi: 10.1681/ASN.2017020143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons M, Saffrich R, Reiser J, Mundel P. Directed membrane transport is involved in process formation in cultured podocytes. J Am Soc Nephrol 10: 1633–1639, 1999. [DOI] [PubMed] [Google Scholar]
- 31.Sistani L, Dunér F, Udumala S, Hultenby K, Uhlén M, Betsholtz C, Tryggvason K, Wernerson A, Patrakka J. Pdlim2 is a novel actin-regulating protein of podocyte foot processes. Kidney Int 80: 1045–1054, 2011. doi: 10.1038/ki.2011.231. [DOI] [PubMed] [Google Scholar]
- 32.Sjölinder M, Uhlmann J, Ponstingl H. DelGEF, a homologue of the Ran guanine nucleotide exchange factor RanGEF, binds to the exocyst component Sec5 and modulates secretion. FEBS Lett 532: 211–215, 2002. doi: 10.1016/S0014-5793(02)03677-3. [DOI] [PubMed] [Google Scholar]
- 33.Sjölinder M, Uhlmann J, Ponstingl H. Characterisation of an evolutionary conserved protein interacting with the putative guanine nucleotide exchange factor DelGEF and modulating secretion. Exp Cell Res 294: 68–76, 2004. doi: 10.1016/j.yexcr.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 34.Su X, Lin Z, Lin H. The biosynthesis and biological function of diphthamide. Crit Rev Biochem Mol Biol 48: 515–521, 2013. doi: 10.3109/10409238.2013.831023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34a.Tian X, Kim JJ, Monkley SM, Gotoh N, Nandez R, Soda K, Inoue K, Balkin DM, Hassan H, Son SH, Lee Y, Moeckel G, Calderwood DA, Holzman LB, Critchley DR, Zent R, Reiser J, Ishibe S. Podocyte-associated talin1 is critical for glomerular filtration barrier maintenance. J Clin Invest 124: 1098–1113, 2014. doi: 10.1172/JCI69778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voskarides K, Damianou L, Neocleous V, Zouvani I, Christodoulidou S, Hadjiconstantinou V, Ioannou K, Athanasiou Y, Patsias C, Alexopoulos E, Pierides A, Kyriacou K, Deltas C. COL4A3/COL4A4 mutations producing focal segmental glomerulosclerosis and renal failure in thin basement membrane nephropathy. J Am Soc Nephrol 18: 3004–3016, 2007. doi: 10.1681/ASN.2007040444. [DOI] [PubMed] [Google Scholar]
- 36.Wang L, Shi Y, Ju P, Liu R, Yeo SP, Xia Y, Owlanj H, Feng Z. Silencing of diphthamide synthesis 3 (Dph3) reduces metastasis of murine melanoma. PLoS One 7: e49988, 2012. doi: 10.1371/journal.pone.0049988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei C, El Hindi S, Li J, Fornoni A, Goes N, Sageshima J, Maiguel D, Karumanchi SA, Yap HK, Saleem M, Zhang Q, Nikolic B, Chaudhuri A, Daftarian P, Salido E, Torres A, Salifu M, Sarwal MM, Schaefer F, Morath C, Schwenger V, Zeier M, Gupta V, Roth D, Rastaldi MP, Burke G, Ruiz P, Reiser J. Circulating urokinase receptor as a cause of focal segmental glomerulosclerosis. Nat Med 17: 952–960, 2011. doi: 10.1038/nm.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Winograd-Katz SE, Fässler R, Geiger B, Legate KR. The integrin adhesome: from genes and proteins to human disease. Nat Rev Mol Cell Biol 15: 273–288, 2014. doi: 10.1038/nrm3769. [DOI] [PubMed] [Google Scholar]
- 39.Wrobel JK, Choi JJ, Xiao R, Eum SY, Kwiatkowski S, Wolff G, Spangler L, Power RF, Toborek M. Selenoglycoproteins attenuate adhesion of tumor cells to the brain microvascular endothelium via a process involving NF-κB activation. J Nutr Biochem 26: 120–129, 2015. doi: 10.1016/j.jnutbio.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang Y, Guo L, Blattner SM, Mundel P, Kretzler M, Wu C. Formation and phosphorylation of the PINCH-1-integrin linked kinase-alpha-parvin complex are important for regulation of renal glomerular podocyte adhesion, architecture, and survival. J Am Soc Nephrol 16: 1966–1976, 2005. doi: 10.1681/ASN.2004121112. [DOI] [PubMed] [Google Scholar]
- 41.Zenker M, Aigner T, Wendler O, Tralau T, Müntefering H, Fenski R, Pitz S, Schumacher V, Royer-Pokora B, Wühl E, Cochat P, Bouvier R, Kraus C, Mark K, Madlon H, Dötsch J, Rascher W, Maruniak-Chudek I, Lennert T, Neumann LM, Reis A. Human laminin beta2 deficiency causes congenital nephrosis with mesangial sclerosis and distinct eye abnormalities. Hum Mol Genet 13: 2625–2632, 2004. doi: 10.1093/hmg/ddh284. [DOI] [PubMed] [Google Scholar]
- 42.Zhang F, Zhao Y, Han Z. An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J Am Soc Nephrol 24: 191–197, 2013. doi: 10.1681/ASN.2012080769. [DOI] [PMC free article] [PubMed] [Google Scholar]