Abstract
Clinical trials using adult stem cells to regenerate damaged heart tissue continue to this day1,2 despite ongoing questions of efficacy and a lack of mechanistic understanding of the underlying biologic effect3. The rationale for these cell therapy trials is derived from animal studies that show a modest but reproducible improvement in cardiac function in models of cardiac ischemic injury4,5. Here we examined the mechanistic basis for cell therapy in mice after ischemia/reperfusion (I/R) injury, and while heart function was enhanced, it was not associated with new cardiomyocyte production. Cell therapy improved heart function through an acute sterile immune response characterized by the temporal and regional induction of CCR2+ and CX3CR1+ macrophages. Intra-cardiac injection of 2 distinct types of adult stem cells, freeze/thaw-killed cells or a chemical inducer of the innate immune response similarly induced regional CCR2+ and CX3CR1+ macrophage accumulation and provided functional rejuvenation to the I/R-injured heart. This selective macrophage response altered cardiac fibroblast activity, reduced border zone extracellular matrix (ECM) content, and enhanced the mechanical properties of the injured area. The functional benefit of cardiac cell therapy is thus due to an acute inflammatory-based wound healing response that rejuvenates the mechanical properties of the infarcted area of the heart.
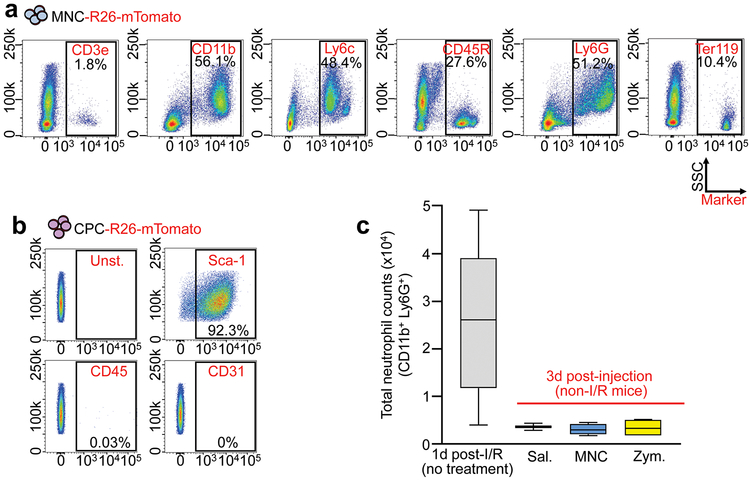
Initial animal studies with adult stem cells reported improved heart function through new cardiomyocyte formation by transdifferentiation of the injected cells6,7. However, adult stem cell transdifferentiation was not observed in later studies4,5,8 and clinical trials using adult stem cells in patients with acute myocardial infarction (MI) injury or decompensated heart failure have been indeterminate1,9. Hence the mechanistic basis for cell therapy remains unclear, although a paracrine hypothesis has been proposed10. Here we focused on 2 primary adult stem cell-types: fractionated bone marrow mononuclear cells (MNCs), as extensively used in human clinical trials2, and cardiac mesenchymal cells that express the receptor tyrosine kinase c-Kit, originally termed cardiac progenitor cells (CPCs)7,10. We also examined the effect of injecting zymosan, a non-cellular and potent activator of the innate immune response11. Isolated MNCs were a heterogeneous cell population consisting of all major hematopoietic lineages although monocytes and granulocytes were the most predominant (Extended Data Fig. 1a). CPCs expressed mesenchymal cell surface markers but were negative for markers of hematopoietic or endothelial cells (Extended Data Fig. 1b).
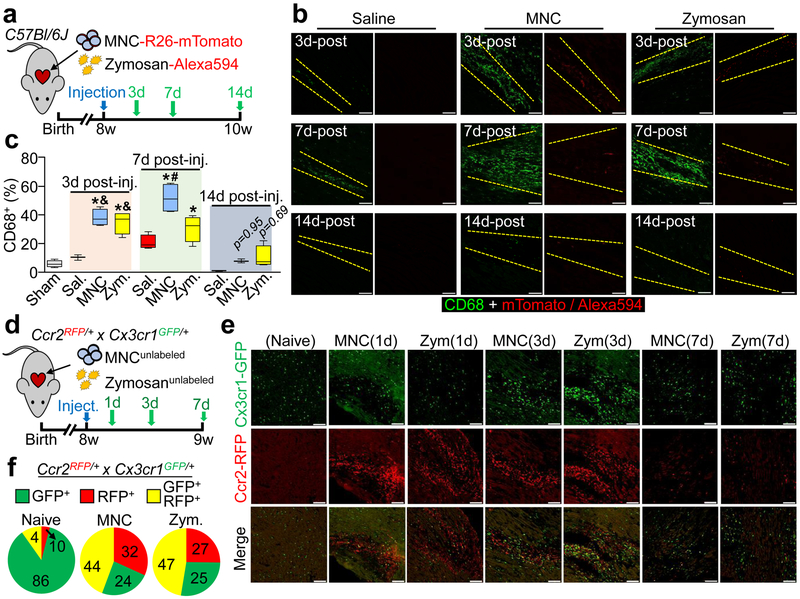
Uninjured 8-week-old male and female C57Bl/6J mice received intra-cardiac injection of either strain-matched MNCs, zymosan or saline (Fig 1a). Histological foci of acute inflammation were observed within areas of cell or zymosan injection, as examined by confocal microscopy from heart sections 3 days, 7 days, or 2 weeks post-injection (Fig. 1b). Activated CD68+ macrophages were significantly increased within the area of injection at 3 and 7 days, with a diminishing effect by 2 weeks as the cells or zymosan were cleared (Fig. 1b, c). No differences in neutrophil levels were observed from dissociated hearts at 3 days (Extended Data Fig. 1c).
Figure 1 ∣. Cardiac cell injection causes local inflammation with accumulation of distinct macrophage subtypes.
a, Experimental scheme using 8-week-old male and female C57Bl/6J mice subjected to intra-cardiac injection of strain-matched bone marrow mononuclear cells (MNC), Alexa Fluor 594-conjugated zymosan (Zym.) or sterile saline (Sal.). Sham animals received thoracotomy but no intra-cardiac injection. MNCs were isolated from Rosa26-mTomato mice on the C57Bl/6J background. b, Representative confocal immunohistochemistry micrographs of hearts showing activated CD68 macrophages (green) or the injected MNCs or zymosan (red). Dashed lines show injection sites. Images are from a minimum of 18 histological sections per mouse heart assessed from n=3 mice (Sal/3 days, Sal/14 days, MNC/14 days) or n=4 mice (all other groups). Scale bars = 100 μm. c, Quantitation of CD68+ cells as a percentage of total cells (DAPI+) imaged at areas of injection from the groups described in (b). Sample sizes for all groups are listed above in (b). At 3 days; *p<0.0001 versus sham, &p=0.0003 (MNC versus saline) or &p=0.0015 (Zym. versus saline). At 7 days; *p<0.0001 (MNC versus sham) or *p=0.0005 (Zym. versus sham), #p<0.0001 (MNC versus saline). Significance determined by one-way ANOVA with Tukey’s post-hoc test. Data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively). d, Experimental scheme using 8-week-old male and female Ccr2-RFP × Cx3cr1-GFP knock-in mice to simultaneously visualize CCR2+ and CX3CR1+ macrophage subtypes after injection of MNCs or zymosan. e, Representative confocal micrographs from cardiac histological sections from MNC or zymosan-injected mice, versus naïve (uninjected) controls (minimum of 30 sections assessed per mouse heart from n=2 naïve control mice and n=3 MNC or n=3 zymosan-injected mice), showing endogenous RFP and GFP immunofluorescence from CCR2+ or CX3CR1+ cells, respectively, at the injection site over a 7-day time course. Scale bars = 100 μm. f, Distribution of CCR2+ and CX3CR1+ macrophage subtypes in hearts at 3 days post-injection. Pie charts reflect the proportion of RFP (CCR2+) or GFP (CX3CR1+) expressing cells, as well as CCR2+ CX3CR1+ double-positive (yellow) cells detected by flow cytometry, as a percent of total macrophages identified by staining for F4/80 and CD64. Data are from n=6 MNC and n=6 zymosan-injected mice and n=2 naïve (uninjected) mice.
To further profile the induction of macrophages due to MNCs or zymosan injection, we used Ccr2-RFP12 and Cx3cr1-GFP13 knock-in mice to broadly distinguish major macrophage subtypes in the heart14,15. We delivered unlabeled MNCs or zymosan into 8-week-old Ccr2-RFP × Cx3cr1-GFP mice by intra-cardiac injection (Fig. 1d). Uninjured (non-injected) adult hearts showed GFP+ (CX3CR1+) macrophages throughout the myocardium while RFP+ (CCR2+) macrophages were largely absent at baseline (Fig. 1e). After 1 day, MNC and zymosan injected areas of the heart showed a robust accumulation of CCR2+ macrophages while CX3CR1+ macrophages began to expand in the periphery of the injected area (Fig 1e). By 3 days these CX3CR1+ macrophages were also expanded within the injection area along with CCR2+ macrophages, and this began to resolve by 7 days. Flow cytometry analysis from these mice at 3 days also indicated a shift in overall macrophage subtype content from a largely CX3CR1+ CCR2− population in the naïve state to a mix of enhanced numbers of CCR2+ and CCR2+ CX3CR1+ (double-positive) macrophages with either MNC or zymosan injection (Fig. 1f).
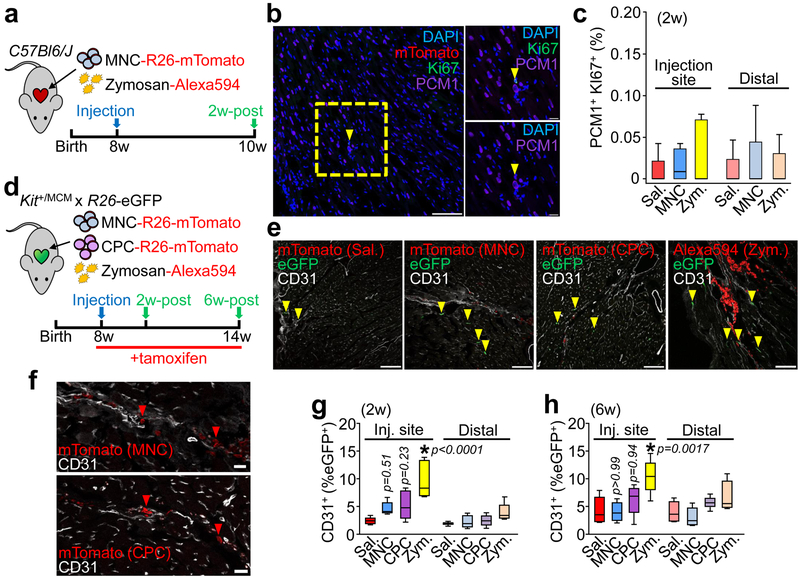
The ability of injected MNCs or zymosan to induce new cardiomyocyte (PCM1+) formation in the heart was also evaluated by immunohistochemistry (Fig. 2a, 2b). No appreciable increase in cardiomyocyte cell cycle activity (Ki67+) was observed versus saline-injected controls, either at areas of injection or distally across the entire tissue (Fig. 2c). Another proposed effect of cell therapy is the activation of endogenous CPCs that we previously determined can contribute to cardiac endothelial cell content after MI injury16. A tamoxifen-inducible KitMerCreMer/+ × Rosa26-eGFP lineage tracing mouse strategy was used to examine endothelial cell formation from endogenous CPCs after cellular or zymosan injection (Fig. 2d, 2e). Importantly, no transdifferentiation of injected MNCs or CPCs into cardiomyocytes or endothelial cells was observed (Fig. 2f). However, 2 and 6 weeks after injection, eGFP+ endothelial cells were significantly increased at the injection sites of zymosan-treated hearts but not with MNC or CPC injection (Fig. 2g, 2h). Zymosan persisted the longest within these hearts while CPCs and MNCs were essentially cleared by 2 weeks (Fig. 2e), potentially explaining why zymosan was more effective. None of the treatments increased c-Kit+-derived endothelial cells in the distal areas of the heart.
Figure 2 ∣. Cell or inflammatory therapy induces endothelial cell but not cardiomyocyte formation.
a, Schematic of experiments performed in panels b and c in 8-week-old male and female C57Bl/6J mice receiving intra-cardiac injection of MNCs, zymosan, or saline and analyzed 2 weeks later. b, Representative cardiac immunohistochemistry for Ki67 (green) and PCM1 (purple) from MNC-injected hearts. DAPI (blue) shows nuclei. Scale bar = 100 μM. A minimum of 45 histological sections were analyzed per mouse heart from n=4 MNC-treated mice or n=5 for all other groups of mice. Yellow box denotes area shown in higher magnification insets on the right. Yellow arrowhead denotes a cardiomyocyte with cell cycle activity. Scale bars for inset images = 10 μm. c, Quantitation of cardiomyocytes with cell cycle activity (PCM1+ Ki67+) as a percentage of all cardiomyocytes imaged (PCM1+) at 2 weeks. Data are from a minimum of 45 cardiac histological sections analyzed per mouse (n=4 MNC; injection site or n=5 for all other groups). d, Schematic of experiments performed in panels (e-h) using c-Kit lineage tracing mice (KitMerCreMer/+ x R26-eGFP) injected with MNCs, CPCs, zymosan or saline, then analyzed 2 weeks or 6 weeks later. Tamoxifen was administered continuously (in chow) starting one day before cell injection. e, Representative confocal immunohistochemistry images from hearts showing CD31+ endothelial cells (white) and injected MNCs, CPCs, or zymosan (red). eGFP (green) shows Kit allele-derived endothelial cells. Yellow arrowheads denote CD31+ endothelial cells that are also eGFP+. Scale bars = 100 μm. f, larger insets of images shown in e, indicating injected MNCs (top) or CPCs (bottom, rotated 90°) with red arrowheads that are negative for CD31 and lack known cardiomyocyte morphology. Scale bars = 20 μm. g,h, Quantitation of percent eGFP+ endothelial cells relative to total endothelial cells counted, either 2 weeks (g) or 6 weeks (h) post-injection. All data in (e-h) are from n=6 (Zym/6 weeks) or n=5 all other groups of mice. All p-values in (g-h) were determined by one-way ANOVA with Tukey’s post-hoc test. All numerical data in this figure (c,g,h) are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively). Data and representative micrographs in e-h are from a minimum of 45 histological sections analyzed per individual mouse heart from the numbers of mice indicated above.
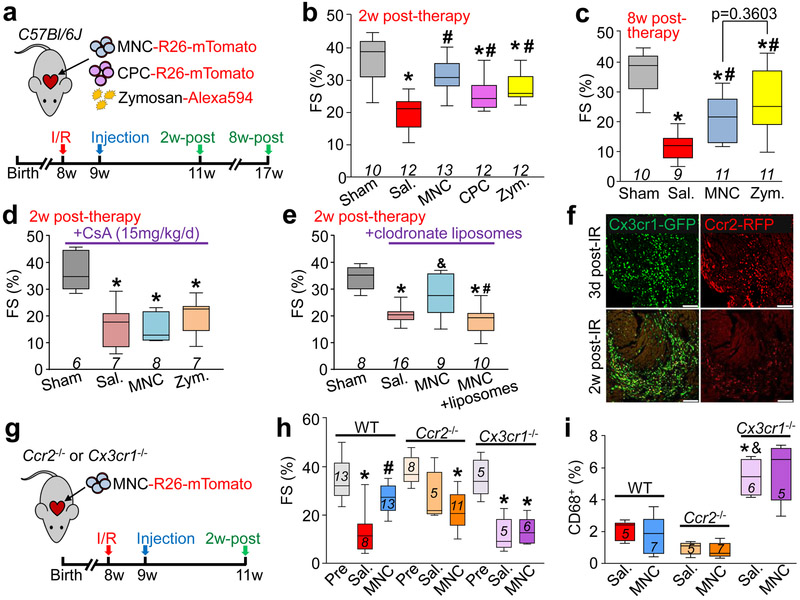
We next injected strain matched MNCs, CPCs, zymosan, or saline on each side of the infarct border zone in C57Bl/6J mice one week post-I/R injury (Fig. 3a). Importantly, cell or zymosan injection into uninjured hearts did not alter LV structure or function (Extended Data Fig. 2a-f). Injection of MNCs, CPCs or zymosan each significantly improved post-I/R cardiac ventricular performance 2w post-injection compared with saline injected controls (Fig. 3b). Of note, while the intra-cardiac injection procedure itself (saline) produced a mild inflammatory response (Extended Data Fig. 3a-e), it was mild and did not improve cardiac function after I/R (Extended Data Fig. 3f-i). Cell or zymosan injection was also associated with improvements in left ventricular end-systolic volume consistent with better cardiac function (LVESV; Extended Data Fig. 4a). In contrast, there was no change in end-diastolic volume (LVEDV; Extended Data Fig. 4b) or heart rate (Extended Data Fig. 4c) across any of the treatment groups 2w post-therapy. Importantly, the observed functional benefit persisted for at least 8w after injection of MNCs or zymosan (Fig. 3c).
Figure 3 ∣. Cell or inflammatory therapy rejuvenates post-I/R heart function.
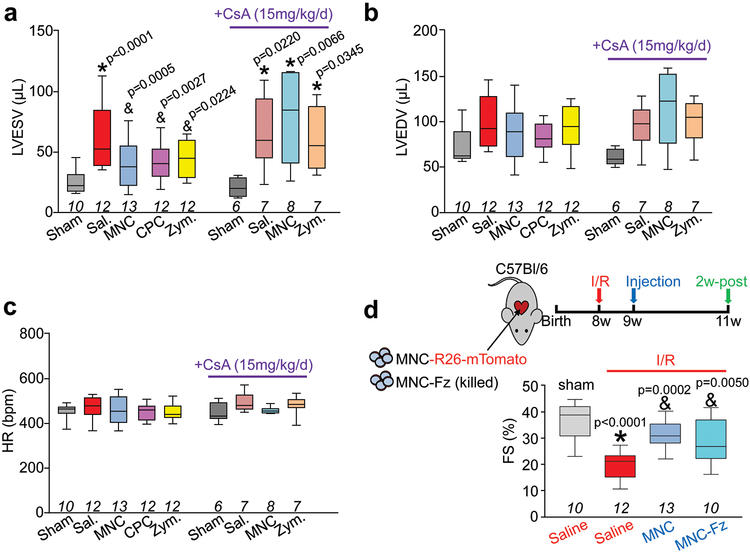
a, Schematic of experiments performed in panels b-c in which 8-week-old male and female C57Bl/6J mice received 120 min of myocardial ischemia followed by reperfusion (I/R) injury then 1 week later intra-cardiac injection of MNCs, CPCs, zymosan, or sterile saline flanking the injury area, followed by analysis 2w or 8 weeks later. b, Fractional shortening (FS) as measured by echocardiography in the groups indicated, 2 weeks post-cell or zymosan therapy. *p<0.0001 (Sal. versus sham) or *p=0.0002 (CPC versus sham) or *p=0.0019 (Zym. versus sham); #p<0.0001 (MNC versus Sal.) or #p=0.0157 (CPC versus Sal.) or #p=0.0019 (Zym. versus Sal.). c, FS at 8 weeks post-cell or zymosan therapy. *p<0.0001 (Sal. versus Sham) or *p=0.0002 (MNC versus Sham) or *p=0.0086 (Zym. versus Sham); #p=0.0194 (MNC versus Sal.) or #p=0.0005 (Zym. versus Sal.). Significance in b-c was determined by one-way ANOVA with Dunnett’s post-hoc test. The same sham group is shown in b and c as these experiments were performed in parallel. d, FS in male and female post-I/R mice that received cyclosporine A (CsA; 15 mg/kg body weight/d) delivered by osmotic minipump, starting one day before MNC, zymosan, or saline injection and continuing for 2 weeks post-injection. *p=0.0004 (Sal. versus Sham) or *p=0.0006 (MNC versus Sham) or *p=0.0018 (Zym. versus Sham), all by one-way ANOVA with Dunnett’s post-hoc test. e, FS in male and female post-I/R mice that received 2 injections of clodronate liposomes delivered intraperitoneally (i.p.) one day before MNC injection and again 5 days after MNC injection. No difference between control and liposome treatment was observed in mice that received intra-cardiac injection of saline post-I/R, so these groups were combined (denoted as Sal.). *p<0.0001 (Sal. versus Sham, or MNC+liposomes versus Sham); &p=0.0276 (MNC versus Sal.); #p=0.0042 (MNC+liposomes versus MNC), all determined by one-way ANOVA with Dunnett’s post-hoc test. f, Confocal micrographs of histological sections at the infarct border zone of hearts from male and female Ccr2-RFP × Cx3cr1-GFP knock-in mice (n=2 mice per group and timepoint with a minimum of 10 sections assessed per mouse heart) at either 3 days or 2 weeks post-I/R. g, Schematic outline of experiments performed in panels (h) and (i) in male and female Ccr2−/− or Cx3cr1−/− mice in the C57Bl/6J background that were subjected to I/R then injected with MNCs or sterile saline 1 week later. h, FS in Ccr2−/− or Cx3cr1−/− mice or wild-type (WT) C57Bl/6J mice 2w post-cell injection, or non-injured mice (Pre). *p<0.0001 (WT; Sal. versus Pre) or *p=0.0004 (Ccr2−/−; MNC versus Pre) or *p= 0.0001 (Cx3cr1−/−; Sal. versus Pre) or *p=0.0002 (Cx3cr1−/−; MNC versus Pre); #p=0.0030 (WT; MNC versus Sal), all by one-way ANOVA with Tukey’s post-hoc test. i, Quantitation of CD68+ cells as a percentage of total cells (DAPI+) imaged at the infarct border zone, 3 weeks post-I/R. *p=0.0001 versus WT/Sal.; &p<0.0001 versus Ccr2−/−/Sal., all by one-way ANOVA with Tukey’s post-hoc test. The number (n) of mice in all experimental groups is indicated below or within the respective graphs. All numerical data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively).
Mice were next treated with high dosage cyclosporine A (CsA), a broad-spectrum immunosuppressant, which abrogated the restorative effects on cardiac function seen with MNC or zymosan injection after I/R injury (Fig. 3d). Macrophages were acutely depleted by administering clodronate liposomes, which similarly abolished the protective effect of MNC injection post-I/R (Fig. 3e). Finally, injected cellular debris from freeze-thaw killed MNCs also improved cardiac function post-I/R (Extended Data Fig. 4d).
Cardiac I/R injury itself is associated with a robust, temporal recruitment of discrete myeloid cell populations17, which we also observed using Ccr2-RFP × Cx3cr1-GFP mice (Fig. 3f). Of note, characterization of Ccr2-RFP × Cx3cr1-GFP mice at baseline by flow cytometry showed that over 90% of the CX3CR1+ CCR2− cells in the heart were tissue resident CD169+ macrophages14, with a minor contribution to dendritic cells and neutrophils (Extended Data Fig. 5a-f). We also employed Ccr2−/− or Cx3cr1−/− gene-targeted mice, and while initial infarct sizes post-I/R were not different among Ccr2−/− or Cx3cr1−/− mice or strain-matched wild-type controls (not shown), Ccr2 deficiency significantly improved cardiac function after I/R (Fig. 3h), consistent with previous reports18,19. Moreover, cell therapy by MNC injection in mice lacking Ccr2 imparted no further functional benefit beyond the improvement seen in these mice post-I/R (Fig.3h). Loss of Ccr2 showed a reduction in overall CD68+ cell content specifically at the border zone of post-I/R hearts with or without cell therapy (Fig. 3i). By comparison, Cx3cr1 null mice showed left ventricular dysfunction after I/R injury that was similar to wild-type controls but these mice no longer benefitted from MNC therapy and showed a much greater total inflammatory response (Fig. 3h, i). This result is consistent with a recent study showing that ablation of CX3CR1+ macrophages increases mortality and peri-infarct fibrosis post-MI in mice15. However, Cx3cr1 null mice still show similar content of GFP+ (expressed from the Cx3cr1-GFP allele) macrophages after permanent occlusion MI injury as compared with control, as well as the same content of activated CD68+ macrophages in the infarct border zone at 3d post-MI (Extended Data Fig. 6a-c). This observation is consistent with past analysis of Cx3cr1 null mice in which monocyte extravasation during peritonitis is unaffected13 and trafficking of tissue macrophages from the yolk sac during development is unaltered20. Given these past observations, the most likely explanation for our results is that CX3CR1 deficiency does not compromise tissue-resident macrophage content, but instead impacts their function, resulting in greater tissue inflammation. Finally, the acute monocyte and neutrophil responses at 3d post-MI were also not different between Cx3cr1 null mice and heterozygous Cx3cr1-GFP controls (Extended Data Fig. 6d). However, significantly greater mortality was observed post-MI in Cx3cr1 null animals (Extended Data Fig. 6e), suggesting that CX3CR1+ cells play an important role in the later stages of infarct maturation (after day 3) and/or in modulation of the fibrotic response, as also recently proposed15
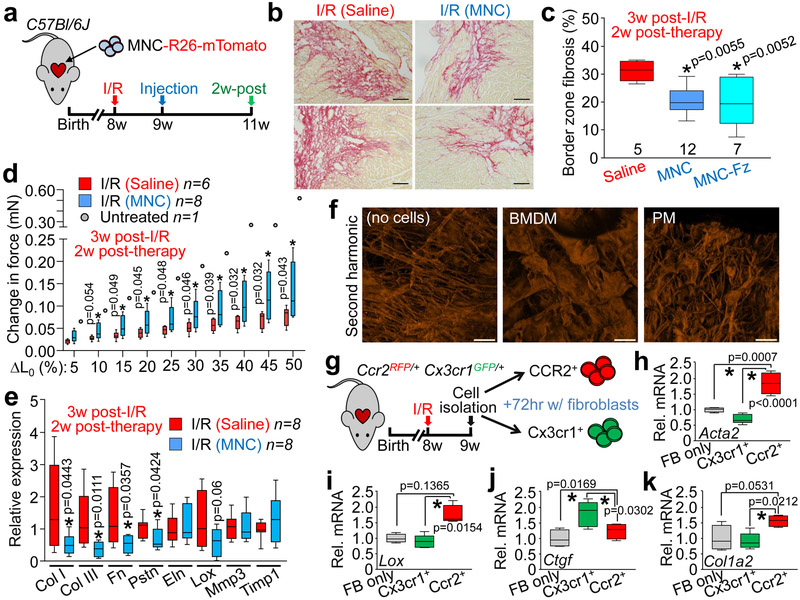
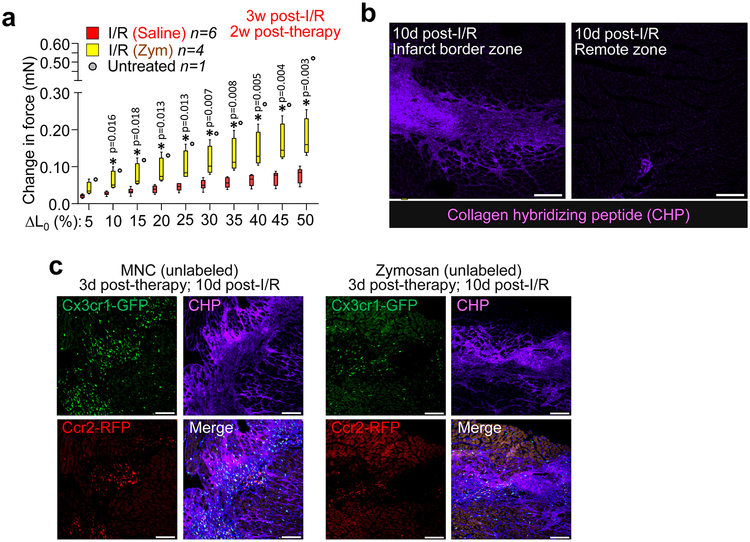
Mechanistically, ECM content in the peri-infarct border zone was significantly decreased with injection of MNCs around the I/R injury area (Fig. 4a, b). This was also observed with injection of non-viable MNCs, suggesting that it was primarily due to immunoreactivity and not active paracrine signaling (Fig. 4c). Remarkably, infarct strips from MNC-injected hearts produced a significantly greater change in passive force over increasing stretch, a profile that was more like the uninjured heart (change in initial length [L0]; Fig. 4d). This profile was also associated with a decrease in gene expression of several ECM and matricellular components in MNC versus saline treated hearts post-I/R injury (Fig. 4e). The force-lengthening assay was also repeated on infarct strips from post-I/R hearts injected with zymosan, which showed an even larger improvement in passive force dynamics compared with either saline or MNC treatment (Extended Data Fig. 7a).
Figure 4 ∣. Cell therapy benefits the mechanical properties of the infarct via remodeling of the extracellular matrix.
a, Schematic outline of experiments performed in panels (b-e). b, Representative picrosirius red-stained cardiac histological images from the infarct border zone of 3 weeks post-I/R mice subjected to MNC or saline injection. Fibrosis is shown in red. Scale bars = 100 μm. c, Quantitation of fibrotic area at the infarct border zone in MNC, freeze thaw-killed MNC, or saline-treated hearts, 3 weeks post-I/R. *p<0.05 by one-way ANOVA with Tukey’s post-hoc test with exact p-values shown in figure. Images in (b) and quantitation in (c) are from n=5 saline-treated, n=12 MNC-treated, or n=7 freeze-killed MNC-treated mice, with a minimum of 20 histological sections assessed from each individual mouse heart. d, Change in passive force generation over increasing stretch-lengthening (percent of L0) in isolated infarct strips from MNC or saline-treated hearts, 3 weeks post-I/R. *p<0.05 versus I/R/Sal. by Student’s 2-tailed t-test (exact p-values are shown in figure). Data from one uninjured control heart (no I/R or cell therapy) is also shown for comparison. e, Gene expression levels by RT-PCR for selected extracellular matrix (ECM) and matrix-associated genes in isolated infarct regions from MNC or saline-treated hearts, 3 weeks post-I/R. *p<0.05 versus I/R/Sal. by Student’s 2-tailed t-test (exact p-values shown in figure). f, Representative confocal micrographs of pre-fabricated collagen patches that were seeded and cultured for 5 days with either bone marrow-derived macrophages (BMDM) or peritoneal macrophages (PM) isolated from wild-type male and female mice, versus cell-free control patches cultured in media. Fluorescence signal is from native type I and II collagen using second harmonic generation microscopy. Scale bars = 100 μm. g, Schematic of experiments using activated cardiac macrophages isolated from post-I/R Ccr2-RFP × Cx3cr1-GFP knock-in mice that were then cultured with isolated cardiac fibroblasts (FB) for 72 hours. h-k, Fibroblast mRNA was used for RT-PCR to assess expression of smooth muscle α-actin (Acta2 / αSMA, h), lysyl oxidase (Lox, i), connective tissue growth factor (Ctgf, j) or collagen 1 alpha 2 (Col1a2, k). For panel 4i: *p<0.05 by Kruskal-Wallis with Dunn’s multiple comparisons test. For all other panels: *p<0.05 by one-way ANOVA with Tukey’s post-hoc test (exact p-values shown in figure). All numerical data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively). Micrographs in f are representative of five different collagen patches seeded with cells pooled from n=4 mice (2 male and 2 female). Data in g-k are from 4-5 replicates generated over fibroblasts isolated from n=10 wild-type mice (6 male and 4 female) and macrophages isolated from n=6 Ccr2-RFP × Cx3cr1-GFP heterozygous knock-in mice (3 male and 3 female).
Bone marrow-derived macrophages (BMDM) or peritoneal macrophages (PM) were next isolated from naïve mice and cultured on pre-fabricated collagen patches followed by second harmonic generation microscopy to examine collagen organization (Fig. 4f). Interestingly, BMDMs and PMs each generated different patterns of collagen reorganization. To extend these observations a collagen hybridizing peptide (CHP)21 reagent was used on injured mouse hearts, which detects immature or denatured collagen and areas of active remodeling (Extended Data Fig. 7b). Hearts from MNC or zymosan treated mice showed CHP reactivity that was coincident with regional localization of CCR2+ versus CX3CR1+ macrophages within the microenvironment of the infarct border zone (Extended Data Fig. 7c). Finally, CCR2+ or CX3CR1+ macrophages were isolated from hearts at 7 days post-I/R (Fig. 4g) and cultured with freshly isolated cardiac fibroblasts for 72 hrs. Gene expression analysis revealed that CCR2+ macrophages increased fibroblast expression of smooth muscle α-actin (Acta2 / αSMA), lysyl oxidase (Lox), and collagen 1 alpha 2 (Col1a2) (Fig. 4h, i, k). In contrast, CX3CR1+ macrophages slightly reduced expression of these genes but increased fibroblast expression of connective tissue growth factor (Ctgf) (Fig. 4j)22. Taken together, these results demonstrate that specific macrophages subtypes mobilized by cell therapy differentially affect the passive mechanical properties of the cardiac infarct area by influencing the activity of cardiac fibroblasts.
As suggested nearly 1 decade ago23, we observed that the acute inflammatory response is a primary beneficial effect underlying cell therapy in the post-MI injured heart. A unique mechanism was identified whereby temporarily stimulating the intrinsic wound healing cascade and select subtypes of macrophages positively impacted the ECM around and within the infarcted region of the heart, such that functional performance was significantly improved. These results are consistent with recent reports demonstrating key functional distinctions between CCR2+ and CX3CR1+ CCR2− macrophages in cardiac wound healing14,15. In conclusion, the data presented here suggest re-evaluation of current and planned cell therapy-based clinical trials to maximize the effects of the most prevalent underlying biologic mechanism of action.
Methods:
Mice
This study was performed entirely in mice, using transgenic mouse models either commercially available or generated as described below. No human subjects or human material was used. All experiments involving mice were approved by the Institutional Animal Care and Use Committee (IACUC) at Cincinnati Children’s Hospital under protocol IACUC2018-0047. All procedures were performed in compliance with institutional and governmental regulations under PHS Animal Welfare Assurance number D16-00068 (A3108-01). The generation and characterization of mice carrying the tamoxifen-inducible MerCreMer recombinase cDNA within the Kit allele (KitMerCreMer/+) and reporter mice carrying the Cre-regulated loxP-stop cassette and eGFP within the Rosa26 gene locus, Rosa26-eGFP (R-GFP) were previously described16. All other mouse strains used were purchased from Jackson Labs, as follows: C57Bl/6J; (#000664), constitutive mTomato expressing mice targeted in the Rosa26 locus for bone marrow mononuclear cell (MNC) or cardiac progenitor cell (CPC) isolation; (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J, #007676), Ccr2 gene-deleted mice; (B6.129S4-Ccr2tm1Ifc/J, # 004999), Cx3cr1 GFP knock-in mice (Cx3cr1 homozygotes are nulls); B6.129P-Cx3cr1tm1Litt/J, #005582), Ccr2 RFP knock-in mice; (B6.129(Cg)-Ccr2tm2.1Ifc/J #017586). Both male and female sexes were used in all experiments, at age ranges indicated in the figures and main text for each experiment. Mice were housed single-sex at a maximum of 4 animals per cage in a specific pathogen free (SPF), temperature-controlled vivarium under a 12 hr light/dark cycle with ad libitum access to food and water. Randomization was not performed because mice are genetically identical, housed together and of the same age ranges and sex ratios.
Preparation of Cell or Inflammatory Therapies
To generate MNCs for injection, whole bone marrow was first isolated by flushing dissected femurs and tibiae of 10-12w-old Rosa26-mTomato expressing mice or C57Bl/6J; mice with 10 mL of sterile Hanks Balanced Salt Solution (HBSS, Fisher Scientific #SH3058801) + 2% bovine growth serum (BGS, Fisher Scientific, #SH3054103) + 2 mM EDTA as previously described24. This suspension was then filtered through a 40 μM mesh strainer (Fisher Scientific #22-363-547), centrifuged at 400 g for 10 min at 4 °C, resuspended in 3 mL of sterile saline, and layered on top of 4 mL of Ficoll Paque Plus (GE Healthcare #17-1440-02). Cells were then centrifuged at 2,500 g for 30 min at 4 °C in a swinging bucket rotor centrifuge without brakes. MNCs were isolated by removal of the resulting thin mononuclear cell layer (second layer from the top). Total MNCs were counted with a hemocytometer, washed twice with sterile saline, and resuspended in sterile saline at a final concentration of either 2.5×106 cells/mL (for injection into uninjured hearts, final dose 50,000 cells) or 7.5×106 cells/mL (for injection into post-I/R hearts, final dose 150,000 cells). A higher dose of cells (MNCs or CPCs, see below) was used in post-I/R hearts to account for greater cell loss in the setting of damaged myocardium and to align with past studies4,5. The full intra-cardiac injection procedure is described in the following section (Mouse Procedures). Cell viability was tested by incubating an aliquot of the MNC suspension with eFluor 450 Fixable Viability Dye (eBioscience #65-0863-18) and found to be over 90% viable at the time of injection. All MNC preparations for injection were combined from an equal number of male and female mice. For experiments that used non-viable MNCs (freeze-thawed; Fz), this final MNC suspension was split into two equal aliquots. One aliquot was placed immediately at −80 °C for 10 min, followed immediately by incubation at 55 °C for 10 min, and this was repeated for a total of 3 freeze-thaw cycles.
To generate CPCs for injection, hearts from 10-12w-old Rosa26-mTomato expressing male and female mice were rapidly excised and briefly rinsed in cold 1X PBS. Single-cell suspensions from these hearts were prepared according to previously published protocols25,26 with minor modifications, as follows. The atria were removed, and the ventricles were minced on ice using surgical scissors into approximately 2 mm pieces (8-10 pieces per mouse heart). Each dissociated ventricle was transferred into 2 mL of digestion buffer in 1 well of a 12-well tissue culture plate. Digestion buffer consisted of 2 mg/mL collagenase type IV (Worthington, #LS004188), 1.2 U/mL dispase II (Roche, #10165859001) and 0.9 mM CaCl2, in 1x HBSS. Tissues were incubated at 37 °C for 20 min with gentle rotation followed by manual trituration 12-15 times with a 10 mL serological pipette, such that all of the tissue pieces were able to pass through the pipette. The tissues were settled by sedimentation and the supernatant was passed through a 40 μM mesh strainer and stored on ice. Two milliliters of fresh digestion buffer was added, followed by 2 additional rounds of incubation, trituration and replacement of supernatant with fresh digestion buffer, except trituration was performed with a 5 mL serological pipette for round 2 and a 1 mL p1000 pipette tip (USA Scientific, #1112-1720) for round 3. The pooled supernatant from the 3 rounds of digestion was washed with sterile PBS and centrifuged at 200 g for 20 min at 4 °C in a swinging bucket rotor centrifuge without brakes. The pellet was resuspended in flow cytometry sorting buffer, consisting of 1x HBSS supplemented with 2% bovine growth serum (BGS) and 2 mM EDTA, and incubated with anti-c-Kit microbeads (Miltenyi Biotec #130-091-224) for 20 min at 4°C with gentle rotation. The suspensions were washed twice with sorting buffer and c-Kit+ cells were enriched via positive selection over Miltenyi Biotec LS columns (#130-042-401) using benchtop magnetic cell separation (MACS) according to manufacturer’s instructions. Cells isolated via positive MACS selection for c-Kit were cultured in DMEM/F12/Glutamax media (Gibco #10565-018) supplemented with 10% fetal bovine serum (Sigma, #F2442), 0.2% insulin/transferrin/selenium (ITS, Lonza, #17-838Z), 20 μg/mL basic recombinant human fibroblast growth factor (Promega, #G5071), 103 U/mL leukemia inhibitor factor (Millipore, #ESG1106), 20 μg/mL epidermal growth factor (Sigma, #E9644) and 1% penicillin-streptomycin (Fisher Scientific, #30-002-CI).
c-Kit+ isolated cells from the heart are referred to as cardiac progenitor cells (CPCs) as previously named, although recent data would suggest that these cells only transdifferentiate into endothelial cells and not cardiac myocytes in vivo3,8,16. c-Kit+ isolated cells were expanded in culture for 12-15 passages before being used for injection, at which point cells were washed 3 times with sterile saline, trypsinized, counted, and resuspended in sterile saline at a final concentration of either 2.5×106 cells/mL (for injection into uninjured hearts, final dose 50,000 cells) or 7.5×106 cells/mL (for injection into post-I/R hearts, final dose 150,000 cells). As with MNCs, suspensions consisted of pooled CPCs from an equal number of male and female mice.
Alexa Fluor-594 conjugated zymosan, or unconjugated zymosan, were purchased from Thermo Fisher (zymosan A [S. cerevisiae] BioParticles, Alexa Fluor 594 conjugate, #Z-23374, zymosan A [S. cerevisiae] BioParticles, unlabeled, # Z2849). A suspension of either 1 mg/mL (for injection into uninjured hearts, final dose 10 μg) or 2 mg/mL (for injection into post-I/R hearts, final dose 20 μg) was prepared for injection in sterile saline according to manufacturer’s instructions. Zymosan dosing was extrapolated from a recent study using injection into neonatal mouse hearts27. A higher dose of zymosan was used in post-I/R hearts to allow comparison to MNC and CPC-treated animals.
Mouse Procedures
To deliver cell or inflammatory therapies by intra-cardiac injection, mice were anesthetized using isoflurane inhalation (to effect), intubated, and a left lateral thoracotomy was performed. A 25 μL gas-tight syringe (Hamilton, #7654-01) fitted with a 33-gauge needle (Hamilton, #7803-05) was used for injections. For experiments in mice without injury, 20 μL of MNCs at a concentration of 2.5×106 cells/mL (50,000 cells total) was injected over 3 regions of the LV (6.7 μL per injection). For experiments with zymosan, 10 μL of a 1 mg/mL suspension was injected over 3 regions of the LV (3.3 μL per injection, 10 μg zymosan total). Injections were equidistant along the anterior wall of the left ventricle (LV), with the needle entering just parallel to the long axis of the ventricle to avoid entering the LV chamber. For experiments with injections occurring after cardiac injury, 2 injections were performed, one on either side of the infarct zone, as follows. Twenty microliters total of MNCs or CPCs at a concentration of 7.5×106 cells/mL (150,000 cells total) was injected (10 μL per injection). For experiments with zymosan, 10 μL total of a 2 mg/mL suspension was injected (5 μL per injection, 20 μg zymosan total). A greater number of cells and higher amount of zymosan were injected into the injured heart compared with the uninjured (naïve) heart because of less retention and greater turnover of the cells or zymosan due to the infarction injury. The sham procedure for intra-cardiac injection consisted of anesthesia, intubation, and thoracotomy as described above. The chest was then immediately closed, and mice recovered.
To induce cardiac injury, we used a modified surgical model of ischemia with reperfusion (I/R) via temporary left coronary artery ligation as previously described28, whereby 120 min of ischemia was used before inducing reperfusion, which gave more complete killing of the ischemic zone and greater reproducibility. After each surgical procedure (I/R or intra-cardiac injection), animals were given post-operative analgesics (buprenorphine, 0.1 mg/kg body weight) and allowed to recover until the experimental time points indicated where mice were then further analyzed, or tissue harvested. For experiments with permanent occlusion MI, the same procedure was performed except the ligature was not released. Randomization was not performed because mice are genetically identical, housed together and of the same age ranges and sex ratios. See final section for discussion of blinding and sample elimination considerations.
Infarct size and area-at-risk post-I/R was determined using triphenyl tetrazolium chloride (TTC) and Evans blue staining as previously described28. In experiments using immunosuppression via cyclosporine A (CsA), mice were anesthetized with 2% isoflurane inhalation to effect, and osmotic minipumps (Alzet, #1002) were implanted subcutaneously on the left lateral side of the mouse. Minipumps were loaded with CsA (Neoral, Novartis, NDC 0078-0274-22) dissolved in Cremophor EL (Sigma, #C5135) such that 15 mg CsA per kg body weight was delivered per day29. In experiments using acute macrophage ablation, mice were administered two doses of 0.2 mL each of clodronate liposomes (Clophosome, #F70101C-N) via intraperitoneal injection, one day before cell therapy and again on day 5 after cell therapy. Liposomes were kept on ice until administration and were rapidly mixed by inversion to deliver a uniform suspension. Control animals were injected with 0.2 mL of sterile saline. In experiments using KitMerCreMer/+ × R26-eGFP genetic lineage tracing mice, tamoxifen was administered as previously described42 via ad libitum feeding with pre-manufactured tamoxifen chow (tamoxifen citrate 40 mg/kg body weight per day, Envigo, #TD.1308603) for the time indicated in each individual experiment. For echocardiographic analysis of cardiac structure and function, mice were anesthetized with 2% isoflurane inhalation to effect and analyzed using a Vevo2100 instrument (VisualSonics) with an 18–38 MHz transducer as previously described30. Randomization was not performed because mice are genetically identical and of the same age ranges. See final section for discussion of blinding and sample elimination considerations.
Histology and Immunohistochemistry
Primary antibodies and dilutions used for immunohistochemistry are listed in Supplementary Table 1. For histological analysis, animals were anesthetized by isoflurane inhalation and sacrificed by cervical dislocation. The chest was opened, and the heart was flushed with cold cardioplegia solution (1 M KCl in 1x PBS) via cardiac apical insertion of a 25-gauge needle. The left atrium was cut to allow drainage of blood from the heart, and animals were briefly perfused with cold fixative (4% paraformaldehyde in sodium phosphate buffer, pH 7.4) through the apex of the heart. Tissues were excised, flushed with fixative, and incubated in cold fixative for 3.5 h at 4 °C with gentle rotation. Tissues were washed 3 times in cold 1x PBS and then cryopreserved by incubation in 30% sucrose in 1x PBS overnight at 4 °C with gentle rotation. Tissues were then embedded in TissueTek optimal cutting temperature (O.C.T) medium (VWR, # 25608-930) and flash-frozen at −80°C. Five-micrometer cryosections were cut using a Leica CM1860 cryostat.
Picrosirius red staining was performed with a kit from Abcam (ab150681) as per manufacturer’s instructions. High magnification images of picrosirius red-stained hearts were captured at 200X magnification using an Olympus BX51 microscope equipped with a single chip color CCD camera (DP70) and DP controller software (Olympus America Inc., v3.1.1.). Border zone fibrosis was quantified as the percentage of picrosirius red-stained area over total tissue area analyzed, as previously described33.
All detection of genetic reporter-driven mTomato, RFP, GFP, or eGFP expression was performed using endogenous fluorescence without antibody labeling. Immunohistochemistry was performed on cardiac cryosections as previously described16,26 with the following modifications (see Supplementary Table 1 for primary antibodies and dilutions. Alexa Fluor fluorochrome-conjugated secondary antibodies were used at a 1:200 dilution for visualization; Life Technologies). For immunohistochemistry using antibodies against PCM-1, antigen retrieval was first performed by incubation with 1% SDS for 5 min at room temperature with gentle rotation. PCM-1 antibody was used to specifically identify cardiomyocytes in heart tissue sections. Histological cardiac sections were washed thoroughly in 1x PBS before proceeding. For immunohistochemistry using the collagen hybridizing peptide (CHP; 3Helix, #BIO300), a stock solution of 15 μm biotin-conjugated CHP was prepared according to manufacturer’s instructions. The solution was heated to 80 °C for 5 min to denature the peptide, as previously described21, followed by rapid cooling on ice and incubation on tissue sections overnight at 4 °C. Sections were then washed and processed for secondary antibody staining with fluorophore-conjugated streptavidin antibody as described above. Confocal microscopy and image acquisition were performed using a Nikon Eclipse Ti inverted microscope equipped with a Nikon A1R confocal running NIS Elements AR 4.50.
Passive Force Measurements
Tissue strips from the infarct region of the left ventricle (or the left ventricular free wall for uninjured hearts) were dissected using a Zeiss Discovery V8 dissection microscope. Tissues were cut into 3 mm (length) × 2 mm (width) strips, and 3-4 strips were cut from each infarct region of the left ventricle. These strips contained scar and a small region of border zone on each end. Tissue strips were maintained in M199 media (Corning Cellgro, 10-060-CV) with no supplementation throughout the procedure of force measurements. Tissue strips were attached to aluminum t-clips (Kem-Mil, #1870) and mounted onto a permeabilized muscle fiber test apparatus (Aurora Scientific, Model: 802D-160-322) initially set to zero tension. Cardiac tissue length was then increased 5% over 50 ms, held for 450 ms, then stretched again from 5% to 50% in intervals of 5% with no period of relaxation, and force was monitored using DMC v600A software (Aurora Scientific). Change in force was calculated as the difference between max force generated after the 50 ms pull and the minimum force achieved after each time period. Minimum force was calculated when the rate of force decay was zero by solving for the derivative of the best fit trend line, which was a 2nd degree polynomial equation.
RT-PCR from Isolated Infarct Tissues
Tissue strips isolated from the infarct region as described above were homogenized with a Precellys 24 homogenizer (Bertin Instruments #03119.200.RD000) and RNA was isolated by using the RNeasy fibrous tissue kit according to the manufacturer’s instructions (Qiagen #74704). One microgram of total RNA was reverse transcribed using random oligo-dT primers and a Verso cDNA synthesis kit (Thermo Fisher Scientific # AB1453A) according to manufacturer's instructions. Real-time PCR was performed using Sso Advanced SYBR Green (BioRad # 1725274) according to the following program: one cycle of 95°C for 10 min, one cycle of 95°C for 15 s, 40 cycles of 95°C for 15 s, 57°C for 10 s, and 62°C for 30s, and one cycle of 62°C for 30s. Gapdh expression was used for normalization. Primer sequences used are included in Supplementary Table 2.
Flow Cytometry and Cell Sorting
For analysis of surface markers on MNCs or CPCs, cells were resuspended in 1x HBSS supplemented with 2% BGS and 2 mM EDTA and incubated with fluorophore or biotin-conjugated primary antibodies (see Supplementary Table 1) for 20 min at 4 °C with gentle rotation. Cells were then washed twice with 1x HBSS. For detection of biotinylated antibodies, cells were incubated with streptavidin-conjugated BV421 (BD Horizon #563259) for 15 min at 4 °C with gentle rotation and then washed twice with 1x HBSS. Samples were analyzed using a BD FACSCanto running BD FACSDiVa V.8.0 software (BD Biosciences) and using the following laser configuration: Blue (488 nm), Yellow-Green (561 nm) and Red (635 nm). Analysis and quantitation were performed using FlowJo V.10 (Tree Star, Inc.).
For flow cytometry analysis of whole-heart cardiac immune cell content, single-cell suspensions were first prepared using enzymatic dissociation and trituration as described above. Animals were anesthetized by 2% isoflurane inhalation to effect and sacrificed by cervical dislocation. Hearts were rapidly excised and briefly rinsed in cold cardioplegic solution (1M KCl in 1x PBS) prior to enzymatic dissociation. The pellet resulting from dissociation was resuspended in 1 mL of red blood cell lysis buffer (150 mM NH4Cl, 10 mM KHCO3, 0.1 mM Na2EDTA) and incubated at room temperature for 5 min. Samples were then centrifuged at 400 g for 10 min at 4 °C and resuspended in 1x HBSS supplemented with 2% BGS and 2 mM EDTA. Cells were incubated with fluorophore-conjugated primary antibodies for 20 min at 4 °C with gentle rotation, washed twice with 1x HBSS and analyzed using a BD LSRFortessa running BD FACSDiVa V.8.0 software (BD Biosciences) and using the following laser configuration: UV (355 nm), Violet (405 nm), Blue (488 nm), Yellow-Green (561 nm) and Red (635 nm) to detect fluorophore-conjugated antibodies and/or endogenous RFP and GFP signal from Ccr2-RFP × Cx3cr1-GFP mice. Analysis and quantitation were performed using FlowJo V.10 (Tree Star, Inc.).
For isolation of cardiac CCR2+ or CX3CR1+ macrophages by fluorescence-activated cell sorting (FACS), hearts from Ccr2-RFP × Cx3cr1-GFP mice at 7 days post-I/R injury were isolated and dissociated to single-cell suspensions as described above for isolation of CPCs, except that the digestion solution was made in DMEM + 2% BGS and 1% penicillin-streptomycin instead of HBSS. Isolated cells were sorted by FACS using a Sony SH800S benchtop cell sorter in a BSL-2 biosafety cabinet. Endogenous RFP and GFP fluorescence were detected using 4 collinear lasers and CCR2+ (RFP+ GFP−) or CX3CR1+ (GFP+ RFP+ or−) cells were sorted into 1.5 mL Eppendorf tubes containing DMEM + 10% BGS and 1% penicillin-streptomycin. Cells were then cultured on isolated cardiac fibroblasts as described below.
Cardiac Fibroblast Isolation
Hearts were excised from ten 8-week-old male and female C57Bl/6J mice and the ventricles and septum were isolated, rinsed in ice-cold PBS and minced into small pieces using sterile micro-scissors. Tissue fragments were digested in DMEM + 2% BGS and 1% penicillin-streptomycin containing type 2 collagenase (100 units/mL; LS004177, Worthington, USA) for 20 minutes at 37 °C under gentle agitation. The digested tissue was triturated repeatedly to promote tissue dissociation. Dense fragments settled for 2 minutes and the supernatant, containing the cardiac fibroblasts, was collected and spun at 1,000 g for 5 minutes. The cell pellet was resuspended in 10 mL of DMEM + 10% BGS and 1% penicillin-streptomycin and kept on ice. This process was repeated 3 times until all the tissue was adequately digested. To remove cardiomyocytes and cell debris, cell suspensions were spun at 300 g, followed by centrifugation of the supernatant at 1,000 g. The final cell pellet containing the cardiac fibroblasts was resuspended in DMEM + 10% BGS and 1% penicillin-streptomycin and pre-incubated on 0.1% gelatin-coated plates for 2 hours to allow fibroblast adherence before replenishment of the cell culture medium.
Cardiac Fibroblast and Macrophage Co-Culture
Isolated cardiac fibroblasts were split into 24-well 0.1% gelatin-coated plates at a seeding density of 15,000 cells/well and allowed to adhere overnight. Macrophage subtypes (CCR2+ and CX3CR1+ cells) isolated as described above were then seeded onto these cardiac fibroblasts at a density of 10,000 macrophages per 15,000 fibroblasts. Control fibroblasts received an equivalent amount of culture media containing no macrophages. Adherence was verified the following day by fluorescence microscopy for RFP or GFP. Cells were isolated after 72 hours later for mRNA quantification.
mRNA Isolation and qRT-PCR from Cultured Cardiac Fibroblasts
Total RNA was purified from cultured cells with TRIzol reagent (Fisher Scientific # 15596018) according to manufacturer’s instructions. Two hundred nanograms of RNA was reverse transcribed to cDNA using the Verso cDNA synthesis kit (Thermo Fisher Scientific # 277.97). Quantitative PCR was performed using SsoAdvanced Universal SYBR Green Supermix (BioRad # 1725274) and assayed in duplicate, according to manufacturer’s instructions in a CFX96 PCR system. Primer sequences are included in Supplementary Table 2. All data were normalized to Gapdh (verified to not deviate between samples).
Macrophage Culture on Fibrillar Collagen Patches
Pre-sterilized resorbable collagen membranes (Ace Surgical Supply #509-3040) were cut into circles of a uniform thickness and a diameter of 6 mm and then placed into 96-well plates. Bone marrow or peritoneal macrophages were isolated as previously described31,32 and cultured in DMEM + 10% BGS and 1% penicillin-streptomycin were then seeded onto the collagen patches at a density of 10,000 cells/patch. Control patches were incubated in culture media without macrophages. Five biological replicates were performed per group. After 5 days in culture, fibrillar collagen assembly was analyzed by second harmonic generation microscopy using a Nikon A1R multiphoton upright confocal microscope equipped with a tunable Coherent Chameleon II TiSapphire IR laser set to 840 nm. Three images were randomly taken per patch and assessed by a blinded investigator to select the representative images for each group.
Statistical Information and Experimental Rigor (blinding)
All statistical tests used and graphical depictions of data (means and error bars, or box and whisker plots) are defined within the figure legends for the respective data panels. Exact n values for all experiments with statistical analysis are included in the figure legends or within the figure itself. For comparisons between 2 groups, unpaired or Student’s 2-tailed t-tests were performed as noted within figure legends. For comparisons between more than two groups, a one-way analysis of variance (ANOVA) with Tukey’s or Dunnett’s post-hoc test was performed as noted within figure legends. p<0.05 was considered as statistically significant. Data in Main Figure 4i used a Kruskal-Wallis test with Dunn’s multiple comparisons test as these data did not follow a normal distribution. All other data were found to follow a normal distribution as determined by the Shapiro-Wilk normality test or Kolmogorov-Smirnov test (α=0.05). For experiments involving I/R surgery, the number of animals that received surgery was determined based on prior experimentation in the lab, which demonstrated a peri-operative surgical mortality rate of 20%. Only animals that did not survive a given surgical procedure or were found at the time of I/R surgery to have incomplete reperfusion (failure of slipknot suture release) were excluded from analysis, otherwise no exclusions occurred. Randomization of mice within a group to receive a given surgical procedure (I/R vs sham) or treatment (saline vs cells vs zymosan) was not needed because the mice were genetically identical and were littermates, although equal sex ratios and age ranges were maintained. Echocardiographic analysis, quantitation of eGFP+ endothelial cells, quantitation of cardiomyocytes in cell cycle, measures of fibrosis, measures of tissue passive force, in vivo and in vitro gene expression analysis, and analysis of collagen organization in culture were conducted by investigators blinded to experimental treatment or procedure. Quantitation of macrophage content by immunohistochemistry was performed using automated fluorescence threshold analysis in NIS Elements 4.50.
Data Availability
All raw data generated or analyzed in this study will be made available from the corresponding author upon reasonable request. More specifically, original source data used to generate graphs in each of the figures and extended data figures are available as Microsoft Excel data sheet files.
Extended Data
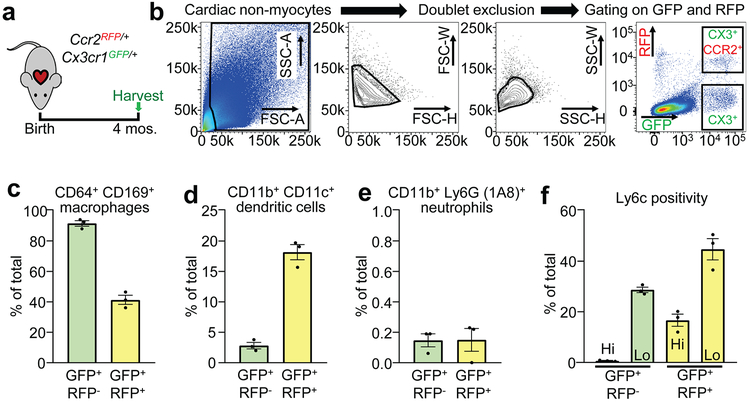
Extended Data Figure 1 ∣. Characterization of cells used in the injection studies and initial neutrophil response to injections.
a, Flow cytometry analysis of MNCs isolated from Rosa26-mTomato mice for intra-cardiac injection. Singlet cells were selected by forward and side scatter properties followed by mTomato-positivity. b, Flow cytometry plots for CPCs immunolabeled with antibodies against mesenchymal, endothelial, or hematopoietic lineages as indicated in the plots. An unstained negative control (Unst.) plot is also shown. Gating was determined versus unstained negative controls. Similar results in a-b were obtained from at least three independent cell preparations. c, Quantitation via flow cytometry of total neutrophil levels in dissociated hearts from MNC, zymosan, or saline-injected male and female C57Bl/6J mice, 3 days post-injection. As a comparison, data from n=5 C57Bl/6J mice isolated 1 day after I/R injury are also shown when neutrophil levels are high. Data are from n=4 (MNC, Zym.) or n=2 (Sal.) mice. Numerical data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively).
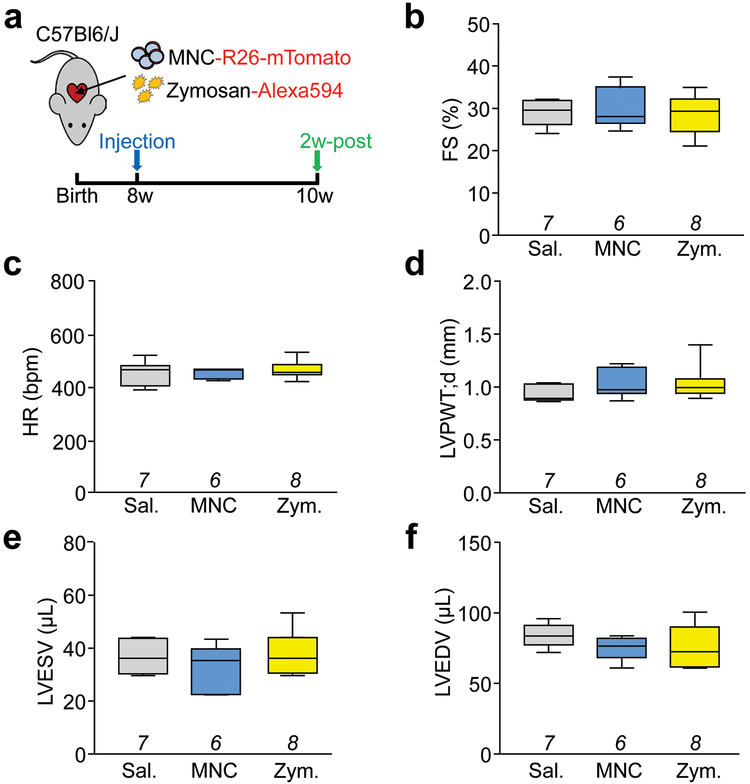
Extended Data Figure 2 ∣. Basal cardiac structure and function following cell or zymosan injection in uninjured mice.
a, Schematic outline of all experiments performed in this figure in which normal 8-week-old male and female C57Bl/6J mice received intra-cardiac injection of mTomato-labeled MNCs, Alexa594-labeled zymosan (Zym.) or sterile saline (Sal.) and were assessed by echocardiography after 2 weeks. b, Echocardiography measured fractional shortening percentage (FS%); c, heart rate (HR) as beats per minute (bpm) under isoflurane anesthesia; d, left ventricular posterior wall thickness in diastole (LVPWT;d) in millimeters; e, left ventricular end-systolic volume (LVESV) in microliters; f, and left ventricular end-diastolic volume (LVEDV) in microliters. All values in b-f were unchanged with injection of MNCs or zymosan versus saline. All numerical data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively). The number (n) of mice for each group in b-f is indicated below each respective box and whisker plot.
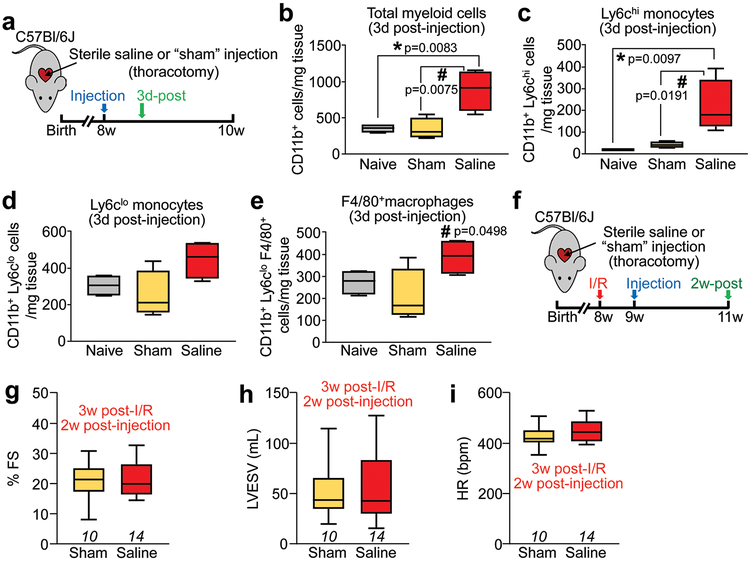
Extended Data Figure 3 ∣. Characterization of inflammatory and functional effects of the intra-cardiac injection protocol.
a, Schematic outline of experiments performed in panels (b-e). Eight-week-old normal (uninjured) male and female C57Bl/6J mice received intra-cardiac injection of sterile saline or a sham procedure in which the heart was exposed by thoracotomy but no intra-cardiac injections were done. Naïve mice without surgery served as an additional control. c-e, Quantitation via flow cytometry of immune cells from enzymatically dissociated hearts from the above groups of mice. Total CD11b+ myeloid cells (b), CD11b+ Ly6chi (c) or CD11b+ Ly6clo (d) monocytes, and CD11b+ F4/80+ macrophages (e) from n=4 mice per group are shown, normalized to starting weight of dissociated tissue. *p<0.05 versus naïve and #p<0.05 versus sham by one-way ANOVA with Tukey’s post-hoc test (exact p-values shown in figure). f, Schematic outline of experiments performed in panels g-i in which 8-week-old male and female C57Bl/6J mice received I/R injury followed by either intra-cardiac injection of saline or thoracotomy (sham) after 1w, and then were assessed by echocardiography 2w later. g, Fractional shortening (FS), h, left ventricular end-systolic volume (LVESV) and i, heart rate as beats per minute (bpm) under isoflurane anesthesia, as measured by echocardiography in the groups indicated. The number (n) of mice for each group is indicated in each graph in g-i. All numerical data in this figure are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively).
Extended Data Figure 4 ∣. Additional echocardiographic parameters and effect of freeze-thaw killed MNCs post-I/R injury.
a, Echocardiography to measure left ventricular end-systolic volume (LVESV) in microliters; b, left ventricular end-diastolic volume (LVEDV) in microliters; c, or heart rate under isoflurane anesthesia in mice that received intra-cardiac injection of MNCs CPCs, zymosan, or sterile saline, 3 week post-I/R. These data were collected in the same group of mice shown in Figures 3b and 3d. Data in (a) were significantly different as assessed by one-way ANOVA with Dunnett’s post-hoc test (exact p-values shown in figure). d, Schematic in which mice received intra-cardiac injection of mTomato-labeled MNCs or freeze-thaw (Fz)-killed MNCs 1 week after I/R and then 2 week later cardiac ventricular fractional shortening (FS%) percentage was measured by echocardiography. *p<0.05 vs Sham/Sal. or &p<0.05 vs I/R/Sal. by one-way ANOVA with Dunnett’s post-hoc test (exact p-values shown in figure). The sham, I/R+Sal. and I/R+MNC groups shown here in (d) are the same data as also shown in Main Figure 3 as these studies were performed in parallel. All numerical data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively). The number (n) of mice for each group is indicated below the respective plot.
Extended Data Figure 5 ∣. Characterization of cell-types labeled from Ccr2-RFP × Cx3cr1-GFP knock-in mice.
a, Schematic of experiments in which hearts from male and female double heterozygous Ccr2-RFP × Cx3cr1-GFP knock-in mice (n=3) were isolated and analyzed by flow cytometry at 4 months of age. b, Representative flow cytometry plots with the gating strategy shown in which singlet cells were first selected by forward and side scatter properties followed by gating on endogenous GFP and RFP fluorescence for either CX3CR1+ CCR2− (GFP+ RFP−) or CX3CR1+ CCR2+ (GFP+ RFP+) cells. c-f, Cells within the GFP+ RFP− or GFP+ RFP+ gates as shown in b were then assessed for surface marker expression via antibodies. c, CD64+ CD169+ macrophages, d, CD11b+ CD11c+ dendritic cells, or e, CD11b+ Ly6G+ neutrophils are shown as a percentage of all GFP+ RFP− or GFP+ RFP+ cells. f, Ly6c positivity in the total GFP+ RFP− or GFP+ RFP+ populations is also shown. All data are represented as the mean +/−SEM from n=3 animals.
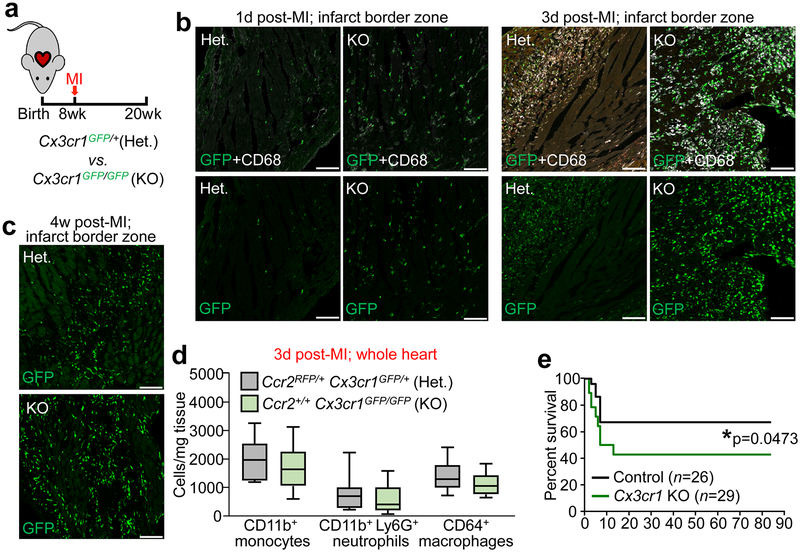
Extended Data Figure 6 ∣. Response of Cx3cr1 global gene-deleted mice to myocardial infarction injury.
a, Schematic of experiments in which 8w-old male and female Cx3cr1-GFP heterozygous (Het.) or Cx3cr1-GFP/GFP homozygous (KO) mice received myocardial infarction (MI) via permanent occlusion of the left coronary artery and were then followed out for 12 weeks. b-c, Representative confocal immunohistochemistry images from hearts of these mice at either 1 day, 3 day, or 4 weeks post-MI, showing endogenous GFP fluorescence from the Cx3cr1 knock-in allele (b,c). Immunohistochemistry for activated CD68 macrophages (white) is also shown in b. Micrographs are representative of n=3 mice per time point. Scale bars = 100 μm. d, Quantitation via flow cytometry of total monocyte, neutrophil, and macrophage levels in dissociated whole hearts from Cx3cr1 Het. or KO mice at 3 days post-MI. Data are from n=11 (Het.) or n=12 (KO) mice and numerical data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively). e, Survival curve for Cx3cr1-GFP/GFP (KO) mice versus controls over 12 weeks following MI injury. Control animals in this experiment included both Cx3cr1-GFP heterozygous and C57Bl/6J animals. The numbers of mice for each group are indicated in the figure. *p=0.0473 by two-sided Gehan-Breslow-Wilcoxon test.
Extended Data Figure 7 ∣. Mechanical and structural improvements in cell therapy or zymosan-treated hearts post-I/R.
a, Change in passive force generation over increasing stretch-lengthening (percent of L0) in isolated infarct strips from zymosan or saline-injected hearts, analyzed 3 weeks after surgical injury (injection of zymosan or saline occurred 2 weeks before harvesting mice). *p<0.05 versus I/R/Saline by unpaired two-tailed t-test. The I/R+Sal. and untreated control data shown here are the same as in Main Figure 4, as these studies were performed in parallel. Numerical data are summarized as box and whisker plots indicating the median value (black bar inside box), 25th and 75th percentiles (bottom and top of box, respectively), and minimum and maximum values (bottom and top whisker, respectively), from the number (n) of mice indicated in the figures. b, Representative confocal micrographs from heart histological sections from Ccr2-RFP × Cx3cr1-GFP mice at 10 days post-I/R showing the infarct border zone versus remote myocardium and labeled with a biotin-conjugated collagen hybridizing peptide (CHP) that detects immature or denatured collagen. CHP labeling was detected with a streptavidin-conjugated Alexa647 secondary antibody (purple). c, Representative confocal micrographs of heart histological sections from the post-I/R border zone of Ccr2-RFP × Cx3cr1-GFP mice that received intra-cardiac injection of MNCs or zymosan at 7 days post-I/R and analyzed after an additional 3 days. Endogenous RFP (red) or GFP (green) fluorescence shows CCR2+ or CX3CR1+ macrophages, respectively. Sections were treated with CHP (purple) as in (b) to visualize immature collagen and areas of active remodeling versus areas where macrophage subtypes differentially localize. Images in b-c are representative over n=2 mice per group. Scale bars = 100 μm.
Supplementary Material
Acknowledgments:
This work was supported by grants from the National Institutes of Health to J.D.M., S.S., and M.N. J.D.M. was supported by the Howard Hughes Medical Institute. R.J.V. was supported by a National Research Service Award from the NIH (F32 HL128083) and a Career Development Award from the American Heart Association (19CDA34670044). All flow cytometric data were acquired using equipment maintained by the Research Flow Cytometry Core in the Division of Rheumatology at Cincinnati Children’s Hospital Medical Center.
Footnotes
Competing Financial Interest:
The authors declare that this work has no associated competing financial interest.
References
- 1.Nguyen PK, Rhee JW & Wu JC Adult Stem Cell Therapy and Heart Failure, 2000 to 2016: A Systematic Review. JAMA Cardiol 1, 831–841, doi: 10.1001/jamacardio.2016.2225 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Aviles F et al. Global position paper on cardiovascular regenerative medicine. Eur Heart J 38, 2532–2546, doi: 10.1093/eurheartj/ehx248 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Epstein JA A Time to Press Reset and Regenerate Cardiac Stem Cell Biology. JAMA Cardiol, doi: 10.1001/jamacardio.2018.4435 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Zwetsloot PP et al. Cardiac Stem Cell Treatment in Myocardial Infarction: A Systematic Review and Meta-Analysis of Preclinical Studies. Circ Res 118, 1223–1232, doi: 10.1161/CIRCRESAHA.115.307676 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Tompkins BA et al. Preclinical Studies of Stem Cell Therapy for Heart Disease. Circ Res 122, 1006–1020, doi: 10.1161/CIRCRESAHA.117.312486 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orlic D et al. Bone marrow cells regenerate infarcted myocardium. Nature 410, 701–705, doi: 10.1038/35070587 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Beltrami AP et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 114, 763–776 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Eschenhagen T et al. Cardiomyocyte Regeneration: A Consensus Statement. Circulation 136, 680–686, doi: 10.1161/CIRCULATIONAHA.117.029343 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.A futile cycle in cell therapy. Nat Biotechnol 35, 291, doi: 10.1038/nbt.3857 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Sanganalmath SK & Bolli R Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ Res 113, 810–834, doi: 10.1161/CIRCRESAHA.113.300219 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pillemer L, Blum L, Pensky J & Lepow IH The requirement for magnesium ions in the inactivation of the third component of human complement (C’3) by insoluble residues of yeast cells (zymosan). J Immunol 71, 331–338 (1953). [PubMed] [Google Scholar]
- 12.Saederup N et al. Selective chemokine receptor usage by central nervous system myeloid cells in CCR2-red fluorescent protein knock-in mice. PLoS One 5, e13693, doi: 10.1371/journal.pone.0013693 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jung S et al. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol Cell Biol 20, 4106–4114 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bajpai G et al. Tissue Resident CCR2− and CCR2+ Cardiac Macrophages Differentially Orchestrate Monocyte Recruitment and Fate Specification Following Myocardial Injury. Circ Res 124, 263–278, doi: 10.1161/CIRCRESAHA.118.314028 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dick SA et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nat Immunol 20, 29–39, doi: 10.1038/s41590-018-0272-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Berlo JH et al. c-kit+ cells minimally contribute cardiomyocytes to the heart. Nature 509, 337–341, doi: 10.1038/nature13309 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nahrendorf M et al. The healing myocardium sequentially mobilizes two monocyte subsets with divergent and complementary functions. J Exp Med 204, 3037–3047, doi: 10.1084/jem.20070885 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaikita K et al. Targeted deletion of CC chemokine receptor 2 attenuates left ventricular remodeling after experimental myocardial infarction. Am J Pathol 165, 439–447, doi: 10.1016/S0002-9440(10)63309-3 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leuschner F et al. Therapeutic siRNA silencing in inflammatory monocytes in mice. Nat Biotechnol 29, 1005–1010, doi: 10.1038/nbt.1989 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stremmel C et al. Yolk sac macrophage progenitors traffic to the embryo during defined stages of development. Nat Commun 9, 75, doi: 10.1038/s41467-017-02492-2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang J et al. In Situ Imaging of Tissue Remodeling with Collagen Hybridizing Peptides. ACS Nano 11, 9825–9835, doi: 10.1021/acsnano.7b03150 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brigstock DR Regulation of angiogenesis and endothelial cell function by connective tissue growth factor (CTGF) and cysteine-rich 61 (CYR61). Angiogenesis 5, 153–165 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Thum T, Bauersachs J, Poole-Wilson PA, Volk HD & Anker SD The dying stem cell hypothesis: immune modulation as a novel mechanism for progenitor cell therapy in cardiac muscle. J Am Coll Cardiol 46, 1799–1802, doi: 10.1016/j.jacc.2005.07.053 (2005). [DOI] [PubMed] [Google Scholar]
Online References for Online Methods:
- 24.Schwanekamp JA, Lorts A, Vagnozzi RJ, Vanhoutte D & Molkentin JD Deletion of Periostin Protects Against Atherosclerosis in Mice by Altering Inflammation and Extracellular Matrix Remodeling. Arterioscler Thromb Vasc Biol 36, 60–68, doi: 10.1161/ATVBAHA.115.306397 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pinto AR et al. Revisiting Cardiac Cellular Composition. Circ Res 118, 400–409, doi: 10.1161/CIRCRESAHA.115.307778 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vagnozzi RJ et al. Genetic Lineage Tracing of Sca-1(+) Cells Reveals Endothelial but Not Myogenic Contribution to the Murine Heart. Circulation 138, 2931–2939, doi: 10.1161/CIRCULATIONAHA.118.035210 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Han C et al. Acute inflammation stimulates a regenerative response in the neonatal mouse heart. Cell Res 25, 1137–1151, doi: 10.1038/cr.2015.110 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaiser RA et al. Targeted inhibition of p38 mitogen-activated protein kinase antagonizes cardiac injury and cell death following ischemia-reperfusion in vivo. J Biol Chem 279, 15524–15530, doi: 10.1074/jbc.M313717200 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Sussman MA et al. Prevention of cardiac hypertrophy in mice by calcineurin inhibition. Science 281, 1690–1693, doi: 10.1126/science.281.5383.1690 (1998). [DOI] [PubMed] [Google Scholar]
- 30.Liu R et al. Cardiac-specific deletion of protein phosphatase 1beta promotes increased myofilament protein phosphorylation and contractile alterations. J Mol Cell Cardiol 87, 204–213, doi: 10.1016/j.yjmcc.2015.08.018 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Goncalves R & Mosser DM The isolation and characterization of murine macrophages. Curr Protoc Immunol Chapter 14, Unit 14 11, doi: 10.1002/0471142735.im1401s83 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies LC, Jenkins SJ, Allen JE & Taylor PR Tissue-resident macrophages. Nat Immunol 14, 986–995, doi: 10.1038/ni.2705 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khalil H et al. Fibroblast-specific TGF-beta-Smad2/3 signaling underlies cardiac fibrosis. J. Clin. Invest. 127 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data generated or analyzed in this study will be made available from the corresponding author upon reasonable request. More specifically, original source data used to generate graphs in each of the figures and extended data figures are available as Microsoft Excel data sheet files.