Abstract
Phenylketonuria (PKU) due to recessively inherited phenylalanine hydroxylase (PAH) deficiency results in hyperphenylalaninemia, which is toxic to the central nervous system. Restriction of dietary phenylalanine intake remains the standard of PKU care and prevents the major neurologic manifestations of the disease, yet shortcomings of dietary therapy remain, including poor adherence to a difficult and unpalatable diet, an increased incidence of neuropsychiatric illness, and imperfect neurocognitive outcomes. Gene therapy for PKU is a promising novel approach to promote lifelong neurological protection while allowing unrestricted dietary phenylalanine intake. In this study, liver-tropic recombinant AAV2/8 vectors were used to deliver CRISPR/Cas9 machinery and facilitate correction of the Pahenu2 allele by homologous recombination. Additionally, a non-homologous end joining (NHEJ) inhibitor, vanillin, was co-administered with the viral drug to promote homology-directed repair (HDR) with the AAV-provided repair template. This combinatorial drug administration allowed for lifelong, permanent correction of the Pahenu2 allele in a portion of treated hepatocytes of mice with PKU, yielding partial restoration of liver PAH activity, substantial reduction of blood phenylalanine, and prevention of maternal PKU effects during breeding. This work reveals that CRISPR/Cas9 gene editing is a promising tool for permanent PKU gene editing.
Keywords: phenylketonuria, phenylalanine, gene therapy, gene editing, gene correction, CRISPR/Cas9, phenylalanine hydroxylase, homology directed repair
Introduction
Phenylketonuria (PKU), caused by recessively inherited phenylalanine hydroxylase (PAH) deficiency (OMIM: 261600), is one of the most common inborn errors of metabolism (IEMs) and results in hyperphenylalaninemia and neurotoxic effects of phenylalanine upon the brain.1 PAH is a cytosolic hepatic homo-tetramer that metabolizes L-phenylalanine (Phe) into L-tyrosine.2 Without PAH function, chronic untreated hyperphenylalaninemia causes severe neurological damage, leading to intellectual disability, psychological disorders, and seizures.1,3 It is important to note that human clinical PKU phenotypes form a disease continuum, ranging from mild hyperphenylalaninemia (blood Phe = 120–600 μM) to mild PKU (blood Phe = 600–1200 μM), to the most severe, classical PKU (cPKU) (blood Phe > 1200 μM), depending upon the amount of residual liver PAH activity in the patient. Due to the severity of largely preventable neurological symptoms, all infants born in medically advanced countries undergo newborn screening for hyperphenylalaninemia and are placed on dietary treatment early in life.4
The current standard of care for PKU is lifelong dietary restriction of Phe intake, which requires supplementation with medical foods lacking Phe but containing critical nutrients, including amino acids other than Phe, along with vitamins and minerals that individuals with PKU are unable to retrieve from the severely restricted diet.5, 6, 7, 8 Impaired access to specialized medical foods is prevalent in the United States due to inconsistent insurance coverage, particularly for adults. Inconsistent access to medical foods is associated with chronic hyperphenylalaninemia, cognitive and behavioral symptoms, and functional disability.9 Federal legislative efforts to guarantee uniform insurance coverage of medical foods across state boundaries are ongoing through the proposed Medical Nutrition Equity Act. Though the diet is successful at preventing the major manifestations of this disease, cognitive and behavioral abnormalities remain prevalent in the treated PKU population and may occur even in individuals who successfully maintain blood Phe concentrations between 120 and 360 μM, the recommended treatment range.8 These abnormalities can include lower IQ, executive functioning deficits, and psychiatric disorders (anxiety and depression), with the incidence of deficits increasing with increased blood Phe concentration.10, 11, 12
A recent US Food and Drug Administration (FDA)-approved enzyme substitution therapy (pegvaliase) for adults with uncontrolled hyperphenylalaninemia on a diet has revealed dramatic improvements in blood Phe management13, 14, 15, 16 but is associated with a significant incidence of immune-mediated hypersensitivity reactions against the foreign protein. Additionally, pegvaliase is not approved for use in individuals less than 18 years of age. Pegvaliase therapy requires daily injections, while gene therapy has the potential to correct the disease in a single treatment. In a survey performed by the National PKU Alliance (NPKUA), a national non-profit organization committed to unite, inform, and support people with PKU, over 85% of the community were interested in gene therapy administered as a one-time infusion that would alleviate the lifelong limitations, burdens, and consequences of PKU.9 Preclinical studies using adeno-associated virus (AAV)-mediated gene addition therapy in the Pahenu2/enu2 mouse have shown robust efficacy, but the effectiveness is limiting in neonatal mice.17,18 This loss of therapeutic efficacy is multifactorial, including the loss of AAV episomes as liver cells divide, the lack of selective advantage for PAH+ liver cells, and the high therapeutic threshold of PKU (~10% hepatocyte correction to lower blood Phe).19, 20, 21 In pursuit of a more permanent gene therapy, this study aimed to correct the mutation causing PKU in Pahenu2/enu2 mice using a targeted, integrating AAV approach. We utilized the CRISPR/Cas9 nuclease system22,23 to induce a double-strand break (DSB) near the Pahenu2 mutation and to enhance the opportunity for homologous recombination with the provided repair template harboring the wild-type (WT) sequence. This correction is expected to be permanent even if the targeted hepatocyte divides and its genome is replicated. The barrier impeding efficacy with this approach is the ability to achieve a sufficient number of correctly gene-edited hepatocytes to produce a physiologically relevant improvement in Phe clearance in the absence of any selective growth advantage for PAH+ cells. To address this challenge, a non-homologous end joining (NHEJ) inhibitor shown to facilitate homology-directed repair (HDR) was evaluated as a means of increasing the frequency of successful gene repair in our system.24
The Pahenu2/enu2 mouse model of cPKU is ideal for this proof-of principle study, as it contains a single missense mutation in the Pah gene associated with hyperphenylalaninemia. The pathologic c.835T > C missense mutation alters amino-acid position 263 from a phenylalanine into a serine, disrupting proper function of the catalytic domain of PAH. Pahenu2/enu2 homozygotes exhibit all of the symptoms of classical untreated human PKU, including a blood phenylalanine concentration > 2,000 μM while consuming normal mouse chow, hypopigmentation, and associated neuropathology, resulting in cognitive and behavioral deficits.25, 26, 27
Results
Experimental Design
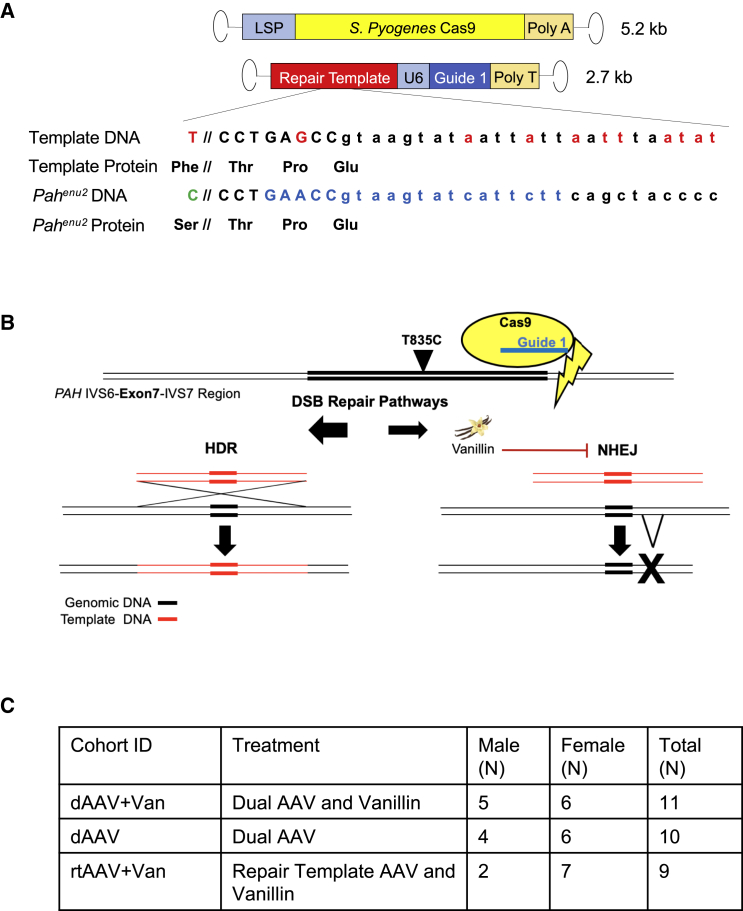
Gene repair occurs through homologous recombination and is greatly enhanced by DSBs, which can be mediated by the Streptococcus pyogenes CRISPR/Cas9 system. In order to correct the Pahenu2 mutation in this fashion, three components must be delivered to hepatocytes by the liver-tropic AAV serotype 8 (AAV8): the Cas9 enzyme, a single-guide RNA (sgRNA) with homology to sequences near the target mutation, and a repair template containing homology to the target allele except for a thymine at position c.835, the WT sequence (Figure 1).22 Due to the limiting packaging capacity of AAV, this system must be manipulated into two AAV vectors in a dual-AAV approach.23 Here, one recombinant AAV genome contained the S. pyogenes Cas9 coding sequence with expression driven by a liver-specific promoter (LSP) designated P3 (minimal transthyretin promoter [TTRmin], coupled to a de novo designed hepatocyte-specific cis-regulatory module 8, [HS-CRM8]),28,29 and a second vector expressed a validated guide RNA 46 bp downstream of the c.835 mutation site under the expression control of the human U6 RNA polymerase III (pol III) promoter. This second vector also contained a 2-kb fragment of the WT mouse Pah genomic sequence (GRCm38: 87569274–87571296) flanking the Pahenu2 mutation site with purposefully introduced synonymous and intronic mutations that alter the PAM and guide sequence, thereby preventing re-cutting by Cas9 as well as facilitating downstream sequence analyses (Figure 1A). Expression of Cas9 and the guide RNA was expected to result in DNA DSBs 46 bp downstream of the Pahenu2 mutation. Innate cellular DNA repair mechanisms could alternately repair the DSBs by NHEJ, yielding either a perfectly repaired Pahenu2 sequence or the introduction of small insertions or deletions (indels) through alternative end joining (Alt-EJ) or by HDR using the Pah genomic DNA fragment from the second AAV vector as a repair template (Figure 1B). The latter, desired outcome would restore the nucleotide at cDNA position 835 to a thymine and lead to the functional correction of PAH activity in the edited hepatocyte. In an effort to maximize the frequency of HDR in preference over NHEJ, vanillin, a potent NHEJ-inhibitor previously shown to increase HDR in a gene-targeting AAV system,24 was co-administered with the viral vectors in some mice. Animals were re-dosed with drug as adolescents due to volume restraints in neonates and to provide an opportunity for additional correction.
Figure 1.
Design of In Vivo CRISPR/Cas9 Gene-Editing Strategy in Pahenu2 Mice
(A) Dual AAV constructs. The Cas9-expressing rAAV2/8 vector genome was 5.2 kb in length and contained the S. pyogenes Cas9 gene driven by a transthyretin-based liver-specific promoter (LSP). The second rAAV2/8 vector genome was 2.7 kb in length and contained a 2-kb repair template flanking 1 kb in either direction of the enu2 missense mutation, as well as guide 1 driven by a human U6 promoter. The repair template sequence pictured below the vector genomes contained the wild-type Pah exon 7 sequence with correction of the Pahenu2 mutation, as well as purposefully introduced synonymous and intronic mutations (denoted in red) to hinder Cas9 cutting of corrected alleles. Uppercase denotes exonic Pah sequence, and lowercase indicates intronic sequence. The wild-type protein sequence of the repair template demonstrates correction of the Ser263Phe and a synonymous mutation at Pro279. The Pahenu2 allele below reveals the c.835T > C mutation in green and the guide targeted sequence in blue. The symbol “//” indicates 46-bp DNA and 15-aa separation. (B) Diagram of expected Cas9-mediated DNA cutting and cellular repair. Guide 1, indicated in blue, directs Cas9 (yellow oval) to induce a double-strand break (DSB) (indicated by the lightning bolt) 46 bp downstream of the c.835T > C mutation (black arrow) in the Pahenu2 allele; double lines indicate double-stranded DNA (dsDNA), where thick lines are exons and thin lines are introns. Potential DSB intracellular pathways that the cell may utilize to repair the Cas9 DSB are depicted below. The repair template, indicated in red, may be incorporated into the genome by homology-directed repair (HDR), pictured on the left. Alternatively, non-homologous end joining (NHEJ) restores the mutant allele (not shown); or alternate end joining (Alt-EJ), pictured at the right, results in a small insertion or deletion (indel, indicated by the bold X). Either NHEJ or Alt-EJ fails to incorporate the repair template. Vanillin is a potent NHEJ inhibitor (indicated by the smaller arrow at the right) that directs DSB repair toward HDR (indicated by the larger arrow at the left). (C) Table of experimental animal cohorts. Three experimental animal cohorts were tested in this study. The cohort with dual AAV plus vanillin (dAAV+Van) received both viral vectors plus vanillin, with a total of 11 animals treated: 5 males and 6 females. The dual-AAV (dAAV) cohort received both viruses but no vanillin, with a total of 10 animals treated: 4 males and 6 females. The cohort with repair template vector plus vanillin (rtAAV+Van) received only the repair template virus with vanillin, with a total of 9 animals treated: 2 males and 7 females.
Three animal cohorts were tested in this study. In two cohorts, dual AAV (dAAV) and dAAV plus vanillin (dAAV+Van), all animals received both AAV vectors, and the latter also received vanillin injections. In the third cohort, repair template AAV plus vanillin (rtAAV+Van), animals received the repair template vector only, without the Cas9-expressing vector, but did receive vanillin (Figure 1C). All animals were screened for and tested positive for the presence of appropriate viral DNA according to treatment group in liver tissue via PCR post-harvest.
Therapeutic Efficacy
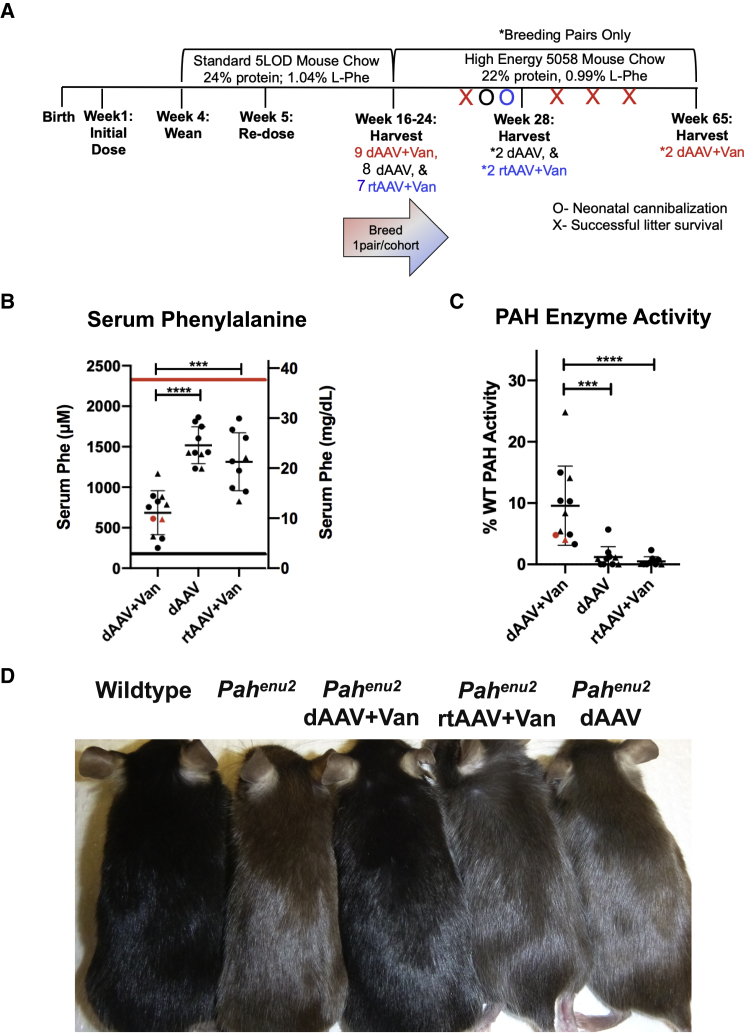
Single male and female mice from each treatment group were reserved for breeding experiments (Figure 2A) and allowed to survive up to 65 weeks of age. The remaining animals were euthanized at 16–24 weeks of age for characterization and tissue harvest. At the time of euthanasia, the coat color on all dAAV+Van-treated animals was indistinguishable from that of heterozygous or WT animals, while animals in the dAAV and rtAAV+Van groups had minimal, if any, coat color darkening, suggesting that substantial reduction of blood phenylalanine concentration had occurred in dAAV+Van-treated animals only (Figure 2D).
Figure 2.
Efficacy of In Vivo CRISPR/Cas9 Gene Editing in Pahenu2 Mice
(A) Experimental timeline. Pahenu2/enu2 mice were born, administered the initial dose of drug in the first week of life, weaned at 4 weeks, and placed on standard mouse chow. At week 5, mice were re-dosed with drug. Between 16 and 24 weeks, mice from each cohort were harvested, and one breeding pair per treatment group was placed on high-energy mouse chow and allowed to breed. By 28 weeks, one successful litter was produced by the dAAV+Van pair, while the dAAV and rtAAV+Van pairs did not successfully breed. At this point, the dAAV and rtAAV+Van breeding pairs were euthanized for tissue collection. The dAAV+Van breeder pair produced three more successful litters and were euthanized at 65 weeks. (B) Serum phenylalanine. The graph shows serum phenylalanine levels of dAAV+Van, dAAV, and rtAAV+Van animals at time of euthanasia. The left y axis indicates serum Phe in micromol/l (Mmicromol/l, while the right y axis indicates serum Phe in micrograms per deciliter. The red line indicates the mean serum phenylalanine concentration of an unrelated cohort of untreated male and female Pahenu2/enu2 animals combined consuming standard mouse chow.27 The black line indicates the upper limit of serum Phe levels in wild-type mice (180 μM or 3 mg/dL). The x axis is separated by treatment group, in the order of dAAV+Van, dAAV, and rtAAV+Van from left to right, whisker plots depict mean serum phenylalanine ± 2 SD, and each dot on the graph indicates an individual animal. Males are depicted with triangles and females with circles. Red indicates 65-week-old animals, and black indicates 16- to 24-week-old animals. The average Phe levels were 685 μM, 1,518 μM, and 1,314 μM, respectively; and ranges were 252–1,168 μM, 1,231–1,863 μM, and 827–1,850 μM, respectively. Intergroup comparisons (depicted as brackets above the treatment groups) revealed significant differences between dAAV+Van and dAAV (p < 0.0001, ∗∗∗∗) and between dAAV+Van and rtAAV+Van (p = 0.0001, ∗∗∗). (C) PAH enzyme activity. The graph shows the PAH enzyme activity levels of dAAV+Van, dAAV, and rtAAV+Van animals at time of euthanasia. The y axis indicates percent wild-type PAH activity; the x axis is separated by treatment group in the order of dAAV+Van, dAAV, and rtAAV+Van from left to right, whisker plots depict mean liver PAH activity ± 2 SD, and each dot on the graph indicates an individual animal. Males are depicted with triangles, and females are depicted with circles. Red indicates 65-week-old animals, and black indicates 16- to 24-week-old animals. The average activity was 9.5%, 1.2%, and 0.5%, respectively, and ranges were 3.3%–25%, 0%–5.7%, and 0%–2.3%, respectively. Intergroup comparison reveals a significant difference between dAAV+Van and dAAV (p = 0.0002, ∗∗∗) and between dAAV+Van and rtAAV+Van (p < 0.0001, ∗∗∗∗). (D) Coat color at euthanasia. Photograph of coat colors of mice from each treatment group at 16–24 weeks of age in comparison to wild-type or untreated Pahenu2/enu2 mice. Mice are lined up from left to right as wild-type, untreated Pahenu2/enu2, dAAV+Van, rtAAV+Van, and dAAV.
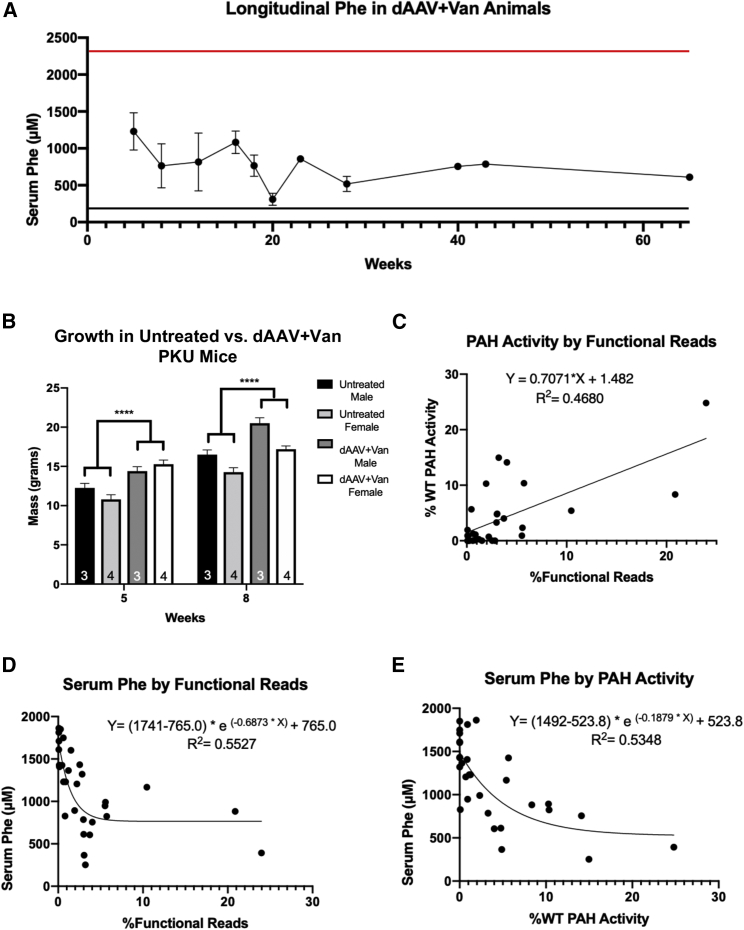
Mean serum phenylalanine concentration at euthanasia was individually variable but approximately 2-fold lower, on average, in dAAV+Van-treated mice (mean ± SEM: 685.2 ± 81.79 μM; range = 252–1,168 μM) in comparison to either dAAV-treated animals (1,518 ± 72.04 μM; range = 1,231–1,863 μM) or rtAAV+Van-treated animals (1,314 ± 119.2 μM; range = 827–1,850 μM). Statistical analysis by one-way ANOVA revealed an overall significant difference in mean serum Phe among the three treatment groups, F(2, 27) = 24.11, p < 0.0001 (Figure 2B). A post hoc intergroup comparison revealed significant differences between dAAV+Van and the other two groups but no differences between dAAV and rtAAV+Van. There appeared to be no significant additional effects from the second AAV administration in any treatment group. The decrease in serum Phe was sustained through 65 weeks in the two animals in the dAAV+Van cohort that were allowed to survive to that age (shown later in Figure 4A).
Figure 4.
Efficacy of In Vivo CRISPR/Cas9 Gene Editing in dAAV+Van-Treated Animals
(A) Longitudinal Phe in dAAV+Van animals. Plot of long-term (up to 65 weeks) serum phenylalanine concentration in the dAAV+Van cohort mice. The red line denotes the mean serum phenylalanine concentration of an unrelated cohort of untreated male and female combined Pahenu2/enu2 animals consuming standard mouse chow.27 The black line indicates the upper limit of serum Phe concentration in wild-type C57BL/6 mice (180 μM). For the time points for which Phe measurements in multiple mice were available, the data are presented as mean ± SEM. Data at 40–65 weeks are means of the two dAAV+Van animals, one male and one female, that remained a breeding pair until euthanasia. (B) Improved growth in dAAV+Van-treated animals. Three untreated age-matched males and three untreated age matched females were compared to dAAV+Van-treated animals. A three-way ANOVA comparing age, treatment group (control versus treated), and sex showed statistically significant differences between these animals, albeit low sample sizes, F(1, 10) = 10.44, p = 0.0090. Post hoc intergroup comparisons revealed consistent significant differences between each treated and untreated group of either sex (p < 0.0001) as depicted by the brackets with asterisks (∗∗∗∗). (C) Correlation between liver PAH activity and functional Pah exon 7 reads in dAAV+Van-treated mice. Plot of percent wild-type liver PAH enzyme activity on y axis versus the percent functional Pah exon 7 reads on the x axis with linear regression analysis. Each point is a single dAAV+Van mouse at euthanasia. Graph shows percent functional Pah exon 7 reads on the x axis and percent wild-type PAH enzyme activity on the y axis. Each point represents a dAAV+Van animal at time of euthanasia. (D) Correlation between serum Phe and percent functional Pah exon 7 reads in dAAV+Van-treated animals. Plot of serum Phe on the y axis versus percent functional Pah exon 7 reads on the x axis. Each point represents a dAAV+Van animal at time of euthanasia. The data were fit with a non-weighted, non-linear exponential one-phase decay using least-squares regression. (E) Correlation between serum Phe and liver PAH enzyme activity in dAAV+Van-treated animals. Plot of serum Phe on the y axis versus percent wild-type liver PAH enzyme activity on the x axis. Each point represents a dAAV+Van animal at time of euthanasia. The data were fit with a non-weighted, non-linear exponential one-phase decay using least-squares regression.
Mean liver phenylalanine hydroxylase activity at euthanasia was, again, individually variable and approximately 10-fold higher in dAAV+Van-treated mice (mean percent WT PAH activity ± SEM: 9.6% ± 1.9%; range = 3.3%–24.8%) in comparison to either dAAV-treated animals (1.2% ± 0.5%; range = 0%–5.7%) or rtAAV+Van-treated animals (0.5% ± 0.3; range = 0%–2.3%). One-way ANOVA revealed an overall significant difference in mean PAH enzyme activity among the three treatment groups, F(2, 27) = 16.02, p < 0.0001 (Figure 2B). A post hoc intergroup comparison revealed significant differences between dAAV+Van and the other two groups but no differences between dAAV and rtAAV+Van.
On-Target Genetic Analysis
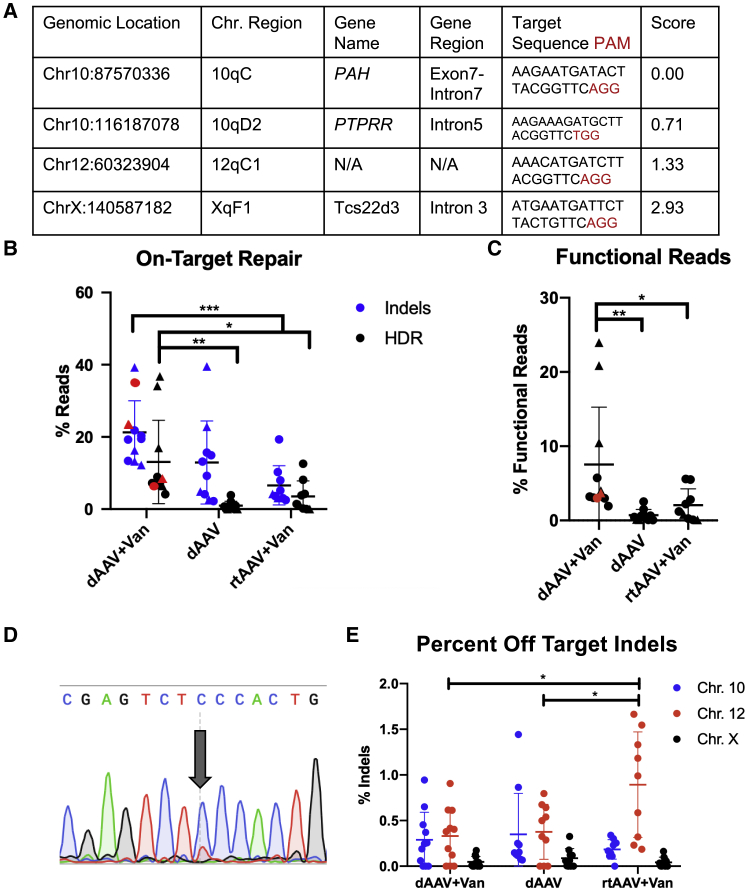
PCR-based next-generation sequencing (NGS) was applied to on- and off-target genomic regions of treated mice (Figure 3A). In order to avoid amplifying and sequencing residual episomal or non-targeted integrations of AAV genomes containing the repair template, we used a nested PCR approach on genomic DNA isolated from treated mouse liver to amplify the on-target genomic region. To produce our first amplicon, a 1.3-kb fragment spanning Pah exon 7, we used a forward primer that was homologous to sequences in Pah intron 6 but outside the 5′ end of the repair template; this fragment was subjected to a second amplification using nested primers containing convenient indexing adapters to yield a 350-bp product. The products of these PCR reactions were indexed with unique barcodes and submitted for NGS on the Illumina MiSeq platform.
Figure 3.
On- and Off-Target CRISPR/Cas9 DNA Analyses
(A) Table of Top Four Cutting Sites for SpCas9+Guide 1 in the Mouse Genome. Two in silico tools, Cas-OFFinder and COSMID, were used to identify the top four SpCas9-Guide1 cutting sites in the mouse genome. Locations of cutting sites are described in the first four columns, followed by target sequence (black) and PAM (red) in the next column. The last column indicates the COSMID score, in which the lowest, 0, indicated the highest probability of cutting (the target site) and ranged to over 40, with the lowest probability of cutting. The top three off-target sites located on chr 10, chr 12, and chr X were scored as 0.17, 1.33, and 2.93, respectively, as indicated in the last column. (B) On-target repair. The graph shows percent reads of indels (blue) or “full” HDR (including exonic and intronic sequences) (black) in dAAV+Van, dAAV, and rtAAV+Van animals at time of euthanasia. The y axis indicates percent reads of overall NGS reads; the x axis is separated by treatment group in order of dAAV+Van, dAAV, and rtAAV+Van from left to right, whisker plots depict the mean percent reads ± 2 SD, and each dot on the graph indicates an individual animal. Males are depicted with triangles, and females are depicted with circles. Red indicates animals at 65 weeks, and black indicates animals at 16–24 weeks. The average percentages of indel reads were 21.27%, 12.92%, and 6.58%, respectively, with ranges of 12.25%–39.29%, 2.2%–39.54%, and 2.52%–19.34%, respectively. The average percentages of HDR reads were 13.06%, 0.96%, and 3.51%, respectively, and ranges were 4.13%–36.78%, 0%– 3.90%, and 0%–12.57%, respectively. Two-way ANOVA revealed a significant difference between treatment groups, F(2, 54) = 12.87, p < 0.0001; and DNA repair, F(1, 54) = 13.11, p = 0.0006. Brackets above the treatment groups depict post hoc intergroup statistical comparisons. ∗ - p < 0.05; ∗∗ - p < 0.01, ∗∗∗ - p < 0.001, ∗∗∗∗ - p < 0.0001 (C) Functional reads. The graph indicates functional reads of exonic-only NGS reads in dAAV+Van, dAAV, and rtAAV+Van animals at time of euthanasia. The y axis indicates the percentages of functional Pah-exon7 NGS reads. The x axis is separated by treatment group in order of dAAV+Van, dAAV, and rtAAV+Van from left to right, whisker plots depict the mean percent functional reads ± 2 SD, and each dot on the graph indicates an individual animal. Males are depicted with triangles, and females are depicted with circles. Red indicates 65-week-old animals, and black indicates 16- to 24-week-old animals. The averages of functional reads were 7.54%, 0.70%, and 2.06%, respectively; and ranges were 1.94%–23.97%, 0.11%–2.54%, and 0.08%–5.58%, respectively. One-way ANOVA revealed significant difference of treatment groups, F(2, 27) = 5.805, p = 0.0080; with a p value of 0.0093 between dAAV+Van and dAAV and a p value of 0.048 between dAAV+Van and rtAAV+Van. Brackets above the treatment groups depict post hoc intergroup statistical comparisons. ∗ - p < 0.05; ∗∗ - p < 0.01. (D) Sanger sequencing of dAAV+Van enu2 allele correction. The Sanger sequencing chromatogram shows low levels of correction in liver genomic DNA, as indicated with the large arrow pointing to the Pahenu2 c.835T > C (indicated with “C”) site that contains a small peak of corrected (indicated with “T”) DNA. (E) Percent off-target indels. The graph shows the percentage of overall NGS reads containing indels in the top three off-target regions identified in Figure 3A. The y axis shows the percentage of indels; the x axis is separated by treatment groups in the order of dAAV+Van, dAAV, and rtAAV+Van from left to right, with further nested separation of each chromosomal region in order of highest likelihood of cutting to lowest; chr 10, chr 12, and chr X, for each treatment group. Each animal is represented by a blue, a red, and a black dot, indicating chr 10, chr 12, and chr X, respectively. Whisker plots depict the mean percent off target indels ± 2 SD. The average percentages of indels for chr 10 were 0.29%, 0.35%, and 0.18%, respectively; for chr 12, they were 0.33%, 0.38%, and 0.89%, respectively; and for chr X, they were 0.05%, 0.09%, and 0.04%, respectively. The ranges of percent indels for chr 10 were 0%–0.94%, 0.07%–1.44%, and 0%–0.34%, respectively; for chr 12, they were 0%–0.91%, 0%–0.80%, and 0.19%–1.66%, respectively; and for chr X, they were 0%–0.17%, 0%–0.33%, and 0%–0.16%, respectively. One-way ANOVA of each chromosomal region between treatment groups revealed significant difference in only the chr 12 region, F(2, 27) = 5.709, p = 0.0086; with a p value of 0.012 between rtAAV+Van and dAAV+Van and a p value of 0.0248 between rtAAV+Van and dAAV. Brackets above the treatment groups depict post hoc intergroup statistical comparisons. ∗ - p < 0.05.
We define NHEJ as DNA repair of a DSB that results in perfect restoration of the original sequence without correction of the Pahenu2 mutation, an event that cannot be detected experimentally. Repair that results in a deletion of 1 bp or greater in size but retains the c.835C Pahenu2 mutation is defined as Alt-EJ, and on-target HDR was defined as detecting exonic and intronic genomic sequences that were consistent with 98% identity to the repair template having been appropriately incorporated into the amplicon in the expected orientation. The percentages of NGS reads with Alt-EJ (indels) or properly targeted HDR are displayed in Figure 3B. A two-way ANOVA revealed a significant difference between treatment groups, F(2, 54) = 12.87, p < 0.0001; and DNA repair, F(1, 54) = 13.11, p = 0.0006. The percentage of reads with indels in dAAV+Van-treated animals (mean ± SEM: 21.27% ± 2.63%; range = 12.25%–39.29%) was approximately 2- to 3-fold higher than that in either the dAAV group (12.92% ± 3.64%; range = 2.21–39.54) or the rtAAV+Van group (6.59% ± 1.81%; range = 2.52–19.34). There was no statistical difference in the percentage of indels between dAAV+Van and dAAV or between dAAV and rtAAV+Van, but there was between dAAV+Van and rtAAV+Van, with a p value of 0.0007. The percentage of reads with properly targeted HDR in dAAV+Van-treated animals (mean percentage ± SEM: 13.06% ± 11.54%; range = 4.13–36.78) was significantly higher than that in either the dAAV group (0.96% ± 1.23%; range = 0–3.90) or the rtAAV+Van group (3.51% ± 4.32%; range = 0.01–12.57). There was no statistical difference in HDR between dAAV- and rtAAV+Van-treated animals.
NGS data were further analyzed for functional cDNA, in which NGS reads were trimmed to contain only the exon 7 sequence and evaluated for the ability to encode a functional enzyme (Figure 3C), again, revealing a similar significant difference among treatment groups, F(2, 27) = 5.805, p = 0.008 by one-way ANOVA. NGS analysis of dAAV+Van-treated animals, again, revealed an approximately 4-fold increase in the percentage of functional Pah cDNA in the dAAV+Van-treated group (mean ± SEM: 7.54% ± 2.33%; range = 1.94%–24.00%) in comparison to the dAAV group (0.70% ± 0.25%; range = 0.07%–2.54%) or the rtAAV+Van group (2.07% ± 0.73%; range = 0.08%–5.58%).
Correction of the c.835T > C mutation back to thymine by Sanger sequencing in a small portion of genomic liver DNA from treated animals could only be detected in dAAV+Van-treated animals (Figure 3D).
Off-Target Genetic Analysis
Possible sites of off-target Cas9-mediated genotoxicity were selected for NGS analysis using the in silico COSMID tool, which used the guide 1 sequence to determine the top 45 off-target cutting sites in the mouse genome, scored for likelihood of cutting where 0 = highest confidence cutting and 40.7 = lowest confidence cutting (Figure S2). The top three sites selected for further investigation were chromosome chr 10:116187078, chr12:60323904, and chrX:140587182, with scores of 0.17, 1.33, and 2.93, respectively (Figure 3A). PCR-based NGS of liver genomic DNA from treated animals revealed minimal off-target SpCas9 cutting in all three chromosomal regions, with none accumulating to more than 2% of reads (Figure 3E). Surprisingly, a one-way ANOVA of each chromosomal region between treatment groups revealed a significant difference in only the chr 12 region, F(2, 27) = 5.71, p = 0.0086; with an approximately 2-fold higher percentage of indels in the rtAAV+Van-treated group (mean percentage of indels ± SEM: 0.89% ± 0.19%; range = 0.19%–1.66%) than either treatment group containing the SpCas9 enzyme, dAAV+Van (0.33% ± 0.09%; range = 0%–0.91%) or dAAV (0.38% ± 0.09%; range = 0%–0.80%). Despite this significant increase in indels on chr 12 in the rtAAV+Van treatment group—which cannot be easily explained, as this cohort did not receive any Cas9 enzyme—these data show minimal off-target cutting by SpCas9 at the three locations investigated in these animals. While this was not an exhaustive investigation of all off-target indels in the whole genome, this therapy seems safe for genomic integrity in the three highest predicted off-target cutting sites.
Correlations between Functional Pah Genomes, Liver PAH Activity, and Mean Serum Phe Concentrations
The amount of PAH activity measured in liver homogenates from dAAV+Van mice is linearly related to the percentage of functional PAH reads restored by CRISPR/Cas9-mediated exon 7-directed DSB and successful HDR (Figure 4C). The relationships between serum Phe and the percentage of functional PAH reads (Figure 4D) or between serum Phe and liver PAH activity (Figure 4E) fit best to a one-phase exponential decay model, which is consistent with the known first-order Michaelis-Menten kinetics of the PAH reaction.
Adverse Effects
No adverse effects of treatment were seen in any cohort. The overall health and development of the animals in the dAAV and rtAAV+Van treatment groups were consistent with that expected for untreated Pahenu2/enu2 mice, including hypopigmentation and impaired growth. The animals in the dAAV+Van treatment group showed improved health in comparison to the expected Pahenu2/enu2 development with dark coat pigmentation and improved growth (Figure 4B). Mice were euthanized at ages ranging from 16 to 65 weeks of age; the gross macroscopic appearance of all internal organs was normal at necropsy in all mice. There was no gross evidence of liver neoplasia in any animal, though molecular markers of hepatocellular carcinoma were not evaluated for low-grade carcinogenic transformation in this study.
Successful Breeding of Gene-Corrected Pahenu2 Mice
One pair (a single male and a single female) of gene-corrected Pahenu2/enu2 mice from the dAAV+Van treatment group were allowed to survive past 24 weeks to test both the duration of response and evaluate the breeding potential of gene-corrected mice. Although hyperphenylalaninemic mice are fertile, female Pahenu2/enu2 mice cannibalize all their offspring soon after birth due to a powerful maternal PKU effect, as well as congenital defects in the pups.30 Typically, the Pahenu2 colony is maintained through Pahenu2/enu2 male × Pahenu2/+ female crosses to avoid the maternal PKU effect. The efficacy of dAAV+Van upon serum Phe in both the male and female Pahenu2/enu2 mice was durable through 65 weeks of life (Figure 4A). This breeding pair utilized provided nesting materials to build well-constructed nests, a behavior not typically seen in hyperphenylalaninemic Pahenu2/enu2 mice (Figure S3).27 Furthermore, and impressively, this breeding pair yielded four liveborn litters with a total of 26 progeny (12 males and 14 females), which the dam successfully delivered and nursed through weaning without any losses. All of the progeny were hypopigmented, hyperphenylalaninemic, and genotyped with toe-tissue-extracted DNA to be consistent with homozygosity of the Pahenu2 mutation. While germline cells of breeders were not directly tested, these results support that there was no germline transmission of the CRISPR/Cas9-mediated gene correction from dAAV+Van-treated mice to their offspring.
In contrast, the breeding pairs from the dAAV and rtAAV+Van treatment groups that were allowed to survive past 24 weeks to evaluate the breeding potential in control treatment groups were both unsuccessful. All animals, two males and two females, were hypopigmented and hyperphenylalaninemic. Each pair, one dAAV and one rtAAV+Van, gestated to term, but neonates were instantly cannibalized upon birth. Neither of these breeding pairs utilized nesting materials to build nests and remained hypopigmented and hyperphenylalaninemic until harvest at 28 weeks.
Discussion
This study reveals promise for the utilization of the CRISPR/Cas9 system followed by HDR of the AAV-supplied donor DNA to achieve in vivo liver-directed gene correction for PKU. The permanent nature of editing mutations within the genome removes the concern for temporary effectiveness as seen in traditional AAV-mediated gene addition therapy for PKU. Our prior attempts with AAV-mediated, liver-directed gene addition in neonatal mice resulted in the rapid clearance of AAV episomes and only brief periods of liver PAH expression (data not shown). This result has been reported by other investigators following administration of liver-directed AAV vectors to juvenile animals in the absence of any selective growth advantage for random AAV integration events.31,32 For the first time, we have achieved liver-directed gene therapy in neonatal mice that persisted beyond adolescence into adult mice that was associated with substantial, albeit partial, correction of hyperphenylalaninemia for up to at least 6 months of age (at the time when most animals were euthanized for tissue harvest) and impressively up to 65 weeks in the case of a single breeder pair. Importantly, all animals in the dAAV+Van treatment group were improved in classification from severe, classical PKU to either mild PKU or mild hyperphenylalaninemia. If this dAAV+Van regimen were applied to humans suffering from classical PKU, it would greatly improve the overall quality of life by allowing increased dietary phenylalanine intake, improved maternal PKU outcomes, and possibly improved behavior and cognition, even in the case with the lowest correction (4.1% HDR, 3.3% PAH enzyme activity, 1,168 μM blood Phe on an unrestricted diet). This treatment approach was not associated with any notable acute toxicity or any long-term adverse effects. In fact, the treatment was associated with complete reversal of the abnormal phenotypes associated with chronic hyperphenylalaninemia in Pahenu2/enu2 mice, including impaired growth and hypopigmentation, despite the fact that serum phenylalanine concentrations were not fully corrected to levels typical of WT mice. Although most animals did not achieve phenylalanine levels in the recommended recommended therapeutic range of 120–360 μM, this is the first gene therapy approach for PKU that has demonstrated lifelong effectiveness from early infancy in a PKU mouse model.
Furthermore, AAV-delivered, CRISPR/Cas9-mediated, liver-directed gene correction was associated with the prevention of maternal PKU syndrome in Pahenu2/enu2 mice. Elevated maternal blood phenylalanine in humans during pregnancy causes microcephaly, intrauterine growth retardation, congenital heart disease, and postnatal developmental disability, the so-called maternal PKU syndrome.33 Hyperphenylalaninemic Pahenu2/enu2 dams gestate to term but invariably cannibalize their progeny.30 The single breeder pair from the dAAV+Van group successfully produced four litters (Figure 2A) without any observed pregnancy losses, an outcome that is unprecedented in our laboratory, although we and others have had prior limited success with breeding Pahenu2/enu2 dams following treatment of hyperphenylalaninemia applied to adult mice.34
CRISPR/Cas9-mediated genome editing is widely used to introduce DSBs in DNA and to facilitate the deletion of functional genes in cultured cells and in whole organisms, but correction of mutant genes following Cas9-mediated DSB is entirely dependent upon the innate DNA repair activity of the host cell.35 NHEJ, in most cases, is the default repair mechanism used to repair the DSB; this blunt end joining can result in full restoration of the original sequence (an event that cannot be detected experimentally), leaving the mutant gene intact. Alternatively, some 3′ clipping of the free ends may occur prior to annealing of the break (so-called Alt-EJ) or healing of the break with incorporation of a repair template through HDR.35,36 The former, less well-understood repair mechanism, Alt-EJ, yields a small deletion at the site of the DSB. If NHEJ or Alt-EJ occurs following Cas9-mediated cleavage of a mutant gene target, a functional open reading frame will not be restored. Only HDR with the incorporation of a repair template harboring the normal functional sequence will yield a functional open reading frame, but under typical conditions, the desired HDR is a rare event. In our experiment, following administration of both AAV vectors carrying the repair template and expressing the Cas9 reagents (but without the NHEJ inhibitor vanillin), the mean percentage of functional Pah open reading frames recovered following HDR was fewer than 1%. This repair frequency is insufficient to cause any appreciable effect upon the PKU phenotype, although it may be sufficient for treatment for some disorders such as hemophilia, where secreted protein from even a small population of hepatocytes can be therapeutic.37 In diseases and model systems where corrected cells enjoy a selective growth advantage, a small number of gene-edited hepatocytes can expand under selective pressure to a therapeutically relevant population. Such results have been reported for the treatment of murine fumarylacetoacetate hydrolase deficiency, a model of human tyrosinemia type 1.38 Unfortunately, PAH-expressing hepatocytes do not enjoy a selective growth advantage over PAH-negative cells.20 Only when we treated the mice with vanillin, a potent NHEJ inhibitor, did the frequency of HDR increase to a level that became physiologically relevant in this model.
Our results were somewhat less robust than those of the Wilson laboratory, who used a similar dAAV system in the murine ornithine transcarbamylase (OTC) model, OTC spfash, and achieved approximately 10% gene correction in a disease that does not enjoy a selective growth advantage for corrected cells without use of NHEJ inhibitors. These conflicting findings could be due to differences in either the initial efficiency of Cas9-mediated double-strand breakage or the efficiency of on-target HDR using the repair template. In our study, only 14% of Pah alleles in dAAV-treated mice demonstrated molecular evidence of genomic alterations (HDR or indels due to Alt-EJ combined) in comparison to 41% overall genomic alteration in the OTC study. This suggests that the efficiency of Cas9-mediated double-strand breakage was much lower in our experiment. This could be attributed to differences in the total dose of AAV vectors delivered or the ratio of Cas9-expressing to repair template vectors administered between the two experiments, both of which could have affected the efficiency of Cas9-mediated DSB formation. Alternatively, it is also possible that perfect NHEJ following Cas9 administration resulted in the restoration of the original pathogenic sequence in a majority of cleaved alleles, a result that cannot be detected experimentally. When we added the NHEJ inhibitor vanillin to dAAV, the mean frequency of total genomic alteration increased to 34% (21% indels and 13% HDR). This result supports our hypothesis that the initial frequency of Cas9-mediated DSBs was likely similar to that of the previously published OTC experiment but that the majority of Cas9-mediated DSBs were perfectly repaired by NHEJ in dAAV-treated mice rather than by Alt-EJ or HDR. The precise reason for the difference in the efficiency of on-target HDR is yet unknown. Both the repair template in our experiment and the OTC template contained multiple mismatches in comparison to their genomic targets; these differences could impair HDR efficiency. It is possible that nuanced design differences between the two templates or innate differences in the genomic target regions could have contributed to increased heteroduplex rejection, D-loop instability during HDR, or mismatch-repair-mediated antirecombination in our study.39 While the dAAV on-target HDR remained low, the addition of vanillin improved the on-target HDR efficiency to a level similar to that achieved in the OTC study.
The therapeutic effectiveness of this gene-editing approach was highly variable between individual mice. One source of variability could be the technically challenging facial vein injection. While we were able to measure correlations between the percentage of functional open reading frames, liver PAH activity, and final blood Phe concentration, some mice still exhibited higher than expected final blood Phe concentrations for the percentage of functional open reading frames and corresponding liver PAH activity achieved in those animals. Typically, in mice treated with AAV-mediated gene addition, restoring liver PAH activity to greater than 10% of WT activity is sufficient to fully correct blood Phe levels to normal.17 These discordant results could be attributed to the number of PAH-expressing cells, which is also a critical factor in total body Phe flux. PAH activity must be expressed in at least 10% of hepatocytes for complete normalization of blood Phe concentrations; restriction of PAH activity to a smaller population of hepatocytes (i.e., lower transduction frequency) significantly limits Phe clearance.40 We hypothesize that, in this experiment, HDR occurred in a small population of evenly dispersed hepatocytes (likely fewer than 10% of hepatocytes). Given that hepatocytes are polyploid41, 42, 43 (the genetic complement can range from diploid [2n] to octoploid [8n] and even higher [128n]), a small number of cells with many edited genomes per cell could be responsible for the relatively high number of functional genomic reads and PAH activity that we have measured. The therapeutic effect may be limited by total Phe transport capacity into the restricted cell population rather than functional enzyme availability. Alternatively, it is possible that gene correction occurred predominantly within peri-central venous hepatocytes, a zone within the hepatic acinus that is not typically responsible for substantial phenylalanine hydroxylation; this is a function normally accomplished by periportal hepatocytes.44 In this study, we performed systemic injections of AAV8 vectors, which, in adult mice, leads to primarily peri-central venous transduction but may lead to mixed periportal and peri-central venous transduction in newborn mice.45 Unfortunately, we are unable to directly evaluate the percentage or location of gene-corrected hepatocytes in treated liver by immunohistology, because the Pahenu2/enu2 model produces abundant, albeit inactive, PAH protein; no available antibody is able to distinguish between active and inactive PAH protein. Further studies using single-cell omics would be informative for understanding the effects of this therapy at the level of individual cells.
While the Pahenu2/enu2 mouse recapitulates human cPKU phenotypically, this isogenic model does not capture the wide range of almost 1,000 known pathogenic variants in the PAH gene.46 One limitation of CRISPR/Cas9-mediated gene editing as we have used it here or as others have used a CRISPR/Cas9 base-editor fusion47 is that a different guide RNA—and, therefore, one of the two AAV vectors—would have to be redesigned for each patient with a novel mutation, or at least there would need to be unique reagents for each exon of the Pah gene. Future development of CRISPR/Cas9 gene editing should be designed to be broadly applicable to all human PKU genotypes and should aim to drive Phe levels down to WT levels. While this study was a beginning for CRISPR/Cas9 gene editing for PKU, further studies are needed before in vivo gene-editing translation into human medicine.
Materials and Methods
Animal Husbandry
Animal care and experimentation were performed in accordance with the guidelines of the Department of Comparative Medicine, Oregon Health & Science University, and the NIH Guide for the Care and Use of Laboratory Animals. C57BL/6-Pahenu2/enu2 mice, which are homozygous for a missense c.835T > C (p.F263S) mutation in exon 7 of the murine Pah gene, are completely deficient in liver PAH activity; are, consequently, hyperphenylalaninemic on an unrestricted diet; and are a representative animal model of human PKU.48 Neonatal Pahenu2/enu2 mice for these experiments were generated through breeding of Pahenu2/enu2 sires to Pahenu2/+ dams. Genotyping for the presence of the Pahenu2 mutation was performed by a TaqMan qPCR assay. All mice were fed tap water and standard mouse chow (LabDiet Picolab Rodent Diet 5LOD, St. Louis, MO, USA) ad libitum, providing approximately 24% protein and 1.04% L-Phe by weight, except the breeders that received high-energy chow (LabDiet Rodent High Energy Diet 5058, St. Louis, MO, USA), providing approximately 22% protein and 0.99% L-Phe. Given that adult mice consume approximately 5 g chow per day, daily L-Phe intake was estimated to be approximately 50 mg/day. The animals were housed under a standard 12-h:12-h on-off light cycle. All surgical procedures were carried out with inhaled isoflurane general anesthesia to minimize pain and discomfort.
Guide Design and Validation
The sequence flanking c.835T > C was entered into the MIT Guide Design tool. Three guides were chosen for validation using the cell-culture-based Surveyor system and revalidated with the in vitro Takara Guide-It Kit (Figure S1).
Viral Production
Plasmid AAV2 LSP-Cas9 was graciously supplied by the Grompe laboratory, and the AAV2 repair template with guide plasmid was synthesized by GeneScript. Plasmids were confirmed for overall integrity, and intact Inverted Terminal Repeats (ITRs) via RE screen using Taq1 and Ahd1, respectively. The OHSU Molecular Virology Support Core produced large-scale preparations of recombinant rAAV2/8 viruses, referenced as rAAV2/8_LSP_SpCas9 and AAV2/8_Ex7RepairTemp_U6G1, using standard triple plasmid transfection procedures into cultured HEK293 cells and purified by iodixanol gradient ultracentrifugation.49 AAV titers were determined using ITR-based qPCR analysis.
Vector Administration and Vanillin Treatment
Post-natal day 3 Pahenu2 mice were injected through the facial vein with 2.1 × 1011 vg AAV2/8_LSP_SpCas9 and 3.3 × 1011 vg AAV2/8_Ex7RepairTemp_U6G1 in a single injection, with a total volume of 10 μL and administered daily intraperitoneal injections of 100 mg/kg vanillin for 7 days, according to treatment designation. Mice were weaned at 4 weeks of age. Between 5 and 8 weeks of age, mice underwent a retro-orbital bleed to collect plasma using heparinized capillary tubes followed by a tail vein injection of 2.1 × 1012 vg AAV2/8_LSP_SpCas9 and 3.3 × 1012 vg AAV2/8_Ex7RepairTemp_U6G1 in a single injection, with a total volume of 100 μL and administered daily intraperitoneal injections of 100 mg/kg vanillin for 7 days, according to treatment designation.
Euthanasia and Tissue Harvest
Animals were sedated using inhaled isoflurane anesthesia. Whole blood was collected by cardiac puncture and allowed to clot in an Eppendorf tube, and serum was separated by centrifugation. The mice were then euthanized by exsanguination and perfused with 20 mL normal saline via the left cardiac ventricle to clear blood from the liver.
Serum Phenylalanine
Serum phenylalanine was determined using an established fluorometric protocol.50
Liver PAH Enzyme Activity
PAH enzyme activity was determined using an established radioactive chromatography assay,51 with modifications.52
Liver DNA Extraction
Separate liver biopsies, approximately 7 mg each, were collected from four different lobes for each animal and collectively put through the QIAGEN DNeasy Blood & Tissue Kit according to the manufacturer’s protocol.
Screening for Viral DNA
Liver DNA was screened for the presence of viral DNA by standard bench-top PCR using primers specific to each transgene. A portion of Cas9 sequence was amplified from the Cas9 vector (forward primer: 5′-CAGCCAGGAAGAGTTCTACAAG-3′; reverse primer: 5′-CATTCCCTCGGTCACGTATTT-3′), and the human U6 sequence was amplified from the repair template vector (forward primer: 5′-GAGGGCCTATTTCCCATGATT-3′; reverse primer: 5′-TGTTTCGTCCTTTCCACAAGATA-3′).
On-Target PCR Amplicon Preparation
The QIAGEN LongRange PCR Kit was used according to the manufacturer’s protocol on genomic liver DNA using a forward primer targeting genomic DNA 58 bp upstream of the 5′ end of the repair template sequence, 5′-AGTTACTGTCGTTTGCAATGCCGC-3′, and a reverse primer, 5′-GCACAGTAGCCACTAATTCTCTCCTTAG-3′, located 426 bp downstream of the C836T mutation within the repair template region. A secondary nested PCR was performed using the 5′-GGGTTGTAGTCTCTCTGGATTTACCA-3′ forward primer and 5′-GCACAGTAGCCACTAATTCTCTCCTTAG-3′ reverse primer with Invitrogen Platinum Taq DNA Polymerase according to the manufacturer’s recommendations.
On- and Off-Target NGS
Off-target sites were selected using two in silico tools: Cas-OFFinder and COSMID.53,54 Three guides present in both algorithms scored with the highest probability of off-target cutting were selected: Chr10:116187094 (COSMID score, 0.17), Chr12:60323919 (COSMID score, 1.33), and ChrX:140587188 (COSMID score, 2.93), as well as the target site Chr10: 87570342 (COSMID score, 0.00). COSMID-designed Illumina primers were ordered with adaptor sequences developed by the O’Roak lab. The four regions were PCR amplified for each animal with the Invitrogen Platinum Taq DNA Polymerase PCR Kit using genomic liver DNA according to the manufacturer’s protocol. Products were run on a 1.2% agar gel and purified with the QIAGEN PCR Purification Kit according to the manufacturer’s protocol. A secondary PCR was performed to index each animal with a unique barcode using primers developed by the O’Roak lab using the Invitrogen Platinum Taq DNA Polymerase PCR Kit for 6 cycles. Products were run on a 1.2% gel and purified with the QIAGEN QIAquick PCR Purification Kit. Samples were quantified using the Epoch Microplate Spectrophotometer and pooled in 1:1 ratios for each region. Pooled samples were then quantified with the Qubit 4 Fluorometer and diluted to 2-nM concentrations. Each region was then combined in a 1:1 ratio in 20 μL and sent to the Molecular Technologies Core at Oregon Health and Science University. Samples were prepared with the V2 500 kit and run on the Illumina MiSeq instrument that downloaded raw data in FASTQ format to Illumina Base Space.
MiSeq Sequencing Data Analysis
FASTQ files generated by NGS were split by index using Python, paired-end reads were merged with PEAR v0.9.6,55 and common primer sequences were trimmed using Cutadapt v1.1856 as previously described.57 Reads were split into each region for each animal by match to the first 25 bp in each amplicon. Data were analyzed in CRISPResso using the Pahenu2 allele as the reference sequence for the target region and mouse reference for each off-target region; indels were called with >1-bp indel mismatch to the reference, and HDR was called with a 98% identity to the repair template sequence.58 In addition, on-target data were run for percent match to the functional coding sequence of the PAH exon 7 region.
Sanger Sequencing
The purified on-target PCR product described earlier was quantified using the Epoch Microplate Spectrophotometer and sent for standard Sanger sequencing with the 5′-GGGTTGTAGTCTCTCTGGATTTACCA-3′ forward primer on an ABI 3130XL sequencer at the Vollum DNA Services Core at Oregon Health & Science University.
Author Contributions
D.Y.R. performed the experiments, analyzed data, and wrote the manuscript. S.R.W. performed the surgical procedures and assisted with mouse breeding, molecular techniques, and experimental design. S.D. assisted with mouse breeding and molecular techniques. S.N. assisted with experimental design and reagents. T.L.M. assisted with NGS preparation and bioinformatic analyses. M.G. assisted with reagents, experimental design, analyses, data interpretation, and manuscript editing and oversaw D.Y.R. as a dissertation advisory committee member. C.O.H. assisted with funding, experimental design, analyses, data interpretation, and manuscript editing and was primary mentor to D.Y.R. during graduate school.
Acknowledgments
D.Y.R. and the experiments were funded by the National PKU Alliance, USA. S.R.W., S.D., and C.O.H. were funded by Na National Institutes of Health (USA) grant RO1 NS080866. We would like to acknowledge the O’Roak laboratory for supplying us with NGS expertise and reagents. We would also like to express our gratitude to the National PKU Alliance for supporting this research.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtm.2019.12.004.
Supplemental Information
References
- 1.Blau N. Genetics of phenylketonuria: then and now. Hum. Mutat. 2016;37:508–515. doi: 10.1002/humu.22980. [DOI] [PubMed] [Google Scholar]
- 2.Flydal M.I., Martinez A. Phenylalanine hydroxylase: function, structure, and regulation. IUBMB Life. 2013;65:341–349. doi: 10.1002/iub.1150. [DOI] [PubMed] [Google Scholar]
- 3.Cazzorla C., Bensi G., Biasucci G., Leuzzi V., Manti F., Musumeci A., Papadia F., Stoppioni V., Tummolo A., Vendemiale M. Living with phenylketonuria in adulthood: the PKU ATTITUDE study. Mol. Genet. Metab. Rep. 2018;16:39–45. doi: 10.1016/j.ymgmr.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blau N., Shen N., Carducci C. Molecular genetics and diagnosis of phenylketonuria: state of the art. Expert Rev. Mol. Diagn. 2014;14:655–671. doi: 10.1586/14737159.2014.923760. [DOI] [PubMed] [Google Scholar]
- 5.Lichter-Konecki U., Vockley J. Phenylketonuria: current treatments and future developments. Drugs. 2019;79:495–500. doi: 10.1007/s40265-019-01079-z. [DOI] [PubMed] [Google Scholar]
- 6.Vockley J., Andersson H.C., Antshel K.M., Braverman N.E., Burton B.K., Frazier D.M., Mitchell J., Smith W.E., Thompson B.H., Berry S.A., American College of Medical Genetics and Genomics Therapeutics Committee Phenylalanine hydroxylase deficiency: diagnosis and management guideline. Genet. Med. 2014;16:188–200. doi: 10.1038/gim.2013.157. [DOI] [PubMed] [Google Scholar]
- 7.Singh R.H., Cunningham A.C., Mofidi S., Douglas T.D., Frazier D.M., Hook D.G., Jeffers L., McCune H., Moseley K.D., Ogata B. Updated, web-based nutrition management guideline for PKU: An evidence and consensus based approach. Mol. Genet. Metab. 2016;118:72–83. doi: 10.1016/j.ymgme.2016.04.008. [DOI] [PubMed] [Google Scholar]
- 8.Greene C.L., Longo N. National Institutes of Health (NIH) review of evidence in phenylalanine hydroxylase deficiency (phenylketonuria) and recommendations/guidelines for therapy from the American College of Medical Genetics (ACMG) and Genetics Metabolic Dietitians International (GMDI) Mol. Genet. Metab. 2014;112:85–86. doi: 10.1016/j.ymgme.2014.03.005. [DOI] [PubMed] [Google Scholar]
- 9.Brown C.S., Lichter-Konecki U. Phenylketonuria (PKU): a problem solved? Mol. Genet. Metab. Rep. 2015;6:8–12. doi: 10.1016/j.ymgmr.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding X.Q., Fiehler J., Kohlschütter B., Wittkugel O., Grzyska U., Zeumer H., Ullrich K. MRI abnormalities in normal-appearing brain tissue of treated adult PKU patients. J. Magn. Reson. Imaging. 2008;27:998–1004. doi: 10.1002/jmri.21289. [DOI] [PubMed] [Google Scholar]
- 11.Feldmann R., Osterloh J., Onon S., Fromm J., Rutsch F., Weglage J. Neurocognitive functioning in adults with phenylketonuria: report of a 10-year follow-up. Mol. Genet. Metab. 2019;126:246–249. doi: 10.1016/j.ymgme.2018.12.011. [DOI] [PubMed] [Google Scholar]
- 12.Stone W.L., Basit H., Los E. StatPearls. StatPearls Publishing; 2019. Phenylketonuria.https://www.ncbi.nlm.nih.gov/books/NBK535378/ [Google Scholar]
- 13.Gupta S., Lau K., Harding C.O., Shepherd G., Boyer R., Atkinson J.P., Knight V., Olbertz J., Larimore K., Gu Z. Association of immune response with efficacy and safety outcomes in adults with phenylketonuria administered pegvaliase in phase 3 clinical trials. EBioMedicine. 2018;37:366–373. doi: 10.1016/j.ebiom.2018.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harding C.O., Amato R.S., Stuy M., Longo N., Burton B.K., Posner J., Weng H.H., Merilainen M., Gu Z., Jiang J., Vockley J., PRISM-2 Investigators Pegvaliase for the treatment of phenylketonuria: a pivotal, double-blind randomized discontinuation phase 3 clinical trial. Mol. Genet. Metab. 2018;124:20–26. doi: 10.1016/j.ymgme.2018.03.003. [DOI] [PubMed] [Google Scholar]
- 15.Longo N., Dimmock D., Levy H., Viau K., Bausell H., Bilder D.A., Burton B., Gross C., Northrup H., Rohr F. Evidence- and consensus-based recommendations for the use of pegvaliase in adults with phenylketonuria. Genet. Med. 2019;21:1851–1867. doi: 10.1038/s41436-018-0403-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thomas J., Levy H., Amato S., Vockley J., Zori R., Dimmock D., Harding C.O., Bilder D.A., Weng H.H., Olbertz J., PRISM investigators Pegvaliase for the treatment of phenylketonuria: results of a long-term phase 3 clinical trial program (PRISM) Mol. Genet. Metab. 2018;124:27–38. doi: 10.1016/j.ymgme.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 17.Harding C.O., Gillingham M.B., Hamman K., Clark H., Goebel-Daghighi E., Bird A., Koeberl D.D. Complete correction of hyperphenylalaninemia following liver-directed, recombinant AAV2/8 vector-mediated gene therapy in murine phenylketonuria. Gene Ther. 2006;13:457–462. doi: 10.1038/sj.gt.3302678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ding Z., Harding C.O., Rebuffat A., Elzaouk L., Wolff J.A., Thöny B. Correction of murine PKU following AAV-mediated intramuscular expression of a complete phenylalanine hydroxylating system. Mol. Ther. 2008;16:673–681. doi: 10.1038/mt.2008.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mingozzi F., High K.A. Therapeutic in vivo gene transfer for genetic disease using AAV: progress and challenges. Nat. Rev. Genet. 2011;12:341–355. doi: 10.1038/nrg2988. [DOI] [PubMed] [Google Scholar]
- 20.Hamman K., Clark H., Montini E., Al-Dhalimy M., Grompe M., Finegold M., Harding C.O. Low therapeutic threshold for hepatocyte replacement in murine phenylketonuria. Mol. Ther. 2005;12:337–344. doi: 10.1016/j.ymthe.2005.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Embury J.E., Charron C.E., Martynyuk A., Zori A.G., Liu B., Ali S.F., Rowland N.E., Laipis P.J. PKU is a reversible neurodegenerative process within the nigrostriatum that begins as early as 4 weeks of age in Pah(enu2) mice. Brain Res. 2007;1127:136–150. doi: 10.1016/j.brainres.2006.09.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu P.D., Lander E.S., Zhang F. Development and applications of CRISPR-Cas9 for genome engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Y., Wang L., Bell P., McMenamin D., He Z., White J., Yu H., Xu C., Morizono H., Musunuru K. A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat. Biotechnol. 2016;34:334–338. doi: 10.1038/nbt.3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paulk N.K., Loza L.M., Finegold M.J., Grompe M. AAV-mediated gene targeting is significantly enhanced by transient inhibition of nonhomologous end joining or the proteasome in vivo. Hum. Gene Ther. 2012;23:658–665. doi: 10.1089/hum.2012.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shedlovsky A., McDonald J.D., Symula D., Dove W.F. Mouse models of human phenylketonuria. Genetics. 1993;134:1205–1210. doi: 10.1093/genetics/134.4.1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fiori E., Oddi D., Ventura R., Colamartino M., Valzania A., D’Amato F.R., Bruinenberg V., van der Zee E., Puglisi-Allegra S., Pascucci T. Early-onset behavioral and neurochemical deficits in the genetic mouse model of phenylketonuria. PLoS ONE. 2017;12:e0183430. doi: 10.1371/journal.pone.0183430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Winn S.R., Scherer T., Thöny B., Ying M., Martinez A., Weber S., Raber J., Harding C.O. Blood phenylalanine reduction corrects CNS dopamine and serotonin deficiencies and partially improves behavioral performance in adult phenylketonuric mice. Mol. Genet. Metab. 2018;123:6–20. doi: 10.1016/j.ymgme.2017.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuah M.K., Petrus I., De Bleser P., Le Guiner C., Gernoux G., Adjali O., Nair N., Willems J., Evens H., Rincon M.Y. Liver-specific transcriptional modules identified by genome-wide in silico analysis enable efficient gene therapy in mice and non-human primates. Mol. Ther. 2014;22:1605–1613. doi: 10.1038/mt.2014.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nair N., Rincon M.Y., Evens H., Sarcar S., Dastidar S., Samara-Kuko E., Ghandeharian O., Man Viecelli H., Thöny B., De Bleser P. Computationally designed liver-specific transcriptional modules and hyperactive factor IX improve hepatic gene therapy. Blood. 2014;123:3195–3199. doi: 10.1182/blood-2013-10-534032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald J.D., Dyer C.A., Gailis L., Kirby M.L. Cardiovascular defects among the progeny of mouse phenylketonuria females. Pediatr. Res. 1997;42:103–107. doi: 10.1203/00006450-199707000-00016. [DOI] [PubMed] [Google Scholar]
- 31.Cunningham S.C., Spinoulas A., Carpenter K.H., Wilcken B., Kuchel P.W., Alexander I.E. AAV2/8-mediated correction of OTC deficiency is robust in adult but not neonatal Spf(ash) mice. Mol. Ther. 2009;17:1340–1346. doi: 10.1038/mt.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang L., Bell P., Lin J., Calcedo R., Tarantal A.F., Wilson J.M. AAV8-mediated hepatic gene transfer in infant rhesus monkeys (Macaca mulatta) Mol. Ther. 2011;19:2012–2020. doi: 10.1038/mt.2011.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Platt L.D., Koch R., Hanley W.B., Levy H.L., Matalon R., Rouse B., Trefz F., de la Cruz F., Güttler F., Azen C., Friedman E.G. The international study of pregnancy outcome in women with maternal phenylketonuria: report of a 12-year study. Am. J. Obstet. Gynecol. 2000;182:326–333. doi: 10.1016/s0002-9378(00)70219-5. [DOI] [PubMed] [Google Scholar]
- 34.Zeile W.L., McCune H.C., Musson D.G., O’Donnell B., O’Neill C.A., Tsuruda L.S., Zori R.T., Laipis P.J. Maternal phenylketonuria syndrome: studies in mice suggest a potential approach to a continuing problem. Pediatr. Res. 2018;83:889–896. doi: 10.1038/pr.2017.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceccaldi R., Rondinelli B., D’Andrea A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016;26:52–64. doi: 10.1016/j.tcb.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang H.H.Y., Pannunzio N.R., Adachi N., Lieber M.R. Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 2017;18:495–506. doi: 10.1038/nrm.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohmori T., Nagao Y., Mizukami H., Sakata A., Muramatsu S.I., Ozawa K., Tominaga S.I., Hanazono Y., Nishimura S., Nureki O., Sakata Y. CRISPR/Cas9-mediated genome editing via postnatal administration of AAV vector cures haemophilia B mice. Sci. Rep. 2017;7:4159. doi: 10.1038/s41598-017-04625-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yin H., Xue W., Chen S., Bogorad R.L., Benedetti E., Grompe M., Koteliansky V., Sharp P.A., Jacks T., Anderson D.G. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat. Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tham K.C., Kanaar R., Lebbink J.H.G. Mismatch repair and homeologous recombination. DNA Repair (Amst.) 2016;38:75–83. doi: 10.1016/j.dnarep.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 40.Hamman K.J., Winn S.R., Harding C.O. Hepatocytes from wild-type or heterozygous donors are equally effective in achieving successful therapeutic liver repopulation in murine phenylketonuria (PKU) Mol. Genet. Metab. 2011;104:235–240. doi: 10.1016/j.ymgme.2011.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duncan A.W., Taylor M.H., Hickey R.D., Hanlon Newell A.E., Lenzi M.L., Olson S.B., Finegold M.J., Grompe M. The ploidy conveyor of mature hepatocytes as a source of genetic variation. Nature. 2010;467:707–710. doi: 10.1038/nature09414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duncan A.W. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin. Cell Dev. Biol. 2013;24:347–356. doi: 10.1016/j.semcdb.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Kurinna S., Stratton S.A., Coban Z., Schumacher J.M., Grompe M., Duncan A.W., Barton M.C. p53 regulates a mitotic transcription program and determines ploidy in normal mouse liver. Hepatology. 2013;57:2004–2013. doi: 10.1002/hep.26233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trefts E., Gannon M., Wasserman D.H. The liver. Curr. Biol. 2017;27:R1147–R1151. doi: 10.1016/j.cub.2017.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bell P., Wang L., Gao G., Haskins M.E., Tarantal A.F., McCarter R.J., Zhu Y., Yu H., Wilson J.M. Inverse zonation of hepatocyte transduction with AAV vectors between mice and non-human primates. Mol. Genet. Metab. 2011;104:395–403. doi: 10.1016/j.ymgme.2011.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Himmelreich N., Shen N., Okun J.G., Thiel C., Hoffmann G.F., Blau N. Relationship between genotype, phenylalanine hydroxylase expression and in vitro activity and metabolic phenotype in phenylketonuria. Mol. Genet. Metab. 2018;125:86–95. doi: 10.1016/j.ymgme.2018.06.011. [DOI] [PubMed] [Google Scholar]
- 47.Villiger L., Grisch-Chan H.M., Lindsay H., Ringnalda F., Pogliano C.B., Allegri G., Fingerhut R., Häberle J., Matos J., Robinson M.D. Treatment of a metabolic liver disease by in vivo genome base editing in adult mice. Nat. Med. 2018;24:1519–1525. doi: 10.1038/s41591-018-0209-1. [DOI] [PubMed] [Google Scholar]
- 48.McDonald J.D., Bode V.C., Dove W.F., Shedlovsky A. Pahhph-5: a mouse mutant deficient in phenylalanine hydroxylase. Proc. Natl. Acad. Sci. USA. 1990;87:1965–1967. doi: 10.1073/pnas.87.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kleven M.D., Gomes M.M., Wortham A.M., Enns C.A., Kahl C.A. Ultrafiltered recombinant AAV8 vector can be safely administered in vivo and efficiently transduces liver. PLoS ONE. 2018;13:e0194728. doi: 10.1371/journal.pone.0194728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McCaman M.W., Robins E. Fluorimetric method for the determination of phenylalanine in serum. J. Lab. Clin. Med. 1962;59:885–890. [Google Scholar]
- 51.Ledley F.D., Hahn T., Woo S.L. Selection for phenylalanine hydroxylase activity in cells transformed with recombinant retroviruses. Somat. Cell Mol. Genet. 1987;13:145–154. doi: 10.1007/BF01534694. [DOI] [PubMed] [Google Scholar]
- 52.Harding C.O., Wild K., Chang D., Messing A., Wolff J.A. Metabolic engineering as therapy for inborn errors of metabolism--development of mice with phenylalanine hydroxylase expression in muscle. Gene Ther. 1998;5:677–683. doi: 10.1038/sj.gt.3300653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bae S., Park J., Kim J.S. Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cradick T.J., Qiu P., Lee C.M., Fine E.J., Bao G. COSMID: a web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol. Ther. Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang J., Kobert K., Flouri T., Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics. 2014;30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. 2011;17:10–12. [Google Scholar]
- 57.Mighell T.L., Evans-Dutson S., O’Roak B.J. A saturation mutagenesis approach to understanding PTEN lipid phosphatase activity and genotype-phenotype relationships. Am. J. Hum. Genet. 2018;102:943–955. doi: 10.1016/j.ajhg.2018.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pinello L., Canver M.C., Hoban M.D., Orkin S.H., Kohn D.B., Bauer D.E., Yuan G.C. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.