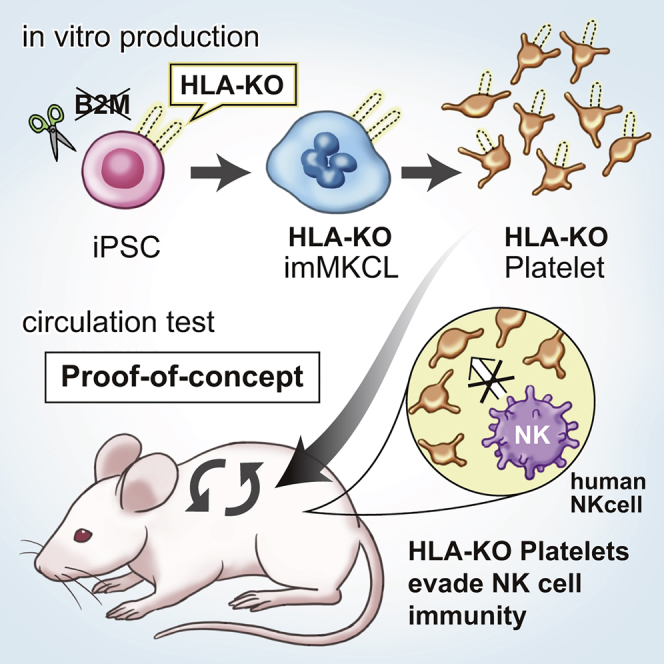
Summary
The ex vivo production of platelets depleted of human leukocyte antigen class I (HLA-I) could serve as a universal measure to overcome platelet transfusion refractoriness caused by HLA-I incompatibility. Here, we developed human induced pluripotent cell-derived HLA-I-deficient platelets (HLA-KO iPLATs) in a clinically applicable imMKCL system by genetic manipulation and assessed their immunogenic properties including natural killer (NK) cells, which reject HLA-I downregulated cells. HLA-KO iPLATs were deficient for all HLA-I but did not elicit a cytotoxic response by NK cells in vitro and showed circulation equal to wild-type iPLATs upon transfusion in our newly established Hu-NK-MSTRG mice reconstituted with human NK cells. Additionally, HLA-KO iPLATs successfully circulated in an alloimmune platelet transfusion refractoriness model of Hu-NK-MISTRG mice. Mechanistically, the lack of NK cell-activating ligands on platelets may be responsible for evading the NK cell response. This study revealed the unique non-immunogenic property of platelets and provides a proof of concept for the clinical application of HLA-KO iPLATs.
Keywords: platelet, megakaryocyte, iPSC, HLA class I, natural killer cell, regenerative medicine, imMKCL, platelet transfusion, refractoriness, MSTRG mice, IL-15
Graphical Abstract
Highlights
-
•
Clinically applicable iPSC-derived HLA class I knockout platelets (HLA-KO iPLATs)
-
•
HLA-KO iPLATs do not elicit NK cell activation in vitro
-
•
HLA-KO iPLATs circulate comparably with wild type in human NK cell-reconstituted mice
-
•
HLA-KO iPLATs circulate competently in alloimmune PTR model mice
T cells and antibodies or NK cells reject HLA-I-incompatible or HLA-I-deficient cells, respectively. Sugimoto and colleagues produced HLA-I null platelets from iPSCs in a clinically applicable system and found that platelets can circulate even in human NK cell-reconstituted mice. Potentially, universal HLA-KO platelets could treat patients suffering from platelet transfusion refractoriness caused by HLA-I incompatibility.
Introduction
Platelet transfusion is an essential treatment for patients with thrombocytopenia (Szczepiorkowski and Dunbar, 2013, Estcourt et al., 2017). The standard blood source for transfusion has been blood donors for many decades, and guidelines have been set to optimize transfusion conditions. However, this donation system requires large efforts to keep supply-demand balance due to the short shelf life of the platelet products (i.e., 4 days in Japan and 5 days in the United States), the anticipated absolute shortage in the near future in countries with aging societies, and the risk of blood-borne infection. Moreover, alloimmune platelet transfusion refractoriness (allo-PTR) is observed in approximately 5%–15% of patients who receive platelet transfusion, with the most dominant cause being the production of alloantibodies against human leukocyte antigen class I (HLA-I), which includes A, B, and C antigens (Pavenski et al., 2013, Saito et al., 2002, Stanworth et al., 2015). In such HLA-I-mediated allo-PTR, transfused platelets are immediately rejected, except for HLA-I-compatible platelets. However, the need to select compatible donors limits the supply, with the most difficult cases being rare HLA-I types.
Human induced pluripotent stem cells (iPSCs) have been extensively studied as an ex vivo source for producing human cells and tissues (Karagiannis and Eto, 2016), and iPSC-derived platelets have the potential to resolve the aforementioned issues in current transfusion systems (Sugimoto and Eto, 2017). They can be produced without donor dependency and with good manufacturing practice from pathogen-free assured master cells devoid of blood-borne infections. As an expandable master cell source for platelets, we previously established immortalized megakaryocyte progenitor cell lines (imMKCLs) from human iPSCs, whereby the selectively qualified iPSC clone-derived imMKCLs can be prepared beforehand (Nakamura et al., 2014). To create imMKCLs, in the megakaryocyte (MK)-lineage differentiation from iPSCs, three doxycycline (DOX)-inducible transgenes, c-MYC, BCL-XL, and BMI1, are introduced. In the presence of DOX, imMKCLs proliferate, whereas depletion of DOX from the culture medium leads to the maturation of imMKCLs to produce platelets (which we call iPLATs). We recently succeeded at producing the clinically required order of competent iPLATs by maturing imMKCLs under turbulent flow conditions consisting of optimal rages of turbulent energy and shear stress (Ito et al., 2018). From the perspective of alloimmune compatibility, the ideal platelet product is autologous. Because iPSCs can be autologous, so too can iPLATs, but autologous products have high cost and are of variable quality. Therefore, the production of HLA-I homologous platelets as an off-the-shelf product from a clinically applicable iPSC library is under consideration (Gourraud et al., 2012, Turner et al., 2013). However, due to high variability of the HLA-I gene, there is still a need to prepare a wide line-up of HLA-I types.
Platelets depleted of HLA-I can potentially be used as a universally effective transfusion measure for HLA-I-mediated allo-PTR. For depletion of HLA-I, genetic manipulation of the HLA-I complex molecule β2-microglobulin (B2M) has been shown to be effective (Gras et al., 2013, Riolobos et al., 2013, Feng et al., 2014). The knockout procedure can completely deplete HLA-I expression for all A, B, C, and E antigens (“HLA-KO”), but this effect can also lead to the activation of human natural killer (NK) cells, which commit cytotoxic activity against cells downregulated of HLA-I (Lanier, 2008, Vivier et al., 2011, Long et al., 2013). Therefore, strategies to overexpress single-chain HLA-E fused with B2M (Gornalusse et al., 2017) or to disrupt HLA-A/B but retain HLA-C (Xu et al., 2019) have been proposed. However, allo-PTR due to anti-HLA-C has been reported (Saito et al., 2002). Furthermore, whether HLA-KO platelets activate NK cells remains unaddressed, including in vivo models with highly reconstituted human NK cells in circulation. In the present study, we produced HLA-KO iPLATs by knocking out B2M using the CRISPR/Cas9 method in our clinically applicable imMKCL system and evaluated their in vitro functionality and immunogenicity to NK cells. We also succeeded in establishing humanized mice with a high reconstitution of human NK cells by using MSTRG mice injected with interleukin-15 (IL-15) ligand and IL-15 receptor (Hu-NK-MSTRG mice) and assessed the circulation of HLA-KO iPLATs in vivo.
Results
HLA-Null Platelets Were Successfully Produced from a β2-Microglobulin-Knockout imMKCL
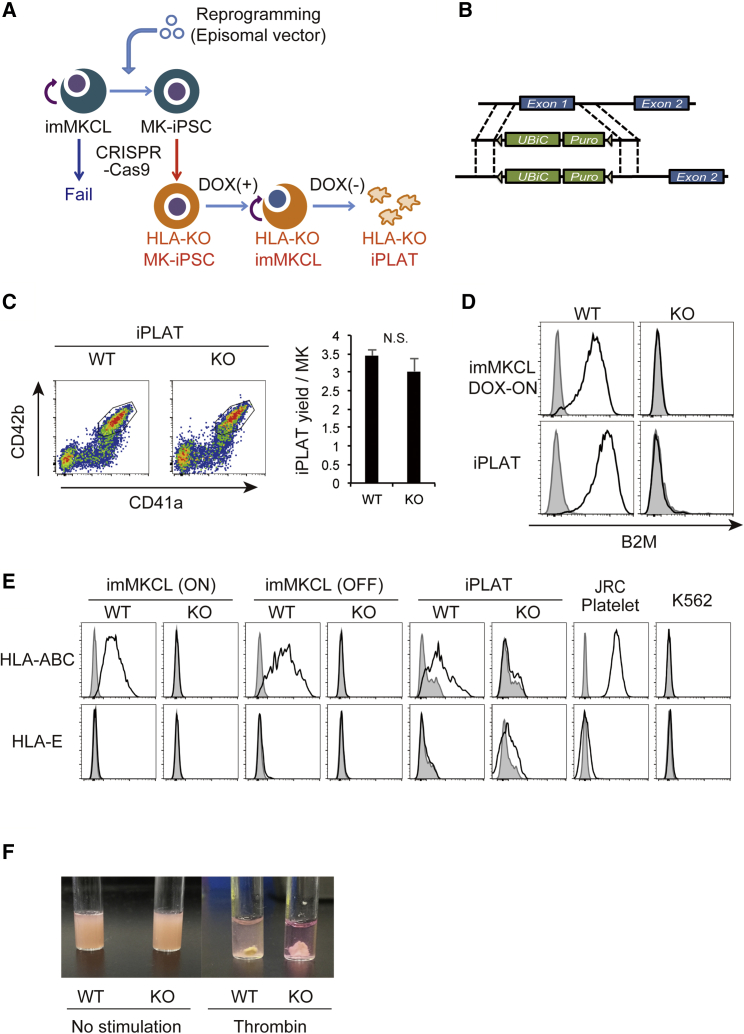
To produce platelets without HLA-I expression, we sought to create HLA-I-depleted imMKCLs by knocking out the B2M gene. Because we did not succeed in genome editing the imMKCLs, we adopted the “re-reprogramming” method (Seo et al., 2018), whereby imMKCLs are first reprogrammed to iPSCs (MK-iPSCs) and then subjected to B2M knockout using CRISPR/Cas9 technology (Figures 1A and 1B). Here, we used already established imMKCLs, which are highly proliferative and have high iPLAT production capacity, as the starting material, assuring the derivation of high-quality imMKCLs with the B2M-KO trait. These B2M-knockout MK-iPSCs bear the DOX-inducible c-MYC, BCL-XL, and BMI1 transgenes of the original imMKCLs and were reinduced to imMKCLs (HLA-KO imMKCLs) and expanded in MK-differentiating medium including DOX (Figure 1A).
Figure 1.
Production of HLA-KO iPLATs by Knocking Out β2-Microglobulin in imMKCL
(A) Schema of the HLA-KO platelet production procedure. Knockout of β2-microglobulin (B2M) by CRISPR/Cas9 failed in imMKCL. Therefore, imMKCL was first re-reprogrammed to secondary iPSCs (MK-iPSC), in which B2M was knocked out. MK-iPSCs were then reinduced to imMKCL (HLA-KO imMKCL) in the presence of doxycycline (DOX) and, after expansion, matured to release iPLATs in DOX-OFF condition.
(B) The targeting strategy of knocking out B2M by replacing exon 1 to a UBiC promoter-regulated puromycin-resistant gene for HLA-I nullification.
(C–E) Flow-cytometry analysis of the generated CD41a+CD42b+ iPLATs and their yield (C), and the cell-surface expression of B2M (D) and of HLA-ABC and HLA-E (E) on imMKCLs, iPLATs, JRC platelets, and K562 cells. Gray histograms in (D) and (E) represent no staining control.
(F) Clot retraction assay of iPLATs.
WT, wild type; KO, HLA-KO; JRC, Japanese Red Cross; N.S., not significant. Data are representative of three independent experiments with error bars representing the mean ± SEM. See also Figure S1.
The production of CD41a+CD42b+ iPLATs from HLA-KO imMKCLs was comparable with the wild-type (WT) counterpart (Figure 1C). HLA-KO iPLATs were confirmed to lack the surface expression of B2M and HLA-I molecules (Figures 1D and 1E). The cell-surface characteristics of HLA-KO iPLATs were comparable with those of WT iPLATs, donor platelets provided from the Japanese Red Cross Society (JRC), and peripheral blood platelets from healthy donors, as shown by the levels of human platelet antigens (HPAs) (Figure S1A). The cell size and ultrastructure of HLA-KO iPLATs were comparable with those of WT iPLATs (Figures S1B and S1C), which have a similar ultrastructure to JRC platelets but are slightly larger, as reported previously (Ito et al., 2018). The in vitro functionality of HLA-KO iPLATs was also comparable, as shown by the low level of Annexin V binding and high level of hallmarks of in vitro platelet activation, namely, PAC-1 binding and CD62P expression upon stimulation (Figures S1D–S1F). Finally, HLA-KO iPLATs and WT iPLATs were comparable for clotting (Figure 1F). These data indicate that the knockout procedure did not affect the production efficiency or function of iPLATs.
NK Cells Do Not Show Cytotoxic Response against iPLATs Regardless of HLA-I Expression
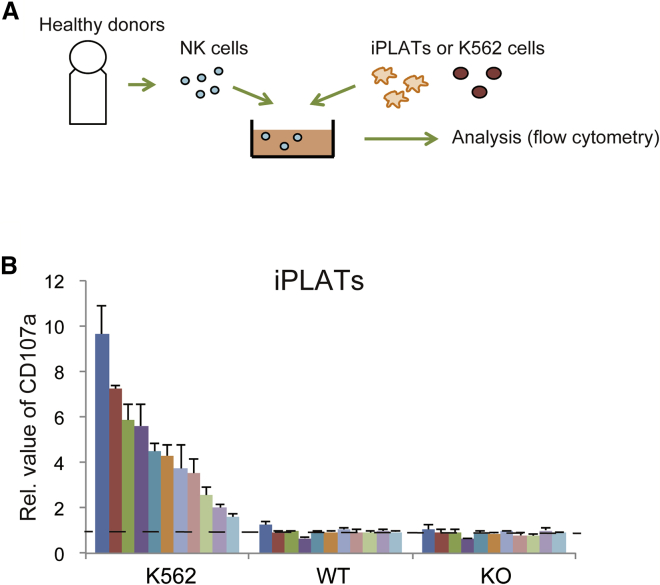
To assess whether iPLATs of HLA-KO phenotype preferentially elicit a cytotoxic response by NK cells, we performed co-culture assays in vitro (Figure 2A). NK cells separated from the peripheral blood mononuclear cells (PBMCs) of 11 healthy donors (Table 1) were each co-cultured with iPLATs for 6 h. The response of the NK cells was assessed by measuring the expression of CD107a, which reflects the release of cytotoxic granules from NK cells (Alter et al., 2004). The expression of CD107a was highly enhanced against K562 cells, a typical positive control leukemic cell line used for cytotoxic assays, but just baseline level against WT and HLA-KO iPLATs (Figures 2B and S2A). We also tested the interferon-γ (IFN-γ) secretion from NK cells, finding the response, like CD107a, to be high against K562 cells, but baseline level against iPLATs (Figure S2B). Furthermore, Annexin V binding on iPLATs was not observed, suggesting no cellular damage to iPLATs (Figure S2C). Thus, the degranulation activity of NK cells was not induced by iPLATs regardless of HLA-I expression.
Figure 2.
In Vitro Cytotoxic Assay of NK Cells Co-cultured with iPLATs or imMKCLs
(A) Schema of the in vitro cytotoxic assay in vitro. NK cells and target cells were co-cultured with 1,000 U/mL human IL-2 for 6 h and then analyzed.
(B) Flow-cytometry analysis of CD107a expression on NK cells from 11 healthy volunteers co-cultured with K562 cells, WT, or HLA-KO iPLATs. WT, wild type; KO, HLA-KO. Data are the mean ± SEM of three independent experiments.
See also Figure S2.
Table 1.
HLA Class I Profiles of the imMKCL and NK Cell Donors (A–K)
| HLA-A | HLA-B | HLA-C | C1 | C2 | |
|---|---|---|---|---|---|
| imMKCL | 02:01 | 15:01 | 02:02 | + | |
| 02:01 | 15:01 | 03:03 | + | ||
| A | 02:01 | 40:01 | 03:04 | + | |
| 24:02 | 15:01 | 03:03 | + | ||
| B | 02:01 | 07:02 | 03:04 | + | |
| 33:03 | 40:01 | 07:02 | + | ||
| C | 24:02 | 07:02 | 07:02 | + | |
| 33:03 | 58:01 | 03:02 | + | ||
| D | 11:01 | 51:01 | 15:02 | + | |
| 24:02 | 40:02 | 03:04 | + | ||
| E | 33:03 | 44:03 | 14:03 | + | |
| 11:01 | 15:01 | 04:01 | + | ||
| F | 02:01 | 15:01 | 04:01 | + | |
| 26:03 | 44:03 | 08:01 | + | ||
| G | 24:02 | 39:01 | 07:02 | + | |
| 26:03 | 35:01 | 03:03 | + | ||
| H | 24:02 | 51:01 | 01:02 | + | |
| 31:01 | 35:01 | 03:03 | + | ||
| I | 24:02 | 54:01 | 01:02 | + | |
| 30:01 | 56:01 | 07:02 | + | ||
| J | 11:01 | 15:01 | 04:01 | + | |
| 31:01 | 56:01 | 07:02 | + | ||
| K | 24:02 | 15:01 | 04:01 | + | |
| 31:01 | 35:01 | 03:03 | + |
iPLATs Do Not Express NK Cell-Activation Molecules
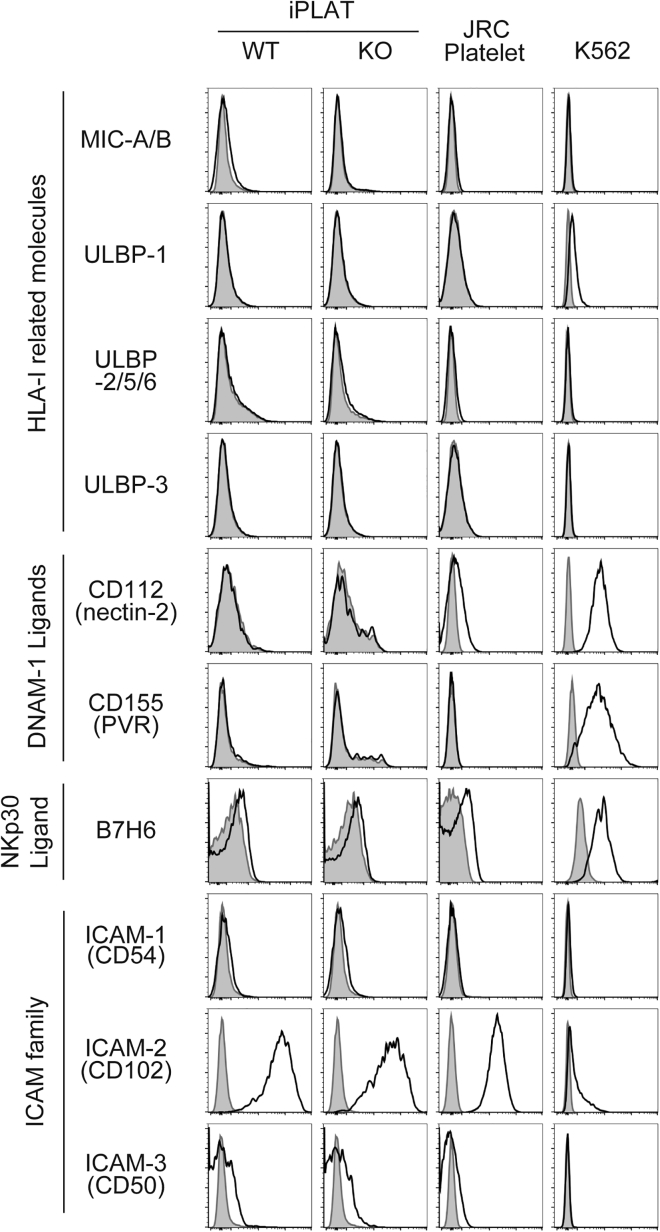
The balance between activation and inhibitory signals regulates NK cell activity (Lanier, 2008, Vivier et al., 2011, Long et al., 2013). Therefore, we compared the expression pattern of ligands for these signals on iPLATs. The major inhibitory ligands of NK cells, HLA-I molecules, suppress NK cell activation through killer cell immunoglobulin-like receptors (KIR). HLA-ABC was expressed on iPLATs without B2M knockout as well as on blood donor-derived platelets from the JRC, but not on B2M-knockout iPLATs, and had low expression on K562 cells (Figure 1E).
MIC-A, MIC-B, and ULBP family proteins are HLA-I-related molecules that are induced under stress situations such as virus infection, and act as activation ligands for NK cells (Bauer et al., 1999, Wu et al., 1999). The expression of MIC-A/B and ULBP proteins was not observed in any of the cells tested, except for ULBP-1 on K562 cells (Figure 3). Similarly, the DNAM-1 ligands CD112 and CD155 and NKp30 ligand B7H6, which are other activation ligands of NK cells (Shibuya et al., 1996, Bottino et al., 2003, Long et al., 2013, Vivier et al., 2011), were expressed on K562 cells but not on platelets (Figure 3). Other Nectin family proteins, including CD111 (nectin-1), CD113 (nectin-3) (Martinet and Smyth, 2015), and CD48, a 2B4 ligand (Nakajima et al., 1999), were not expressed (Figure S3). Among the adhesion molecule intercellular adhesion molecule (ICAM) family proteins, which are ligands for LFA-1 on NK cells (Barber et al., 2004, Long et al., 2013), ICAM-2 was expressed in all cells, but not differently between platelets and K562 cells (Figure 3).
Figure 3.
Platelets Do Not Express NK Cell-Activating Molecules
Representative histogram of the flow-cytometry analysis of HLA-I-related molecules, DNAM-1 ligands, B7H6 (NKp30 ligand), and ICAM family molecules for iPLATs, JRC platelets, and K562 cells. For iPLATs, both WT and HLA-KO types were analyzed. Gray histograms represent isotype control. WT, wild type; KO, HLA-KO. Data are representative of three independent experiments.
See also Figure S3.
The major inhibitory ligands are HLA-I, but there are other inhibitory ligands for NK cells. Cadherin family proteins are inhibitory ligands for KLRG1 receptor on NK cells (Ito et al., 2006), but E-, N-, and R-cadherin were not expressed (Figure S3). Additionally, PD-L1 and PD-L2 modulate NK cell activity through the receptor PD-1 (Benson et al., 2010), and proliferating cell nuclear antigen (PCNA) and lectin-like transcript-1 (LLT1) modulate NK cell activity through NKp44 (Rosental et al., 2011) and NKR-P1A (Rosen et al., 2005), respectively. However, none of these ligands were expressed on platelets (Figure S3). Therefore, while K562 cells may activate NK cells through ULBP-1, DNAM-1 ligands, and B7H6, platelets do not specifically express ligands that inhibit or activate NK cells.
HLA-KO iPLATs Can Circulate in Humanized Mice with Abundant Human NK Cells and Anti-HLA-I Antibodies
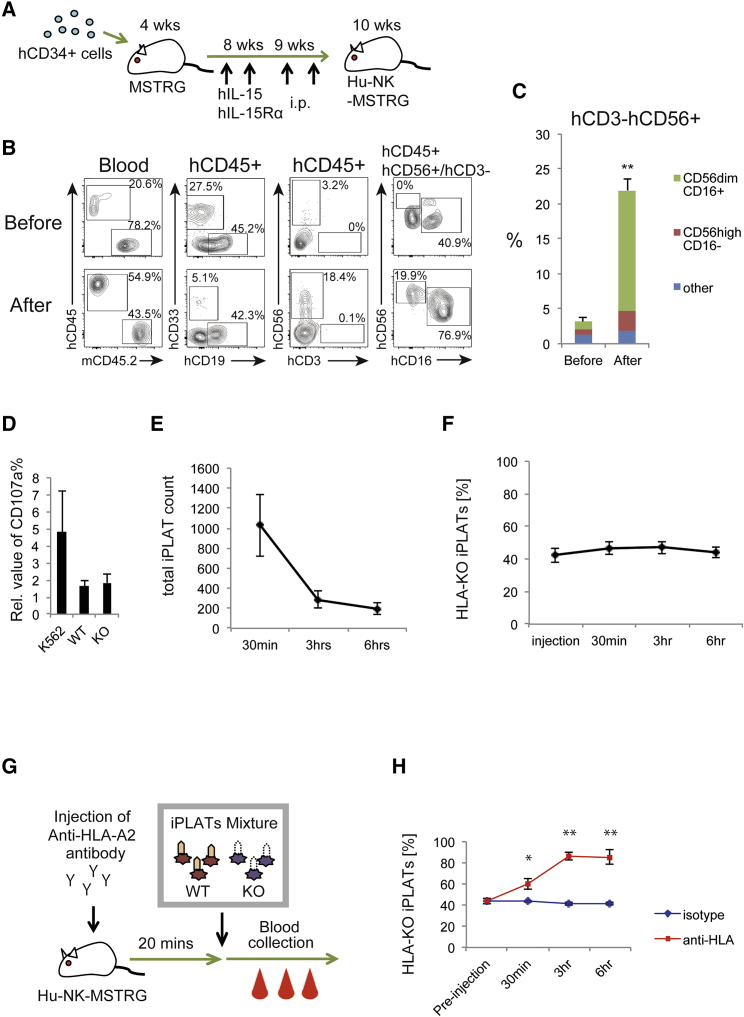
Next, we assessed whether HLA-KO iPLATs are rejected by NK cells in vivo. To generate mice with abundant human circulating NK cells, we transplanted MSTRG mice that express human macrophage colony-stimulating factor (M-CSF), thrombopoietin (TPO), and signal regulatory protein α (SIRPα) on Rag2−/−Il2rg−/− background (to enhance human hematopoietic cell engraftment) (Rongvaux et al., 2014, Saito et al., 2016) with human CD34+ cells derived from cord blood, then intraperitoneally administered a human IL-15 and IL-15R-α mixture at 8 and 9 weeks to promote NK cell reconstitution (Figure 4A). By this method, MSTRG mice showed a high frequency of human NK cells in circulation, with the majority being CD56dimCD16+ mature NK cells (hereafter called Hu-NK-MSTRG mice; Figures 4B, 4C, and S4A). NK cells separated from the spleen of Hu-NK-MSTRG mice showed CD107a upregulation when co-cultured with K562 cells but not with iPLATs (Figure 4D), indicating that the NK cells are competent for cytotoxic function. We further confirmed the in vivo functionality of human NK cells in the mice by infusing a mixture of WT and HLA-KO human iPSC-derived hematopoietic cells. The ratio of HLA-KO was lower in the lungs of Hu-NK-MSTRG mice than in non-humanized MSTRG mice, indicating that the NK cells are competent for cytotoxic function against HLA-deficient cells in vivo as well (Figure S4B).
Figure 4.
HLA-KO iPLATs Successfully Circulated in a Humanized Mouse Model with High NK Cell Reconstitution
(A) Schema of the establishment of NK cell-reconstituted humanized mice (Hu-NK-MSTRG mice). Four-week-old MSTRG mice were transplanted with cord blood-derived human CD34+ cells. At 8 and 9 weeks old, the mice were intraperitoneally administered recombinant human IL-15 and IL-15 receptor α. At 10 weeks old, the mice were checked for the reconstitution of human hematopoietic cells and underwent transfusion experiments (n = 8).
(B and C) Representative flow-cytometry scatterplots of circulating human blood cells in the mice (B) and the percentage of circulating human CD56+CD3− cells (C) (n = 4).
(D) Human NK cells were purified from Hu-NK-MSTRG mice and subjected to an in vitro cytotoxic assay against K562 cells and iPLATs. CD107a expression on NK cells was analyzed by flow cytometry (n = 3).
(E and F) The number of total circulating hCD41a+CD42b+ iPLATs (E) and percentage of HLA-KO type among circulating iPLATs (F) in Hu-NK-MSTRG mice at pre-transfusion and 30 min, 3 h, and 6 h after transfusion (n = 8).
(G) Schema of the allo-PTR model mice. Hu-NK-MSTRG mice were injected with mouse anti-HLA-A2 antibody 20 min prior to the transfusion of iPLATs.
(H) The percentage of HLA-KO type among circulating iPLATs in Hu-NK-MSTRG mice treated with anti-HLA-A2 antibody (allo-PTR mice) or the isotype control at pre-transfusion and 30 min, 3 h, and 6 h after transfusion (n = 3 or 4 in each group).
WT, wild type; KO, HLA-KO. Data are representative of three or more independent experiments with error bars representing the mean ± SEM. ∗p < 0.05 and ∗∗p < 0.01. See also Figure S4.
To assess the circulation capacity of HLA-KO iPLATs, we transfused a mixture of WT and HLA-KO iPLATs labeled with TAMRA dye to Hu-NK-MSTRG mice and compared the ratio between the WT and HLA-KO types in the blood circulation at various time points (Figure S4C). Circulation of the transfused iPLATs was observed up to 6 h post transfusion (Figure 4E). Importantly, the ratio of HLA-KO was stable from pre-injection to up to 6 h (Figure 4F). This result indicates that HLA-KO iPLATs have the same circulating capability as WT iPLATs even in the presence of human NK cells.
In a separate experiment, 20 min prior to the transfusion of iPLATs, mouse anti-HLA-A2 antibody was injected into Hu-NK-MSTRG mice to create an HLA-I-mediated allo-PTR model (Gras et al., 2013) (Figure 4G). Similar to the kinetics in patients with anti-HLA-I antibody-mediated allo-PTR transfused with incompatible donor-derived platelets, the circulation of HLA-WT iPLATs was less at 30-min transfusion, and only HLA-KO iPLATs were essentially observed after 3 h (Figures 4G, 4H, and S4D). These data suggest that HLA-KO iPLATs are an effective source for alloimmune PTR, which is mostly mediated by anti-HLA-I antibodies.
Discussion
We have recently proposed a clinically applicable system to produce transfusion-relevant numbers of iPLATs from imMKCLs by using a turbulent flow-based bioreactor and adding an aryl hydrocarbon receptor antagonist and Rho-associated protein kinase inhibitor for imMKCL maturation and iPLAT release (Ito et al., 2018). iPLATs produced in this system have shown comparable function with blood donor-derived platelets and are devoid of tumorigenicity upon irradiation, a precaution also taken for usual transfusion products to eliminate the risk of transfusion-related graft-versus-host disease. Based on this system, this study shows that HLA-KO iPLATs were successfully produced from HLA-KO imMKCLs (Figures 1 and S1), and further found the unique property of HLA-KO iPLATs to evade NK cell-mediated and anti-HLA-I antibody-mediated immune responses from in vitro and in vivo assays (Figures 2, 3, 4, and S2–S4). Thus, a functional, safe, and universal HLA-I iPLAT product can be supplied, resolving the risk of compatible-donor shortage in HLA-I-mediated allo-PTR. Because one HLA-KO iPLAT line can be applied for all HLA-I types, the cost will be significantly lower than had we prepared autologous iPLATs or a wide line-up of iPLATs with various HLA-I types from an iPSC library (Gourraud et al., 2012, Turner et al., 2013).
The nullification of HLA-I on platelets has been achieved before by knocking out or knocking down B2M to enable platelets to evade anti-HLA-I alloresponses (Gras et al., 2013, Feng et al., 2014, Borger et al., 2016). While the gene knockout procedure is capable of achieving complete deletion of the HLA-I expression, it may not inhibit the activation of NK cells through the HLA-I receptor, KIR, thus leading to rejection of the allografts (Ichise et al., 2017). A short hairpin RNA (shRNA)-based mRNA knockdown procedure was reported to leave a low expression level of HLA-I, which has the potential advantage of suppressing the NK cell response against HLA-I nullified cells (Wiegmann et al., 2014). Other approaches to evade NK cell response include the overexpression of single-chain HLA-E fused with B2M (Gornalusse et al., 2017), knockout of the HLA-A and HLA-B loci while retaining HLA-C (Xu et al., 2019), and the overexpression of CD47 (Deuse et al., 2019). However, it has not been addressed whether platelets nullified with all HLA-I molecules elicit an NK cell response, and thus it remains unknown which approach is most suitable. In our current study, we found that platelets do not elicit cytotoxic responses by NK cells even when HLA-I is completely depleted (Figures 2 and S2). Our data show that the lack of NK cell-activating receptor ligands such as DNAM-1 ligands and the NKp30 ligand B7H6 on iPLATs may be partly involved in this effect (Figures 3 and S3).
We also looked into various ligands and adhesion molecules that could affect NK cell activation, such as non-DNAM-1 ligand Nectin family molecules, including Nectin-1 and Nectin-3, ICAM family proteins, which are ligands for LFA-1 on NK cells (Barber et al., 2004, Long et al., 2013), CD48, which binds 2B4 receptor for activation (Nakajima et al., 1999), and cadherins, which bind KLRG1 receptor to inhibit NK function via ITAM (immunoreceptor tyrosine-based activation) signaling (Ito et al., 2006, Li et al., 2009) and also have a functional role in aggregation and thrombus formation (Dunne et al., 2012). Other inhibitory ligands, such as LLT1, PD-L1, PD-L2, and PCNA (Aldemir et al., 2005, Rosen et al., 2005, Benson et al., 2010, Pesce et al., 2017, Rosental et al., 2011), were also analyzed. However, we did not find a specific lack of activating molecules or specific expression of inhibitory molecules on the platelets. We speculate that the inert response may depend on the non-nuclear properties of platelets, since erythrocytes, which are also anucleate and do not express HLA-I, are not attacked by NK cells, except in specific circumstances such as Plasmodium infection (Chen et al., 2014). Further studies are required, but elucidating how iPLATs evade NK cells could contribute to understanding other immune phenomena such as tumor immune evasion.
For in vivo evaluation, we adopted MSTRG mice transplanted with human cord blood cells (Figures 4 and S4). MSTRG mice have an enhanced reconstitution of human hematopoiesis by knocking in genes encoding human M-CSF, TPO, and SIRPα into the Rag2−/−Il2rg−/− background (Rongvaux et al., 2014, Saito et al., 2016). By further injecting a mixture of human IL-15 ligand and receptor, as previously reported (Huntington et al., 2009), we succeeded in establishing “Hu-NK-MSTRG” mice, which reconstituted high levels of human NK cells. Our data showed that the reconstituted human NK cells in Hu-NK-MSTRG mice have cytotoxic activity similar to that of NK cells in human peripheral blood (Figures 4D and S4B). We further set up an allo-PTR model by injecting mouse anti-HLA antibody. HLA-KO iPLAT transfusion to this model suggested that HLA-KO iPLATs could circulate upon transfusion to human subjects, including allo-PTR patients with anti-HLA-I alloantibodies. Several groups have previously reported the establishment of humanized mice with a high reconstitution of NK cells and have tested the in vivo NK cell functionality (Strowig et al., 2010, Kübler et al., 2014, Rongvaux et al., 2014, Katano et al., 2015, Herndler-Brandstetter et al., 2017). However, these reports show the in vivo rejection of only tumorigenic HLA-deficient cells, while this report showed this in non-tumor HLA-deficient cells, thus providing a pre-clinical model for HLA-depleted cell therapies. Moreover, there are no reports on platelet circulation in humanized mice with a high reconstitution of NK cells and anti-HLA-I antibodies.
There has been a debate about whether platelets capture circulating B2M to express functional HLA-I (Gouttefangeas et al., 2000). With regard to our model, although human blood cells that express B2M were in circulation, we did not observe a difference in the ratio of HLA-KO and WT iPLATs, and their HLA expression level remained unchanged during the assessment. Based on these observations, we conclude that iPLATs do not readily upregulate HLA-I expression through the adoption of circulating B2M.
The production of anti-HPA alloantibodies is the second major cause of allo-PTR and also a primary cause of fetal/neonatal alloimmune thrombocytopenia and post-transfusion purpura. However, HPA knockout is not appropriate due to the existence of antibody-recognition sites on crucial functional platelet molecules, such as CD61 (ITGB3), CD41 (ITGA2B), CD42b, CD36, and CD29 (Curtis and McFarland, 2014, Hayashi and Hirayama, 2015). As such, HPA conversion using the CRISPR/Cas9 system has been proposed (Zhang et al., 2016). Considering that a substantial portion of patients with anti-HPA antibodies also carry anti-HLA-I antibody, HLA-KO iPLATs may be an ideal platform for further gene editing of HPA.
In conclusion, our study shows a proof of concept that HLA-KO iPLATs can be used as universal HLA-I-type platelets. This study should lead to the clinical application of HLA-KO iPLATs for resolving the shortage risk of HLA-I-compatible donors in allo-PTR conditions and contribute to the cost reduction of iPLATs. In addition, we also found that the unique non-immunogenic property of HLA-depleted platelets does not activate NK cells. Owing to their capacity to evade alloimmune responses and their safe and uniform product profile by sterile production from quality-assured imMKCL master cells, HLA-KO iPLATs may also lead to new standardized platelet-based therapies such as injection for tissue regeneration and drug-delivery systems.
Experimental Procedures
Study Approval
Donor-derived human platelets were provided by the JRC. Human platelets were used in compliance with the Guidelines on the Use of Donated Blood in R&D from the Ministry of Health, Labour and Welfare of Japan. The collection and usage of peripheral blood from healthy volunteers, the collection of cord blood from healthy volunteers, and animal experiments were approved by the Ethical Committee of Kyoto University and Kumamoto University. All experiments using human samples were conducted in accordance with the Declaration of Helsinki.
Cell Culture
imMKCLs were established from human iPSCs as previously reported (Nakamura et al., 2014). imMKCLs were cultured in IMDM medium (Sigma) with L-glutamine (25030-081; Thermo Fisher Scientific), Insulin-transferrin-selenium (41400-045; Thermo Fisher), 50 μg/mL ascorbic acid (A4544; Sigma-Aldrich), and 450 μM 1-thioglycerol (M6145; Sigma-Aldrich). DOX-ON proliferation condition: imMKCLs were cultured with 15% fetal bovine serum (FBS), 50 ng/mL recombinant human thrombopoietin (rhTPO: PeproTech), 50 ng/mL recombinant human stem cell factor (rhSCF: R&D Systems), and 5 μg/mL DOX-inducible transgenes (c-MYC, BIM1, and BCL-XL). DOX-OFF production of iPLATs condition: the medium contained 10% human plasma (Cosmo Bio #12250210, Japan Blood Products Organization), 10 U heparin sodium (#(01)14987476163428; Yoshido), 50 ng/mL rhSCF, 200 ng/mL TA-316 (Nissan Chemical), 15 μM KP-457 (Kaken Pharmaceutical), 0.5 μM GNF-351 (Calbiochem), and 0.5 μM Y39983 (MedChemExpress). imMKCLs were cultured with this medium for 6 days in 125- or 250-mL Corning Erlenmeyer cell-culture flasks (#431143 and #431144; Sigma-Aldrich) in shaking conditions using Lab-Therm shakers (Kuhner) (Ito et al., 2018).
MK-iPSCs were established from imMKCLs by re-reprogramming (Seo et al., 2018) and cultured with the mouse C3H10T1/2 cell line in DMEM F-12 medium (Sigma) with MEM non-essential amino acids solution (100×) (11140050; Thermo Fisher), penicillin-streptomycin-glutamine (100×) (10378016; Thermo Fisher), 20% KnockOut Serum Replacement (10828028; Thermo Fisher), 0.1 mM 2-mercaptoethanol (Sigma), and 5 ng/mL basic fibroblast growth factor (R&D Systems).
K562 cells were cultured with RPMI-1640 medium (Sigma), 10% FBS, and 100× penicillin-streptomycin-glutamine (10378016; Thermo Fisher).
Establishment of a β2-Microglobulin-Knockout imMKCL
Exon 1 of B2M was selected as the knockout target. The sequence for homologous recombination was constructed as 5′-arms-loxP-Ubic-Puror-loxP-3′-arms. The sequence of the 5′-arm was Fw 5′-TGG CGG CCG CTC TAG ACC CTT TGT CTT CCA GTG TCT-3′, and Rv 5′-ATT ATA CGA AGT TAT GCC CGC ATG CTG TCA GCT TCA GGA ATG-3′; and the 3′-arm was Fw 5′-ATA CGA AGT TAT GCA ACA GGG TTT CAC CGT GTT AG-3′, and Rv 5′-GCT TGA TAT CGA ATT CGA GGC ACA GTA CAT CTT GGA-3′. This cassette was integrated into pBluescript KS(+) vector using In-Fusion reaction to construct the targeting vector plasmid. The single-guide RNA (sgRNA) expression vector was designed as Guide Fw 5′-TCA TGC CAT CCG TAA GAT GC-3′ and Guide Rv 5′-CTA AAA CTA GCT GTG CTC GC-3′, and inserted into pHL-H1-ccdB-mEF1a-RiH vector using In-Fusion reaction to construct the sgRNA expression vector pHL-H1-B2M-sgRNA-mEF1a-RiH.
MK-iPSC line #11 (MKiPS#11) was established from imMKCLs by re-reprogramming (Seo et al., 2018). imMKCLs at the DOX-ON proliferation stage were transfected with an episomal vector carrying reprogramming factors (OCT3/4 with shRNA of TP53, SOX2, KLF4, and MYCL) by using Human Stem Cell Nucleofecto Kit 2 (VPH-5022; Lonza). The transfected imMKCLs were co-cultured with the C3H10T1/2 cell line as feeder cells for 3–4 weeks. iPSC-like colonies were picked up as MK-iPSCs. For gene manipulation, MKiPS#11 was placed into feeder-free culture condition: iMatrix-511 (892012; MTR)-coated dishes in StemFit AK03N medium (Ajinomoto, Tokyo, Japan). For establishment of B2M-KO imMKCL, the sgRNA expression vector, the targeting vector, and the CRISPR/Cas9 expression vector (pHL-UbicP-SphcCas9-iP-A) were transfected into 0.8 × 106 MK-iPSCs by electroporation. MK-iPSCs were cultured with 10 μM Y27632 (Wako) for 1 day and selected with 1 μg/mL puromycin from day 3. Seven days after the electroporation, individual colonies were picked. After genotyping each clone by using PCR primer 1, 5′-ACG AAA TGG CGG CAC CTT AT-3′, primer 2, 5′-CCC GTC CTA AAA TGT CCT TC-3′, primer 3, 5′-CTG CAA GAA CTC TTC CTC AC-3′, and primer 4, 5′-TGG CGC CAT GAT AGC TCA AA-3′, an adequately targeted clone was selected (HLA-KO MK-iPSCs). HLA-KO MK-iPSCs were placed into the C3H10T1/2 cell co-culture condition. For differentiation to imMKCLs, HLA-KO MK-iPSCs were cultured by the Sac method (Nakamura et al., 2014). HLA-KO MK-iPSCs were cultured in differentiation medium from day 0 to day 14. After day 14, HLA-KO hematopoietic progenitor cells were collected from the Sac-like structure and cultured in imMKCL medium with 50 ng/mL rhSCF, 50 ng/mL rhTPO, and 5 μg/mL DOX. The DOX-induced transgenes were expressed, and HLA-KO imMKCLs were obtained.
Flow Cytometry
The following antibodies were used: from BD Biosciences, hCD42b-APC (clone HIP1: 551061), HLA-ABC-FITC (clone G46–2.6: 557348), β2-microglobulin-FITC (clone TU99; 551338), hCD3-FITC (clone SP34: 556611), hCD56-PerCP/Cy5.5 (clone B159: 560842), hCD107a-BV421 (clone H4A3: 562623), and hCD49b-PE (clone 12F1: 555669); from R&D systems, ULBP-1-PE (clone 170818; FAB1380P), ULBP-2/5/6-APC (clone 165903; FAB1298A), ULBP-3-PE (clone 166510; FAB1517P), hOCIL/CLEC2d(LLT1)-APC (clone 402659; FAB3480A), and B7H6-PE (clone 875001; FAB7144P); from BioLegend, hCD41a-APC (clone HIP8; 303710), hCD42b-PE (clone HIP1; 303906), hCD112-PE (clone TX31; 337409), hCD155-APC (clone SKII.4; 337617), MIC-A/B-PE (clone 6D4; 320906), HLA-E-PE (clone 3D12; 342603), ICAM-1-Pacific Blue (clone HA58; 353109), ICAM-2-PE (clone CBR-IC2/2; 328505), ICAM-3-APC (clone CBR-IC3/1; 330011), hCD19-APC/Cy7 (clone HIB19; 302218), hCD45-PE/Cy7 (clone HI30; 304016), hCD3-FITC (clone UCHT1; 300406), mCD45.2-PE (clone 104; 109808), hCD56-BV421 (clone HCD56; 318328), hCD16-BV510 (clone 3G8; 560918), hCD324 (E-Cadherin)-APC (clone 67A4; 324107), hCD36-APC/Cy7 (clone 5-271; 336213), hCD29-PE (clone TS2/16; 303003), hCD109-PE (clone W7C5; 323305), hCD61-FITC (clone VI-PL2; 336403), PD-L1-PE (clone 29E.2A3; 329705), PD-L2-PE (clone 24F.10C12; 329605), hCD48-PE (clone BJ40; 336707), h/m/rPCNA-PE (clone PC10; 307908), and hCD111-PE (clone R1.302; 340404); from eBioscience, hCD33-APC (clone WM-53; 17-0338-41) and hCD41a-efluor 450 (clone HIP8; 48-0419-42); from Santa Cruz Biotechnology, N/R-cadherin-Alexa Fluor 647 (clone H-4; sc-271386) and hCD42c-PE (clone F-11; sc-377129 PE); and from MBL International, hCD113-Alexa Fluor 488 (clone N3.12.4; K0224-A48).
For staining, cells were incubated with antibodies in PBS(−) (Sigma) with 3% FBS for 30 min at 4°C, except platelets at room temperature. The data were obtained with a FACS Verse, FACS Aria IIIu, or FACS Canto II (BD Biosciences), and analyzed by FlowJo 10.4 (FlowJo).
Clot Retraction Assay
iPLATs were suspended in IMDM medium with 20% human plasma. iPLATs were then mixed with 2 U/mL thrombin and incubated at 37°C for 2 h (Hirata et al., 2017).
NK Cell Co-culture Assay
PBMCs were isolated from whole blood by density gradient centrifugation using Ficoll-Paque PREMIUM (GE Healthcare) from healthy volunteers. PBMCs were incubated with CD4, CD14, and CD19 MicroBeads (130-045-101, 130-050-201, 130-050-301; Miltenyi Biotec) at 4°C for 30 min. After washing, MACS Columns (Miltenyi Biotec) were used for the isolation. A mixture of CD4+, CD14+, and CD19+ cells was used as negative control cells. CD4-CD14-CD19 cells were incubated with CD3 and CD8 MicroBeads (130-050-101, 130-045-201; Miltenyi Biotec) at 4°C for 30 min, and cells negatively separated by MACS columns were used as the NK cell population. For the CD107a expression assay, 1 × 106 cells/mL human NK cells were co-cultured with target cells at an E/T ratio of 1:1 in 200 μL of RPMI-1640 medium with 10% pooled human serum (inactivated) (12181450; Cosmo Bio), 2 mM L-glutamine, 100 U/mL penicillin, 0.1 mg/mL streptomycin (basal medium) with 1,000 U/mL recombinant human IL-2, and 1/200 volume of CD107a-BV421 for 6 h at 37°C in a 5% CO2 incubator in a 96-well plate. After incubation, the plate was centrifuged, and each well was resuspended in 100 μL of PBS with 2% FBS with anti-CD56-PerCP-Cy5.5 and anti-CD3-FITC antibodies. After 30-min incubation, the samples were analyzed by flow cytometry. The cytotoxic activities of NK cells were evaluated by measuring CD107a expression and normalized as follows: relative value of CD107a = (CD107a positive [%] of NK cells with target cells)/(CD107a positive [%] of NK cells alone). For Annexin V binding of the target cells, the E/T ratio was 8:1, 4:1, or 0.25:1 in basal medium with 1,000 U/mL recombinant human IL-2. K562 cells were labeled by the CellTrace CFSE Cell Proliferation Kit (Thermo Fisher Scientific). After incubation, the plate was centrifuged, and each well was resuspended in PBS(−) with 10× Binding Buffer (556454; BD Biosciences), Annexin V, and antibodies for hCD41a and hCD42b. After 30-min incubation, the samples were analyzed by flow cytometry.
Mouse Model
Human CD34+ cells (1 × 106) were isolated from human cord blood using the CD34+ Cell Isolation Kit (Miltenyi Biotec) and intravenously injected into MSTRG (CSF1h/h SIRPAtg THPOh/h Rag2−/− Il2rg−/−) mice irradiated with 2.5 Gy at 6 weeks old. Four weeks after the transplantation, the reconstitution of human cells was confirmed by flow-cytometric analysis. Recombinant human IL-15 (0.5 μg) (#200-15; PeproTech) and 1.0 μg of recombinant human IL-15 R alpha Fc chimera protein (#7195-IR-050; RSD) in 200 μL of PBS were intraperitoneally injected at days 0, 3, 7, and 14 at 4 weeks post transplantation. The reconstitution of human NK cells was confirmed by flow-cytometric analysis. For the allo-PTR mouse model, 3 μg/body weight (g) of anti-HLA-A2 immunoglobulin G (#0131HA; One Lambda) in 100 μL of PBS was injected into Hu-NK-MSTRG mice 20 min before platelet transfusion.
Human NK Cell Isolation from Mouse Spleen
The spleens of mice were minced and filtered through 40-μm cell strainers to make single-cell suspensions. The suspensions were treated with ammonium-chloride-potassium lysis buffer to lyse erythrocytes. CD56-positive NK cells were then isolated using anti-hCD56 microbeads (#130-050-401; Miltenyi Biotec) and positively selected through LS columns (#130-122-729; Miltenyi Biotec).
Transfusion Assay
For the platelet analysis, WT and HLA-KO iPLATs were mixed and labeled with 5′-TAMRA (C6121; Thermo Fisher Scientific). iPLATs (5–8 × 107 in 100 μL of bicarbonate buffer) were intravenously injected into the mice. Blood samples (10 μL) were collected at 30 min, 3 h, and 6 h after the transfusion.
Statistical Analysis
All data are presented as means ± SEM and were analyzed by GraphPad Prism 5 (GraphPad Software). Statistically significant differences were determined by one-way ANOVA, Tukey-Kramer test for multiple comparisons, and two-tailed t test for pairwise comparisons. p values are indicated in the figures as ∗p < 0.05 and ∗∗p < 0.01.
Author Contributions
D.S. designed and performed the experiments, evaluated the data, and wrote the manuscript; C.F., I.S., Y.H., A.S., M.A., N.H., H.X., T.M., and K.F. designed and performed the experiments; N.Y. designed and established the HLA-KO imMKCLs and iPLATs; S.N. supported the gene manipulation and iPLAT production; A.H. provided guidance on the CRISPR/Cas9 experiments; M.G.M. and H.T. provided guidance and established humanized mice models; N.S. provided guidance on the data interpretation and wrote the manuscript; N.S. and K.E. managed the overall project, contributed to the data interpretation, and edited the manuscript.
Acknowledgments
The authors thank Dr. Sayuri Nakata, Dr. Akinori Yuzuriha, Toshie Kusunoki, and Dr. Keiji Hirota for assisting with experiments, Dr. Shin Kaneko for providing an experimental protocol, Dr. Peter Karagiannis for critical reading of the manuscript, and Misaki Ouchida for the visual abstract. This work was supported in part by Highway Program for Realization of Regenerative Medicine (JP17bm0504008, K.E.), Practical Applications of Regenerative Medicine (JP17bk0104039, K.E.), Projects for Technological Development (JP19bm0404037h0002, N.S. and K.E.), Core Center for iPS Cell Research (JP17bm0104001, N.S. and K.E.) from Japan Agency for Medical Research and Development (AMED), by Grant-in-Aid for scientific research (15H03005, K.E.) from Japan Society for the Promotion of Science, by Initiative for Accelerating Regulatory Science in Innovative Drug, Medical Device, and Regenerative Medicine from Ministry of Health, Labour and Welfare (MHLW) (K.E.), by Japan Student Services Organization scholarship (D.S.), and by Japanese Government (Monbukagakusho: MEXT) scholarship (C.F.).
H.X., S.N., A.H., K.E., and N.S. have applied for patents related to this paper. K.E. is a founder of Megakaryon and a member of its scientific advisory board without salary; K.F. is an employee of Otsuka Pharmaceuticals; this work was supported in part by grants from Megakaryon and Otsuka Pharmaceuticals; the interests of K.E. were reviewed and are managed by Kyoto University in accordance with its conflict-of-interest policies.
Published: December 26, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.11.011.
Contributor Information
Koji Eto, Email: kojieto@cira.kyoto-u.ac.jp.
Naoshi Sugimoto, Email: naoshi.sugi@cira.kyoto-u.ac.jp.
Supplemental Information
References
- Aldemir H., Prod’homme V., Dumaurier M.-J., Retiere C., Poupon G., Cazareth J., Bihl F., Braud V.M. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J. Immunol. 2005;175:7791–7795. doi: 10.4049/jimmunol.175.12.7791. [DOI] [PubMed] [Google Scholar]
- Alter G., Malenfant J.M., Altfeld M. CD107a as a functional marker for the identification of natural killer cell activity. J. Immunol. Methods. 2004;294:15–22. doi: 10.1016/j.jim.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Barber D.F., Faure M., Long E.O. LFA-1 contributes an early signal for NK cell cytotoxicity. J. Immunol. 2004;173:3653–3659. doi: 10.4049/jimmunol.173.6.3653. [DOI] [PubMed] [Google Scholar]
- Bauer S., Groh V., Wu J., Steinle A., Phillips J.H., Lanier L.L., Spies T. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science. 1999;285:727–729. doi: 10.1126/science.285.5428.727. [DOI] [PubMed] [Google Scholar]
- Benson D.M., Bakan C.E., Mishra A., Hofmeister C.C., Efebera Y., Becknell B., Baiocchi R.A., Zhang J., Yu J., Smith M.K. The PD-1/PD-L1 axis modulates the natural killer cell versus multiple myeloma effect: a therapeutic target for CT-011, a novel monoclonal anti–PD-1 antibody. Blood. 2010;116:2286–2294. doi: 10.1182/blood-2010-02-271874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borger A.-K., Eicke D., Wolf C., Gras C., Aufderbeck S., Schulze K., Engels L., Eiz-Vesper B., Schambach A., Guzman C.A. Generation of HLA-universal iPSC-derived megakaryocytes and platelets for survival under refractoriness conditions. Mol. Med. 2016;22:274–285. doi: 10.2119/molmed.2015.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino C., Castriconi R., Pende D., Rivera P., Nanni M., Carnemolla B., Cantoni C., Grassi J., Marcenaro S., Reymond N. Identification of PVR (CD155) and Nectin-2 (CD112) as cell surface ligands for the human DNAM-1 (CD226) activating molecule. J. Exp. Med. 2003;198:557–567. doi: 10.1084/jem.20030788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Amaladoss A., Ye W., Liu M., Dummler S., Kong F., Wong L.H., Loo H.L., Loh E., Tan S.Q. Human natural killer cells control Plasmodium falciparum infection by eliminating infected red blood cells. Proc. Natl. Acad. Sci. U S A. 2014;111:1479–1484. doi: 10.1073/pnas.1323318111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis B.R., McFarland J.G. Human platelet antigens—2013. Vox Sang. 2014;106:93–102. doi: 10.1111/vox.12085. [DOI] [PubMed] [Google Scholar]
- Deuse T., Hu X., Gravina A., Wang D., Tediashvili G., De C., Thayer W.O., Wahl A., Garcia J.V., Reichenspurner H. Hypoimmunogenic derivatives of induced pluripotent stem cells evade immune rejection in fully immunocompetent allogeneic recipients. Nat. Biotechnol. 2019;37:252–258. doi: 10.1038/s41587-019-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunne E., Spring C.M., Reheman A., Jin W., Berndt M.C., Newman D.K., Newman P.J., Ni H., Kenny D. Cadherin 6 has a functional role in platelet aggregation and thrombus formation. Arterioscler. Thromb. Vasc. Biol. 2012;32:1724–1731. doi: 10.1161/ATVBAHA.112.250464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estcourt L.J., Birchall J., Allard S., Bassey S.J., Hersey P., Kerr J.P., Mumford A.D., Stanworth S.J., Tinegate H. Guidelines for the use of platelet transfusions. Br. J. Haematol. 2017;176:365–394. doi: 10.1111/bjh.14423. [DOI] [PubMed] [Google Scholar]
- Feng Q., Shabrani N., Thon J.N., Huo H., Thiel A., Machlus K.R., Kim K., Brooks J., Li F., Luo C. Scalable generation of universal platelets from human induced pluripotent stem cells. Stem Cell Rep. 2014;3:817–831. doi: 10.1016/j.stemcr.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gornalusse G.G., Hirata R.K., Funk S.E., Riolobos L., Lopes V.S., Manske G., Prunkard D., Colunga A.G., Hanafi L.-A., Clegg D.O. HLA-E-expressing pluripotent stem cells escape allogeneic responses and lysis by NK cells. Nat. Biotechnol. 2017;35:765–772. doi: 10.1038/nbt.3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gourraud P.-A., Gilson L., Girard M., Peschanski M. The role of human leukocyte antigen matching in the development of multiethnic “haplobank” of induced pluripotent stem cell lines. Stem Cells. 2012;30:180–186. doi: 10.1002/stem.772. [DOI] [PubMed] [Google Scholar]
- Gouttefangeas C., Diehl M., Keilholz W., Hörnlein R.F., Stevanović S., Rammensee H.-G. Thrombocyte HLA molecules retain nonrenewable endogenous peptides of megakaryocyte lineage and do not stimulate direct allocytotoxicity in vitro. Blood. 2000;95:3168–3175. [PubMed] [Google Scholar]
- Gras C., Schulze K., Goudeva L., Guzman C.A., Blasczyk R., Figueiredo C. HLA-universal platelet transfusions prevent platelet refractoriness in a mouse model. Hum. Gene Ther. 2013;24:1018–1028. doi: 10.1089/hum.2013.074. [DOI] [PubMed] [Google Scholar]
- Hayashi T., Hirayama F. Advances in alloimmune thrombocytopenia: perspectives on current concepts of human platelet antigens, antibody detection strategies, and genotyping. Blood Transfus. 2015;13:380–390. doi: 10.2450/2015.0275-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndler-Brandstetter D., Shan L., Yao Y., Stecher C., Plajer V., Lietzenmayer M., Strowig T., de Zoete M.R., Palm N.W., Chen J. Humanized mouse model supports development, function, and tissue residency of human natural killer cells. Proc. Natl. Acad. Sci. U S A. 2017;114:E9626–E9634. doi: 10.1073/pnas.1705301114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirata S., Murata T., Suzuki D., Nakamura S., Jono-Ohnishi R., Hirose H., Sawaguchi A., Nishimura S., Sugimoto N., Eto K. Selective inhibition of ADAM17 efficiently mediates glycoprotein ibalpha retention during ex vivo generation of human induced pluripotent stem cell-derived platelets. Stem Cells Transl Med. 2017;6:720–730. doi: 10.5966/sctm.2016-0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huntington N.D., Legrand N., Alves N.L., Jaron B., Weijer K., Plet A., Corcuff E., Mortier E., Jacques Y., Spits H., Di Santo J.P. IL-15 trans-presentation promotes human NK cell development and differentiation in vivo. J. Exp. Med. 2009;206:25–34. doi: 10.1084/jem.20082013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichise H., Nagano S., Maeda T., Miyazaki M., Miyazaki Y., Kojima H., Yawata N., Yawata M., Tanaka H., Saji H. NK cell alloreactivity against KIR-ligand-mismatched HLA-haploidentical tissue derived from HLA haplotype-homozygous iPSCs. Stem Cell Rep. 2017;9:1–15. doi: 10.1016/j.stemcr.2017.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M., Maruyama T., Saito N., Koganei S., Yamamoto K., Matsumoto N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J. Exp. Med. 2006;203:289–295. doi: 10.1084/jem.20051986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Nakamura S., Sugimoto N., Shigemori T., Kato Y., Ohno M., Sakuma S., Ito K., Kumon H., Hirose H. Turbulence activates platelet biogenesis to enable clinical scale ex vivo production. Cell. 2018;174:636–648.e18. doi: 10.1016/j.cell.2018.06.011. [DOI] [PubMed] [Google Scholar]
- Karagiannis P., Eto K. Ten years of induced pluripotency: from basic mechanisms to therapeutic applications. Development. 2016;143:2039–2043. doi: 10.1242/dev.138172. [DOI] [PubMed] [Google Scholar]
- Katano I., Takahashi T., Ito R., Kamisako T., Mizusawa T., Ka Y., Ogura T., Suemizu H., Kawakami Y., Ito M. Predominant development of mature and functional human NK cells in a novel human IL-2-producing transgenic NOG mouse. J. Immunol. 2015;194:3513–3525. doi: 10.4049/jimmunol.1401323. [DOI] [PubMed] [Google Scholar]
- Kübler A., Woiterski J., Witte K.-E., Bühring H.-J., Hartwig U.F., Ebinger M., Oevermann L., Mezger M., Herr W., Lang P. Both mature KIR+ and immature KIR− NK cells control pediatric acute B-cell precursor leukemia in NOD.Cg-Prkdcscid IL2rgtmWjl/Sz mice. Blood. 2014;124:3914–3923. doi: 10.1182/blood-2014-05-572743. [DOI] [PubMed] [Google Scholar]
- Lanier L.L. Up on the tightrope: natural killer cell activation and inhibition. Nat. Immunol. 2008;9:495–502. doi: 10.1038/ni1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hofmann M., Wang Q., Teng L., Chlewicki L.K., Pircher H., Mariuzza R.A. Structure of natural killer cell receptor KLRG1 bound to E-cadherin reveals basis for MHC-independent missing self recognition. Immunity. 2009;31:35–46. doi: 10.1016/j.immuni.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long E.O., Kim H.S., Liu D., Peterson M.E., Rajagopalan S. Controlling natural killer cell responses: integration of signals for activation and inhibition. Annu. Rev. Immunol. 2013;31:227–258. doi: 10.1146/annurev-immunol-020711-075005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinet L., Smyth M.J. Balancing natural killer cell activation through paired receptors. Nat. Rev. Immunol. 2015;15:243–254. doi: 10.1038/nri3799. [DOI] [PubMed] [Google Scholar]
- Nakajima H., Cella M., Langen H., Friedlein A., Colonna M. Activating interactions in human NK cell recognition: the role of 2B4-CD48. Eur. J. Immunol. 1999;29:1676–1683. doi: 10.1002/(SICI)1521-4141(199905)29:05<1676::AID-IMMU1676>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Takayama N., Hirata S., Seo H., Endo H., Ochi K., Fujita K.-I., Koike T., Harimoto K.-I., Dohda T. Expandable megakaryocyte cell lines enable clinically applicable generation of platelets from human induced pluripotent stem cells. Cell Stem Cell. 2014;14:535–548. doi: 10.1016/j.stem.2014.01.011. [DOI] [PubMed] [Google Scholar]
- Pavenski K., Rebulla P., Duquesnoy R., Saw C.L., Slichter S.J., Tanael S., Shehata N. Efficacy of HLA-matched platelet transfusions for patients with hypoproliferative thrombocytopenia: a systematic review. Transfusion. 2013;53:2230–2242. doi: 10.1111/trf.12175. [DOI] [PubMed] [Google Scholar]
- Pesce S., Greppi M., Tabellini G., Rampinelli F., Parolini S., Olive D., Moretta L., Moretta A., Marcenaro E. Identification of a subset of human natural killer cells expressing high levels of programmed death 1: a phenotypic and functional characterization. J. Allergy Clin. Immunol. 2017;139:335–346.e3. doi: 10.1016/j.jaci.2016.04.025. [DOI] [PubMed] [Google Scholar]
- Riolobos L., Hirata R.K., Turtle C.J., Wang P.-R., Gornalusse G.G., Zavajlevski M., Riddell S.R., Russell D.W. HLA engineering of human pluripotent stem cells. Mol. Ther. 2013;21:1232–1241. doi: 10.1038/mt.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rongvaux A., Willinger T., Martinek J., Strowig T., Gearty S.V., Teichmann L.L., Saito Y., Marches F., Halene S., Palucka A.K. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol. 2014;32:364–372. doi: 10.1038/nbt.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen D.B., Bettadapura J., Alsharifi M., Mathew P.A., Warren H.S., Lanier L.L. Cutting edge: lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J. Immunol. 2005;175:7796–7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- Rosental B., Brusilovsky M., Hadad U., Oz D., Appel M.Y., Afergan F., Yossef R., Rosenberg L.A., Aharoni A., Cerwenka A. Proliferating cell nuclear antigen is a novel inhibitory ligand for the natural cytotoxicity receptor NKp44. J. Immunol. 2011;187:5693–5702. doi: 10.4049/jimmunol.1102267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Ota S., Seshimo H., Yamazaki Y., Nomura S., Ito T., Miki J., Ota M., Fukushima H., Maeda H. Platelet transfusion refractoriness caused by a mismatch in HLA-C antigens. Transfusion. 2002;42:302–308. doi: 10.1046/j.1537-2995.2002.00051.x. [DOI] [PubMed] [Google Scholar]
- Saito Y., Ellegast J.M., Rafiei A., Song Y., Kull D., Heikenwalder M., Rongvaux A., Halene S., Flavell R.A., Manz M.G. Peripheral blood CD34(+) cells efficiently engraft human cytokine knock-in mice. Blood. 2016;128:1829–1833. doi: 10.1182/blood-2015-10-676452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo H., Chen S.J., Hashimoto K., Endo H., Nishi Y., Ohta A., Yamamoto T., Hotta A., Sawaguchi A., Hayashi H. A β1-tubulin-based megakaryocyte maturation reporter system identifies novel drugs that promote platelet production. Blood Adv. 2018;2:2262–2272. doi: 10.1182/bloodadvances.2018019547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya A., Campbell D., Hannum C., Yssel H., Franz-Bacon K., McClanahan T., Kitamura T., Nicholl J., Sutherland G.R., Lanier L.L., Phillips J.H. DNAM-1, a novel adhesion molecule involved in the cytolytic function of T lymphocytes. Immunity. 1996;4:573–581. doi: 10.1016/s1074-7613(00)70060-4. [DOI] [PubMed] [Google Scholar]
- Stanworth S.J., Navarrete C., Estcourt L., Marsh J. Platelet refractoriness—practical approaches and ongoing dilemmas in patient management. Br. J. Haematol. 2015;171:297–305. doi: 10.1111/bjh.13597. [DOI] [PubMed] [Google Scholar]
- Strowig T., Chijioke O., Carrega P., Arrey F., Meixlsperger S., Rämer P.C., Ferlazzo G., Münz C. Human NK cells of mice with reconstituted human immune system components require preactivation to acquire functional competence. Blood. 2010;116:4158–4167. doi: 10.1182/blood-2010-02-270678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto N., Eto K. Platelet production from induced pluripotent stem cells. J. Thromb. Haemost. 2017;15:1717–1727. doi: 10.1111/jth.13736. [DOI] [PubMed] [Google Scholar]
- Szczepiorkowski Z.M., Dunbar N.M. Transfusion guidelines: when to transfuse. Hematol. Am. Soc. Hematol. Educ. Program. 2013;2013:638–644. doi: 10.1182/asheducation-2013.1.638. [DOI] [PubMed] [Google Scholar]
- Turner M., Leslie S., Martin N.G., Peschanski M., Rao M., Taylor C.J., Trounson A., Turner D., Yamanaka S., Wilmut I. Toward the development of a global induced pluripotent stem cell library. Cell Stem Cell. 2013;13:382–384. doi: 10.1016/j.stem.2013.08.003. [DOI] [PubMed] [Google Scholar]
- Vivier E., Raulet D.H., Moretta A., Caligiuri M.A., Zitvogel L., Lanier L.L., Yokoyama W.M., Ugolini S. Innate or adaptive immunity? The example of natural killer cells. Science. 2011;331:44–49. doi: 10.1126/science.1198687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiegmann B., Figueiredo C., Gras C., Pflaum M., Schmeckebier S., Korossis S., Haverich A., Blasczyk R. Prevention of rejection of allogeneic endothelial cells in a biohybrid lung by silencing HLA-class I expression. Biomaterials. 2014;35:8123–8133. doi: 10.1016/j.biomaterials.2014.06.007. [DOI] [PubMed] [Google Scholar]
- Wu J., Song Y., Bakker A.B., Bauer S., Spies T., Lanier L.L., Phillips J.H. An activating immunoreceptor complex formed by NKG2D and DAP10. Science. 1999;285:730–732. doi: 10.1126/science.285.5428.730. [DOI] [PubMed] [Google Scholar]
- Xu H., Wang B., Ono M., Kagita A., Fujii K., Sasakawa N., Ueda T., Gee P., Nishikawa M., Nomura M. Targeted disruption of HLA genes via CRISPR-Cas9 generates iPSCs with enhanced immune compatibility. Cell Stem Cell. 2019;24:1–13. doi: 10.1016/j.stem.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Zhang N., Zhi H., Curtis B.R., Rao S., Jobaliya C., Poncz M., French D.L., Newman P.J. CRISPR/Cas9-mediated conversion of human platelet alloantigen allotypes. Blood. 2016;127:675–680. doi: 10.1182/blood-2015-10-675751. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.