Abstract
Precise timing in hormone release from the hypothalamus, the pituitary and ovary is critical for fertility. Hormonal release patterns of the reproductive axis are regulated by a feedback loop within the hypothalamic-pituitary-gonadal (HPG) axis. The timing and rhythmicity of hormone release and tissue sensitivity in the HPG axis is regulated by circadian clocks located in the hypothalamus (suprachiasmatic nucleus, kisspeptin and GnRH neurons), the pituitary (gonadotrophs), the ovary (theca and granulosa cells), the testis (Leydig cells), as well as the uterus (endometrium and myometrium). The circadian clocks integrate environmental and physiological signals to produce cell endogenous rhythms generated by a transcriptional-translational feedback loop of transcription factors that are collectively called the “molecular clock”. This review specifically focuses on the contribution of molecular clock transcription factors in regulating hormone release patterns in the reproductive axis, with an emphasis on the female reproductive system. Specifically, we discuss the contributions of circadian rhythms in distinct neuronal populations of the female hypothalamus, the molecular clock in the pituitary and its overall impact on female and male fertility.
Keywords: Circadian rhythms, hypothalamic-pituitary-gonadal axis, clock genes, gene transcription, hormone release, suprachiasmatic nucleus, ovary, gonadotropin-releasing hormone, kisspeptin, estrogen
Introduction
Reproduction is tightly controlled by circadian (rhythms of ~24h) and seasonal rhythms. In seasonal breeders, changes in day length are linked to transitions in hormone release patterns within the reproductive axis. This allows for the fine-tuning of reproductive status to the time of year in which there is the highest chance of survival of the offspring and the mother. Hormone release patterns in seasonal breeders are well studied and will not be addressed here (For reviews on seasonal changes in reproductive status see (Ebenhöh and Hazlerigg 2013, Simonneaux, Bahougne et al. 2017). What is less studied, is the role of cell endogenous circadian rhythms in hormone release patterns of the reproductive axis. Timed hormone release within the hypothalamic-pituitary-gonadal (HPG) axis is required for normal fertility. This review will focus on the contributions of molecular clock transcription factors in proper hormone release and target tissue sensitivity within the reproductive axis with an emphasis on females.
The reproductive axis
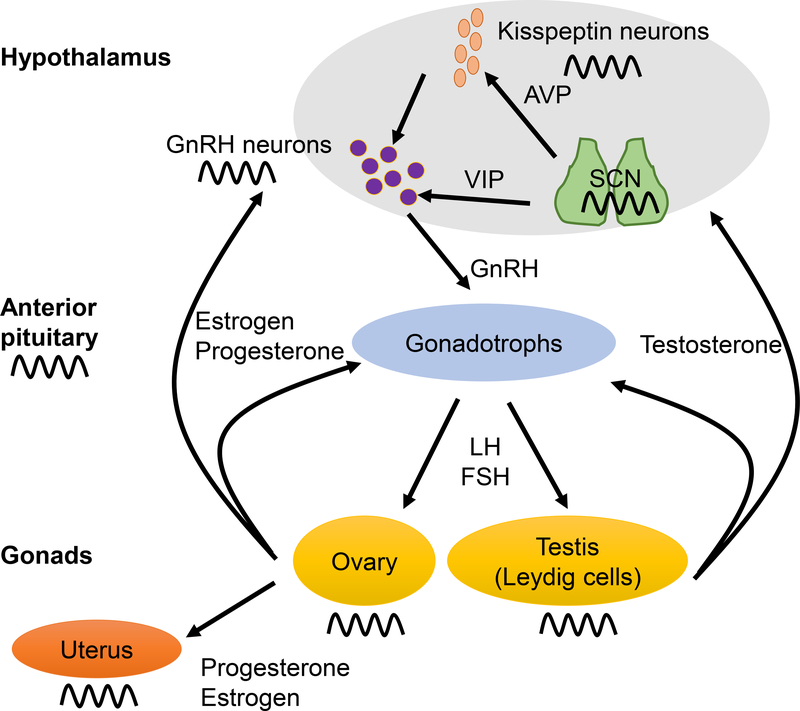
Reproduction is regulated by the HPG axis. At the apex of this axis are two hypothalamic neuronal populations required for fertility, the kisspeptin neurons and the gonadotropin-releasing hormone (GnRH) neurons (Estrada, Clay et al. 2006, Adachi, Yamada et al. 2007, Jacobi, Martin et al. 2007, Kauffman, Park et al. 2007, Herbison 2008, Smith 2008, Robertson, Clifton et al. 2009) (Figure 1). Kisspeptin neurons are sexually dimorphic, where kisspeptin expressing neurons located in the anterior ventral periventricular area (AVPV) are involved in the LH surge, and account for a larger neuronal population in females than males. The second hypothalamic kisspetin neuron population important in reproduction is in the arcuate nucleus. The arcuate nucleus kisspeptin neurons are integrating metabolic status information to the HPG axis and are key in generating pulsatile GnRH release (Smith, Cunningham et al. 2005, Smith, Popa et al. 2006, Adachi, Yamada et al. 2007, Kauffman, Gottsch et al. 2007, Clarkson, Boon et al. 2009, Desroziers, Mikkelsen et al. 2010, Gill, Wang et al. 2010, Lehman, Merkley et al. 2010, Lehman, Hileman et al. 2013, Herbison 2018, Dulka and Moenter 2019, McQuillan, Han et al. 2019, Padilla, Perez et al. 2019). The primary role of kisspeptin neurons in reproduction is to stimulate GnRH neurons to release GnRH peptide into the hypophyseal-portal system, promoting pituitary gonadotrophs to release follicle-stimulating hormone (FSH) and luteinizing hormone (LH) into the bloodstream (Irwig, Fraley et al. 2004, Han, Gottsch et al. 2005, Messager, Chatzidaki et al. 2005, Gottsch, Clifton et al. 2006). LH and FSH act on the gonads to promote gametogenesis and sex steroid production. Sex steroids (testosterone, progesterone and estrogen), in turn feed back to the gonadotrophs and the hypothalamus thereby regulating hormone release patterns (Figure 1). The fine-tuning of these hormone release patterns combined with optimal tissue sensitivity is obtained through the molecular circadian clock (Figure 2). This cell-endogenous clock has been linked to every aspect of reproductive physiology including ovarian steroidogenesis, follicular development (Chen, Zhao et al. 2013, Chen, Zhao et al. 2013), ovulation (Sellix, Yoshikawa et al. 2010, Mereness, Murphy et al. 2015), development of the gametes (Johnson, Lim et al. 2002, Morse, Cermakian et al. 2003, Amano, Matsushita et al. 2009), fertilization (Johnson, Lim et al. 2002), implantation (Greenhill 2014, Liu, Johnson et al. 2014, Xu, Li et al. 2016), as well as pregnancy and parturition (Gamble, Resuehr et al. 2013, Miller and Takahashi 2013). Manipulations (environmental or genetic) that disrupt or alter clock function globally or in specific tissues of the reproductive tract lead to irregular patterns of hormone secretion and infertility (Kennaway, Boden et al. 2012, Mereness, Murphy et al. 2015). Conversely, reproductive pathophysiology is associated with irregular clock function in tissues of the HPG axis, including the ovary (Sellix, Murphy et al. 2013, Mereness, Murphy et al. 2015).
Figure 1. Circadian rhythms exist throughout the reproductive axis.
Fertility is regulated by the hypothalamic-pituitary-gonadal axis. The presence of a functional molecular clock, composed of clock transcription factors including BMAL1, CRY1/2/3, CLOCK and PER1/2 has been identified at all levels of the reproductive axis (indicated by the sinusoidal symbol). The absence of core clock genes, such as Per, Cry, Bmal1 and Clock are all associated with impaired fertility. To align peripheral clocks to the time-of-day, a time signal from the SCN is transmitted to peripheral clocks. At the level of the hypothalamus the SCN projects directly onto kisspeptin and GnRH neurons to modulate their activity. Increased release of GnRH is mandatory for the LH surge, and the combination of SCN-afferents on GnRH neurons with increased estrogen levels and kisspeptin signaling allows for correct GnRH neuron firing. Abbreviations: AVP: arginine vasopressin, Bmal1: aryl hydrocarbon receptor nuclear translocator like 1, Clock: circadian locomotor output cycles kaput, Cry: cryptochrome, FSH: follicle stimulating hormone, GnRH: gonadotropin-releasing hormone, LH: luteinizing hormone, Per: period circadian clock, SCN: suprachiasmatic nucleus, VIP: vasoactive intestinal peptide.
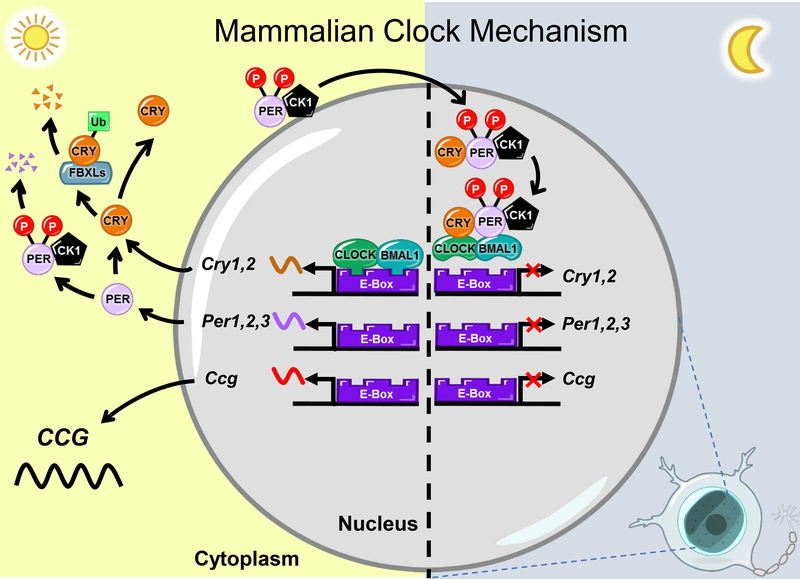
Figure 2. The molecular clock generates cell-endogenous circadian rhythms and circadian gene expression of clock-controlled genes (CCGs).
Cell endogenous circadian rhythms are generated by a fine-tuned transcription-translation feedback loop within nucleated cells. The core transcription factors generating these circadian rhythms are the transcription factors CLOCK and BMAL1. CLOCK and BMAL1 form a heterodimer and bind to E-boxes in regulatory regions to promote the expression of clock genes, including Per1/2/3 and Cryl/2, as well as clock controlled genes (Ccg containing E-boxes in their promoters. Per1/2/3 and Cryl/2, along with the Ccgs’ translocate to the nucleus for translation. To generate a circadian rhythm in gene expression, PER1/2/3 and CRY1/2 proteins form a complex and return to the nucleus to inhibit transcription by binding to and inactivating the CLOCK:BMAL1 complex. The stability of PER1/2/3 is regulated by CK1δ, CK1ε, and CK2, enzymes that phosphorylate PERs and promote their degradation. The stability of CRY1/2 is regulated by ubiquitination by FBXL3 and FBXL21. Abbreviations: Bmal1: aryl hydrocarbon receptor nuclear translocator like 1, CCG: clock-controlled genes, CK: casein kinase, Clock: circadian locomotor output cycles kaput, Cry: cryptochrome, FBXL: F-box proteins and Per: period. Adapted with permission from UC San Diego BioClock Studio.
The molecular clock generates circadian rhythms in reproductive tissues
The role of circadian rhythms is to align tissue function to the environmental day-night cycle. Most cells have an endogenous circadian timekeeper, referred to as their “molecular clock” (Figure 2). The molecular clock is an auto-regulatory transcription-based feedback loop consisting of a group of transcriptional regulators referred collectively to as core clock genes. In mammals, the core loop includes the transcriptional activator, Brain and muscle arntl-like1 (BMAL1) and its binding partner Circadian locomotor output cycles kaput (CLOCK), a histone acetyltransferase that drives expression of several genes, including the repressors Period (Per1,2,3) and Cryptochrome (Cry1,2). PER and CRY in a negative feedback loop act as potent repressors of BMAL1:CLOCK-dependent transcription. BMAL1:CLOCK and PER:CRY are considered to be the core clock genes. The pace of the molecular clock is fine-tuned by numerous factors, where key regulators include casein kinases (CK1δ, CK1ε, CK2). These enzymes phosphorylate PER1/2/3 and promote PER1/2/3 degradation, in contrast, the formation of a large complex containing PERs, CRY s, and other proteins prevent their degradation. The stability of CRY1/2 is regulated by F-box proteins (FBXL3, FBXL21). FBXLs are enzymes that add ubiquitin chains to their target proteins and promote their degradation. Ubiquitination by FBXL3, results in fast degradation of the CRYs, whereas ubiquitination by FBXL21, results in slow degradation of CRYs (Figure 2) (Lowrey and Takahashi 2011, Buhr and Takahashi 2013, Minami, Ode et al. 2013, Stojkovic, Wing et al. 2014). Silencing or genetic deletion of any of the core clock transcription factors impairs the generation of the circadian rhythm within a cell (Hogenesch and Ueda 2011, Lowrey and Takahashi. Of all the core clock genes, deletion of Bmal1 most dramatically impacts both cellular and behavioral circadian rhythms, and therefore is the most studied molecular clock transcription factor (Welsh, Logothetis et al. 1995, Abraham, Granada et al. 2010, Ko, Yamada et al. 2010, Evans, Pan et al. 2011). In addition to regulating their own transcription, molecular clock transcription factors drive expression of clock-controlled genes (CCG), generating a ~24h expression pattern of tissue specific genes (Koike, Yoo et al. 2012). Except for sperm, all other reproductive tissues have consistently been described to possess a functional molecular clock. In the following sections the role of these core clock transcription factors in hormone release and HPG axis function will be discussed.
The contributions of circadian rhythms in distinct neuronal populations of the female hypothalamus
Circadian rhythms in the suprachiasmatic nucleus (SCN)
The SCN plays two distinct roles in circadian control of physiological functions. First, the SCN translates light-information to the rest of the body to align peripheral circadian rhythms to the time of day (Figure 1). Second, neurons within the SCN utilize core clock genes (Figure 2) combined with a tight intercellular communication network (Liu, Welsh et al. 2007, Abraham, Granada et al. 2010, Welsh, Takahashi et al. 2010, Lowrey and Takahashi 2011, Evans, Pan et al. 2012, Bedont, LeGates et al. 2014, Hatori, Gill et al. 2014), to generate a synchronized and sustained circadian rhythm that persist in absence of a daily light signal, together creating a strong buffer capacity against deviations of the ~24h rhythm (Welsh, Logothetis et al. 1995, Abraham, Granada et al. 2010, Ko, Yamada et al. 2010, Evans, Pan et al. 2012, Hastings, Maywood et al. 2019). This buffer effect can mask the contribution of genes in the function of specific SCN neuron populations creating difficulties in elucidating the contribution of single genes in respect to SCN output and function. Despite this compensation mechanism it has been thoroughly established that a daily signal from the SCN times the LH surge promoting ovulation (Legan and Karsch 1975, Brown-Grant and Raisman 1977, Wiegand and Terasawa 1982, Abrahamson and Moore 2001, Christian and Moenter 2010). For the interested reader numerous outstanding reviews discussing the circadian timing of the LH surge are available (de la Iglesia and Schwartz 2006, Miller and Takahashi 2012). In this review we are specifically focusing on the role of clock genes in this circadian timing. Despite great efforts, it remains elusive which neuronal population(s) of the SCN drive the daily signal synchronizing HPG axis function. Mice which are full body knock out (KO) of core clock genes, including Per1/2, Bmal1, and ClockΔ19 (the ClockΔ19 is a dominant negative Clock mutant mouse) are all associated with impaired fertility (Chappell, White et al. 2003, Boden and Kennaway 2004, Miller, Olson et al. 2004, Dolatshad, Campbell et al. 2006, Alvarez, Hansen et al. 2008, Pilorz and Steinlechner 2008, Ratajczak, Boehle et al. 2009, Boden, Varcoe et al. 2010). Intriguingly, while the Bmal1KO (Chu, Zhu et al. 2013), and ClockΔ19 mutant (Van Der Horst, Muijtjens et al. 1999, Bae, Jin et al. 2001, Miller, Olson et al. 2004) both have an undetectable LH surge, these mice still undergo ovulation and can become pregnant. This shows that while the circadian system is essential in gating the LH surge, it does not appear to be required for successful ovulation. This is consistent with the maintained ovulation in transgenic mice which lack LH pulsatility (Kumar and Sait 2011), suggesting that under certain circumstances, ovulation can occur without the classical LH surge. Despite evidence the reproductive system can support ovulation in absence of a detectable LH surge and SCN signaling, under most circumstances a daily timing signal arising in the SCN is necessary for the LH surge and ovulation. Two studies recently asked if the circadian timing signal for the LH surge was localized within arginine vasopressin (AVP) neurons of the SCN (Figure 1). To do this, they used an estrogen induced surge model, where the LH surge is observed around activity onset (ZT12). Interestingly, in C57BL6/CBA mice with the Bmal1flox/flox allele conditionally deleted in AVP neurons (Bmalfl/fl:Avpcre) the LH surge did not occur at ZT12, but was mis-timed (advanced or delayed) (Bittman 2019), whereas Bmalfl/fl:Avpcre on a C57BL6 genetic background had a comparable surge to controls (Tonsfeldt, Cui et al. 2019). One important distinction between these two studies were the used mouse strains; C57BL6 mice do not secrete endogenous melatonin (Tonsfeldt, Cui et al. 2019), whereas C57BL6/CBA mice are expected to produce melatonin, although this was not verified in this study (Ebihara, Marks et al. 1986, Goto, Oshima et al. 1989, Bittman 2019). This is an important distinction, as melatonin is a hormone secreted exclusively at night, and which plays an important role in communicating timing information to tissues throughout the body and the brain (Hardeland, Pandi-Perumal et al. 2006, Gorman In Press). Interestingly, in melatonin deficient mice (C57BL6), the LH surge was abolished in triple transgenic mice with the Bmalfl/fl allele deleted in both AVP and vasoactive intestinal peptide (VIP) neurons (Tonsfeldt, Cui et al. 2019). The loss of a detectable LH surge in the Bmalfl/fl :Avpcre:Vipcre mice was not caused by loss of Bmal1 in VIP neurons alone, as Bmalfl/fl:Vipcre females maintained an intact LH surge and normal fertility (Tonsfeldt, Cui et al. 2019). Interestingly, mice mutant for VIP (VipKO), an SCN neuropeptide modulating GnRH neuron activity (Vijayan, Samson et al. 1979, Samson, Burton et al. 1981, Alexander, Clifton et al. 1985, Kimura, Mitsugi et al. 1987, Van der Beek, Van Oudheusden et al. 1994, van der Beek, Horvath et al. 1997, Horvath, Cela et al. 1998, Beek, Swarts et al. 1999, Smith, Jennes et al. 2000, Kriegsfeld, Silver et al. 2002, Christian and Moenter 2008, Christian and Moenter 2008, Piet, Dunckley et al. 2016) have female subfertility due to decreased SCN cell synchrony (Loh, Kuljis et al. 2014), confirming that SCN pacemaker function, particularly SCN VIP output, is central to maintaining normal reproductive function and estrous cyclicity. It should be noted that some important considerations need to be taken into account in many of the cited studies. One is that conditional KO mouse models come with certain limits such as inefficient flox-allele deletion and ectopic cre-allele expression which can impact the studied phenotypes (Hoffmann, Larder et al. 2019, Pandolfi, Tonsfeldt et al. 2019, Pandolfi, Breuer et al. 2019). In addition, both AVP and VIP are expressed outside the SCN, including the liver, in smooth and striate muscle, as well as the gastric system, although the degree of Avpcre and Vipcre-allele targeting to these additional tissues is not well described. This potential Avpcre and Vipcre allele expression in peripheral tissues raises the possibility of Bmal1fl/fl allele deletion in non SCN cells which might contribute to the observed phenotypes. Further, the discrepancies in the LH surge in the two Bmalfl/fl:Avpcre studies (Bittman 2019, Tonsfeldt, Cui et al. 2019) suggest that the melatonin deficient C57BL6 mice might have adapted to their melatonin deficient state, potentially rendering the LH surge more robust. Another likely possibility is that melatonin plays a role in promoting the correct timing of the LH surge, as it is well established that melatonin is important in regulating reproductive status and the HPG axis in seasonal breeders (Karsch, Bittman et al. 1984, Chemineau, Pelletier et al. 1988, Revel, Masson-Pevet et al. 2009, Gorman In Press).
Circadian rhythms in GnRH neurons
The role of circadian rhythms in relation to GnRH neuron function has primarily been studied in females, specifically in relation to the LH surge due to the central role of GnRH neurons as “peripheral and central signal integrators” adapting the GnRH pulse frequency to balance LH and FSH release from the pituitary (Figure 1) (Knobil 1980, Gillespie, Chan et al. 2003, Tsutsumi and Webster 2009, Zhao and Kriegsfeld 2009, Hickok and Tischkau 2010, Tonsfeldt, Goodall et al. 2011, Williams, Jarjisian et al. 2011, Hoffmann, Trang et al. 2016, Clarkson, Han et al. 2017, Herbison 2018, Pandolfi, Tonsfeldt et al. 2019). To prepare GnRH neurons for the timed release of GnRH to drive the LH surge, a combination of SCN-afferents on GnRH neurons with increased estrogen levels and kisspeptin stimulation are required (Everett and Sawyer 1950, van der Beek, Horvath et al. 1997, Kriegsfeld, Silver et al. 2002, Barbacka-Surowiak, Surowiak et al. 2003, Semaan and Kauffman 2010, Tonsfeldt, Goodall et al. 2011, Dror, Franks et al. 2013, Kriegsfeld 2013, Miller and Takahashi 2013). In addition, the molecular clock within GnRH neurons drive the rhythmic expression of the kisspeptin receptor (Kiss1R), GnRH (Gnrh1) and GnRH peptide release (the latter was assessed in vitro using immortalized GnRH neurons) (Chappell, White et al. 2003, Chappell, White et al. 2003, Gillespie, Chan et al. 2003, Olcese, Domagalski et al. 2003, Zhao and Kriegsfeld 2009, Hickok and Tischkau 2010, Hickok and Tischkau 2010, Williams, Jarjisian et al. 2011, Matagne, Kim et al. 2012). Although both GnRH and KISS1R are indispensable for maintaining LH and FSH levels required for fertility (Gibson, Kasowski et al. 1994, Singh and Handelsman 1996, Ma, Dong et al. 2004, Foster, Jackson et al. 2006, Lapatto, Pallais et al. 2007, Tonsfeldt, Goodall et al. 2011, Wierman, Kiseljak-Vassiliades et al. 2011, Choe, Kim et al. 2013), in vivo studies in transgenic mice with disrupted circadian rhythms support a minor role of the molecular clock within neurons outside the SCN in relation to timing GnRH release and Kiss1R expression. GnRH neurons in the Bmal1KO have increased sensitivity to kisspeptin input, as evident from enhanced LH release in response to a kisspeptin challenge (Tonsfeldt, Schoeller et al. 2019). This increase in kisspeptin sensitivity was not associated with detectable changes in the transcription levels of hypothalamic Kiss1R, Gnrh1, or pituitary release of LH in response to GnRH, and the mice had normal gonadal status, supporting maintained hormone release patterns (Tonsfeldt, Schoeller et al. 2019). Despite these seemingly minor molecular changes in the Bmal1KO hypothalamus, these mice do not present a timed LH surge (Tonsfeldt, Schoeller et al. 2019). This suggests the existence of some fundamental differences in the sensitivity and function of GnRH neurons in the Bmal1KO mouse. What these changes in GnRH neurons are remains unknown. Condition KO of the Bmalflox/flox allele in GnRH neurons (Bmalfl/fl:Gnrhcre) appear to blunt the induced LH surge in C57BL6/CBA mice (Bittman 2019), but not in C57BL6 mice (Tonsfeldt, Schoeller et al. 2019). Both studies agreed that the timing of the LH increase/surge was comparable to controls. This discrepancy might be contributed to mouse strain, or caused by the use of two different Gnrh-cre alleles, which we have previously shown to target GnRH neurons at slightly different rates, where the Gnrh-cre mouse used in the Tonsfeldt et al. study targets ~100% of GnRH neurons, but also has ectopic expression in the SCN (Bittman 2019, Hoffmann, Larder et al. 2019, Pandolfi, Tonsfeldt et al. 2019, Pandolfi, Breuer et al. 2019), although this ectopic expression does not significantly impact SCN regulated locomotor output in Bmalfl/fl:Gnrhcre males (Pandolfi, Tonsfeldt et al. 2019). On the other hand, the Gnrh-cre allele used in the Bittman study, targets −95–100°% of GnRH neurons, and does not target the SCN. The potential incomplete deletion of Bmal1 within GnRH neurons due to <100% targeting, or inefficient flox-allele deletion might impact the results, as it is well established that only a small handful of GnRH neurons are required to maintain HPG axis function (Herbison, Porteous et al. 2007, Hoffmann, Pandolfi et al. 2018, Tonsfeldt, Schoeller et al. 2019, Hoffmann, Tamrazian et al. 2014).
Circadian rhythms in kisspeptin neurons
The kisspeptin neuron populations most extensively studied in regards to fertility are located within the hypothalamus in the anteroventral periventricular nucleus (AVPV) as well as the arcuate nucleus. Both these two kisspeptin neuron populations are regulated by sex-steroid feedback, where the AVPV kisspeptin neurons mediate the positive feedback promoting the LH surge, whereas the arcuate nucleus kissppetin neuron population mediates negative feedback (Moenter, Caraty et al. 1990, Watson, Colston et al. 1995, Gu and Simerly 1997, Chappell and Levine 2000, Petersen, Ottem et al. 2003, Wintermantel, Campbell et al. 2006, Christian, Glidewell-Kenney et al. 2008, Glanowska, Venton et al. 2012, Wang, Vanacker et al. 2019). Both of these two neuronal populations have a functional molecular clock (Smarr, Gile et al. 2013, Padilla, Perez et al. 2019). Within the AVPV kisspeptin neurons, the circadian expression of Bmal1 is not responsive to estrogen, whereas absence of estrogen (ovariectomy) disrupts circadian expression of both Kiss1 and the transcript for the AVP receptor (Smarr, Gile et al. 2013). This has led to the hypothesis that the AVPV kisspeptin neurons function as a coincidence detector for high estrogen and SCN output, thus timing the LH surge (Wintermantel, Campbell et al. 2006, Robertson, Clifton et al. 2009, Williams III, Jarjisian et al. 2010, Smarr, Morris et al. 2012, Smarr, Gile et al. 2013, Chassard, Bur et al. 2015). This hypothesis is partially supported by the finding that conditional knock-out mice on a C57BL6/CBA genetic background with disrupted circadian rhythms within kisspeptin neurons (Bmalfl/fl:Kisscre mice) have a mistimed LH surge, and often present with two LH peaks in a ~12h period (Bittman 2019). This contrasts with a comparable study in the same transgenic mouse model (Bmalfl/fl:Kisscre mice), with the difference of these mice being on a C57BL6 genetic background, which presented with a normal LH surge (ZT4 and ZT12 were evaluated), normal sex hormone levels, and intact fertility (Tonsfeldt, Schoeller et al. 2019). Together these studies suggest the timing of the LH surge as well as normal LH and FSH release patterns from the pituitary occurs relatively independent of the molecular clock within kisspeptin neurons, and again suggest melatonin might play a role in timing of the surge. Another possibility is that the conditional KO mice (Bmalfl/fl:Kisscre mice) experience some genetic compensation. This is supported by a recent study where targeted silencing of kisspeptin neurons within the adult arcuate nucleus caused impaired estrous cycling, indicating abnormal hormone release in the HPG axis (Padilla, Perez et al. 2019). Interestingly, these mice also presented with abnormal circadian activity patterns (Padilla, Perez et al. 2019), a phenotype not observed in mice with conditional deletion of Bmal1 in kisspeptin neurons (Tonsfeldt, Schoeller et al. 2019). In summary, the current literature supports that circadian rhythms in arcuate nucleus kisspeptin neurons might be involved in the control of hypothalamic hormonal release, however more work is required to determine the exact contribution of the molecular clock in AVPV and arcuate nucleus kisspeptin neurons.
The molecular clock in the pituitary
The gonadotroph cells within the pituitary translate GnRH pulses into a balanced secretion of FSH and LH (Figure 1) (Tsutsumi and Webster 2009, Glanowska, Burger et al. 2014). As such it seems obvious that cellular circadian rhythms would be critical in modulating time-of-day specific sensitivity of gonadotrophs to GnRH, sex steroids and/or play a central role in Fshb and Lhb transcription. However, despite the presence of a functional molecular clock in gonadotrophs (Chu, Zhu et al. 2013), conditional deletion of Bmal1 in this cell population only causes a slight increase in LH levels in proestrus, and increased FSH in estrus together resulting in modest irregularities in estrous cycles, without impacting fertility (Chu, Zhu et al. 2013). Together this indicates that the molecular clock in gonadotrophs play a minor role in regulating these cells sensitivity to input (such as GnRH and sex steroids) as well as the production and secretion of LH and FSH.
The molecular clock in uterine function
In mammals, including humans, labor preferentially occur during the rest phase of the day, which translates into labor at night in diurnal species, and labor during the day in nocturnal species (Bosc 1990). The mechanisms timing labor to the rest phase of the day varies dependent on the species, but include signals from the fetus (Mesiano and Jaffe 1997), the maternal and fetal SCN (Reppert, Henshaw et al. 1987) and the uterus (Ratajczak, Asada et al. 2012). In diurnal primates, the nocturnal onset of labor coincides with increased sensitivity of the pregnant uterus to oxytocin and melatonin, two hormones driving pregnancy associated uterine contractions (Hirst, Haluska et al. 1993, Olcese, Lozier et al. 2013, Olcese 2014). The daily change in uterine sensitivity to oxytocin and melatonin suggest a role of circadian rhythms in adapting uterine sensitivity to stimuli (Sharkey, Puttaramu et al. 2009). Studies in rats and mice have shown that the non-pregnant uterus possesses a functional molecular clock (Nakamura, Sellix et al. 2008, Loh, Kuljis et al. 2014), and that pregnancy is associated with increased expression of clock genes and a prolonged period of uterine circadian rhythms (unpublished data). Although based on a limited literature, the molecular clock in the uterus appear to be involved in embryo implantation as well as parturition (Gamble, Resuehr et al. 2013, Miller and Takahashi 2013). Conditional deletion of Bmal1 in the uterine smooth muscle, the myometrium, causes mis-timed labor onset (Ratajczak, Asada et al. 2012), whereas disrupted molecular function in early pregnancy impairs embryo implantation (Ratajczak, Boehle et al. 2009, Greenhill 2014, Liu, Johnson et al. 2014, Xu, Li et al. 2016, Li, Chen et al. 2017, Lv, Wang et al. 2019). It remains unknown what genetic pathways are regulated by the molecular clock in the uterine endometrium and myometrium, and how molecular clock gene transcriptional targets might change during pregnancy potentially leading to changes in the time of day of receptor expression enhancing uterine sensitivity to hormones during the rest phase of the day (Hirst, Haluska et al. 1993, Sharkey, Puttaramu et al. 2009, Beesley, Lee et al. 2015). Some promising candidates as regulators of uterine circadian rhythms are the sex steroids estrogen (Nakamura, Sellix et al. 2008) and progesterone (Rubel, Lanz et al. 2012), which directly modulate the circadian clock in the uterus (He, Hirata et al. 2007). What drives the change in circadian period in the uterus during pregnancy is unknown, and more research is required to understand how uterine sensitivity to hormones change in both the non-pregnant and pregnant state (Perrin, Segall et al. 2006, Nakamura, Sellix et al. 2010).
The molecular clock and ovarian function
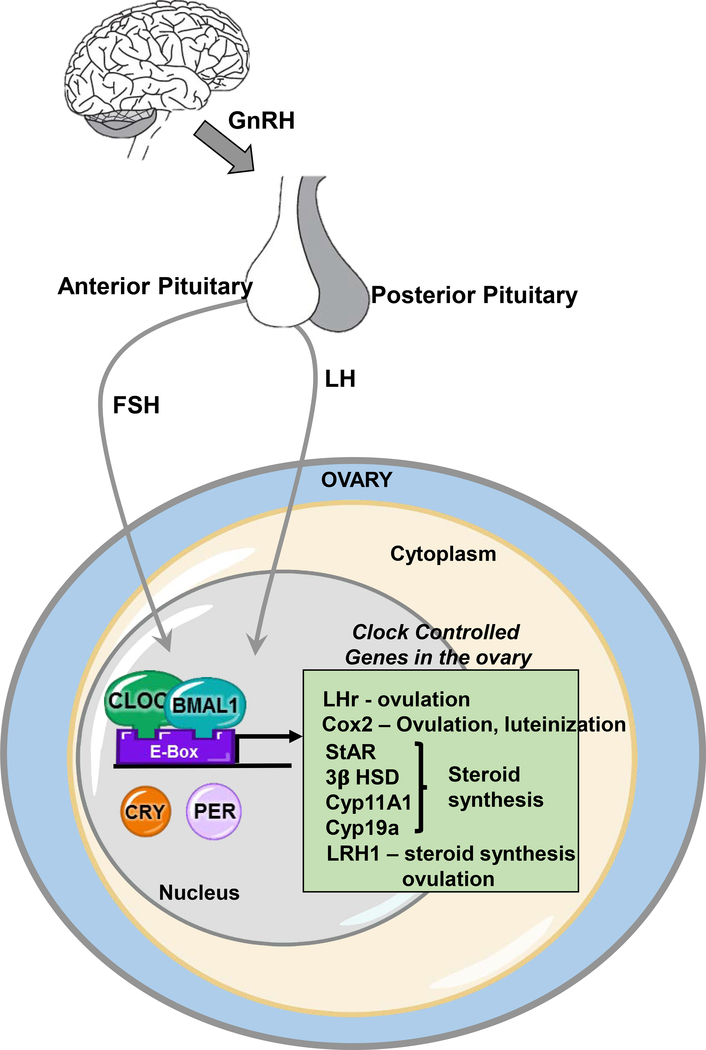
While gonadotropins clearly influence the timing and amplitude of expression of the core clock genes in the ovarian cells (Figures 1 and 3), in recent years, a number of studies have begun to focus on understanding the physiological significance of the ovarian clock. The bulk of our current knowledge about the ovarian clock and its function come from studies in granulosa and luteal cells from rodent models. The current theory is that the clock in follicular cells regulates the timing of sensitivity and/or prepares the preovulatory follicle to coincide with the LH surge and cause ovulation (Sellix 2015). It is well known that the LH surge induces the expression of a large number of genes in the ovary (Richards, Russell et al. 1998, Richards 2001). It is believed that this LH-induced ovarian gene expression is mediated through the circadian clock. For example, the expression of cyclooxegenase-2 (COX2), a rate-limiting enzyme in prostaglandin synthesis is a clock-controlled gene (CCG) regulated by BMAL1:CLOCK through E-box elements on its promoter (Figure 3) (Sirois, Sayasith et al. 2004). Prostaglandins are critical for successful ovulation (Sugimoto, Inazumi et al. 2015). Interestingly, it has been reported that prostaglandin endoperoxide synthase-1 (Ptgs2) and LH receptor (Lhcgr) gene expression show strong circadian rhythms in luteinized rat granulosa cells disrupted by siRNA-mediated knockdown of Bmal1 (Chen, Zhao et al. 2013). Moreover, siRNA-mediated knockdown of Per2 or Clock in granulosa cells causes downregulation of LH receptor expression (Shimizu, Hirai et al. 2011), indicating that the expression of LH receptor is directly regulated by the circadian clock. Given the importance of COX-2 and LH receptor in ovulation, it can be said that the regulation of Cox-2 and LH receptor expression by the circadian timing system is one of the underlying mechanisms by which the ovarian clock prepares the granulosa cells for the LH surge and subsequent transition to a functional corpus luteum (Figure 3). Another possible CCG in the ovary is liver receptor homolog 1 / CYP7A promoter binding factor (LRH-1) that was first discovered in the liver as an orphan nuclear receptor (Nitta, Ku et al. 1999). In the liver, LRH-1 directly interacts with CLOCK and has a synergistic effect on BMAL1:CLOCK mediated transcription (Oiwa, Kakizawa et al. 2007). In the ovary, LRH-1 is expressed in granulosa cells (Kim, Havelock et al. 2005) and is involved in steroid biosynthesis by inducing the expression of cytochrome 450 side chain cleavage enzyme (CYP11A1) (Kim, Havelock et al. 2005) and is essential for ovulation (Duggavathi, Volle et al. 2008).
Figure 3. Regulation of the ovarian clock by gonadotropins.
To maintain optimum ovarian function and normal fertility, the hypothalamus and the pituitary maintain a synchrony with the ovarian clock via endocrine signals involving FSH and LH. The gonadotropins, through regulation of core clock genes like Per and Bmal1, regulate the ovarian circadian clock which in turn controls the expression and timing of expression of genes critical for normal ovarian physiology (Clock controlled genes). Abbreviations: LHr: luteinizing hormone receptor, Cox2: cyclooxygenase 2, StAR: steroidogenic acute regulatory protein, 3b HSD: 3-beta hydroxysteroid dehydrogenase, Cyp11A: cholesterol side-chain cleavage enzyme, Cyp19a: aromatase.
While granulosa cells have been the focus of most studies with respect to the ovarian clock, a recent study (Mereness, Murphy et al. 2016) used granulosa- and theca-specific Bmal1 conditional knockout mouse models to determine that the circadian clock function in theca cells is also critical for maintaining the normal rhythms of gene expression necessary for ovarian sensitivity to gonadotropins and timing of ovulation. In this study, the rhythmic expression of the LH receptor was disrupted in the theca- and not the granulosa cell-specific Bmal1 knockout mice causing decreased ovarian sensitivity to LH (Mereness, Murphy et al. 2016).
The circadian clock in the ovary also plays a major role in regulating steroid biosynthesis. In mature granulosa cells (Chen, Zhao et al. 2013), some of the key steroidogenic genes like aromatase (CYP19a), steroidogenic acute regulatory protein (StAR), 3beta-hydroxysteroid dehydrogenase (3ß-HSD) and 11 ß-hydroxylase are clock controlled genes (Chen, Zhao et al. 2013). The rhythmic pattern of expression of these genes is severely affected with disruption of the circadian clock (Chen, Zhao et al. 2013). Moreover, global (Ratajczak, Boehle et al. 2009) and ovarian steroidogenic cell-specific (Liu, Johnson et al. 2014) Bmal1 knockout mouse models show decreases in progesterone levels due to disruption of StAR gene expression that leads to implantation failure. Given the regulatory role of the circadian clock ovarian steroid hormone synthesis, it is now being realized that the circadian clock may also play a major role in the HPG axis feedback loop.
Modulation of the ovarian circadian clock by gonadotropins and ovarian steroids
Evidence for an active circadian oscillator in ovarian cells comes from the phasic sensitivity towards gonadotrophins in granulosa and theca cells of the ovary (Yoshikawa, Sellix et al. 2009). This concept is further supported by the rhythmic expression of the LH receptor in the ovary (Figure 3)(Chen, Zhao et al. 2013), which confers ovarian sensitivity to the LH surge around activity onset (Sellix, Yoshikawa et al. 2010). Moreover, core clock genes (Bmal1, Clock, Per1/2) and their transcripts are rhythmically expressed in the granulosa, theca, and luteal cells (Fahrenkrug, Georg et al. 2006, Karman and Tischkau 2006, He, Hirata et al. 2007). In the ovary, as other tissues, the peak phases of Bmal1 and Per2 are in anti-phase (Figure 2), and indicative of a self-sustaining circadian oscillator (Karman and Tischkau 2006). For normal ovarian physiology and female fertility, the circadian clock in the ovary maintains synchrony with the hypothalamus and the pituitary (Figure 1 and 3). In the last decade, a number of studies have postulated that this synchronization in large part is carried by endocrine cues. Some of the strongest evidence in support of this idea comes from studies utilizing a hypophysectomized immature rat model where it has been shown that hypophysectomy causes disruption of the ovarian clock function which can be rescued by exogenous hCG (human Chorionic Gonadotropin)(Karman and Tischkau 2006). Moreover, in vitro studies with granulosa cells from mouse (Yoshikawa, Sellix et al. 2009), rat (He, Hirata et al. 2007) and hen (Zhang, Lai et al. 2017) have reported that LH and FSH treatment cause changes in the rhythm and amplitude of the ovarian circadian timing system (He, Hirata et al. 2007, Yoshikawa, Sellix et al. 2009, Zhang, Lai et al. 2017). However, the effectivity of LH and/or FSH in regulating the circadian timing system may vary among different species. For example, in a hen model (Zhang, Lai et al. 2017), while it has been shown that LH elicits a significant response from the circadian timing system in granulosa cells from mature follicles, a combination of LH and FSH is much more effective than LH or FSH alone (Zhang, Lai et al. 2017). In contrast, another study in the chicken ovary reported that LH is the primary regulator of the ovarian clock genes (Tischkau, Howell et al. 2011). In rodents, (Chu, Yoshida et al. 2011) while FSH alone can significantly induce Per2 expression in granulosa cells of preantral follicles, the effect of LH on the circadian timing system occurs only after FSH stimulation, likely owing to the expression of LH receptor. The same study (Chu, Yoshida et al. 2011) reported that luteal regression by apoptosis causes significant dysregulation of Per2 circadian oscillation. Consequently, it has been suggested that in the ovary, the circadian clockwork alters with respect to follicular development, luteinization, and apoptosis, and expression of Bmal1 acts as a “molecular-switch” of the circadian oscillation (Chu, Yoshida et al. 2011). With respect to the ovarian clock, the current understanding is that the rhythmic expression of clock genes is limited to mature granulosa cells and luteal cell (He, Hirata et al. 2007, He, Hirata et al. 2007, Chu, Yoshida et al. 2011, Chu, Misawa et al. 2012)
Another major question, that is less understood is, what is the underlying mechanism by which gonadotrophins modulate the circadian timing system in the ovary? A plausible suggested mechanism involves gonadotropin-induced signaling and transcription factors that regulate the expression of core clock genes. For example, in avian granulosa cells, LH-induced cAMP, p38 mitogen-activated protein kinases (p38MAPK) and extracellular signal-regulated kinases 1 and 2 (ERK1/2) pathways have been shown to affect transcript levels of clock genes (Li, Zhang et al. 2014). Similarly, in rat granulosa cells, promoter analysis revealed that Per1 expression is mediated by the cAMP response element binding protein (He, Hirata et al. 2007). Moreover, in rat granulosa cells, FSH induces the development of the circadian timing system through the expression of gap junction protein, Connexin 43-dependent pathway (Chen, Zhao et al. 2013).
In addition to direct effects, gonadotropins may also influence the ovarian clock through ovarian steroids. Studies in rats have shown that the ovarian circadian clock is influenced by the estrous cycle, and it has been hypothesized that this may be in parts due to the fluctuating steroid hormone levels (Nakamura, Sellix et al. 2010). For example, in the ovary, during estrus, the amplitude of Per1 mRNA expression declines and its phase is advanced relative to proestrus and is recovered by diestrus (Nakamura, Sellix et al. 2010). However, to date, there is no evidence of a direct effect of estrogen or progesterone on the ovarian circadian system. Androgens are the only steroid hormones reported to directly influence the ovarian circadian system (Mereness, Murphy et al. 2015). Given the importance of the circadian timing system in normal female fertility and that high levels of androgen is associated with pathophysiological conditions like poly-cystic ovary syndrome (PCOS), a number of studies have proposed that androgen-induced dysregulation of the ovarian clock may be an underlying mechanism of PCOS (Sellix, Murphy et al. 2013, Mereness, Murphy et al. 2015). While the effects of gonadotropins in regulating the ovarian clock are now well established, the direct role of steroids affecting the circadian timing system in the ovary and its effect in normal and pathophysiological conditions remain poorly understood.
Circadian rhythms in the male reproductive tract
Understanding the role of the molecular clock in male reproductive function, and specifically within the testis, lags behind the large body of work aimed at understanding its role in female reproductive function. It is well established that timed hormone release patterns and sperm maturation are critical in male fertility (recently reviewed by(Bittman 2016), however the contribution of core clock genes relation to hormone release and male fertility have so far proven to be minor (Alvarez, Chen et al. 2003, Bittman, Doherty et al. 2003, Morse, Cermakian et al. 2003, Bebas, Goodall et al. 2009). For example, Bmal1KO males show a modest reduction in testosterone levels which might slightly reduce sperm production (Alvarez, Hansen et al. 2008, Schoeller, Clark et al. 2016). However the infertility of these mice are not caused by impaired sex hormone levels or a reduction in sperm production, but rather the disruption of the brain circuit driving male sexual behavior (Schoeller, Clark et al. 2016). This is consistent with findings in both ClockΔ19 mutant males and Per1/2KO mice (Dolatshad, Campbell et al. 2005, Alvarez, Hansen et al. 2008) where male fertility and circulating hormone levels are comparable to controls. Although the testis contains all the constituents of the molecular clock (Alvarez, Hansen et al. 2008), these transcription factors only appear to generate detectable circadian rhythms in Leydig cells (Bittman 2016). Thus, it seems that except for Leydig cells, the remaining cells in the testis as well as sperm are exceptions in the reproductive axis of cells and tissues possessing strong circadian rhythms (Mongrain, Ruan et al. 2008, Klose, Grote et al. 2011, Bittman 2016). In the future it will be of interest to determine if clock genes are important in correct sperm production and maturation, and it will be interesting to determine the potential contribution of circadian rhythms in the function of the epididymis.
Impact of disrupted circadian rhythms in humans
Absence of the SCN or disrupted rhythm generation induced by shift work or genetic mutations has important health consequences (Kennaway, Boden et al. 2012, Williams and Kriegsfeld 2012). Today, it is estimated that 50–70 million Americans suffer from disrupted circadian rhythms, which are associated with a large number of serious medical illnesses including cardiovascular disease, obesity, decreased cognitive capacities, impaired fertility and increased incidence of miscarriages and infertility (Nurminen 1998, Baker and Driver 2007, Mahoney 2010). As the timing and synchronization of hormonal release and tissue sensitivity plays an essential role in fertility, the desynchrony between the SCN and the rest of the HPG axis is emerging as a potential contributor to various pathophysiological conditions related to human fertility. Indeed, the relationship between shift work and changes in hormonal function is especially evident in female shift workers, who display increased reports of altered menstrual cycles (Labyak, Lava et al. 2002, Chung, Yao et al. 2005, Mahoney 2010, Fernandez, Marino et al. 2016, Stock, Knight et al. 2019), which are accompanied by changes in follicular stage length (Knutsson 2003, Lohstroh, Chen et al. 2003, Mahoney 2010, Attarchi, Darkhi et al. 2013) and FSH concentrations (Lohstroh, Chen et al. 2003, Mahoney 2010). However, how these reproductive changes impact sex-steroid levels, and cellular circadian rhythms needs further investigation.
Conclusion
It is now well established that the circadian timing system not only plays a critical role in regulating normal functioning of the HPG axis, its disruption has also been associated with a number of pathophysiological conditions. What needs to be understood is the interactions of the circadian system with gonadotrophins and steroid hormones and its downstream physiological effects. The molecular mechanism of how the clock is regulated in the SCN or how the circadian timing system fine tunes the pulsatile release of gonadotropins in the pituitary, the underlying mechanism by which LH and FSH regulates the circadian clock in the ovary or the effects of steroids on the circadian system under normal and pathophysiological conditions, are some of the questions that needs to be addressed in the future.
Highlights.
Clock genes generate circadian rhythms in the hypothalamic-pituitary-gonadal axis
Clock genes are required for proper reproductive axis function and fertility
Circadian rhythms drive daily changes in tissue sensitivity to hormones
Hormones and steroids in turn regulate the circadian timing system
Acknowledgments
Funding
H.M.H. was in part supported by Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number R00HD084759, the USDA National Institute of Food and Agriculture Hatch project MICL1018024, MSU AgBioResearch and by the March of Dimes Grant no 5-FY19-111. A.S. was in part supported by Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under Award Number R01HD086062-01A1, the USDA National Institute of Food and Agriculture Hatch project MICL02561 and MSU AgBioResearch.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abraham U, et al. (2010). “Coupling governs entrainment range of circadian clocks.” Mol Syst Biol 6: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abrahamson EE and Moore RY (2001). “Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections.” Brain Res 916(1–2): 172–191. [DOI] [PubMed] [Google Scholar]
- Adachi S, et al. (2007). “Involvement of anteroventral periventricular metastin/kisspeptin neurons in estrogen positive feedback action on luteinizing hormone release in female rats.” Journal of Reproduction and Development: 0701090061–0701090061. [DOI] [PubMed] [Google Scholar]
- Alexander MJ, et al. (1985). “Vasoactive intestinal polypeptide effects a central inhibition of pulsatile luteinizing hormone secretion in ovariectomized rats.” Endocrinology 117(5): 2134–2139. [DOI] [PubMed] [Google Scholar]
- Alvarez J, et al. (2003). “Non-cyclic and developmental stage-specific expression of circadian clock proteins during murine spermatogenesis.” Biology of reproduction 69(1): 81–91. [DOI] [PubMed] [Google Scholar]
- Alvarez J, et al. (2008). “The circadian clock protein BMAL1 is necessary for fertility and proper testosterone production in mice.” Journal of biological rhythms 23(1): 26–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano T, et al. (2009). “Expression and functional analyses of circadian genes in mouse oocytes and preimplantation embryos: Cry1 is involved in the meiotic process independently of circadian clock regulation.” Biology of reproduction 80(3): 473–483. [DOI] [PubMed] [Google Scholar]
- Attarchi M, et al. (2013). “Characteristics of menstrual cycle in shift workers.” Global journal of health science 5(3): 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae K, et al. (2001). “Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock.” Neuron 30(2): 525–536. [DOI] [PubMed] [Google Scholar]
- Baker FC and Driver HS (2007). “Circadian rhythms, sleep, and the menstrual cycle.” Sleep Med 8(6): 613–622. [DOI] [PubMed] [Google Scholar]
- Barbacka-Surowiak G, et al. (2003). “The involvement of suprachiasmatic nuclei in the regulation of estrous cycles in rodents.” Reprod Biol 3(2): 99–129. [PubMed] [Google Scholar]
- Bebas P, et al. (2009). “Circadian clock and output genes are rhythmically expressed in extratesticular ducts and accessory organs of mice.” The FASEB Journal 23(2): 523–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedont JL, et al. (2014). “Lhx1 controls terminal differentiation and circadian function of the suprachiasmatic nucleus.” Cell Rep 7(3): 609–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beek E v. d., et al. (1999). “Reproductive Neuroendocrinology-Central Administration of Antiserum to Vasoactive Intestinal Peptide Delays and Reduces Luteinizing Hormone and Prolactin Surges in Ovariectomized, Estrogen-Treated.” Neuroendocrinology 69(4): 227–237. [DOI] [PubMed] [Google Scholar]
- Beesley S, et al. (2015). “Circadian clock regulation of melatonin MTNR1B receptor expression in human myometrial smooth muscle cells.” Molecular human reproduction 21(8): 662–671. [DOI] [PubMed] [Google Scholar]
- Bittman EL (2016). “Timing in the testis.” Journal of biological rhythms 31(1): 12–36. [DOI] [PubMed] [Google Scholar]
- Bittman EL (2019). “Circadian Function in Multiple Cell Types Is Necessary for Proper Timing of the Preovulatory LH Surge.” Journal of biological rhythms: 0748730419873511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittman EL, et al. (2003). “Period gene expression in mouse endocrine tissues.” American Journal of Physiology-Regulatory, Integrative and Comparative Physiology 285(3): R561–R569. [DOI] [PubMed] [Google Scholar]
- Boden MJ and Kennaway DJ (2004). “Reproductive Consequences of Circadian Dysfunction: Fertility in the Bmal1 null mouse.” Reprod. Fertil. Dev 16(9): 280–280. [Google Scholar]
- Boden MJ, et al. (2010). “Reproductive biology of female Bmal1 null mice.” Reproduction 139(6): 1077–1090. [DOI] [PubMed] [Google Scholar]
- Bosc M (1990). “Photoperiodic regulation of the time of birth in rats: involvement of circadian endogenous mechanisms.” Physiology & behavior 48(3): 441–446. [DOI] [PubMed] [Google Scholar]
- Brown-Grant K and Raisman G (1977). “Abnormalities in reproductive function associated with the destruction of the suprachiasmatic nuclei in female rats. “ Proceedings of the Royal Society of London. Series B. Biological Sciences 198(1132): 279–296. [DOI] [PubMed] [Google Scholar]
- Buhr ED and Takahashi JS (2013). “Molecular components of the Mammalian circadian clock.” Handb Exp Pharmacol(217): 3–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappell PE and Levine JE (2000). “Stimulation of gonadotropin-releasing hormone surges by estrogen. I. Role of hypothalamic progesterone receptors.” Endocrinology 141(4): 1477–1485. [DOI] [PubMed] [Google Scholar]
- Chappell PE, et al. (2003). “Circadian gene expression regulates pulsatile gonadotropin-releasing hormone (GnRH) secretory patterns in the hypothalamic GnRH-secreting GT1–7 cell line.” Journal of Neuroscience 23(35): 11202–11213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassard D, et al. (2015). “Evidence for a putative circadian kiss-clock in the hypothalamic AVPV in female mice.” Endocrinology 156(8): 2999–3011. [DOI] [PubMed] [Google Scholar]
- Chemineau P, et al. (1988). “Photoperiodic and melatonin treatments for the control of seasonal reproduction in sheep and goats.” Reproduction Nutrition Développement 28(2B): 409–422. [DOI] [PubMed] [Google Scholar]
- Chen H, et al. (2013). “FSH induces the development of circadian clockwork in rat granulosa cells via a gap junction protein Cx43-dependent pathway.” American journal of physiology. Endocrinology and metabolism 304(6): E566–575. [DOI] [PubMed] [Google Scholar]
- Chen H, et al. (2013). “Downregulation of core clock gene Bmal1 attenuates expression of progesterone and prostaglandin biosynthesis-related genes in rat luteinizing granulosa cells.” American journal of physiology. Cell physiology 304(12): C1131–1140. [DOI] [PubMed] [Google Scholar]
- Choe HK, et al. (2013). “Synchronous activation of gonadotropin-releasing hormone gene transcription and secretion by pulsatile kisspeptin stimulation.” Proceedings of the National Academy of Sciences 110(14): 5677–5682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian CA, et al. (2008). “Classical estrogen receptor a signaling mediates negative and positive feedback on gonadotropin-releasing hormone neuron firing.” Endocrinology 149(11): 5328–5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian CA and Moenter SM (2008). “Vasoactive intestinal polypeptide can excite gonadotropin-releasing hormone neurons in a manner dependent on estradiol and gated by time of day.” Endocrinology 149(6): 3130–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian CA and Moenter SM (2010). “The neurobiology of preovulatory and estradiol-induced gonadotropin-releasing hormone surges.” Endocrine reviews 31(4): 544–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu A, et al. (2013). “Global but not gonadotrope-specific disruption of Bmal1 abolishes the luteinizing hormone surge without affecting ovulation.” Endocrinology 154(8): 2924–2935. [DOI] [PubMed] [Google Scholar]
- Chu G, et al. (2012). “Contribution of FSH and triiodothyronine to the development of circadian clocks during granulosa cell maturation.” Am J Physiol Endocrinol Metab 302(6): E645–653. [DOI] [PubMed] [Google Scholar]
- Chu G, et al. (2011). “Alterations of circadian clockworks during differentiation and apoptosis of rat ovarian cells.” Chronobiol Int 28(6): 477–487. [DOI] [PubMed] [Google Scholar]
- Chung F-F, et al. (2005). “The associations between menstrual function and life style/working conditions among nurses in Taiwan.” Journal of occupational health 47(2): 149–156. [DOI] [PubMed] [Google Scholar]
- Clarkson J, et al. (2009). “Postnatal development of an estradiol-kisspeptin positive feedback mechanism implicated in puberty onset.” Endocrinology 150(7): 3214–3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson J, et al. (2017). “Definition of the hypothalamic GnRH pulse generator in mice.” Proceedings of the National Academy of Sciences 114(47): E10216–E10223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Iglesia HO and Schwartz WJ (2006). “Minireview: timely ovulation: circadian regulation of the female hypothalamo-pituitary-gonadal axis.” Endocrinology 147(3): 1148–1153. [DOI] [PubMed] [Google Scholar]
- Desroziers E, et al. (2010). “Mapping of kisspeptin fibres in the brain of the pro-oestrous rat.” Journal of neuroendocrinology 22(10): 1101–1112. [DOI] [PubMed] [Google Scholar]
- Dolatshad H, et al. (2005). “Developmental and reproductive performance in circadian mutant mice.” Human reproduction 21(1): 68–79. [DOI] [PubMed] [Google Scholar]
- Dror T, et al. (2013). “Analysis of multiple positive feedback paradigms demonstrates a complete absence of LH surges and GnRH activation in mice lacking Kisspeptin signaling.” Biol Reprod 88(6): 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggavathi R, et al. (2008). “Liver receptor homolog 1 is essential for ovulation.” Genes Dev 22(14): 1871–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulka E and Moenter S (2019). “SAT-412 Effects of the Ovary and Dihydrotestosterone on Firing Rate of GnRH Neurons in Prenatally Androgenized Mice.” Journal of the Endocrine Society 3(Supplement_1): SAT–412. [Google Scholar]
- Ebenhöh O and Hazlerigg D (2013). “Modelling a molecular calendar: the seasonal photoperiodic response in mammals.” Chaos, Solitons & Fractals 50: 39–47. [Google Scholar]
- Ebihara S, et al. (1986). “Genetic control of melatonin synthesis in the pineal gland of the mouse.” Science 231(4737): 491–493. [DOI] [PubMed] [Google Scholar]
- Estrada K, et al. (2006). “Elevated KiSS-1 expression in the arcuate nucleus prior to the cyclic preovulatory gonadotrophin-releasing hormone/lutenising hormone surge in the ewe suggests a stimulatory role for kisspeptin in oestrogen-positive feedback.” Journal of neuroendocrinology 18(10): 806–809. [DOI] [PubMed] [Google Scholar]
- Evans JA, et al. (2012). “Cryl−/− circadian rhythmicity depends on SCN intercellular coupling.” J Biol Rhythms 27(6): 443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett JW and Sawyer CH (1950). “A 24-hour periodicity in the “LH-release apparatus” of female rats, disclosed by barbiturate sedation.” Endocrinology 47(3): 198–218. [DOI] [PubMed] [Google Scholar]
- Fahrenkrug J, et al. (2006). “Diurnal rhythmicity of the clock genes Per1 and Per2 in the rat ovary.” Endocrinology 147(8): 3769–3776. [DOI] [PubMed] [Google Scholar]
- Fernandez RC, et al. (2016). Fixed or rotating night shift work undertaken by women: implications for fertility and miscarriage Seminars in reproductive medicine, Thieme Medical Publishers. [DOI] [PubMed] [Google Scholar]
- Foster DL, et al. (2006). “Programming of GnRH feedback controls timing puberty and adult reproductive activity.” Mol Cell Endocrinol 254–255: 109–119. [DOI] [PubMed] [Google Scholar]
- Gamble KL, et al. (2013). “Shift work and circadian dysregulation of reproduction.” Frontiers in endocrinology 4: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson MJ, et al. (1994). “Continuous gonadotropin-releasing hormone infusion stimulates dramatic gonadal development in hypogonadal female mice.” Biol. Reprod 50(3): 680–685. [DOI] [PubMed] [Google Scholar]
- Gill JC, et al. (2010). “Reproductive hormone-dependent and-independent contributions to developmental changes in kisspeptin in GnRH-deficient hypogonadal mice.” PloS one 5(7): e11911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JM, et al. (2003). “Expression of circadian rhythm genes in gonadotropin-releasing hormone-secreting GT1–7 neurons.” Endocrinology 144(12): 5285–5292. [DOI] [PubMed] [Google Scholar]
- Glanowska KM, et al. (2014). “Development of gonadotropin-releasing hormone secretion and pituitary response.” Journal of Neuroscience 34(45): 15060–15069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanowska KM, et al. (2012). “Fast scan cyclic voltammetry as a novel method for detection of real-time gonadotropin-releasing hormone release in mouse brain slices.” Journal of Neuroscience 32(42): 14664–14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorman M (In Press). “Temporal organization of melatonin signalling in mammals.” Molecular and cellular endocrinology. [DOI] [PubMed] [Google Scholar]
- Goto M, et al. (1989). “Melatonin content of the pineal gland in different mouse strains.” Journal of pineal research 7(2): 195–204. [DOI] [PubMed] [Google Scholar]
- Gottsch M, et al. (2006). “Kisspepeptin-GPR54 signaling in the neuroendocrine reproductive axis.” Molecular and cellular endocrinology 254: 91–96. [DOI] [PubMed] [Google Scholar]
- Greenhill C (2014). “Circadian clock involved in embryo implantation.” Nature Reviews Endocrinology 10(12). [DOI] [PubMed] [Google Scholar]
- Gu G and Simerly R (1997). “Projections of the sexually dimorphic anteroventral periventricular nucleus in the female rat.” Journal of Comparative Neurology 384(1): 142–164. [PubMed] [Google Scholar]
- Han S-K, et al. (2005). “Activation of gonadotropin-releasing hormone neurons by kisspeptin as a neuroendocrine switch for the onset of puberty.” Journal of Neuroscience 25(49): 11349–11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland R, et al. (2006). “Melatonin.” The international journal of biochemistry & cell biology 38(3): 313–316. [DOI] [PubMed] [Google Scholar]
- Hastings MH, et al. (2019). “The mammalian circadian timing system and the suprachiasmatic nucleus as its pacemaker.” Biology 8(1): 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori M, et al. (2014). “Lhx1 maintains synchrony among circadian oscillator neurons of the SCN.” eLife 3: e03357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He PJ, et al. (2007). “The disruption of circadian clockwork in differentiating cells from rat reproductive tissues as identified by in vitro real-time monitoring system.” J Endocrinol 193(3): 413–420. [DOI] [PubMed] [Google Scholar]
- He PJ, et al. (2007). “Gonadotropic regulation of circadian clockwork in rat granulosa cells.” Mol Cell Biochem 302(1–2): 111–118. [DOI] [PubMed] [Google Scholar]
- He PJ, et al. (2007). “Up-regulation of Per1 expression by estradiol and progesterone in the rat uterus.” J Endocrinol 194(3): 511–519. [DOI] [PubMed] [Google Scholar]
- Herbison AE (2008). “Estrogen positive feedback to gonadotropin-releasing hormone (GnRH) neurons in the rodent: the case for the rostral periventricular area of the third ventricle (RP3V).” Brain research reviews 57(2): 277–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbison AE (2018). “The gonadotropin-releasing hormone pulse generator.” Endocrinology 159(11): 3723–3736. [DOI] [PubMed] [Google Scholar]
- Herbison AE, et al. (2007). “Gonadotropin-releasing hormone neuron requirements for puberty, ovulation, and fertility.” Endocrinology 149(2): 597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickok JR and Tischkau SA (2010). “In vivo circadian rhythms in gonadotropin-releasing hormone neurons.” Neuroendocrinology 91(1): 110–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst JJ, et al. (1993). “Plasma oxytocin and nocturnal uterine activity: maternal but not fetal concentrations increase progressively during late pregnancy and delivery in rhesus monkeys.” American journal of obstetrics and gynecology 169(2): 415–422. [DOI] [PubMed] [Google Scholar]
- Hoffmann HM, et al. (2019). “Differential CRE Expression in Lhrh-cre and GnRH-cre Alleles and the Impact on Fertility in Otx2-Flox Mice.” Neuroendocrinology 108(4): 328–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, et al. (2018). “Haploinsufficiency of Homeodomain Proteins Six3, Vax1, and Otx2 Causes Subfertility in Mice via Distinct Mechanisms.” Neuroendocrinology 108(4): 328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, et al. (2016). “Deletion of Vax1 from gonadotropin-releasing hormone (GnRH) neurons abolishes GnRH expression and leads to hypogonadism and infertility.” Journal of Neuroscience 36(12): 3506–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann HM, et al. (2014).” Heterozygous deletion of ventral anterior homeobox (vaxl) causes subfertility in mice.” Endocrinology. 155(10):4043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenesch JB and Ueda HR (2011). “Understanding systems-level properties: timely stories from the study of clocks.” Nat Rev Genet 12(6): 407–416. [DOI] [PubMed] [Google Scholar]
- Horvath TL, et al. (1998). “Gender-specific apposition between vasoactive intestinal peptide-containing axons and gonadotrophin-releasing hormone-producing neurons in the rat.” Brain Res 795(1–2): 277–281. [DOI] [PubMed] [Google Scholar]
- Irwig MS, et al. (2004). “Kisspeptin activation of gonadotropin releasing hormone neurons and regulation of KiSS-1 mRNA in the male rat.” Neuroendocrinology 80(4): 264–272. [DOI] [PubMed] [Google Scholar]
- Jacobi JS, et al. (2007). “17-Beta-estradiol directly regulates the expression of adrenergic receptors and kisspeptin/GPR54 system in GT1–7 GnRH neurons.” Neuroendocrinology 86(4): 260–269. [DOI] [PubMed] [Google Scholar]
- Johnson MH, et al. (2002). “Circadian clockwork genes are expressed in the reproductive tract and conceptus of the early pregnant mouse.” Reprod.Biomed.Online 4(2): 140. [DOI] [PubMed] [Google Scholar]
- Karman BN and Tischkau SA (2006). “Circadian clock gene expression in the ovary: Effects of luteinizing hormone.” Biol Reprod 75(4): 624–632. [DOI] [PubMed] [Google Scholar]
- Karsch FJ, et al. (1984). Neuroendocrine basis of seasonal reproduction Proceedings of the 1983 Laurentian Hormone Conference, Elsevier. [Google Scholar]
- Kauffman AS, et al. (2007). “Sexual differentiation of Kiss1 gene expression in the brain of the rat.” Endocrinology 148(4): 1774–1783. [DOI] [PubMed] [Google Scholar]
- Kauffman AS, et al. (2007). “The kisspeptin receptor GPR54 is required for sexual differentiation of the brain and behavior.” Journal of Neuroscience 27(33): 8826–8835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KKennaway DJ, et al. (2012). “Circadian rhythms and fertility.” Molecular and cellular endocrinology 349(1): 56–61. [DOI] [PubMed] [Google Scholar]
- Kim JW, et al. (2005). “The orphan nuclear receptor, liver receptor homolog-1, regulates cholesterol side-chain cleavage cytochrome p450 enzyme in human granulosa cells.” J Clin Endocrinol Metab 90(3): 1678–1685. [DOI] [PubMed] [Google Scholar]
- Kimura F, et al. (1987). “Effects of preoptic injections of gastrin, cholecystokinin, secretin, vasoactive intestinal peptide and PHI on the secretion of luteinizing hormone and prolactin in ovariectomized estrogen-primed rats.” Brain Res 410(2): 315–322. [DOI] [PubMed] [Google Scholar]
- Klose M, et al. (2011). “Temporal control of spermatogenesis is independent of the central circadian pacemaker in Djungarian hamsters (Phodopus sungorus).” Biology of reproduction 84(1): 124–129. [DOI] [PubMed] [Google Scholar]
- Knobil E (1980). The neuroendocrine control of the menstrual cycle Proceedings of the 1979 Laurentian Hormone Conference, Elsevier. [Google Scholar]
- Knutsson A (2003). “Health disorders of shift workers.” Occupational medicine 53(2): 103–108. [DOI] [PubMed] [Google Scholar]
- Ko CH, et al. (2010). “Emergence of noise-induced oscillations in the central circadian pacemaker.” PLoS Biol 8(10): e1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, et al. (2012). “Transcriptional architecture and chromatin landscape of the core circadian clock in mammals.” Science 338(6105): 349–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegsfeld LJ (2013). “Circadian regulation of kisspeptin in female reproductive functioning.” Adv Exp Med Biol 784: 385–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegsfeld LJ, et al. (2002). “Vasoactive intestinal polypeptide contacts on gonadotropin-releasing hormone neurones increase following puberty in female rats.” J Neuroendocrinol 14(9): 685–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P and Sait SF (2011). “Luteinizing hormone and its dilemma in ovulation induction.” Journal of human reproductive sciences 4(1): 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labyak S, et al. (2002). “Effects of shiftwork on sleep and menstrual function in nurses.” Health care for women international 23(6–7): 703–714. [DOI] [PubMed] [Google Scholar]
- Lapatto R, et al. (2007). “Kiss1−/− mice exhibit more variable hypogonadism than Gpr54−/− mice.” Endocrinology 148(10): 4927–4936. [DOI] [PubMed] [Google Scholar]
- Legan SJ and Karsch FJ (1975). “A daily signal for the LH surge in the rat.” Endocrinology 96(1): 57–62. [DOI] [PubMed] [Google Scholar]
- Lehman MN, et al. (2013). Neuroanatomy of the kisspeptin signaling system in mammals: comparative and developmental aspects Kisspeptin signaling in reproductive biology, Springer: 27–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman MN, et al. (2010). “Anatomy of the kisspeptin neural network in mammals.” Brain Res 1364: 90–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, et al. (2017). “Altered expression of miRNAs in the uterus from a letrozole-induced rat PCOS model.” Gene 598: 20–26. [DOI] [PubMed] [Google Scholar]
- Li L, et al. (2014). “Cooperation of luteinizing hormone signaling pathways in preovulatory avian follicles regulates circadian clock expression in granulosa cell.” Mol Cell Biochem 394(1–2): 31–41. [DOI] [PubMed] [Google Scholar]
- Liu AC, et al. (2007). “Intercellular coupling confers robustness against mutations in the SCN circadian clock network.” Cell 129(3): 605–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, et al. (2014). “Loss of BMAL1 in ovarian steroidogenic cells results in implantation failure in female mice.” Proceedings of the National Academy of Sciences 111(39): 14295–14300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh DH, et al. (2014). “Disrupted Reproduction, Estrous Cycle, and Circadian Rhythms in Female Mice Deficient in Vasoactive Intestinal Peptide.” J Biol Rhythms. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohstroh PN, et al. (2003). “Bone resorption is affected by follicular phase length in female rotating shift workers.” Environmental health perspectives 111(4): 618–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL and Takahashi JS (2011). “Genetics of circadian rhythms in Mammalian model organisms.” Adv Genet 74: 175–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv S, et al. (2019). “Impaired decidualization caused by downregulation of circadian clock gene BMAL1 contributes to human recurrent miscarriage.” Biology of reproduction. [DOI] [PubMed] [Google Scholar]
- Ma X, et al. (2004). “Targeted disruption of luteinizing hormone beta-subunit leads to hypogonadism, defects in gonadal steroidogenesis, and infertility.” Proc. Natl. Acad. Sci. USA 101(49): 17294–17299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahoney MM (2010). “Shift work, jet lag, and female reproduction.” Int J Endocrinol 2010: 813764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matagne V, et al. (2012). “Thyroid transcription factor 1, a homeodomain containing transcription factor, contributes to regulating periodic oscillations in GnRH gene expression.” J Neuroendocrinol 24(6): 916–929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuillan HJ, et al. (2019). “GnRH pulse generator activity across the estrous cycle of female mice.” Endocrinology 160(6): 1480–1491. [DOI] [PubMed] [Google Scholar]
- Mereness AL, et al. (2016). “Conditional Deletion of Bmal1 in Ovarian Theca Cells Disrupts Ovulation in Female Mice.” Endocrinology 157(2): 913–927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mereness AL, et al. (2015). “Developmental programming by androgen affects the circadian timing system in female mice.” Biol Reprod 92(4): 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesiano S and Jaffe RB (1997). “Developmental and functional biology of the primate fetal adrenal cortex.” Endocrine reviews 18(3): 378–403. [DOI] [PubMed] [Google Scholar]
- Messager S, et al. (2005). “Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54.” Proceedings of the National Academy of Sciences 102(5): 1761–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH, et al. (2004). “Circadian clock mutation disrupts estrous cyclicity and maintenance of pregnancy.” Current biology 14(15): 1367–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BH and Takahashi JS (2013). “Central circadian control of female reproductive function.” Frontiers in endocrinology 4: 195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami Y, et al. (2013). “Mammalian circadian clock: the roles of transcriptional repression and delay.” Handb Exp Pharmacol(217): 359–377. [DOI] [PubMed] [Google Scholar]
- Moenter SM, et al. (1990). “The estradiol-induced surge of gonadotropin-releasing hormone in the ewe.” Endocrinology 127(3): 1375–1384. [DOI] [PubMed] [Google Scholar]
- Mongrain V, et al. (2008). “Clock-dependent and independent transcriptional control of the two isoforms from the mouse Rorygene.” Genes to Cells 13(12): 1197–1210. [DOI] [PubMed] [Google Scholar]
- Morse D, et al. (2003). “No circadian rhythms in testis: Period1 expression is clock independent and developmentally regulated in the mouse.” Molecular endocrinology 17(1): 141–151. [DOI] [PubMed] [Google Scholar]
- Nakamura TJ, et al. (2010). “Influence of the estrous cycle on clock gene expression in reproductive tissues: effects of fluctuating ovarian steroid hormone levels.” Steroids 75(3): 203–212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura TJ, et al. (2008). “Estrogen directly modulates circadian rhythms of PER2 expression in the uterus.” American Journal of Physiology-Endocrinology and Metabolism 295(5): E1025–E1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitta M, et al. (1999). “CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene.” Proc Natl Acad Sci U S A 96(12): 6660–6665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurminen T (1998). “Shift work and reproductive health.” Scand J Work Environ Health 24 Suppl 3: 28–34. [PubMed] [Google Scholar]
- Oiwa A, et al. (2007). “Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1.” Biochem Biophys Res Commun 353(4): 895–901. [DOI] [PubMed] [Google Scholar]
- Olcese J (2014). “Circadian clocks and pregnancy.” Frontiers in endocrinology 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olcese J, et al. (2003). “Expression and regulation of mPer1 in immortalized GnRH neurons.” Neuroreport 14(4): 613–618. [DOI] [PubMed] [Google Scholar]
- Olcese J, et al. (2013). “Melatonin and the circadian timing of human parturition.” Reproductive Sciences 20(2): 168–174. [DOI] [PubMed] [Google Scholar]
- Padilla SL, et al. (2019). “Kisspeptin neurons in the arcuate nucleus of the hypothalamus orchestrate circadian rhythms and metabolism.” Current biology 29(4): 592–604. e594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi EC, et al. (2019). “The Homeodomain Transcription Factors Vax1 and Six6 Are Required for SCN Development and Function.” Molecular Neurobiology. Nov 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi EC, et al. (2019). “Deletion of the Homeodomain Protein Six6 from GnRH Neurons Decreases GnRH Gene Expression Resulting in Infertility.” Endocrinology. 160(9):2151–2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin JS, et al. (2006). “The expression of the clock protein PER2 in the limbic forebrain is modulated by the estrous cycle.” Proceedings of the National Academy of Sciences 103(14): 5591–5596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen SL, et al. (2003). “Direct and indirect regulation of gonadotropin-releasing hormone neurons by estradiol.” Biology of reproduction 69(6): 1771–1778. [DOI] [PubMed] [Google Scholar]
- Piet R, et al. (2016). “Vasoactive intestinal peptide excites GnRH neurons in male and female mice.” Endocrinology 157(9): 3621–3630. [DOI] [PubMed] [Google Scholar]
- Pilorz V and Steinlechner S (2008). “Low reproductive success in Per1 and Per2 mutant mouse females due to accelerated ageing?” Reproduction 135(4): 559. [DOI] [PubMed] [Google Scholar]
- Ratajczak CK, et al. (2012). “Generation of myometrium-specific Bmal1 knockout mice for parturition analysis.” Reproduction, Fertility and Development 24(5): 759–767. [DOI] [PubMed] [Google Scholar]
- Ratajczak CK, et al. (2009). “Impaired steroidogenesis and implantation failure in Bmal1−/− mice.” Endocrinology 150(4): 1879–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reppert SM, et al. (1987). “The circadian-gated timing of birth in rats: disruption by maternal SCN lesions or by removal of the fetal brain.” Brain Res 403(2): 398–402. [DOI] [PubMed] [Google Scholar]
- Revel FG, et al. (2009). “Melatonin controls seasonal breeding by a network of hypothalamic targets.” Neuroendocrinology 90(1): 1–14. [DOI] [PubMed] [Google Scholar]
- Richards JS (2001). “Perspective: The ovarian follicle--A perspective in 2001.” Endocrinology 142(6): 2184–2193. [DOI] [PubMed] [Google Scholar]
- Richards JS, et al. (1998). “Molecular mechanisms of ovulation and luteinization.” Mol Cell Endocrinol 145(1–2): 47–54. [DOI] [PubMed] [Google Scholar]
- Robertson JL, et al. (2009). “Circadian regulation of Kiss1 neurons: implications for timing the preovulatory gonadotropin-releasing hormone/luteinizing hormone surge.” Endocrinology 150(8): 3664–3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubel CA, et al. (2012). “Research resource: Genome-wide profiling of progesterone receptor binding in the mouse uterus.” Mol Endocrinol 26(8): 1428–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson WK, et al. (1981). “Vasoactive intestinal peptide stimulates luteinizing hormone-releasing hormone release from median eminence synaptosomes.” Regulatory peptides 2(4): 253–264. [DOI] [PubMed] [Google Scholar]
- Schoeller EL, et al. (2016). “Bmal1 is required for normal reproductive behaviors in male mice.” Endocrinology 157(12): 4914–4929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellix MT (2015). “Circadian clock function in the mammalian ovary.” J Biol Rhythms 30(1): 7–19. [DOI] [PubMed] [Google Scholar]
- Sellix MT, et al. (2013). “Excess androgen during puberty disrupts circadian organization in female rats.” Endocrinology 154(4): 1636–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellix MT, et al. (2010). “A circadian egg timer gates ovulation.” Curr Biol 20(6): R266–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semaan SJ and Kauffman AS (2010). “Sexual differentiation and development of forebrain reproductive circuits.” Curr Opin Neurobiol 20(4): 424–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharkey JT, et al. (2009). “Melatonin synergizes with oxytocin to enhance contractility of human myometrial smooth muscle cells.” The Journal of Clinical Endocrinology & Metabolism 94(2): 421–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, et al. (2011). “Circadian Clock genes Per2 and clock regulate steroid production, cell proliferation, and luteinizing hormone receptor transcription in ovarian granulosa cells.” Biochem Biophys Res Commun 412(1): 132–135. [DOI] [PubMed] [Google Scholar]
- Simonneaux V, et al. (2017). “Daily rhythms count for female fertility.” Best Practice & Research Clinical Endocrinology & Metabolism 31(5): 505–519. [DOI] [PubMed] [Google Scholar]
- Singh J and Handelsman DJ (1996). “Neonatal administration of FSH increases Sertoli cell numbers and spermatogenesis in gonadotropin-deficient (hpg) mice.” J. Endocrinol 151(1): 37–48. [DOI] [PubMed] [Google Scholar]
- Sirois J, et al. (2004). “Cyclooxygenase-2 and its role in ovulation: a 2004 account.” Hum Reprod Update 10(5): 373–385. [DOI] [PubMed] [Google Scholar]
- Smarr BL, et al. (2013). “Oestrogen-independent circadian clock gene expression in the anteroventral periventricular nucleus in female rats: possible role as an integrator for circadian and ovarian signals timing the luteinising hormone surge.” J Neuroendocrinol 25(12): 1273–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smarr BL, et al. (2012). “The dorsomedial suprachiasmatic nucleus times circadian expression of Kiss1 and the luteinizing hormone surge.” Endocrinology 153(6): 2839–2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JT (2008). “Kisspeptin signalling in the brain: steroid regulation in the rodent and ewe.” Brain research reviews 57(2): 288–298. [DOI] [PubMed] [Google Scholar]
- Smith JT, et al. (2005). “Regulation of Kiss1 gene expression in the brain of the female mouse.” Endocrinology 146(9): 3686–3692. [DOI] [PubMed] [Google Scholar]
- Smith JT, et al. (2006). “Kiss1 neurons in the forebrain as central processors for generating the preovulatory luteinizing hormone surge.” Journal of Neuroscience 26(25): 6687–6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MJ, et al. (2000). “Localization of the VIP2 receptor protein on GnRH neurons in the female rat.” Endocrinology 141(11): 4317–4320. [DOI] [PubMed] [Google Scholar]
- Stock D, et al. (2019). “Rotating night shift work and menopausal age.” Human reproduction 34(3): 539–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stojkovic K, et al. (2014). “A central role for ubiquitination within a circadian clock protein modification code.” Frontiers in molecular neuroscience 7: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto Y, et al. (2015). “Roles of prostaglandin receptors in female reproduction.” J Biochem 157(2): 73–80. [DOI] [PubMed] [Google Scholar]
- Tischkau SA, et al. (2011). “The luteinizing hormone surge regulates circadian clock gene expression in the chicken ovary.” Chronobiol Int 28(1): 10–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonsfeldt KJ, et al. (2019). The Contribution of Bmal1 Expression in the Suprachiasmatic Nucleus to Female Fertility. ENDOExpo. [Google Scholar]
- Tonsfeldt KJ, et al. (2011). “Oestrogen induces rhythmic expression of the Kisspeptin-1 receptor GPR54 in hypothalamic gonadotrophin-releasing hormone-secreting GT1–7 [DOI] [PMC free article] [PubMed]
- Tonsfeldt KJ, et al. (2019). “The Contribution of the Circadian Gene Bmal1 to Female Fertility and the Generation of the Preovulatory Luteinizing Hormone Surge.” J Endocr Soc 3(4): 716–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi R and Webster NJ (2009). “GnRH pulsatility, the pituitary response and reproductive dysfunction.” Endocrine journal 56(6): 729–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Beek E, et al. (1994). “Preferential induction of c-fos immunoreactivity in vasoactive intestinal polypeptide-innervated gonadotropin-releasing hormone neurons during a steroid-induced luteinizing hormone surge in the female rat.” Endocrinology 134(6): 2636–2644. [DOI] [PubMed] [Google Scholar]
- van der Beek EM, et al. (1997). “Evidence for a direct neuronal pathway from the suprachiasmatic nucleus to the gonadotropin-releasing hormone system: combined tracing and light and electron microscopic immunocytochemical studies.” J. Comp. Neurol 384(4): 569–579. [DOI] [PubMed] [Google Scholar]
- Van Der Horst GT, et al. (1999). “Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms.” Nature 398(6728): 627. [DOI] [PubMed] [Google Scholar]
- Vijayan E, et al. (1979). “Vasoactive intestinal peptide: evidence for a hypothalamic site of action to release growth hormone, luteinizing hormone, and prolactin in conscious ovariectomized rats.” Endocrinology 104(1): 53–57. [DOI] [PubMed] [Google Scholar]
- Wang L, et al. (2019). “Genetic dissection of the different roles of hypothalamic kisspeptin neurons in regulating female reproduction.” eLife 8: e43999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson E, et al. (1995). “LH and progesterone concentrations during diestrus in the mare and the effect of hCG.” Theriogenology 43(8): 1325–1337. [Google Scholar]
- Welsh DK, et al. (1995). “Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms.” Neuron 14: 697–706. [DOI] [PubMed] [Google Scholar]
- Welsh DK, et al. (2010). “Suprachiasmatic nucleus: cell autonomy and network properties.” Annu Rev Physiol 72: 551–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegand SJ and Terasawa E (1982). “Discrete lesions reveal functional heterogeneity of suprachiasmatic structures in regulation of gonadotropin secretion in the female rat.” Neuroendocrinology 34(6): 395–404. [DOI] [PubMed] [Google Scholar]
- Wierman ME, et al. (2011). “Gonadotropin-releasing hormone (GnRH) neuron migration: initiation, maintenance and cessation as critical steps to ensure normal reproductive function.” Front. Neuroendocrinol 32(1): 43–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WP 3rd, et al. (2011). “Circadian control of kisspeptin and a gated GnRH response mediate the preovulatory luteinizing hormone surge.” Endocrinology 152(2): 595–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams WP 3rd and Kriegsfeld LJ (2012). “Circadian control of neuroendocrine circuits regulating female reproductive function.” Frontiers in endocrinology 3: 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermantel TM, et al. (2006). “Definition of estrogen receptor pathway critical for estrogen positive feedback to gonadotropin-releasing hormone neurons and fertility.” Neuron 52(2): 271–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, et al. (2016). “Loss of Bmal1 decreases oocyte fertilization, early embryo development and implantation potential in female mice.” Zygote 24(5): 760–767. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, et al. (2009). “Timing of the ovarian circadian clock is regulated by gonadotropins.” Endocrinology 150(9): 4338–4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, et al. (2017). “Rhythmic expression of circadian clock genes in the preovulatory ovarian follicles of the laying hen.” PLoS One 12(6): e0179019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S and Kriegsfeld LJ (2009). “Daily changes in GT1–7 cell sensitivity to GnRH secretagogues that trigger ovulation.” Neuroendocrinology 89(4): 448–457. [DOI] [PMC free article] [PubMed] [Google Scholar]