Summary
In mammals, LINE-1 (L1) retrotransposons constitute between 15% and 20% of the genome. Although only a few copies have retained the ability to retrotranspose, evidence in brain and differentiating pluripotent cells indicates that L1 retrotransposition occurs and creates mosaics in normal somatic tissues. The function of de novo insertions remains to be understood. The transdifferentiation of mouse embryonic fibroblasts to dopaminergic neuronal fate provides a suitable model for studying L1 dynamics in a defined genomic and unaltered epigenomic background. We found that L1 elements are specifically re-expressed and mobilized during the initial stages of reprogramming and that their insertions into specific acceptor loci coincides with higher chromatin accessibility and creation of new transcribed units. Those events accompany the maturation of neuronal committed cells. We conclude that L1 retrotransposition is a non-random process correlating with chromatin opening and lncRNA production that accompanies direct somatic cell reprogramming.
Keywords: LINE-1 elements, cell transdifferentiation, cell plasticity
Highlights
-
•
L1 activation accompanies induced dopaminergic neuron maturation
-
•
L1 inhibition impairs the transdifferentiation potential of MEFs
-
•
L1 retrotransposition creates a lineage-specific genetic mosaicism
-
•
L1 insertions correlates with open chromatin and lncRNA transcription
In this article, Orlando and colleagues show that LINE-1 elements are reactivated and retrotransposed upon transdifferentiation of MEF cells into induced dopaminergic neurons (iDA). The inhibition of LINE-1 elements impairs the transdifferentiation potential of MEF cells. LINE-1 retrotransposition in post-mitotic iDA cells is lineage specific and correlates with chromatin opening and lncRNA production in intergenic genomic regions.
Introduction
L1 retrotransposable elements (TEs) make up 15%–20% of the mammalian genomic DNA (Deininger and Batzera, 2002, de Koning et al., 2011). The mouse genome contains about 500,000 L1 copies, but only approximately 3,000 have retained the property to drive their expansion via a copy-paste mechanism (Mouse Genome Sequencing Consortium et al., 2002). Active elements (∼6.5 kb) are usually silenced by various mechanisms (DiGiacomo et al., 2013, Imbeault et al., 2017, Matsui et al., 2010, Van Meter et al., 2014), as they would be potentially disruptive by affecting genome structure and stability. However, recent reports indicate that L1 reactivation occurs in early development, before germ layer differentiation, and in the adult brain, revealing genomic mosaicism for L1 content in the soma (Coufal et al., 2009, Kano et al., 2009, Muotri et al., 2005, Perrat et al., 2013, Upton et al., 2015). In brain, L1 retrotransposition preferentially hits neuronal expressed genes, and L1-related structural variations include L1 de novo insertions as well as deletions of large fragments of genomic DNA (Erwin et al., 2016, Perrat et al., 2013, Upton et al., 2015).
L1 mobilization might influence gene expression, contributing to phenotype variation (Britten and Davidson, 1971, Chuong et al., 2016, Faulkner et al., 2009, Fort et al., 2014, Glinsky, 2015, Han and Boeke, 2005). In fact, somatic L1 retrotransposition has been extensively correlated to several pathogenic processes, such as neurological diseases, autoimmune disorders, and cancer (Bundo et al., 2014, Coufal et al., 2011, Guffanti et al., 2014, Muotri et al., 2010, Reilly et al., 2013, Thomas et al., 2017). Recent reports have shown that, even if rare (Evrony et al., 2012, Evrony et al., 2016), L1 retrotransposition occurs in the adult hippocampus, and its inhibition impairs long-term memory formation (Bachiller et al., 2017). In addition, L1 copy number variation (CNV) has been reported in a mouse model to be correlated with induced early-life stress (Bedrosian et al., 2018). Although the functional significance of this phenomenon remains to be elucidated, altogether this evidence indicates that L1 retrotransposition-induced genomic mosaicism occurs and influences brain function. The question remains open whether somatic L1 activation would be just a spurious event or a part of specific differentiation or more in general of developmental programs (Chuong et al., 2016).
In order to address this question, we used a post-mitotic somatic cell transdifferentiation model for direct conversion of mouse primary embryonic fibroblasts (MEFs) into induced neurons of the dopaminergic lineage (iDAs) (Caiazzo et al., 2011). Direct transdifferentiation converts a terminally differentiated cell into another one without pluripotency reinstatement (iPS), thus avoiding genome-wide heterochromatin erasure, aberrant retrotransposons reactivation, and preserving global epigenetic transcriptional regulation and genome integrity (Castro-Diaz et al., 2014, Friedli et al., 2014, Gkountela et al., 2015, Grow et al., 2015, Kunarso et al., 2010, Wissing et al., 2012). Using whole-genome sequencing (WGS), we report evidence for L1 reactivation and CNV upon cell identity conversion and a conserved and specific de novo insertion site profile involving iDA expressed gene loci. Further, RNA sequencing (RNA-seq) and assay for transposase-accessible chromatin sequencing (ATAC-seq) analysis revealed that L1 recipient loci show a more accessible chromatin in the proximity of L1 somatic insertion sites concomitant also with increased non-coding RNA (ncRNA) production. Finally, inhibition of L1 dynamics impaired iDA cell maturation, indicating a correlation between L1 reactivation and cell lineage conversion.
Results
L1 Retrotransposition Occurs during Transdifferentiation of Mouse Embryonic Fibroblasts into Dopaminergic Neurons
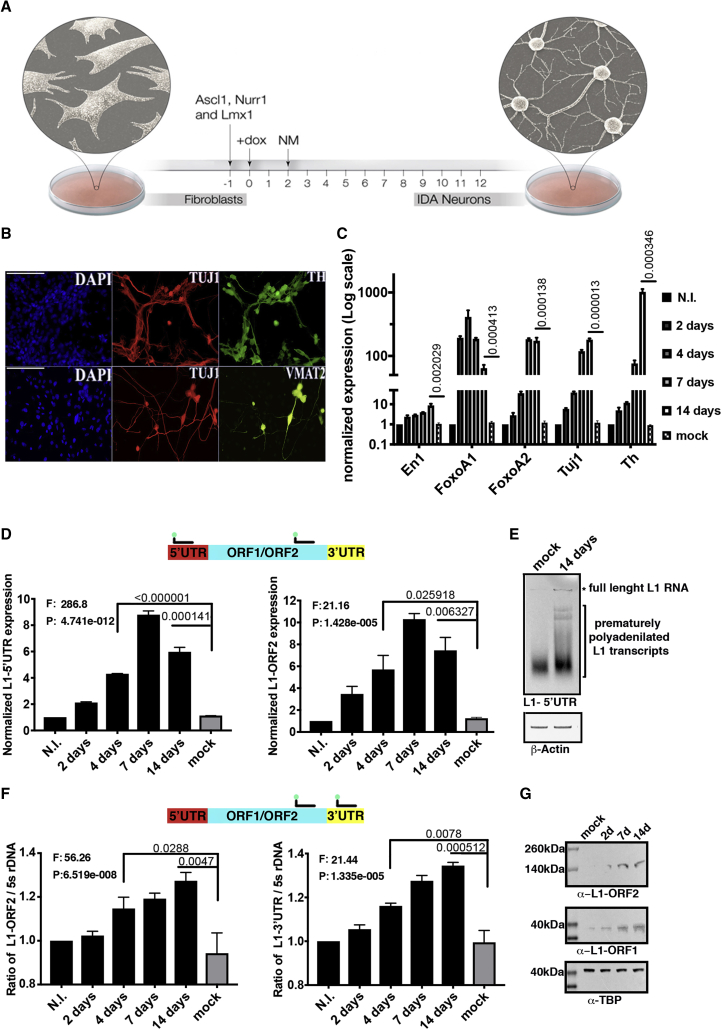
To verify L1 activity during cell transdifferentiation, we adopted a well-characterized protocol for direct conversion of post-mitotic MEFs to iDAs (Caiazzo et al., 2011) obtained by overexpression of three specific transcription factors (Nurr1, Ascl1, and Lmx1a) (Figure 1A). To optimize both production and isolation of fully transdifferentiated cells, we used MEFs carrying an enhanced green fluorescent protein (EGFP) reporter cassette under the control of tyrosine hydroxylase (Th) promoter. Reprogramming efficiency and cell conversion were confirmed by both immunofluorescence and real-time qPCR expression analysis of En1, FoxoA1, FoxoA2, Tuj1, and Th genes (Figures 1B and 1C).
Figure 1.
L1 Dynamics Occurs during MEF Reprogramming into iDA Cells
(A) Schematic representation of MEF transdifferentiation to iDA neurons. Expression of Ascl1, Nurr1, and Lmx1a is induced with doxycycline (dox) 24 h after infection. At 48 h post-induction, culture medium is replaced with a neuronal-inducing medium (NM).
(B) Immunofluorescent assay for principal neural commitment (TH, TUJ1, and VMAT2) and dopaminergic neuron markers (TH).
(C) Expression levels of iDA-specific transdifferentiation markers. Data are represented as mean with SEM. N = 6. T Test results are showed in the plot.
(D) Expression levels of full-length (5′UTR) and truncated (L1-ORF2) L1 elements during MEF transdifferentiation. RNA expression was measured in non-infected (N.I.) cells, mock cells and after 2, 4, 7, 14 days upon transdifferentiation. SEM and one-way ANOVA (F and p value) are indicated; t test is also showed in the plot. n = 3.
(E) Northern blot analysis of poly(A) + LINE-1 RNA. Levels of b-actin RNA have been used as loading control.
(F) Amount of L1-ORF2 and L1-3′ UTR DNA, normalized on 5s rDNA content during transdifferentiation. SEM and one-way ANOVA are indicated. n = 6. See also Figure S1.
(G) Western blot analysis of LINE-1 ORF2 and L1-ORF1 protein production. Levels of TBP protein have been used as loading control.
The expression of active L1 elements was measured by a multiplex TaqMan qPCR assay using an FAM-labeled probe specific for the L1 5′ UTR or L1-ORF2 region (Bedrosian et al., 2018), together with a VIC-labeled probe, for the detection of TBP mRNA as an internal normalizer (Figure 1D). We confirmed the production of full-length LINE-1 RNA in iDA cells by northern blot using a probe complementary to the 0–600 base pair (bp) region of the L1 5′ UTR transcript (Figures 1E and S1B-left) (Deininger and Belancio, 2016). Further, by western blot analysis, we verified the production of the ORF2 protein encoded by the polycistronic L1 full-length transcript on MEF and iDA cells. Total protein extracts were probed using a custom-made antibody specific for mouse L1-ORF2 and L1-ORF1 proteins. Consistently with the L1 RNA expression profile, the ORF2 protein signal is detectable from day 7, further increasing in fully transdifferentiated iDA cells (Figures 1F and S1A).
The relative genome content of all L1 elements (both full length and 5′-truncated) was measured by a second set of TaqMan assays specific for the L1-ORF2 or L1- 3′ UTR DNA sequences (Bedrosian et al., 2018). Time-course experiments revealed a specific trend of reactivation and increase in CNV during cell conversion, starting from day 4 (Figures 1F and S1B-right).
Then, we measured the L1 CNV in TH+ iDA cells and residual TH− cells; only the genome of fully transdifferentiated cells displayed a higher amount of L1 elements. In parallel, we compared the number of copies of L1s and other retrotransposable elements, such as active and autonomous intracisternal A-type particle (IAP) elements and non-autonomous SINE B1 and B2 elements. Notably, during transdifferentiation, only L1s and not other potentially mobile elements are expanding in the genome (Figures S1C and S1D).
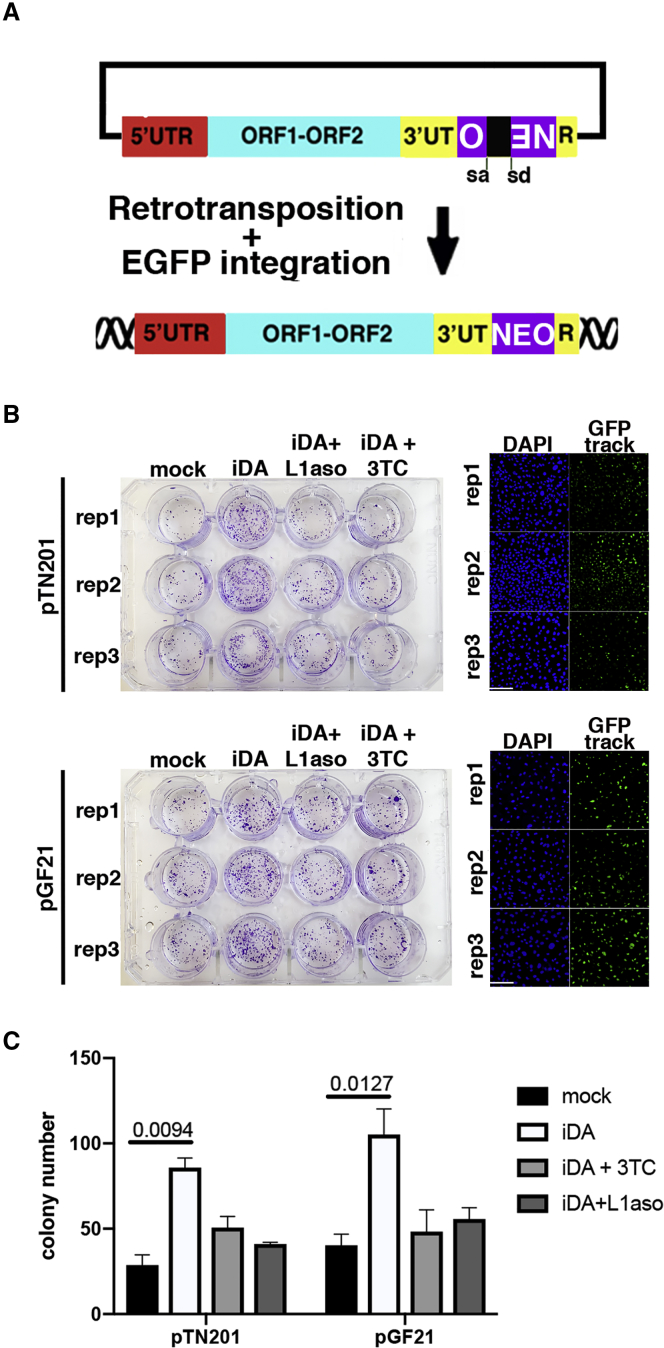
Finally, to prove that transdifferentiating post-mitotic cells are compatible and support the retrotransposition machinery, we overexpressed two different exogenous mouse L1 element (pGF21 and pTN201 plasmids in wild-type MEF cells, which were generously provided by Prof. John Goodier) with an antisense oriented neomycin resistance cassette interrupted by an intron inside the 3′ UTR region (MacIa et al., 2017, Ostertag et al., 2000) (Figure 2A). We verified the integration of the exogenous L1 element by selecting neomycin-resistant cells. Neomycin-resistant iDA colonies were counted through a crystal violet staining assay (Figures 2B and 2C). Indeed, transdifferentiated iDA cells show a higher rate of pGF21 and pTN201 retrotransposition, confirming the L1 reactivation trend detected by qPCR and western blot.
Figure 2.
L1-mNEOMYCINi Retrotransposition Assay
(A) LINE1 retrotransposition assay with pGF21 and pTN201 L1-Neo reporter plasmids. The scheme depicts the rationale of the assay.
(B) Retrotransposition of an engineered exogenous LINE1-Neomycin sequences in the genome has been evaluated by drug selection and crystal violet colony screening as in (MacIa et al., 2017). Plasmid expressing LINE1-Neomycin have been transfected in 3 independent preparation of MEF cells before induction of transdifferentiation. Electroporation efficiency is showed in the picture using a GFP track vector.
(C) Quantification of neomycin-resistant cell colonies from crystal violet assay. n = 3, SEM and t test are indicated.
Block of L1 Dynamics Affects the Efficiency of iDA Cell Transdifferentiation
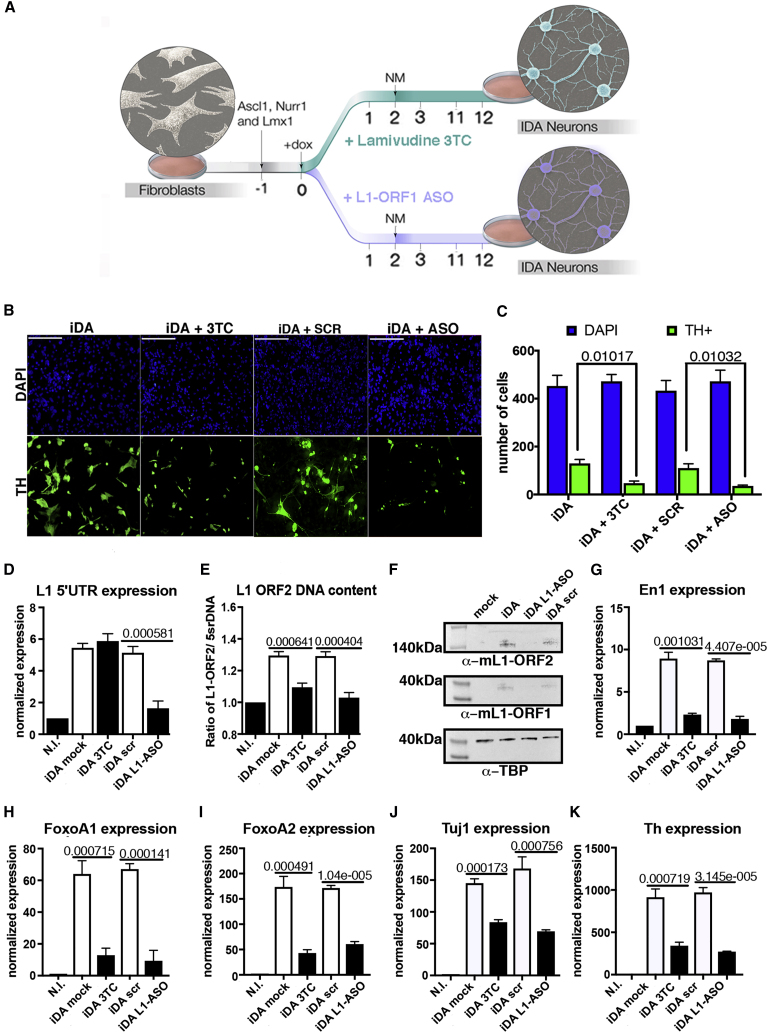
To investigate L1 reactivation events during the acquisition of in vitro-induced dopaminergic neuronal phenotype, we treated the fibroblasts undergoing conversion with either lamivudine (3TC), which blocks reverse transcriptase enzymes essential for L1 retrotransposition (Wood et al., 2016), or 2-deoxy-2-fluoroarabinonucleic acid-modified antisense oligonucleotides (F-ANA ASO) to knock down full-length L1 RNA (Bachiller et al., 2017, Thomas et al., 2017) (Figure 3A).
Figure 3.
LINE-1 Inhibition Severely Affects Fibroblast Transdifferentiation
(A) Schematic representation of the transdifferentiation protocol under treatment with reverse transcriptase inhibitor Lamivudine(3TC) and LINE-1 anti sense oligonucleotide (L1-ASO) treatment.
(B) Immunofluorescent assay for transdifferentiation efficiency (Th marker in green) in iDA cells treated with Lamivudine or L1-ASO compared to mock (1×PBS) or scramble ASO respectively. Nuclei were counterstained with DAPI (blue).
(C) Number of transdifferentiated Th+ cells (green) upon 3TC or L1-ASO treatment. The plot shows the quantification of 6 fields from 3 independent experiments. Scale bar= 100 µm.
(D) Expression levels of L1 full-length elements during MEF transdifferentiation. RNA expression was measured in non-infected cells (N.I.) and after 14 days of differentiation. SEM and two-tailed t test are indicated. n = 6.
(E) Copy Number of L1 full-length elements during MEF transdifferentiation. L1 CNV was measured in non-infected cells (N.I.) and after 14 days of differentiation. SEM and two-tailed t test are indicated. n = 6.
(F) Western Blot analysis of L1 ORF2 and L1-ORF1 protein production in wt and L1-ASO treated iDA cells. Level of TBP protein have been used as loading control.
(G) Expression levels of En1 gene in not infected (N.I.) cells and after 14 days of differentiation in 3TC or L1 ASO treated cells. SEM and two-tailed t test are indicated; n = 6.
(H) Expression levels of FoxoA1 gene in not infected (N.I.) cells and after 14 days of differentiation in 3TC or L1 ASO treated cells. SEM and two-tailed t test are indicated; n = 6.
(I) Expression levels of FoxoA2 gene in not infected (N.I.) cells and after 14 days of differentiation in 3TC or L1 ASO treated cells. SEM and two-tailed t test are indicated; n = 6.
(J) Expression levels of Tuj1 gene in not infected (N.I.) cells and after 14 days of differentiation in 3TC or L1 ASO treated cells. SEM and two-tailed t test are indicated; n = 6.
(K) Expression levels of Th gene in not infected (N.I.) cells and after 14days of differentiation in 3TC or L1 ASO treated cells. SEM and two-tailed t test are indicated; n = 6.
See also Figure S2.
Inhibition of L1 retrotransposition with either 3TC or ASO reduced the number of TH positive cells by 30%–35% (Figures 3B–3F) and deregulated the expression of transcription factors and structural proteins, typical markers of dopaminergic neurons (Figures 3G–3K, S2A, and S2B). These results indicate that suppression of L1 dynamics affected fibroblast conversion into iDA cells, which underscores a supportive role played by L1 retrotransposition in this process. Notably, a TaqMan multiplex qPCR assay on both RNA and genomic DNA showed that FANA ASOs efficiently prevented full-length L1 transcript accumulation and retrotransposition, although lamivudine prevented only the expansion of L1 without affecting L1 expression (Figures 3D–3F). To exclude possible biases due to 3TC toxicity, we controlled cell viability in mock and 3TC-treated cells with propidium iodide-based cell-cycle fluorescence-activated cell sorting analysis, reporting no effects (Figure S2C). Finally, the transfection of an exogenous L1-ASO-resistant mouse active L1 element (pTNC7 plasmid, kindly donated by Edith Heard) partially rescued the transdifferentiation rate in MEF cells treated with L1 ASO (Figure S2D).
Whole-Genome Sequencing Reveals a Specific L1 Insertion Profile in the Genome of iDA Cells
Next, we investigated the consequences at the genome-wide level of L1 reactivation. To map L1 retrotransposition events in the genome of the reprogrammed cells, we performed WGS on parental MEF cells, sorted TH+ iDA neurons and lamivudine-treated TH+ cells. WGS data were filtered with Mobster (Ewing, 2015, Thung et al., 2014) to search for active, non-reference mobile element insertions (MEIs), taking the MEF genome as reference. From this first list (Figure S3A), we isolated the anchors containing L1 elements present uniquely in the iDA cell genome and discarded those shared with the MEF parental cells. Furthermore, we considered only the insertions supported by at least five reads from both the 5′ and 3′ junctions, common to at least two biological replicates and with less than 50-bp of insertion shift between replicates. Further, for each insertion, we verified the presence of target site duplications/deletions, target size duplication sequences, the L1 truncation pattern, the motif of the L1 insertion site, and the allelic frequency (Data S1).
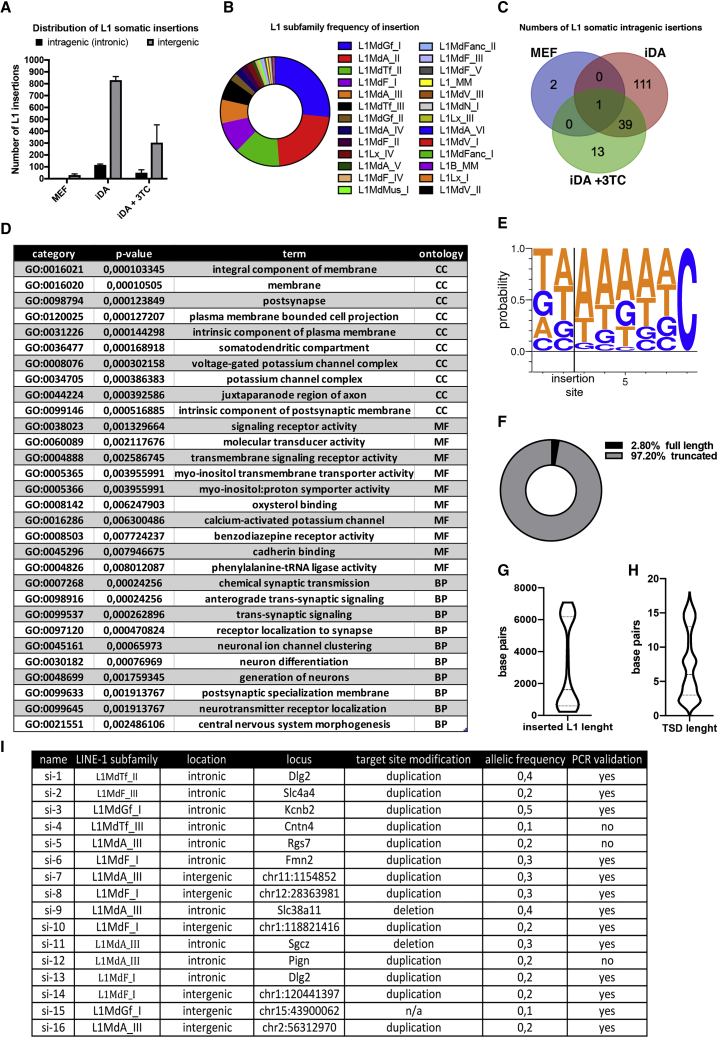
We cataloged about 800 intergenic and 111 intragenic (intronic) insertions unique to the iDA cell genome and undetectable in MEF and 3TC-treated cells; more than the 90% of the insertions were from L1MdGf, L1MdA, L1MdTf, and L1MdF elements (Figures 4A–4C). Most of the insertions were consistent among biological replicates in terms of genomic location, indicating a non-random L1 somatic insertion profile (Figures S3A–S3C). Gene ontology analysis showed that the 111 acceptor genes found in iDA cells were enriched for neuronal functions (Figure 4D), while the few genes receiving insertions in MEFs or 3TC-treated cells are not involved in neuronal functions. This included the Dlg2 neuronal gene, reported in previous studies to be an L1 acceptor gene (Erwin et al., 2016, Muotri et al., 2005). The calculated average frequency per cell was (∼0.3) in line with the detected range shown in other reports (Data S1) (Evrony et al., 2016, Evrony et al., 2012, Paquola et al., 2017, Treiber and Waddell, 2017).
Figure 4.
Analysis of L1 De Novo Insertions in Reprogrammed and Lamivudine-Treated Cells
(A) Whole genome sequencing data of the distribution of L1 somatic insertions supported by more than five reads on each of 5’ and 3’ ends of inserted sites that were consistently inserted in more than two replicates between 0 and 50 base pairs in different replicates. SEM is indicated in the plot. n = 3.
(B) Representation of the frequency of retrotransposition of L1 subfamilies in iDA cells genome.
(C) Venn diagram showing different intragenic L1 de novo insertion rates.
(D) Functional analysis of L1 acceptor genes specific to MEF, iDA and 3TC-treated iDA cells using Gene Ontology.
(E) L1 insertion site motif LogoPlot.
(F) percentage of full length de novo inserted L1s.
(G) Truncation pattern of L1 somatic insertions.
(H) Target Size Duplication (TSD) size distribution. Table showing PCR validation results of 16 L1s somatic insertions.
(I) Selected L1 insertions used for PCR validation.
See also Figures S3 and S4.
For validation, we randomly selected 16 candidate loci of the junction between the 5′ end of the de novo inserted L1 element and the flanking genome. Thirteen of 16 candidate insertions were successfully PCR amplified on DNA from an independent experiment and Sanger sequenced to control the structure of the junction between the L1 and the flanking genome (Figure 4I, Figures S4A and S4B). No residual insertion amplification in both MEF cells and TH+ 3TC-treated cells has been detected (Figures S4A and S4B). In addition, few insertions were further validated through an empty/filled locus amplification screening. The acceptor site containing a retrotransposed L1 element was amplified in each iDA replicate and the heavier locus band Sanger sequenced (Figure S5A). Taken together, these results show that neuronal fate induction is accompanied by L1 retrotransposition, affecting regions of the genome, relevant for neuronal lineage commitment and neuron function.
L1 Retrotransposition Follows the Specificity of Neuronal Gene Expression Profile
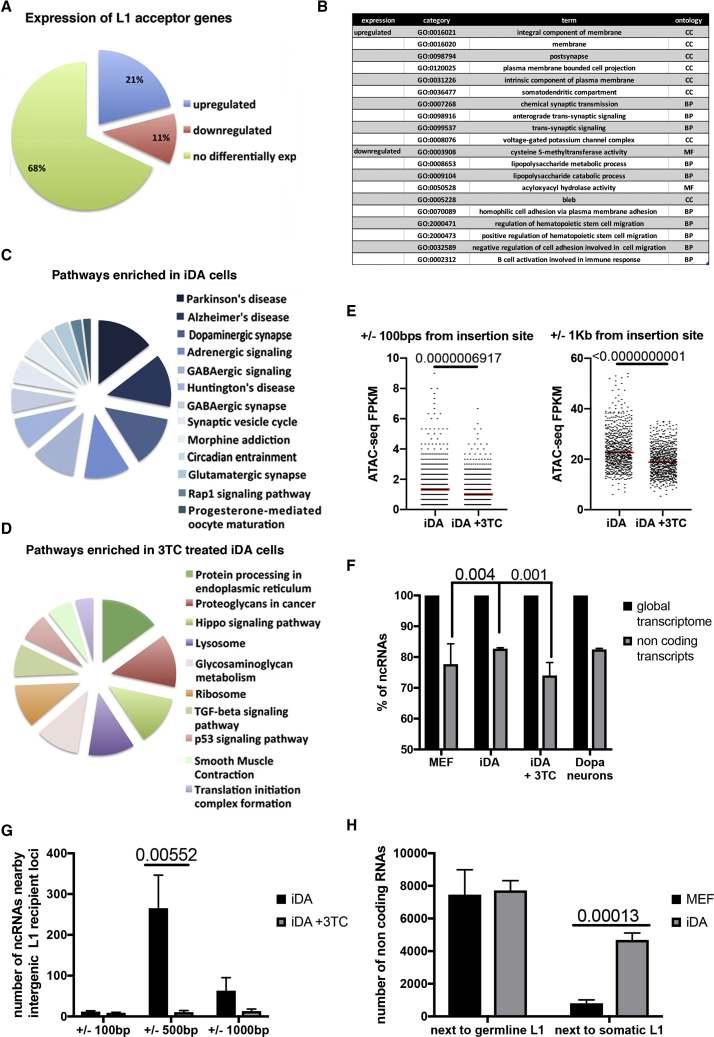
The impact of L1 somatic retrotransposition on the neuronal genetic program remains to be understood. Using RNA-seq analysis of MEFs, iDAs, and 3TC-treated cells, we sought to investigate the connection between the L1 de novo insertion profile and the iDA cell differentiation program. Among all genes that accepted L1 insertions, 32% were differentially expressed in transdifferentiated cells (Figures 5A and 5B). We also looked into the transcriptional behavior of genes proximal to L1 acceptor genes to see if L1 insertions exerted any influence on nearby genes, but we could not find any specific transcriptional pattern (Figure S5B).
Figure 5.
De Novo Insertions Support the Establishment of a Cell-Type-Specific Gene Expression Profile that Is Disrupted upon Lamivudine Treatment
(A) Differential expression analysis of iDA-specific acceptor genes showing up, down, and not differentially expressed. False discovery rate (FDR) of 0.05 and a minimum fold change of 2 on a log2 scale have been considered in the analysis.
(B) Gene Ontology analysis of up- and down-regulated L1 acceptor genes.
(C) RNA-seq pathway analysis of differentially expressed pathways in iDA neurons compared with MEF cells.
(D) RNA-seq pathway analysis of differentially expressed pathways in 3TC-treated iDA neurons compared with mock iDA neurons.
(E) ATAC-seq analysis results at intergenic L1 recipient loci in iDA and 3TC treated cells at different distances from the insertion site; t tests are indicated. n = 3.
(F) Quantification of novel non-coding transcripts produced within ±100, ±500, ±1,000 bp from the somatic L1 element in iDA and 3TC-treated iDA neurons. SEM and t test are indicated. n = 3.
(G) Total amount of long non-coding transcripts produced in MEF and transdifferentiated cells within ±1,000 bp of de novo inserted and germline L1 elements. SEM and t test are indicated in the plot. n = 3.
(H) RNA-seq de novo assembly transcriptome analysis of MEF, iDA, dopaminergic neurons, and 3TC treated iDA cells. The percentage of non-coding transcripts longer than 200 bp has been normalized over the whole transcriptome. SEM and two-tailed t test are indicated in the plot. n = 6.
See also Figure S5.
A comparison between the most significantly represented pathways (Kyoto Encyclopedia of Genes and Genomes database) in iDA neurons and MEF cells reveals that transdifferentiation produces a radical change in cell structures and functions (Figures 5C and S5C). Conversely, L1 inhibition during the transdifferentiation process prevents upregulation of the pathways required for dopaminergic neuron functionality, reflecting incomplete cell conversion (Figures 5D and S5C).
The somatic insertion of an engineered L1 element was shown to increase the expression of the neuron-specific recipient gene (Muotri et al., 2005). A possibility could be specific changes in chromatin accessibility. To test this hypothesis, we used ATAC-seq analysis (Buenrostro et al., 2013) to compare the chromatin structure next to L1 acceptor sites in iDA cells and 3TC-treated cells, which share the same culture conditions and overexpression of the same lineage-specific transcription factors but differ only for L1 insertions profile. Notably, to avoid interference of induced active genes, we selected acceptor loci far from known transcribed units. ATAC-seq analysis showed an increased chromatin accessibility of recipient loci when L1 somatic insertion was present (Figure 5E).
L1 elements might be capable of tuning cell-type-specific expression by acting as regulatory sequences (Glinsky, 2015, Glinsky, 2016) and creating new transcription units (Chuong et al., 2016, Kelley and Rinn, 2012, Kunarso et al., 2010). The finding that L1 somatic insertions in iDA cells correlate with more open chromatin and nearby long non-coding RNA (lncRNA) production is in line with the previously reported connection between retrotransposons and evolution of lineage-specific lncRNAs (Johnson and Guigo, 2014, Kapusta et al., 2013). We performed transcriptome assembly from RNA-seq data, and we measured the percentage of the global long non-coding transcriptome. In iDA cells, compared with MEFs, we detected an increase (8%–10%) of the global non-coding transcriptome consistent with the increase in the L1 number of copies (Figure 5F). Transdifferentiated 3TC-treated cells showed a reduced level of non-coding transcripts. We also compared the novel non-coding transcripts detected in iDA neurons with previously published RNA-seq data of midbrain dopaminergic neurons. Further, iDA and midbrain dopaminergic neurons share an equivalent global amount of long non-coding transcripts (Figure 5F) (PRJNA271250) (Brichta et al., 2015).
To correlate lineage-specific L1 insertions, chromatin accessibility and transcription, we analyzed lncRNA expression at distances as close as 100 bp, 500 bp, and 1 kb around all L1 insertion sites and measured the number of transcripts produced in iDA cells and 3TC-treated cells. We detected de novo or increased production of non-coding RNAs at 500 bp and 1,000 bp in iDA- compared with 3TC-treated cells (Figure 5G). None of the non-coding transcripts contained portions of L1 sequences.
We further investigated the rate of non-coding RNA expression from intergenic regions flanking a de novo inserted L1 element or a germline L1 element. This analysis showed a significantly increased production of long non-coding transcripts around somatic L1 insertions loci in iDA cells compared with the same loci devoid of L1 in MEFs (Figure 5H). In contrast, regions flanking a germline L1 element, present in both MEF and iDA cells, do not show different lncRNA expression upon transdifferentiation (Figure 5H).
We conclude that L1 somatic insertions strongly correlate with both a more open chromatin state of recipient loci and lncRNA production in close proximity to the de novo insertion sites.
Discussion
As L1 reactivation appears to follow brain development, our work addressed the question whether L1 retrotransposition would accompany neuronal cell specialization also in a direct transdifferentiation context. We found that reactivation of the dynamics of L1 elements is elicited in the initial stages of MEF reprogramming and accompanies cell maturation to the complete acquisition of the fate of dopaminergic neurons (Figures 1 and 2). When retrotransposition was prevented, the efficiency of conversion of MEFs to iDA cells was severely impaired, as confirmed by immunofluorescence staining, RT-qPCR analysis of transdifferentiation markers, and RNA-seq-based signaling pathway analysis (Figures 3, 4, and 5).
In order to gain a closer estimate of the de novo insertions, we performed paired-end WGS. The selection of reliable analytical methods for L1 somatic variant identification in the genome has been a debated topic. In particular, doubts have been raised about identification of false-positive events generated by targeted sequencing approaches or single-cell sequencing coupled with a massive whole-genome amplification. The PCR-free WGS approach excluded biases introduced by the capturing of non-specific DNA sequences or PCR amplification. Moreover, the Mobster tool used for WGS data analysis mapped MEIs with a high level of confidence, limiting the number of false positives (Evrony et al., 2016, Ewing, 2015, Thung et al., 2014).
iDA cells showed an accumulation of L1 somatic insertions after transdifferentiation with an average allelic frequency of 0.3 (Figure 4), in line with recently reported studies (Erwin et al., 2016, Evrony et al., 2016). Further, Gene Ontology analysis of the 111 intragenic insertions present only in iDA cells revealed a preference for insertions into lineage-specific active genes, in keeping with previous observations and correlations between L1 retrotransposition and mental disorders (Erwin et al., 2016, Upton et al., 2015). The presence of cell-specific somatic insertions and recurring among individuals has been described recently in Drosophila oocytes for both LTR and LINE-type transposons (Wang et al., 2018).
TEs spreading in intergenic regions and evolution of both regulatory elements and lncRNAs appear to be tightly linked, indicating a key contribution of TEs sequences in influencing somatic gene regulatory networks (Chuong et al., 2016, Han and Boeke, 2005, Johnson and Guigo, 2014, Kapusta et al., 2013, Kelley and Rinn, 2012, Kunarso et al., 2010, Sundaram et al., 2014). As a possible impact of the L1 insertion on gene expression, we integrated the WGS data with RNA-seq data. In the iDA cells, one-third of the genes carrying L1 insertion cells appeared to be differentially expressed (Figures 5A–5D) following the induced phenotypic change of the cell. Conversely, lamivudine-treated cells, despite priming by overexpression of lineage-specific transcription factors, did not accumulate L1 at the same loci and failed to reprogram to dopaminergic cell fate (Figures 2, 3, 4, and S5A; Data S1). Through ATAC-seq analysis, we provide evidence that L1 acceptor sites are more accessible upon insertion than 3TC-treated cells lacking L1 retrotransposition (Figure 5E). The integration of WGS and ATAC-seq with RNA-seq suggests that enhanced chromatin activity due to L1 retrotransposition might elicit the expression of novel non-coding transcripts (Figures 5F–5H) as proposed also in other reports. Future work will shed light on the mechanistic link between L1 reactivation and activation of lineage-specific genetic programs, a theme relevant for direct cell reprogramming-based technologies and their applications.
Experimental Procedures
iDA Cells Reprogramming
MEF transdifferentiation experiments were performed as described in Caiazzo et al. (2011). Third-generation lentiviral vectors containing the coding sequence of Nurr1, Ascl1, and Lmx1a transcription factors under the control of the tetracycline operator were packaged in 293T cells. MEF cells containing the TH-EGFP reporter cassette (25,000 cells/cm2) were infected in MEF medium (DMEM; Gibco) containing 10% fetal bovine serum (Gibco), non-essential amino acids (Gibco), sodium pyruvate, and penicillin/streptomycin (Gibco). At 16–20 h after infection, cells were switched into fresh MEF medium containing doxycycline (2 mg/mL; Sigma). After 48 h, the medium was replaced with serum-free neuronal-inducing medium: DMEM/F12 (Gibco), 25 μg/mL insulin (Sigma), 50 μg/mL transferrin (Sigma), 30 nM sodium selenite, 20 nM progesterone (Sigma), 100 nM putrescine (Sigma), and penicillin/streptomycin (Sigma) containing doxycycline. Lamivudine (3TC, Sigma; 10 mg/mL in PBS), at a final concentration of 150 μM, was added to proliferating fibroblasts after lentiviral infection until the end of transdifferentiation, and the medium was refreshed every 24 h. A mock experiment was performed with an equal volume of PBS. Cell transdifferentiation was induced with doxycycline the day after the initial 3TC treatment.
GFP+ TH-GFP MEF cells were sorted with a BD FACSAria III. Wild-type MEFs were used as negative control for the gating of the cell sorter.
Retrotransposition Assays and Antisense Oligonucleotide Delivery
Primary MEFs were electroporated with plasmid pGF21 and pTN201 (kindly provided by John Goodier, Johns Hopkins University) using a Neon Electroporator (Life Technologies; program: 1,650 V, 30 ms pulse) expressing an active L1 element with the neomycin resistance cassette inside its 3′ UTR (Ostertag et al., 2000). After transfection, confluent cells were transdifferentiated and collected after 14 days of transdifferentiation. Control MEF cells were collected 14 days post transfection. Transfection efficiency was monitored with a GFP track plasmid (Addgene no. 30456).
FANA (2-deoxy-2-fluoroarabinonucleic acid)-modified antisense oligonucleotides specific for four different LINE-1 ORF1 regions was delivered by gymnosis following the manufacturer’s instructions (AUMbiotech). Scramble antisense oligonucleotide was used as control. LINE1 RNA knockdown efficiency was controlled by TaqMan PCR. L1-specific antisense oligonucleotide sequences were as follows: L1-ASO #1, TTG ACC TTT CTC CCT TAC TGC; L1-ASO #2, ATA TGT TAC TTG ACC TTT CTC; L1-ASO #3, TGT AAT TCT GAT AGG CCT TCC; L1-ASO #4, AGT GTC TGT ATA ACA TCT GTC.
DNA Extraction and Quantification
High-molecular-weight genomic DNA was isolated using standard phenol-chloroform-isoamyl alcohol (Sigma-Aldrich) extraction techniques. Extracted DNA size and quality were verified with a NanoDrop spectrophotometer and 0.8% agarose gel electrophoresis. After quantification with Quant-iT PicoGreen dsDNA kit (Thermo Scientific), genomic DNA (gDNA) was diluted to a final concentration of 1 ng/μL RNase (RNase Cocktail, Invitrogen) and Exonuclease 1 (Thermo Scientific) treatment.
RNA Extraction and cDNA Preparation
Total RNA was extracted with an RNeasy Mini kit plus (QIAGWN) according to the manufacturer's instructions. RNA quality and concentration were checked with the NanoDrop spectrophotometer. cDNA was produced from 100 ng of RNA from each sample with a Superscript III first-strand cDNA synthesis system (Thermo Fisher).
Gene Expression Analysis
Real-time PCR analyses were performed with 7900HT Fast Real-Time PCR (Applied Biosystems).
Quantitative PCR was performed using Sybr Select Master mix (Life Technologies) and Ct values were calculated by 7900HT Fast Real-Time PCR RQ manager software (Applied Biosystems). Quantification was normalized as ΔCt between the gene of interest and the housekeeping gene Tbp. Primer sequences used in the analysis were as follows (5′-3′): m-Tbp-fw, CTT CCT GCC ACA ATG TCA CAG; m-Tbp-rev, CCT TTC TCA TGC TTG CTT CTC TG; m-En1-fw, CGC CTG GGT CTA CTG CAC A; m-En1-rev, TCT TCT TTA GCT TCC TGG TGC G; m-Foxoa1-fw, GAA GGG CAT GAG AGC AAC GA; m-Foxoa1-rev, ACA GGG ACA GAG GAG TAG GCC; m-Foxoa2-fw, ACG AGC CAT CCG ACT GGA G; m-Foxoa2-rev, GGC GTT CAT GTT GCT CAC G; m-Tuj1-fw, CAG GCC CGA CAA CTT TAT CT; m-Tuj1-rev, CTC TTT CCG CAC GAC ATC TA; m-Th-fw, CTC ACC TAT GCA CTC ACC CGA; m-Th-rev, GGT CAG CCA ACA TGG GTA CG.
TaqMan PCR for L1 Expression and CNV Analysis
Quantitative PCR experiments were performed on a 7900HT Fast Real-Time PCR (Applied Biosystems) apparatus. L1 genomic content was measured by multiplex qPCR TaqMan assay with probes specific for L1-ORF2 and M-Satellite DNA (SATA) as internal control. In each reaction, two TaqMan probes, labeled with FAM and VIC fluorophores, were combined with 1 ng of gDNA, target-specific primers, and iQ multiplex Powermix (Bio-Rad) in a total volume of 20 μL. Tbp TaqMan premade assay was used as an internal normalizer (Thermo Fisher catalog no. 4333769F). TaqMan probes and primers for LINE-1, 5s rDNA, SINE-B1, SINE-B2, and IAP transposable element expression, and CNVs are described in Bedrosian et al. (2018).
Immunofluorescence Assay
Immunofluorescence assays were performed as described in Caiazzo et al. 2011. Primary antibodies used TH (Millipore ab-152), TUJ1 (Abcam ab-18207, www.abcam.com/beta-III-Tubulin-antibody-ab18207.pdf), VMAT2 (Chemicon, ab1598p). Detection was performed for 40 min at room temperature with the following secondary antibodies: goat-anti-mouse immunoglobulin (Ig)G (Alexa Fluor 594; Thermo Scientific); chicken anti-rabbit IgG (Alexa Fluor 488; Thermo Scientific), dilution 1:1,000. Nuclei were stained with a dilution of 1:5,000 of 5 mg/mL DAPI solution.
A scale bar is indicated in each image, and the dimension of the scale bar is specified in the figure legend.
Western Blot
Total protein extracts were prepared by lysing cells in extraction buffer (HEPES KOH [pH 8.5], NaCl 400 mM, EDTA 0.1 mM, EGTA 0.1 mM, DTT 1 mM, 1× protease inhibitor, SDS 1%). Proteins were separated by electrophoresis on BOLT 4%–12% bis-tris polyacrylamide precast gels in MOPS buffer (Life Technologies). Anti-mLINE1-ORF2 (custom made, Eurogentek) was raised in guinea pigs against the mouse-specific C-term of the LINE-1 ORF2 protein (details and ELISA test available upon request), anti-mouse L1-ORF1 (Abcam EPR21844-108), anti-TBP (Santa Cruz Biotechnology, SC-273). Horseradish peroxidase-conjugated secondary antibodies were revealed with the ECL chemoluminescence kit (Amersham) and signals detected with ChemiDoc (Bio-Rad).
Northern Blot
Total RNA was extracted as before and poly(A)+ transcripts were isolated with a poly(A) spin RNA isolation kit (NEB S1560S). Isolated poly(A)+ RNA was run on a 2% agarose denaturing gel (Thermo Scientific #AM8678). Northern blot was performed using the NorthermMax-Gly Kit (Thermo Scientific #AM1946–#AM8672). Nylon membrane was hybridized at 39°C with 21-nt-long biotinylated probes (designed and produced by Stellaris-LGC Bioscience) complementary to the 0–600 bp region of the mouse L1 5′ UTR. β-Actin predesigned RNA probes were used as loading control following the manufacturer’s instructions (Sigma-Aldrich #11498045910). The signal was revealed using a chemiluminescent Nucleic Acid Detection Kit (Thermo Scientific #89880) and chemiluminescence was detected with ChemiDoc (Bio-Rad)
Genomic DNA Library Preparation, Whole-Genome Sequencing, and Analysis
Genomic libraries were prepared using a PCR-free Illumina TruSeq LT DNA kit. DNA was extracted from MEFs, induced dopaminergic cells transdifferentiated until day 14 (iDA14), and transdifferentiated cells treated with reverse transcriptase inhibitor lamivudine (3TC; Sigma-Aldrich) (iDA14 + 3TC). Sequencing was performed using an Illumina HiSeq2000 sequencer (KAUST Core Lab) (45× minimum coverage). For quality control, we used FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc/) and Trimmomatic v0.38 (http://www.usadellab.org/cms/?page=trimmomatic). Genomic reads were then mapped to the Mus musculus (mm10) reference genome using the BWA v0.7.17 genome aligner (http://bio-bwa.sourceforge.net/) (Langmead et al., 2009, Li and Durbin, 2009) with default parameters. The resulting Bam files were used as input to Mobster v0.1.6 (sourceforge.net/projects/mobst) (Thung et al., 2014) to predict and characterize de novo MEIs and their genomic position as previously reported. Briefly, Mobster searches for candidate active non-reference MEIs using discordant read pairs with at least one uniquely mapped read (anchors for the possible insertion). Mates of the anchoring reads are then mapped to consensus ME sequences. We obtained LINE1 consensus sequences from RepeatMasker v4.0.7 (http://www.repeatmasker.org/) (Smit et al., 2013) and the RepBase database (Jurka et al., 2005) (http://www.girinst.org/). Anchors in the same strand and supportive of the same ME family were clustered together. Then, forward-strand and reverse-strand clusters indicative of the same MEI event are joined. To avoid predicting MEIs already present in the reference, all predictions are filtered with a prediction window within 90 bp of an annotated MEI of the same ME family as the predicted MEI. Mobster results were further filtered to isolate only insertions supported by at least five reads and common between at least two biological repeats for each sample. Karyotype was controlled using Aneufinder with standard parameters.
For downstream validations, we reconstructed a virtual reference genome for each predicted insertion and designed primers across each MEI junction between the 5′ end of the L1 and the flanking genome.
PCR Validation of De Novo LINE1 Insertions
Candidate de novo insertions from WGS were validated by PCR on high-molecular-weight genomic DNA (15–20 kb average size) obtained from MEF cells, iDA cells, and 3TC-treated cells. For this assay, a forward primer was designed on the genomic flanking region and a reverse primer on the 5ʹ end of the de novo inserted LINE1 element to produce amplicons with size between 200 and 300 bp. PCR was performed with Q5 HF 2× master mix (NEB). PCR products were analyzed with standard 2% agarose gel electrophoresis (Bio-Rad). Image acquisition was performed with the ChemiDoc image system (Bio-Rad). PCR fragments were cloned into an entry vector using a TOPO-TA cloning kit (Life Technologies) and Sanger sequenced to verify the sequence of LINE-1 de novo insertion sites. Primers used for validations were as follows (5′-3′): SI-1 FW TGT CGG GCA GAG AGA GCT; SI-1 REV ATT TCC TAA GTT CGG CGG GT; SI-2 FW ACC GTA GTG TCC AGT TTT CAA C; SI-2 REV GAG TCA CGA GTC GAG CGG; SI-3 FW TCC ACA CCA ATA GAC AGC CG; SI-3 REV GAG TCC CGG AGC CAA GAA G; SI-4 FW ATG GCT ACA GTT CCG CAC AT; SI-4 REV CGC ACC CTC TAA CCT GTT CA; SI-5 FW TGT GTG TGT GTC CAA AAT GCA; SI-5 REV AGC CTG CTT CCC TAT GTA CC; SI-6 FW TCT GAG ATG GAC CCT AGG CT; SI-6 REV CGG ACC CCT GTG CTC TCA; SI-7 FW TGT TCT TGG ATG CGA CAC AA; SI-7 REV GGT AGC CTG CTT CCC TAT GT; SI-8 FW AGG CAT CTG TGA ATC TTT CTA CA; SI-8 REV GTT GTC TGG AGC CGA AGA TG; SI-9 FW TGG GTG ATG GTT TGG ATC CT; SI-9 REV ACT TGT GCC CCA GAT CAG G; SI-10 FW AAG GAA GGC TAC TGA TTT GTT TG; SI-10 REV GGT TGT CTG GAG CCG AAG AT; SI-11 FW CCA CAG AAT ACC TAC AAA TGA GC; SI-11 REV AGC CTG CTT CCC TAT GTA CC; SI-12 FW TCC CCT TAC AGC CAA CTT CA; SI-12 REV GGA CTT GTG CCC CAG ATC A; SI-13 FW GGG AGC AGG CAG AAA ATT CA; SI-13 REV CGG ACC CCT GTG CTC TCA; SI-14 FW GAA GGC TAC TGA TTT GTT TGA GT; SI-14 REV GGA AGG TGG CTG GTT GTC T; SI-15 FW CCA CAG AAT ACC TAC AAA TGA GC; SI-15 REV GGC CCG GGT AGC CTG CTT CC; SI-16 FW CCC ACA TCC TCC CAA CAT CT; SI-16 REV ATT CTT GGA GCT GGT GTT GC. For empty/filled PCR validation, primers were designed on the reference DNA to amplify 100 bp flanking the insertion site (empty locus) (Data S1). The band corresponding to the locus with the putative L1 insertion (filled locus) was TA-cloned and Sanger sequenced using M13 forward and M13 reverse primers.
Chromatin Tagmentation
Chromatin tagmentation of MEF cells, iDA cells, and 3TC-treated cells was performed as previously described (Buenrostro et al., 2013). Tagmented chromatin was deep sequenced with an IlluminaHiSeq4000 machine, and data were analyzed using the chipseeker tool with standard parameters.
RNA Library Extraction and Transcriptome Sequencing
Total RNA was isolated from MEF cells, iDA14 cells, and 3TC-treated cells, with three biological replicates each sample. RNA-seq libraries were prepared using an Illumina TruSeq Total RNA library preparation kit (Illumina) following the manufacturer’s instructions. The total RNA library preparation employed a ribosomal removal step using a Ribo-minus kit (Illumina). Ribosomal depleted RNA was then used for downstream library preparation following Illumina's protocol. Quality control and quantification of the libraries were performed using Qubit 3.0 (Life Technologies) and Bioanalyzer Chip RNA 6000 pico (Agilent), and they were sequenced directly using the high-throughput Illumina HiSeq 2500 sequencing system (KAUST Core Lab). The transcriptomic data were aligned to the mm10 reference genome using Tophat v2 (ccb.jhu.edu/software/tophat/index.shtml) and Bowtie v2 (http://bowtie-bio.sourceforge.net/index.shtml). Cufflinks v2.2 (Goff et al., 2013) (http://cole-trapnell-lab.github.io/cufflinks/) was used to assemble and quantify transcript expression.
Transcriptome assembly, mapping, and quantification were done using the Trinity v2.4.0 tool (github.com/trinityrnaseq/trinityrnaseq/wiki) following standard parameters. The transcripts from each assembly were then mapped to virtual references and the list of both chimeric and non-chimeric transcripts were collected for further analysis. The mapped transcripts were then checked with blast against the mm10 reference RNA database for similarity with any existing ncRNAs. To assess the coding potential of Trinity-assembled transcripts, they were mapped to protein coding transcripts and classified as coding or non-coding.
Cuffdiff v2 (http://cole-trapnell-lab.github.io/cufflinks/cuffdiff/) was used for the differential expression analysis; CummeRbund and the R v3.4.1 package were used to visualize the expression of the set of transcripts of interest.
Gene Ontology and pathway analysis were done using GOseq (https://bioconductor.org/packages/release/bioc/html/goseq.html). To compare the compatibility of our transdifferentiated transcriptome with in vivo, we used open-source MEF and dopaminergic data for the mouse genome from NCBI.
Statistical Analysis
In bar plots, values are presented as means and SEM; number of replicates are indicated in the figure legends (at least n = 3). To determine the significance between two mean values, we made comparisons by two-tailed t test. Comparisons among three or more samples were done by one-way ANOVA. For all statistical tests, a 0.05 level of confidence was accepted for significance. All the statistical analyses were performed with GraphPad Prism7 software.
Author Contributions
F.D.V. conceived the study, designed and performed experiments, analyzed the data, and wrote the manuscript. M.T. and M.Celii analyzed all Illumina sequencing data (WGS, RNA-seq and ATAC-seq). S.A. prepared all the libraries for NGS. P.L. prepared all plasmids for Sanger sequencing, V.B., M.Caiazzo, and S.P. collected and provided transdifferentiated MEF cells with TH-GFP reporter cassette. V.O. conceived the study, designed experiments, and wrote the manuscript.
Acknowledgments
We are grateful to Fred Gage (Salk Institute) for the L1-EGFP Plasmid and stimulating discussions. John Goodier for pGF21 and pTN201 plasmids. Edith Heard for pTNC7 plasmid. V.O. is supported by King Abdullah University of Science and Technology (BAS 1037/01-01, CRG URF 126310101). M.C. is supported by Italian Ministry of University and Education grant MIUR FIR 2013 RBFR13LH4X_002. V.B. is supported by Italian Ministry of University and Education, Italian Institute of Technology Seed project, Telethon Foundation, Cariplo Foundation, and Michael Fox Foundation and ERC grant AdERC #340527. Thanks to Dr. Giancarlo Bellenchi for sharing reagents and laboratory facilities.
Published: January 2, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.12.002.
Accession Numbers
All raw Illumina next-generation sequencing data have been deposited at the BioProject database, which is hosted at the NCBI, under the BioProject Database accession number: PRJNA388561. http://www.ncbi.nlm.nih.gov/bioproject/388561.
Supplemental Information
Data S1 contains the complete list of LINE-1 de novo insertions called with Mobster WGS analysis in MEF, iDA and 3TC treated cells.
The table contains the insertions coordinate, Subfamily of L1 inserted, insertion annotation, presence of Target site duplication or deletion, sequence of the TSD, sequences of the genome flanking the insertion site, start/end point of the L1 element inserted, sequence of the assembled virtual reference and allelic frequency of the event in each biological replicate with specified the number of reads supporting the insertion and the total reads from the locus.
References
- Bachiller S., del-Pozo-Martín Y., Carrión Á.M. L1 retrotransposition alters the hippocampal genomic landscape enabling memory formation. Brain Behav. Immun. 2017;64:65–70. doi: 10.1016/j.bbi.2016.12.018. [DOI] [PubMed] [Google Scholar]
- Bedrosian T.A., Quayle C., Novaresi N., Gage F.H. Early life experience drives structural variation of neural genomes in mice. Science. 2018;359:1395–1399. doi: 10.1126/science.aah3378. [DOI] [PubMed] [Google Scholar]
- Brichta L., Shin W., Jackson-Lewis V., Blesa J., Yap E.-L., Walker Z., Zhang J., Roussarie J.-P., Alvarez M.J., Califano A. Identification of neurodegenerative factors using translatome–regulatory network analysis. Nat. Neurosci. 2015;18:1325–1333. doi: 10.1038/nn.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten R.J., Davidson E.H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Buenrostro J.D., Giresi P.G., Zaba L.C., Chang H.Y., Greenleaf W.J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bundo M., Toyoshima M., Okada Y., Akamatsu W., Ueda J., Nemoto-Miyauchi T., Sunaga F., Toritsuka M., Ikawa D., Kakita A. Increased L1 retrotransposition in the neuronal genome in schizophrenia. Neuron. 2014;81:306–313. doi: 10.1016/j.neuron.2013.10.053. [DOI] [PubMed] [Google Scholar]
- Caiazzo M., Dell’Anno M.T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T.D., Menegon A., Roncaglia P., Colciago G. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature. 2011;476:224–227. doi: 10.1038/nature10284. [DOI] [PubMed] [Google Scholar]
- Castro-Diaz N., Ecco G., Coluccio A., Kapopoulou A., Yazdanpanah B., Friedli M., Duc J., Jang S.M., Turelli P., Trono D. Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev. 2014;28:1397–1409. doi: 10.1101/gad.241661.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong E.B., Elde N.C., Feschotte C. Regulatory activities of transposable elements: from conflicts to benefits. Nat. Rev. Genet. 2016;18:71–86. doi: 10.1038/nrg.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal N.G., Garcia-Perez J.L., Peng G.E., Yeo G.W., Mu Y., Lovci M.T., Morell M., O’Shea K.S., Moran J.V., Gage F.H. L1 retrotransposition in human neural progenitor cells. Nature. 2009;460:1127–1131. doi: 10.1038/nature08248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coufal N.G., Garcia-Perez J.L., Peng G.E., Marchetto M.C.N., Muotri A.R., Mu Y., Carson C.T., Macia A., Moran J.V., Gage F.H. Ataxia telangiectasia mutated (ATM) modulates long interspersed element-1 (L1) retrotransposition in human neural stem cells. Proc. Natl. Acad. Sci. U S A. 2011;108:20382–20387. doi: 10.1073/pnas.1100273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deininger P.L., Batzer M.A. Mammalian retroelements. Genome Res. 2002;12:1455–1465. doi: 10.1101/gr.282402. [DOI] [PubMed] [Google Scholar]
- Deininger P., Belancio V.P. Detection of LINE-1 RNAs by northern blot. Methods Mol. Biol. 2016;1400:223–236. doi: 10.1007/978-1-4939-3372-3_15. [DOI] [PubMed] [Google Scholar]
- DiGiacomo M., Comazzetto S., Saini H., DeFazio S., Carrieri C., Morgan M., Vasiliauskaite L., Benes V., Enright A., O’Carroll D. Multiple epigenetic mechanisms and the piRNA pathway enforce LINE1 silencing during adult spermatogenesis. Mol. Cell. 2013;50:601–608. doi: 10.1016/j.molcel.2013.04.026. [DOI] [PubMed] [Google Scholar]
- Erwin J.A., Paquola A.C.M., Singer T., Gallina I., Novotny M., Quayle C., Bedrosian T.A., Alves F.I.A., Butcher C.R., Herdy J.R. L1-associated genomic regions are deleted in somatic cells of the healthy human brain. Nat. Neurosci. 2016;19:1583–1591. doi: 10.1038/nn.4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony G.D., Cai X., Lee E., Hills L.B., Elhosary P.C., Lehmann H.S., Parker J.J., Atabay K.D., Gilmore E.C., Poduri A. Single-neuron sequencing analysis of l1 retrotransposition and somatic mutation in the human brain. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evrony G.D., Lee E., Park P.J., Walsh C.A. Resolving rates of mutation in the brain using single-neuron genomics. Elife. 2016;5:e12966. doi: 10.7554/eLife.12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing A.D. Transposable element detection from whole genome sequence data. Mob. DNA. 2015;6:24. doi: 10.1186/s13100-015-0055-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner G.J., Kimura Y., Daub C.O., Wani S., Plessy C., Irvine K.M., Schroder K., Cloonan N., Steptoe A.L., Lassmann T. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 2009;41:563–571. doi: 10.1038/ng.368. [DOI] [PubMed] [Google Scholar]
- Fort A., Hashimoto K., Yamada D., Salimullah M., Keya C.A., Saxena A., Bonetti A., Voineagu I., Bertin N., Kratz A. Deep transcriptome profiling of mammalian stem cells supports a regulatory role for retrotransposons in pluripotency maintenance. Nat. Genet. 2014;46:558–566. doi: 10.1038/ng.2965. [DOI] [PubMed] [Google Scholar]
- Friedli M., Turelli P., Kapopoulou A., Rauwel B., Castro-Di??z N., Rowe H.M., Ecco G., Unzu C., Planet E., Lombardo A. Loss of transcriptional control over endogenous retroelements during reprogramming to pluripotency. Genome Res. 2014;24:1251–1259. doi: 10.1101/gr.172809.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkountela S., Zhang K.X., Shafiq T.A., Liao W.-W., Hargan-Calvopiña J., Chen P.-Y., Clark A.T. DNA demethylation dynamics in the human prenatal germline. Cell. 2015;161:1425–1436. doi: 10.1016/j.cell.2015.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky G.V. Transposable elements and DNA methylation create in embryonic stem cells human-specific regulatory sequences associated with distal enhancers and noncoding RNAs. Genome Biol. Evol. 2015;7:1432–1454. doi: 10.1093/gbe/evv081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glinsky G.V. Mechanistically distinct pathways of divergent regulatory DNA creation contribute to evolution of human-specific genomic regulatory networks driving phenotypic divergence of Homo sapiens. Genome Biol. Evol. 2016;8:2774–2788. doi: 10.1093/gbe/evw185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grow E.J., Flynn R.A., Chavez S.L., Bayless N.L., Wossidlo M., Wesche D.J., Martin L., Ware C.B., Blish C.A., Chang H.Y. Intrinsic retroviral reactivation in human preimplantation embryos and pluripotent cells. Nature. 2015;522:221–225. doi: 10.1038/nature14308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guffanti G., Gaudi S., Fallon J.H., Sobell J., Potkin S.G., Pato C., Macciardi F. Transposable elements and psychiatric disorders. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. 2014;165:201–216. doi: 10.1002/ajmg.b.32225. [DOI] [PubMed] [Google Scholar]
- Han J.S., Boeke J.D. LINE-1 retrotransposons: modulators of quantity and quality of mammalian gene expression? BioEssays. 2005;27:775–784. doi: 10.1002/bies.20257. [DOI] [PubMed] [Google Scholar]
- Imbeault M., Helleboid P.-Y., Trono D. KRAB zinc-finger proteins contribute to the evolution of gene regulatory networks. Nature. 2017;543:550–554. doi: 10.1038/nature21683. [DOI] [PubMed] [Google Scholar]
- Johnson R., Guigo R. The RIDL hypothesis: transposable elements as functional domains of long noncoding RNAs. RNA. 2014;20:959–976. doi: 10.1261/rna.044560.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kano H., Godoy I., Courtney C., Vetter M.R., Gerton G.L., Ostertag E.M., Kazazian H.H. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 2009;23:1303–1312. doi: 10.1101/gad.1803909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A., Kronenberg Z., Lynch V.J., Zhuo X., Ramsay L.A., Bourque G., Yandell M., Feschotte C. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013;9:e1003470. doi: 10.1371/journal.pgen.1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D., Rinn J. Transposable elements reveal a stem cell-specific class of long noncoding RNAs. Genome Biol. 2012;13:R107. doi: 10.1186/gb-2012-13-11-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Koning A.P.J., Gu W., Castoe T.A., Batzer M.A., Pollock D.D. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7:e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G., Chia N.-Y., Jeyakani J., Hwang C., Lu X., Chan Y.-S., Ng H.-H., Bourque G. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat. Genet. 2010;42:631–634. doi: 10.1038/ng.600. [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:1–10. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIa A., Widmann T.J., Heras S.R., Ayllon V., Sanchez L., Benkaddour-Boumzaouad M., Muñoz-Lopez M., Rubio A., Amador-Cubero S., Blanco-Jimenez E. Engineered LINE-1 retrotransposition in nondividing human neurons. Genome Res. 2017;27:335–348. doi: 10.1101/gr.206805.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T., Leung D., Miyashita H., Maksakova I.A., Miyachi H., Kimura H., Tachibana M., Lorincz M.C., Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927–931. doi: 10.1038/nature08858. [DOI] [PubMed] [Google Scholar]
- Van Meter M., Kashyap M., Rezazadeh S., Geneva A.J., Morello T.D., Seluanov A., Gorbunova V. SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat. Commun. 2014;5:5011. doi: 10.1038/ncomms6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Waterston R.H., Lindblad-Toh K., Birney E., Rogers J., Abril J.F., Agarwal P., Agarwala R., Ainscough R. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Muotri A.R., Chu V.T., Marchetto M.C.N., Deng W., Moran J.V., Gage F.H. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature. 2005;435:903–910. doi: 10.1038/nature03663. [DOI] [PubMed] [Google Scholar]
- Muotri A.R., Marchetto M.C.N., Coufal N.G., Oefner R., Yeo G., Nakashima K., Gage F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature. 2010;468:443–446. doi: 10.1038/nature09544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostertag E.M., Prak E.T., DeBerardinis R.J., Moran J.V., Kazazian H.H., Jr. Determination of L1 retrotransposition kinetics in cultured cells. Nucleic Acids Res. 2000;28:1418–1423. doi: 10.1093/nar/28.6.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquola A.C.M., Erwin J.A., Gage F.H. Insights into the role of somatic mosaicism in the brain. Curr. Opin. Syst. Biol. 2017;1:90–94. doi: 10.1016/j.coisb.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrat P.N., DasGupta S., Wang J., Theurkauf W., Weng Z., Rosbash M., Waddell S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science. 2013;340:91–95. doi: 10.1126/science.1231965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reilly M.T., Faulkner G.J., Dubnau J., Ponomarev I., Gage F.H. The role of transposable elements in health and diseases of the central nervous system. J. Neurosci. 2013;33:17577–17586. doi: 10.1523/JNEUROSCI.3369-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff, L., Trapnell, C., and Kelley, D. (2013). cummeRbund: analysis, exploration, manipulation, and visualization of Cufflinks high-throughput sequencing data. https://rdrr.io/bioc/cummeRbund/
- Smit, A., Hubley, R., and Green, P. (2013). RepeatMasker Open-4.0. 2013-2015.
- Sundaram V., Cheng Y., Ma Z., Li D., Xing X., Edge P., Snyder M.P., Wang T. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 2014;24:1963–1976. doi: 10.1101/gr.168872.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C.A., Tejwani L., Trujillo C.A., Negraes P.D., Herai R.H., Mesci P., Macia A., Crow Y.J., Muotri A.R. Modeling of TREX1-dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell. 2017;21:319–331.e8. doi: 10.1016/j.stem.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thung D.T., de Ligt J., Vissers L.E., Steehouwer M., Kroon M., de Vries P., Slagboom E.P., Ye K., Veltman J.A., Hehir-Kwa J.Y. Mobster: accurate detection of mobile element insertions in next generation sequencing data. Genome Biol. 2014;15:488. doi: 10.1186/s13059-014-0488-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treiber C.D., Waddell S. Resolving the prevalence of somatic transposition in Drosophila. Elife. 2017;6:e28297. doi: 10.7554/eLife.28297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upton K.R., Gerhardt D.J., Jesuadian J.S., Richardson S.R., Sánchez-Luque F.J., Bodea G.O., Ewing A.D., Salvador-Palomeque C., van der Knaap M.S., Brennan P.M. Ubiquitous L1 mosaicism in hippocampal neurons. Cell. 2015;161:228–239. doi: 10.1016/j.cell.2015.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Dou K., Moon S., Tan F.J., Zhang Z.Z. Hijacking oogenesis enables massive propagation of LINE and retroviral transposons. Cell. 2018;174:1082–1094.e12. doi: 10.1016/j.cell.2018.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissing S., Mũoz-Lopez M., Macia A., Yang Z., Montano M., Collins W., Garcia-Perez J.L., Moran J.V., Greene W.C. Reprogramming somatic cells into ips cells activates line-1 retroelement mobility. Hum. Mol. Genet. 2012;21:208–218. doi: 10.1093/hmg/ddr455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood J.G., Jones B.C., Jiang N., Chang C., Hosier S., Wickremesinghe P., Garcia M., Hartnett D.A., Burhenn L., Neretti N. Chromatin-modifying genetic interventions suppress age-associated transposable element activation and extend life span in Drosophila. Proc. Natl. Acad. Sci. U S A. 2016;113:11277–11282. doi: 10.1073/pnas.1604621113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1 contains the complete list of LINE-1 de novo insertions called with Mobster WGS analysis in MEF, iDA and 3TC treated cells.
The table contains the insertions coordinate, Subfamily of L1 inserted, insertion annotation, presence of Target site duplication or deletion, sequence of the TSD, sequences of the genome flanking the insertion site, start/end point of the L1 element inserted, sequence of the assembled virtual reference and allelic frequency of the event in each biological replicate with specified the number of reads supporting the insertion and the total reads from the locus.