Summary
Parkinson's disease (PD) is a complex and highly variable neurodegenerative disease. Familial PD is caused by mutations in several genes with diverse and mostly unknown functions. It is unclear how dysregulation of these genes results in the relatively selective death of nigral dopaminergic neurons (DNs). To address this question, we modeled PD by knocking out the PD genes PARKIN (PRKN), DJ-1 (PARK7), and ATP13A2 (PARK9) in independent isogenic human pluripotent stem cell (hPSC) lines. We found increased levels of oxidative stress in all PD lines. Increased death of DNs upon differentiation was found only in the PARKIN knockout line. Using quantitative proteomics, we observed dysregulation of mitochondrial and lysosomal function in all of the lines, as well as common and distinct molecular defects caused by the different PD genes. Our results suggest that precise delineation of PD subtypes will require evaluation of molecular and clinical data.
Keywords: Parkinson's disease, disease modeling, CRISPR, genome editing, transcriptomics, proteomics, Parkin, DJ1, ATP13A2, human pluripotent stem cells
Highlights
-
•
CRISPR knockin of reporter in TH locus allows live tracking and isolation of DNs
-
•
Large-scale 3D midbrain DN differentiation using spinner flask culture
-
•
Phenotypic comparison of isogenic DNs harboring knockouts of PARKIN, DJ-1, or ATP13A2
-
•
Transcriptomics and quantitative proteomics studies determine common and distinct PD pathways
In this article, Ahfeldt, Rubin, and colleagues model Parkinson's disease (PD) in human pluripotent stem cells by knocking out PARKIN, DJ-1, or ATP13A2. They report increased levels of oxidative stress in all PD lines and death of dopaminergic neurons in the PARKIN-KO. Using transcriptomics and quantitative proteomics approaches they determine common and distinct molecular defects caused by different PD genes.
Introduction
Parkinson's disease (PD) is a chronic and progressive neurodegenerative disorder that disproportionally affects dopaminergic neurons (DNs) in the substantia nigra pars compacta (SNc) of the midbrain (Dauer and Przedborski, 2003, Uhl et al., 1985). Most PD cases are sporadic, and little is known about the cause of their disease. However, approximately 10% of PD patients have a family history of the disease (Thomas and Beal, 2007). Studies of sporadic, as well as familial, PD patients suggest that both are affected by the dysregulation of similar biochemical pathways—most prominently, impairment of protein and mitochondrial homeostasis and oxidative stress (OS) (Chai and Lim, 2013).
A better understanding of the familial forms of PD could help elucidate pathways that are also relevant for the sporadic forms of the disease. Approximately 3%–5% of sporadic PD cases can be linked to mutations in one of six genes. Among these, mutations in the E3 ubiquitin ligase, PARKIN, the PTEN-induced putative kinase 1 (PINK1), the protein deglycase, DJ-1, and the presumptive cation-transporting ATPase 13A2 (ATP13A2) are inherited in an autosomal recessive fashion and cause completely penetrant early-onset PD in homozygous or compound heterozygous carriers (Klein and Westenberger, 2012). PARKIN-, PINK1-, and DJ-1-related forms of PD are clinically indistinguishable from each other in their pathology (Schulte and Gasser, 2011). In contrast, mutations in ATP13A2 cause Kufor-Rakeb syndrome, an atypical presentation of PD involving additional symptoms of dementia, spasticity, and supranuclear gaze palsy (Hampshire et al., 2001, Paisan-Ruiz et al., 2010). The symptomatologies of these recessive mutations suggest that their study in vitro will reveal relevant common, but also distinct, dysregulated pathways for PD.
Advances in gene-editing technology of human pluripotent stem cells (hPSCs) allow studies of familial PD genes compared with isogenic controls. For example, LRRK2 (Reinhardt et al., 2013), SNCA (Ryan et al., 2013), PARKIN (Shaltouki et al., 2015, Tabata et al., 2018), and DJ-1 (Burbulla et al., 2017) mutations have been studied in this way. Although these studies have demonstrated PD-specific phenotypes such as OS, dopamine oxidation, and cell death, we still know little of the shared common or distinct mechanisms that accompany the pathological dysregulation.
Using CRISPR-Cas9 genome editing we developed isogenic loss-of-function models of early-onset autosomal recessive PD (PARKIN−/−, DJ1−/−, and ATP13A2−/−) with the aim of identifying common and distinct elements of each. We combined our isogenic models with a knockin fluorescent reporter at the tyrosine hydroxylase (TH) locus that enabled isolation of large numbers of DNs. We further developed an efficient 3D-spin reactor differentiation protocol to generate DNs on a large-scale in a reproducible fashion that allows studies in organoids/spheres and in a 2D format after dissociation.
These technical advances allowed us to carry out comparative quantitative global proteomic and transcriptomic analyses with the goal of identifying dysregulated pathways that contribute to the development of PD. Our characterization of the three isogenic PD lines revealed increased OS in the basal state in all mutant types of DNs with early specific loss of these neurons in the PARKIN−/− line. Using Ingenuity Pathway Analysis (IPA) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis of transcriptomic and proteomic data, we identified subtype-specific dysregulation of PD-relevant pathways including changes in mitochondrial and lysosomal properties, and apoptosis.
Results
Generation of Isogenic TH Knockin Reporter PD Cell Lines
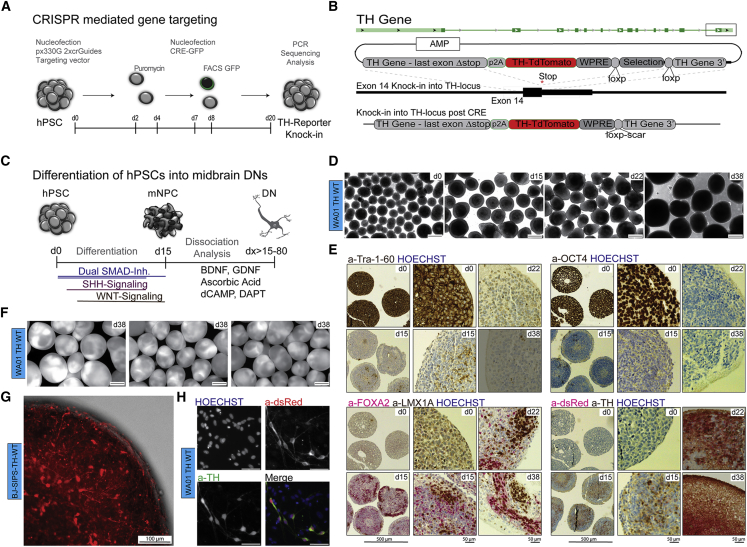
The TH gene, encoding the rate-limiting enzyme in dopamine synthesis, is commonly used in immunocytochemistry experiments to quantify the percentage of DNs derived from hPSCs. We engineered a TH-p2a-Td:Tomato (red fluorophore protein) construct as a reporter for TH expression. We used a CRISPR-Cas9 genome editing strategy that relies on positive selection and CRE-mediated excision of the selection cassette to introduce this fluorescent reporter into the TH gene locus (Figure 1A). The targeting vector retained a largely unaltered endogenous TH gene product (Shaner et al., 2004) (Figures 1B and S1A). We used GFP labeling to enrich cultures that were successfully nucleofected with the CRE-GFP plasmid (Figure S1B). Correct knockin/homologous recombination events were determined through genotyping and DNA sequencing (Figures S1C and S1D). Knockin efficiency across three cell lines, determined by 5′ genotyping PCR, was 60%.
Figure 1.
Spin Culture Differentiation of TH Reporter hPSCs into Midbrain DNs
(A) Experimental scheme depicting the CRISPR-mediated TH reporter knockin strategy.
(B) Donor plasmid containing the targeting vector TH with a 5′TH homology arm followed by a 2A self-cleaving peptide sequence, a WPRE sequence, floxed selection cassette, and 3′ TH homology arm. Genomic locus indicating the targeting area in exon 14 of the TH gene, red star representing the position of the stop codon. Schematic representation of successful targeting post-CRE excision.
(C) Experimental scheme showing the culture conditions in spin culture.
(D) Bright-field images of WA01-WT spheres at time points d0, d15, d22, and d38. Scale bars, 500 μm (4×).
(E) Immunohistochemistry of sectioned organoids/spheres. Each panel organized as (top left, 4× d0; bottom left, 4× d15. Scale bar, 500 μm (4×); top middle, 20× d0; bottom middle, 20× d15; top right, 20× d22; and bottom right, 20× d38. Scale bar, 50 μm (20×) using validated antibodies against: top left panel (TRA-1-60), top right panel (OCT4), bottom left panel (FOXA2 and LMX1A), and bottom right panel (dsRed and TH). Nuclei were counterstained using HOECHST.
(F) Fluorescence images of WA01-WT spheres at d38 showing TH:TdTomato reporter expression. Scale bars, 500 μm, 4× (n = 3 differentiation experiments).
(G) Maximum projection image of d22 sphere from differentiated BJ-SIPS WT TH cells showing TH:TdTomato expression. z stack images acquired using spinning disc confocal CX7 at 10×. Scale bar, 100 μm.
(H) Fluorescence microscopy panel, showing DNs at d38 from WA01-TH-WT cell lines, dissociated on d25 and plated in low density on glial cells. Scale bars, 50 μm, 20× (top left, Hoechst; top right, a-dsRed; bottom left, a-TH [MAB318]; bottom right, merge).
hPSC Differentiation into Midbrain DNs in 3D-Spin Reactors
Culture in 3D-spin reactors is scalable and less variable than standard organoid methods, allowing the production of hundreds of millions of cells per spin reactor (Amit et al., 2011, Qian et al., 2016, Rigamonti et al., 2016). To recapitulate midbrain differentiation (Kriks et al., 2011) we used dual-SMAD inhibition, followed by patterning via modulation of sonic hedgehog (SHH) and WNT signaling (Figure 1C). 2D cultures of hPSCs readily formed spheres after transfer into spin reactors and had a relatively homogeneous appearance. Differentiation of WA01-TH cells was initiated when pluripotent spheres reached an average diameter of 500 μm; that time point was named d0. During differentiation, spheres continuously increased in size. The average sphere diameter was 726 μm at d15, 784 μm at d22, and 1,119 μm at d38 (Figures 1D and S1E, left).
Virtually all cells comprising the spheres at d0 were positive for the pluripotency markers Tra-1-60 and OCT4 (Figure 1E, top). OCT4 and TRA-1-60 expression rapidly decreased and only a few cells were positive at d15. FOXA2 is one of the earliest genes to be expressed in the floor plate of the developing midbrain. After the midbrain patterning at d15, >95% of cells were positive for FOXA2 (Figure 1E, bottom left). Expression of the roof plate marker LMX1A appeared clustered on the edges of the neural progenitor cell (NPC) spheres. LMX1A and FOXA2 expression was absent in pluripotent spheres. Most cells remained FOXA2+ over the course of differentiation, but the staining appeared weaker at later time points. Staining with antibodies against TH and dsRed revealed the presence of a low percentage of TH:TdTomato-positive (TH+) NPCs and early neurons at d15 (Figure 1E, bottom right). The TH percentage rapidly increased up to d22. At d38, about 40% of all cells stained positive for dsRed and TH. Similar differentiation efficiencies were noted in three independent wild-type (WT) hPSC lines that were tested.
The sphere size was also reproducible, as indicated by three independent differentiation experiments (Figures 1F and S1E, right). In differentiating spheres, we observed parallel tracks of axons on the outside, while the inside appeared less organized. To reveal 3D morphology and reporter fluorescence intensity, we took live cell images of d22 spheres derived from BJ-SIPS-TH-WT cells. The maximum intensity projection of 12 images shows bright fluorescent cell bodies with DN morphology, networks of neuronal processes and bright red puncta with regular and spherical appearance (Figure 1G). After differentiation, spheres could be dissociated using a gentle enzymatic digestion protocol.
We sorted cells based on TH:TdTomato expression. Positive neurons were plated on primary mouse glial cells. About 50% of TH+ neurons survived digestion and flow sort, and quickly grew processes. The sorted cells showed the characteristic morphology of hPSC-derived midbrain DNs. The TH:TdTomato reporter appeared very bright in live cultures but was noticeably dimmer after paraformaldehyde fixation. Cells were stained using antibodies against TH and dsRed: 98% of TH+ cells were also positive for dsRed. We did not observe cells that were dsRed+ but not TH+ (Figures 1H and S1F).
Isogenic Cell Lines Carrying Three Distinct PD Mutations
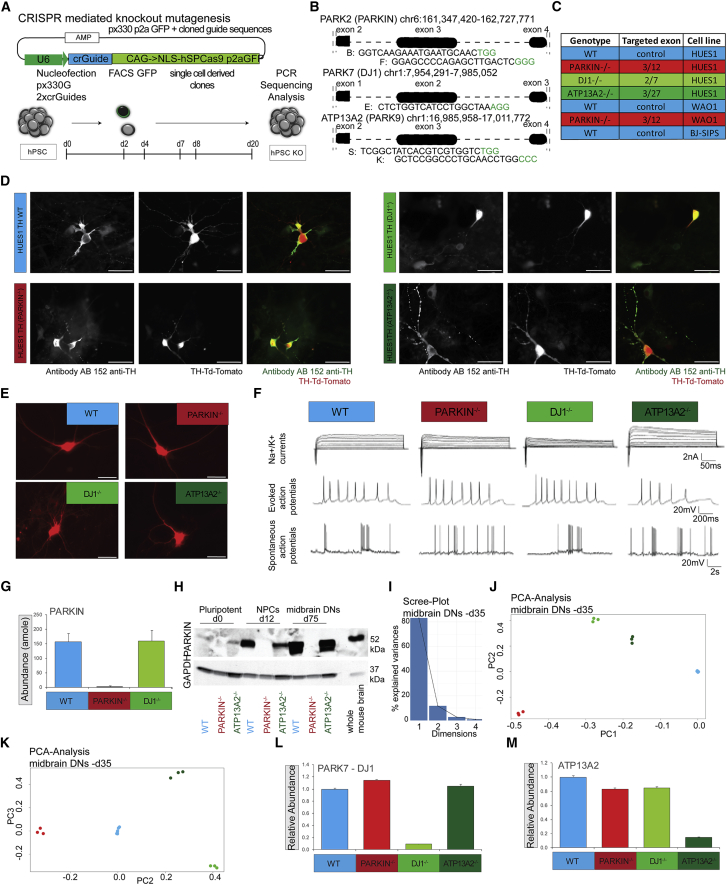
We used CRISPR editing to create isogenic PD lines from WT donor control cell lines (Figure 2A). We created CRISPR guides based on a px330-p2A-GFP variant designed to cause double-strand breaks in the coding regions of three distinct PD loci: PARKIN, DJ-1, and ATP13A2 (Figure 2B). Flow sorting of GFP expression was used to enrich for successfully nucleofected cells (Figure S2A). We generated several PD clones in both HUES1 and WA01 cell lines. Edited clones were identified via PCR genotyping and Sanger sequencing (Figures S2B–S2D). Cell lines used in the study are listed (Figure 2C). We differentiated PD and control cell lines using a 3D-spin protocol. Analyzed clones exhibited a small cytoplasm to nucleus ratio and stained positive for the pluripotency markers TRA-1-60 and OCT4 (Figures S3A and S3B). Normal karyotypes were found in the WA01-TH-WT, BJ-SIPS-TH-WT, as well as WA01-TH-PARKIN clones that were analyzed (Figure S3C). DNs from all isogenic lines were isolated by fluorescence flow cytometry at various times (Figure S3D). Sorted DNs were cultured on mouse glial cells for up to 2 months.
Figure 2.
Generation, and Molecular and Functional Characterization of Knockout Cell Lines in the PD Model
(A) Experimental Scheme depicting the CRISPR knockout mutagenesis strategy.
(B) Graphical display of chromosome positions, targeted exons, and CRISPR sequences.
(C) Table summarizing knockouts indicating targeted exons, and coloring scheme used in this study in isogenic HUES1, WA01 and BJ-SIPS lines: WT, PARKIN−/−, DJ-1−/−, and ATP13A2−/−.
(D) Fluorescence microscopy panel, showing HUES1-derived DNs at d35 from all isogenic reporter cell lines, dissociated on d25 and plated in low density on mouse glial cells. Scale bars, 100 μm, 40× (left, ICC stained with TH antibody AB152; middle, TH:TdTomato reporter fluorophore expression, right merge).
(E) Fluorescence microscopy images of flow sorted TH+ neurons from all isogenic lines 4 weeks post plating on glial cells before electrophysiological analysis. Scale bars, 50 μm.
(F) Whole-cell patch clamp recordings in TH+-labeled DNs derived from all four iPSC lines (n = 6–9 for each line) showing voltage-gated sodium and potassium currents (top row), evoked action potentials (middle), and spontaneous action potentials (bottom).
(G) Quantification of PARKIN abundance in DNs from WT, PARKIN−/−, and DJ-1−/− lines using AQUA peptides.
(H) Western blot of WT, PARKIN−/−, and ATP13A2−/− lines using PARKIN antibody and GAPDH as a control, at three time points, indicated as pluripotent at d0, NPC at d12, midbrain DN at d75, and mouse whole-brain lysate as control.
(I) Scree plot showing the percentage of variances explained by each principal component.
(J) PCA plot of components 1 and 2 at d35.
(K) PCA plot of components 2 and 3 at d35.
(L) Bar chart showing normalized relative abundance of DJ1 protein, WT samples set to 1.
(M) Bar chart showing normalized relative abundance of ATP13A2 protein, WT samples set to 1. Statistical significance was analyzed by one-way ANOVA followed by post hoc test (1% FDR)-Bonferroni-Holm for multiple comparisons.
To ensure that the reporter expression did not change, we co-stained reporter-expressing cells with an anti-TdTomato/RFP antibody. In all analyzed cell lines and clones, we detected strong overlap between the two antibody stains. There were only a few instances of cells that were positive to an anti-TH antibody and not positive using anti-dsRed antibody (Figures 2D and S1F). TdTomato was expressed diffusely throughout the cell body and less strongly within neuronal projections. The TH+ neurons from all isogenic cell lines generally showed multipolar soma, elaborate dendrites, and axons (Figure 2E). Midbrain DNs exhibit two characteristic firing patterns, single spikes and bursting (Shi, 2005). To confirm that the hPSC-derived TdTomato+ neurons were functionally consistent with their putative identification as DNs, we carried out electrophysiological measurements. We detected voltage-gated sodium and potassium currents, evoked repetitive action potentials, and frequent spontaneous potentials in TH:TdTomato DNs derived from both WT and isogenic PD lines (Figure 2F). Furthermore, live cell calcium imaging also showed spontaneous tetrodotoxin-sensitive activity in neurons from all the lines (Figure S3Ei-iii). Thus, all isogenic lines differentiated into functional DNs with electrophysiological properties similar to those described previously (Jiang et al., 2012, Kriks et al., 2011).
To confirm loss of the PARKIN protein we used isotope-labeled peptides (AQUA peptides) as an internal standard to allow absolute quantification of PARKIN in those samples (Ordureau et al., 2018, Stemmann et al., 2001). PARKIN was detected in both the WT and DJ−/− cell lines and was not detectable above background in the PARKIN−/− cell line (Figure 2G). A time course western blot analysis of PARKIN protein expression during differentiation showed complete loss of PARKIN at all developmental time points. PARKIN protein abundance increased substantially during differentiation in both WT and ATP13A2−/− lines (Figures 2H and S3F). We also confirmed loss of PARKIN in several WA01 clones (Figure S3G). In addition, using western blot techniques, we confirmed loss of DJ-1 in several HUES1 clones (Figure S3H).
Proteomics Reveals Global Differences between WT and Isogenic PD Cell Lines
To profile proteome alterations quantitatively in the isogenic PD lines, we used multiplexed tandem mass tag (TMT)-based quantitative mass spectrometry. We performed comparative studies between all cell lines in the pluripotent state as well as in d35 DNs, for which we generated differentiation triplicates. We quantified >7,000 proteins in 4 experiments for all the d35 DN samples. Comparative studies were conducted on a smaller intersection subset of >4,000 high-abundance proteins. All abundances, measured normalized relative abundances and fold changes, including statistical testing, are summarized in Table S1.
Principal-component analysis (PCA) performed on the d35 DN dataset showed strong reproducibility between independent differentiation replicates. A Scree plot shows that most variance in the dataset is represented by three components (Figure 2I). The first, second, and third components explain 83.1%, 11.8%, and 2.6% of the variance, respectively. A PCA plot of components 1 and 2 shows distinct clusters that group samples by genotype, with component 1 separating WT from all isogenic DN lines, while component 2 separates the PARKIN−/− line from the DJ-1−/− and ATP13A2−/− lines (Figure 2J). PCA plots of components 2 and 3 show that component 3 separates the DJ-1−/− and ATP13A2−/− lines (Figure 2K). Three dimensions were sufficient to differentiate all samples by genotype. The variance in the hPSC lines was much smaller. However, loss of PARKIN led to its separation from all other cell lines through component 1 (Figures S4A–S4C). Analysis of differential protein abundance confirmed knockout of the DJ-1 and ATP13A2 proteins (Figures 2L and 2M).
PD Gene Loss Depletes Mitochondrial Proteins and Increases OS in TH+ DNs
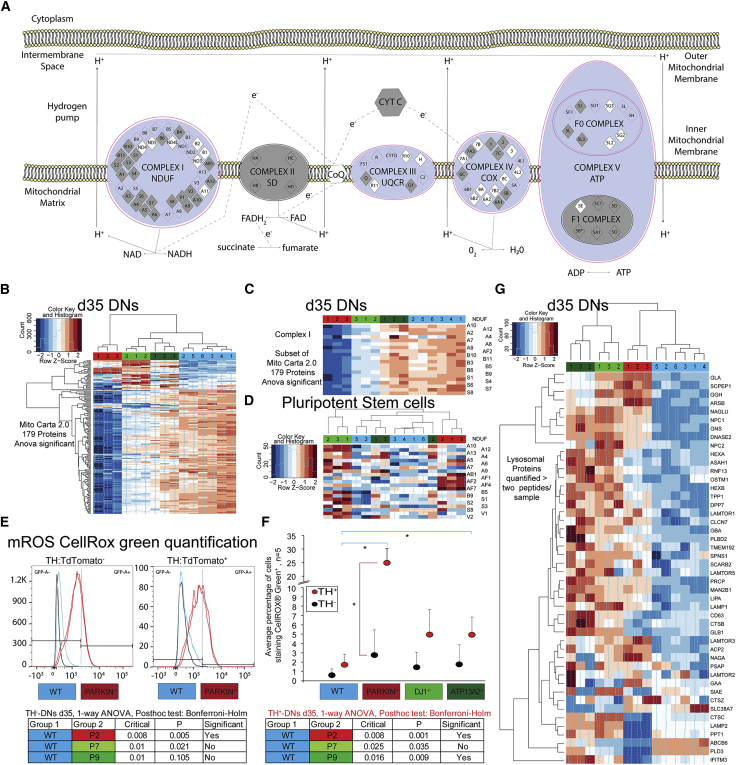
Postmortem brain analyses of PD patients, as well as of PD animal models, show increased OS through mitochondrial dysfunction in DNs as a common feature of PD pathology (Dias et al., 2013). Mitochondrial dysfunction and especially loss of complex I activity have been investigated as part of sporadic and familial PD etiology. We performed IPA on the proteomics data to explore pathways that were dysregulated between WT and PD lines. Canonical IPA showed many dysregulated pathways between WT and PARKIN−/− cell lines. The top 3 most significant pathways in d35 DNs were oxidative phosphorylation, mitochondrial dysfunction, and the sirtuin-signaling pathway (Figure S4D). These pathways were also dysregulated in pluripotent cells, but less significantly.
To determine dysregulation in pluripotent cells, we used significance cutoff adapted to the given dataset (from log2 fold change −0.7/0.7 in d35 DNs to −0.12/0.12 in d0 pluripotent cells). We overlaid the normalized protein abundance as fold changes between d35 DNs from WT and PARKIN−/− lines and mapped the proteins onto the canonical pathway “oxidative phosphorylation” obtained from IPA (Figure 3A). We observed significant depletion of mitochondrial proteins in complexes I, III, and IV. To examine mitochondrial dysfunction in all PD disease lines, we probed for proteins that were detected with at least two unique peptides that were classified as relevant to mitochondrial function (Mito Carta 2.0) and showed dysregulation using an ANOVA test. Heatmap analysis of the 179 dysregulated proteins showed global depletion of mitochondrial proteins in PARKIN−/− lines at d35 of differentiation (Figure 3B). However, subtle global depletion of mitochondrial proteins was noted in the other PD lines as well. We focused on a subset of these proteins, plotting only those that are part of complex I (NADH:ubiquinone oxidoreductase [NDUF] proteins). Complex I proteins were less abundant in the disease lines at d35 (Figure 3C), but not in d0 hPSCs (Figure 3D). Loss of complex I function has been linked to increased OS levels.
Figure 3.
OS, Mitochondrial Dysfunction and Lysosomal Dysregulation Are Shared Phenotypes in all Early-Onset PD DNs
(A) The canonical oxidative phosphorylation pathway created using IPA software. log2 fold changes were plotted. Purple outlines indicate significantly dysregulated proteins/pathways (based on −0.7/0.7 log2 fold change). Red color indicates a positive change or protein enrichment in the PARKIN−/− line (none noted), while blue represents a depletion.
(B, D, G) Heatmap.2 function across all samples using specified gene lists, default clustering and row scaling. Columns represent samples, rows represent genes, and color intensity represents column Z score, where red indicates enriched and blue depleted proteins.
(B) Heatmap analysis of 179 Mito Carta 2.0 proteins with mitochondrial function or localization that shows differential protein abundance between any cell line in d35 DNs.
(C) Subset of Mito Carta 2.0 analysis showing all quantified proteins that are part of the NDUF-complex I in d35 DNs.
(D) Heatmap of all proteins that were quantified with at least two unique peptides that are part of the NDUF-complex I in hPSCs.
(E) Dissociated spheres at d35 were stained with CellROX green to detect mROS production in the basal state. mROS-G+ was quantified in TH− as well as TH+ cells. Histograms of mROS-G positivity overlaid with no staining control, stained WT (blue) and PARKIN−/− (red) lines, respectively.
(F) Averaged mROS percentage, quantified as mROS-G+ using synchronized bisector gates (FloJo, LLC) to divide the x axis into GFP− and GFP+ non-overlapping populations based on the background expression in our unstained control. Statistical significance was analyzed by one-way ANOVA followed by Bonferroni-Holm multiple comparison test (n = 4 independent staining and flow experiments significance indicated in tables [bottom]).
(G) Heatmap analysis of proteins with an annotated lysosomal function that were quantified with at least two unique peptides.
OS might be connected to DN-specific cellular functions such as dopamine metabolism. We found significantly reduced TH protein expression at d35 in independent WA01-derived Parkin−/− cells in line with our observations of reduced numbers of TH+ DNs (Figures S4F and S4G). We measured protein carbonyl content, which is considered a marker of oxidative modification of proteins and, therefore, of OS. We found that the quantity of carbonylated proteins in differentiating hPSCs increased over time and was significantly different between d17 and d35 (Figures S4H–S4J). To test the role of PD mutations in the regulation of mitochondrial reactive oxygen species (mROS), we analyzed the levels of mROS using the live cell dye CellROX Green and the TH:TdTomato reporter in all of the isogenic cell lines following differentiation (Figures 3E and 3F) and quantified the percentage of cells exhibiting high green fluorescence (mROS-G+) in each population. PARKIN−/− cell lines showed significantly elevated levels of mROS in TH+ DNs versus TH− cells (Figure 3F). WT control cell lines showed fewer mROS events than all PD cell lines. This difference was significant for comparison between WT control and ATP13A2−/− cell lines but was most severe as a consequence of loss of PARKIN (Figure 3F, blue arcs). Our data suggest that increased mROS and mitochondrial changes in DNs are part of a shared etiology.
PD Gene Loss Changes Lysosomal Function and Other PD-Relevant Pathways
Dysregulation of the autophagy-lysosomal pathway (ALP) is central to sporadic and familial PD pathology. We hypothesized that changes in protein abundance in the basal state without any perturbation of ALP would be an indicator for dysregulation of lysosomal function. We plotted proteins that were detected with at least two unique peptides and were annotated as part of the lysosomal compartment. Heatmap analysis of the 46 proteins showed global enrichment of lysosomal proteins in both the ATP13A2−/− and DJ-1−/− cell lines. Dysregulation was also present in PARKIN−/−, but hierarchical clustering revealed that the dysregulation was distinct from that of the other PD cell lines (Figure 3G). Thus, autophagy-lysosomal function and OS are commonly dysregulated pathways in PD and similarly appear dysregulated in DNs derived from the isogenic cell lines. It is unclear whether changes in the amount of lysosomal proteins resulted from increased induction or a block in completion of the autophagy process.
PARKIN Loss Causes Cell Death of Early TH+ Neurons
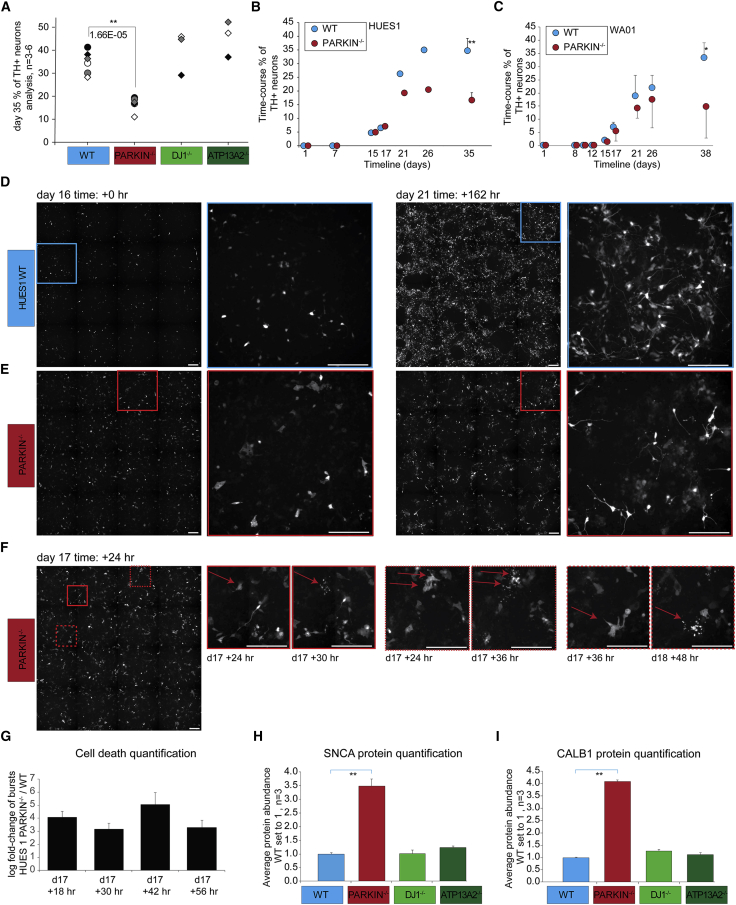
Using IPA comparison analysis, we tested curated pathways for diseases and biological functions that were present in all WT versus PD line comparisons. We found dysregulation of pathways relevant to cellular function and maintenance, cell morphology, nervous system development and function, neurological disease, and cell death and survival (Figure S4E). Cell death and apoptosis can be triggered by excessive generation of free radicals, mitochondrial dysfunction, damage to DNA, or activation of lysosomal proteases. We hypothesized that the increase in OS in early-onset PD mutations could result in increased cell death rates in midbrain DNs in basal culture conditions. We dissociated spheres at different times during differentiation and analyzed the population for the percentage of TH+ cells via fluorescence flow cytometry. The average percentage of TH+ neurons in HUES1 WT, DJ-1−/−, and ATP13A2−/− lines was ∼40% at d35, indicating the absence of substantial changes in differentiation or survival of TH+ neurons in these lines. However, we found that the TH+ fraction was significantly smaller (16.7%) in the PARKIN−/− line at the same time point (Figure 4A). A time course of TH+ neuron emergence showed no significant differences in the onset of reporter expression or the percentage of TH+ cells during the first 17 days of differentiation in WT and PARKIN−/− lines (Figure 4B). We validated this finding using an independent WA01-TH cell line and three PARKIN−/− clones. Differences in TH percentage arose early and were significant at d38 of analysis (Figure 4C). Reduced differentiation or birth of DNs, as well as increased cell death, might have contributed to the smaller numbers of TH+ cells in the PARKIN−/− cell lines.
Figure 4.
Quantification of Cell Death Events in TH+ DNs
(A) Dissociated spheres were analyzed via flow cytometry, TH quantification in isogenic HUES1 lines at d35 showing decreased numbers of TH+ cells in the PARKIN−/− line. Statistical significance p value derived from one-way ANOVA followed by Bonferroni-Holm multiple comparison test (n = 6, 6, 3, and 3 independent differentiation experiments).
(B) TH quantification in WT and PARKIN−/− isogenic lines in a time course experiment in HUES1 line (n = 1 except d35 n = 6).
(C) TH quantification in WT and PARKIN−/− isogenic lines in a time course experiment in WA01 line (d0–d21, WT, n = 3 differentiation replicates; PARKIN−/−, n = 3 independent clones; d26–d38, WT, n = 4 differentiation replicates; PARKIN−/−, n = 5 or 3 independent clones, 2 differentiation replicates; d38, ∗p < 0.05, unpaired two-sided t test).
(D–F) Nikon Biostation CT live cell fluorescence imaging of TH:TdTomato expression during differentiation of WT and PARKIN−/− lines after dissociation at d15. Image acquisition starts 24 h post plating. Images were acquired every 6 h, time points indicated. Squares depict areas of interest that are shown in higher magnification. (D) WT cell line. (E) PARKIN−/− cell line. (F) Representative images of cell death events in the PARKIN−/− cell line. Zoomed images with red arrows indicating cells of interest before and after cell death.
(G) Cell death quantification of burst events in WT and PARKIN−/− cell lines at the indicated time points, shown as log2 fold change between PARKIN−/− and WT cell lines (n = 3 wells per cell line). ∗p < 0.05, unpaired two-sided t test.
(H) Bar chart showing normalized relative abundance of SNCA protein across all samples, WT samples set to 1.
(I) Bar chart showing normalized relative abundance of CALB1 protein across all samples. WT samples set to 1. Statistical significance was analyzed by one-way ANOVA followed by post hoc test (1% FDR) Bonferroni-Holm for multiple comparisons.
To distinguish between reduced birth and increased death, we focused on the critical window between d15 and d21. We conducted a time-lapse experiment using an automated live cell imager (Nikon Biostation, CT) to examine the fate of newly generated TH+ cells, as well as to track these cells over time, with the goal of determining whether the differences were due to differentiation or survival. We plated 50,000 unsorted cells per 0.32 cm2 from HUES1 WT and PARKIN−/− isogenic cell lines on d15. We noted faint TH expression in WT-derived cells with either NPC morphology or immature neuron morphology without apparent neuritic processes (Figure 4D, left—overview and magnification). Over time, WT TH+ cells gained in fluorescence intensity, grew processes, and exhibited a morphology typical of DNs (Figure 4D, right—overview and magnification). However, cells in the PARKIN−/− line behaved differently. At d16, 24 h post-dissociation, many of the faintly TdTomato-expressing cells showed increased vacuolation when compared with WT cells (Figure 4E, left—overview and magnification). At the end of the experiment at d21 the PARKIN−/− DNs exhibited typical morphology, but showed substantially reduced numbers and processes (Figure 4E, right—overview and magnification). During the course of the experiment we observed cell death in TH+ cells in the PARKIN−/− line. Highly vacuolated cells burst and released fluorescent protein particles in small vacuoles that disappeared within 12–24 h (Figure 4F). The phenotype is clearly visible in Video S1. Over the course of the 5-day imaging period, many newly born TH+ DNs disappeared from the culture, after displaying cellular fragmentation (Video S1). We observed significantly fewer such events in the WT cell line. Quantification revealed a 3–5 log fold increase of cell death events in PARKIN−/− TH+ cells (Figure 4G).
Section 1: WT-Well-1 10× magnification. Section 2: WT-Well-1 representative area zoomed in. Section 3: PARKIN−/−-Well-1 10× magnification. Section 4: PARKIN−/−-Well-1 representative area zoomed in. Section 5: WT-Well-2 10× magnification. Section 6: WT-Well-2 representative area zoomed in. Section 7: PARKIN−/−-Well-2 10× magnification. Section 8: PARKIN−/−-Well-2 representative area zoomed in.
Elevated levels of SNCA are sufficient to cause early-onset familial PD in a dose-dependent manner which leads to neuronal dysfunction. We observed significant (>3-fold) protein enrichment of SNCA in d35 PARKIN−/− DNs, but not in the other PD cell lines (Figure 4H). We hypothesized that hPSC-derived DNs represent a heterogeneous group of cells similar to the composition found in the human midbrain and that ventral tegmental area (VTA)-like neurons should be less affected by cell death than SNpc neurons. There are no molecular markers that can clearly delineate SNc from VTA, but calbindin (CALB1) is used in the field for its higher expression in the VTA (Bodea and Blaess, 2015). CALB1 protein abundance was >4-fold upregulated in the PARKIN−/− cell line, but not the other PD lines that did not exhibit cell death (Figure 4I).
Overlapping Dysregulated Genes and Pathways in PARKIN−/− and ATP13A2−/− Cell Lines
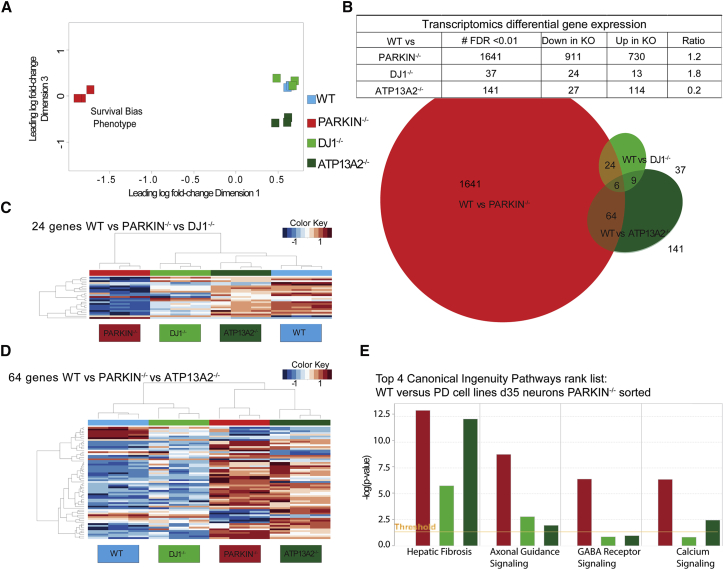
We generated RNA sequencing transcriptomics datasets from all isogenic HUES1 cell lines to identify common and distinct dysregulated genes and networks in the three isogenic PD lines in an unbiased manner, with the goal of grouping similar and distinct forms of PD. To minimize noise and capture signal only from DNs, d35 spheres were dissociated and TH+ cells were purified by flow sorting for TdTomato expression. RNA sequencing profiles were generated in three separate differentiation experiments (n = 3) (Figure S5A). To visualize the relationship between each individual sample in our dataset, we performed unsupervised clustering as a multidimensional scaling plot, in which the distances correspond to leading log fold changes between each pair of RNA samples (Figures 5A and S5B). The plot revealed substantial differences between PARKIN−/− versus all other isogenic lines, which led to the separation by dimension 1. Dimension 2 separated biological replicate two from the other replicates. Dimension 3 separated the ATP13A2−/− line from the WT and DJ-1−/− lines. The PARKIN−/− line is the most dissimilar from all other lines.
Figure 5.
Transcriptomics Analysis of Sorted DNs Highlights Common and Distinct Dysregulation of PD-Relevant Genes and Pathways: HUES1 PD Lines
(A) Multidimensional scaling plot of all isogenic cell lines in which distance corresponds to leading log fold changes between each pair of RNA samples. Dimension 1 separates PARKIN−/− line from all other lines and dimension 3 separates the ATP13A2−/− line from WT and DJ-1−/− lines.
(B) Table showing the number of significant DEGs between WT and isogenic lines (FDR = 0.01), the number of up- and downregulated genes, and the resulting ratio. Overlap analysis as a three-way Venn diagram intersecting DEGs between WT and all isogenic lines.
(C) Heatmap across all samples using the 24 intersecting genes between WT versus PARKIN−/− and WT versus DJ-1−/− analyses.
(D) Heatmap across all samples using the 64 intersecting genes between WT versus PARKIN−/− and WT versus ATP13A2−/− analyses.
(E) Bar chart of d35 global transcriptomics comparisons between WT and all PD cell lines showing select altered disease or biological function pathways determined using IPA curated sets. The y axis shows the significance (–log(p value)) and the orange line shows the significance threshold cutoff of −log(p value = 0.05), sorted by significance in PARKIN−/− versus WT comparison.
We performed differential gene expression analysis and examined the results for overlap of genes and pathways. We identified genes that were differentially regulated by at least 2-fold, with a p value <0.01 between WT and all isogenic PD lines and validated genes via qRT-PCR (Figures 5B, top and S5C; Table S2). We conducted an overlap analysis to determine whether differentially expressed genes (DEGs) between WT and all isogenic disease lines were present in common pathways or were independent from one another (Figure 5B, bottom). Only six genes were differentially regulated in all three isogenic lines. We found 37 genes that were differentially expressed between DJ-1−/− and WT. However, among these, 24 were also differentially expressed in the PARKIN−/− lines. Shared genes were dysregulated in opposite directions, suggesting disparate disease mechanisms in DJ-1−/− compared with PARKIN−/− (Figure 5C). We found no significant dysregulated pathways in our KEGG pathway enrichment analysis of this comparison. In addition, 141 genes were differentially expressed between the ATP13A2−/− and WT lines. Of these, 64 were also differentially expressed between the PARKIN−/− and WT lines. The heatmap analysis demonstrated that in PARKIN−/− and ATP13A2−/− DNs, most expression changes had the same direction when compared with WT or DJ-1−/− DNs (Figure 5D).
To analyze the differential expression results in a network context, we performed IPA and independent KEGG pathway enrichment analysis on all DEGs between WT and isogenic PD lines. We found that the PARKIN−/− and ATP13A2−/− lines showed similar patterns of dysregulation. The two top KEGG pathways found dysregulated in both lines were hsa04512—extracellular matrix (ECM) receptor interaction; and hsa04974—protein digestion and absorption (Figure S5D). IPA of canonical pathways showed dysregulation of hepatic fibrosis, which is based on the dysregulation of ECM proteins, and axonal guidance signaling in all PD lines. Dysregulation of GABA (gamma-aminobutyric acid) receptor signaling was significant in the PARKIN−/− cell line, while calcium signaling was significant in both the PARKIN−/− and ATP13A2−/− cell lines (Figure 5E).
Altered Cellular Functions and DN Compositions in the PARKIN−/− Line
In addition to the commonly dysregulated KEGG pathways, we found several KEGG pathways with direct relevance to DNs that were significantly dysregulated only in the PARKIN−/− line. These pathways included hsa05032, morphine addiction; hsa04726, serotonergic synapse; hsa04727, GABAergic synapse; hsa05030, cocaine addiction; and hsa04080, neuroactive ligand-receptor interaction (Figure S5D). We plotted genes contributing to any addiction pathways in the KEGG analysis (path:hsa05032, path:hsa05031, path:hsa05034, and path:hsa05033) and visualized the contributing genes in a heatmap. The PARKIN−/− line formed a distinct cluster compared with the other isogenic lines, showing dysregulation of many transcripts with prominent roles in dopaminergic function (Figure S5E).
Discussion
In this work, we have established and validated a robust large-scale 3D-spin culture protocol that allows for the derivation of midbrain DNs at high efficiency. We also generated an isogenic in vitro system to study cellular disease processes in WT and mutant DNs carrying severe autosomal recessive PD mutations in either PARKIN, DJ-1, or ATP13A2. We knocked in and validated a TH reporter to isolate relevant cells from our lines. Using our system, we have generated a deep proteomics dataset and analyzed the global protein abundance landscape across WT and PD disease lines. We further generated a transcriptomics dataset of sorted DNs from each of the lines. We found evidence of commonly dysregulated cellular functions involving OS and ALP. Dysregulation of PD-relevant pathways indicate shared and distinct molecular signatures among the isogenic PD cell lines. The PARKIN−/− lines were most distinct from controls and also were most distinct from the other PD lines. Identified shared or specific dysregulated candidate genes may inform efforts to find therapeutic targets and to stratify PD pathology and patients on a molecular level.
3D-Spin Cultures for the Derivation of DNs
Most current protocols to derive midbrain DNs from hPSCs rely on dual-SMAD inhibition, and regulation of SHH and WNT signaling through growth factors or small molecules (Chambers et al., 2009, Kriks et al., 2011). To accommodate high input technologies—such as screening assays and proteomics analyses—we adapted and simplified the existing culture protocols (Chambers et al., 2009, Kriks et al., 2011) for use in large-scale spinner flask bioreactors. Our protocol is technically easy, fully defined, and shows high reproducibility and scalability for deriving large numbers of midbrain DNs. Immunocytochemistry was used to demonstrate midbrain-marker expression in NPCs and DNs. Transcriptomics and proteomics data showed the expected expression of dopaminergic genes and enzymes. Electrophysiology analysis revealed the presence of cells with properties typical of A9 type midbrain DNs, and reporter expression was highly correlated to staining with commercial TH antibodies. TH:TdTomato signal enabled fluorescence-activated cell sorting purification and allowed for live cell tracking and laser confocal microscopy 3D imaging of DN morphology. In addition, we developed a protocol that allows dissociation of 3D-derived neurons for culture on 2D dishes or co-culture of isolated flow sorted neurons on glial cells. Spheres were maintained for up to 75 days, and there was no indication that the cultures cannot be maintained for longer time periods.
Omics Characterization of Isogenic PD Models
Protein stability and degradation are independent of transcriptional activity and strongly contribute to the regulation of protein levels. These processes are widely implicated in neurodegenerative diseases including PD (Caudle et al., 2010, Tai and Schuman, 2008). Here, we provide a comparative global molecular, functional, and phenotypic examination of several familial PD mutations in the same isogenic background. We generated transcriptomics data from sorted DNs and used quantitative proteomics to measure and quantify relative protein abundance in human DNs across our isogenic PD cell lines. PD is a disease of dysregulated proteins and accordingly we have linked PD genetics to emerging phenotypes in the relevant midbrain DNs and have established quantitative proteomics datasets (of >7,000 proteins) to explore genes, networks, and pathways that may contribute to PD etiology. High-confidence proteins were used for comparative studies. This deep quantitative proteomics dataset is a useful resource for the field and revealed common and distinct molecular dysregulation of PD cell lines and demonstrates the importance of studying PD in the relevant cell types.
OS and Mitochondrial Dysfunction Are Shared Phenotypes
There is evidence that OS is a component of the pathophysiology of familial and sporadic forms of PD. mROS are a source of cellular stress and have been examined in the context of PD and its hPSC models (Blesa et al., 2015, Cooper et al., 2012, Csobonyeiova et al., 2016, Dias et al., 2013). IPA of dysregulated proteins highlighted mitochondrial dysfunction, oxidative phosphorylation, and sirtuin-signaling pathways as the three most significant dysregulated pathways in the PARKIN−/− cell line. Despite the presence of antioxidants in the differentiation medium, analysis using flow cytometry revealed significantly increased levels of mROS in the basal state in all mutated lines when compared with the isogenic WT control. This increase was more pronounced in the TH+ cells, demonstrating the cell-type-specific vulnerability of DNs, further highlighting the utility of modeling PD in disease-relevant cells. Reduced activity of complex I, the NADH dehydrogenase complex, in mitochondria has been observed in midbrain tissue of sporadic PD patients (Keeney et al., 2006). Loss of PARKIN caused dysregulation of mitochondrial proteins in hPSCs, but the associated molecular dysregulation and measured phenotypes were significantly higher in the relevant DNs. This effect may be linked to dopamine oxidation, which is thought to initiate a toxic cascade that links OS to lysosomal dysfunction and SNCA aggregation (Burbulla et al., 2017).
The Interplay of Factors that May Explain Midbrain DN Phenotypes and Cell Death
SNCA was the first specific genetic aberration to have been linked to the development of PD (Polymeropoulos et al., 1997), and accumulation of SNCA aggregates and the formation of Lewy bodies are hallmarks of PD. SNCA was more than 3-fold enriched in PARKIN−/− DNs when compared with WT DNs.
PARKIN is broadly expressed throughout the body, including the heart, testis, liver, and kidney, as well as the brain (Kuhn et al., 2004). However, PD patients in general and those carrying loss-of-function PARKIN mutations exhibit the dysfunction and death of midbrain DNs. Parkin protein abundance was lowest in hPSCs and highest in DNs. Knockout mutations in the PARKIN gene resulted in a cell-type-specific phenotype culminating in the selective diminution of TH+ neurons in basal culture conditions. Using live cell time-lapse imaging and in association with a TH reporter we were able to show cellular phenotypes, such as enlarged vacuolated cell bodies and cell death with fragmentation of cellular compartments in PARKIN−/− cell lines, reminiscent of paraptosis, paraptosis-like cell death, necroptosis, and some forms of lysosomal cell death, all of which are associated with strong cytoplasmic vacuolation and which can lead to a cell death phenotype that has been described as bursting (Aits and Jaattela, 2013, Shubin et al., 2016). Cell death occurs at a vulnerable stage, shortly after the onset of TH expression and the first synthesis of dopamine, when the cells are also undergoing major and energy-consuming morphological changes, neuron and neurite growth, and show increased SNCA expression.
Current protocols to derive midbrain DNs result in a heterogeneous mix of non-neuronal cells, DNs, and other neuronal cell types (Marton and Ioannidis, 2018). DNs themselves represent a heterogeneous group, containing SNc and VTA identities, among others (Xia et al., 2016). Midbrain DNs are anatomically separated into SNc and VTA. In PD patients, a subtype of nigral DNs shows enhanced vulnerability, while other populations, such as VTA DNs, are much less affected. Similarly, in our cultures, some, but not all, DNs died. CALB1 is often used as a marker to distinguish VTA from SN and shows higher expression in the VTA. VTA DNs also play a primary role in the reward system and addiction. Both increased CALB1 levels and the analysis of commonly dysregulated KEGG pathways in our PARKIN−/− cell lines point toward a shift in the surviving population and we speculate that hPSC-derived DNs model the specific vulnerabilities of DN subpopulations found in human brains. Pathway analysis of transcriptional data between WT and PARKIN−/− cell lines implicated dysregulation of both GABAergic synapse and reward systems.
Isogenic PD Models Reveal Common and Distinct Disease Pathways
Abnormal protein aggregation, OS/mitochondrial dysregulation and endoplasmic reticulum stress are critical pathological mechanisms involving ALP (Plotegher and Duchen, 2017). Most models of ALP rely on chemical perturbations to initiate or arrest the relevant cellular functions. In our isogenic PD lines, specific PD-associated mutations drove all measured differences. Analyzing proteomics data, we found that DJ1−/− and ATP13A2−/− cell lines showed similar dysregulation, while the PARKIN−/− line shows distinct dysregulation.
Overall, our experimental platform using a knockin fluorescence reporter in the TH locus, together with our results on phenotyping, transcriptomics, and proteomic analyses of isogenic lines, provide an improved understanding of PARKIN, DJ-1, and ATP13A2 function. Our isogenic models allowed us to compare the impact of PD-relevant genes on the differentiation and health of the most vulnerable cell type in PD. Our study demonstrates how isogenic PD hPSCs can be used to understand PD-relevant disease pathology, determine affected pathways, and enhance our knowledge of genetic interactions in PD pathology. We have demonstrated DN-specific disease-relevant phenotypes in the PARKIN−/− line and identified OS as a common pathology and a shared dysregulated pathway in all isogenic cell lines. Conversely, and despite similar clinical presentations, we found evidence for at least two etiological PD subtypes, PARKIN−/− versus the ATP13A2−/−and DJ-1−/− lines, indicating that precise delineation of PD subtypes will require evaluation of both molecular and clinical data.
Experimental Procedures
See further details in the Supplemental Experimental Procedures.
Propagation and Maintenance of hPSCs
hESC line HUES01, from Harvard University (Cowan et al., 2004), was cultured on Matrigel-coated plates (ESC qualified, BD Biosciences) using hESC mTeSR-1 cell culture medium (STEMCELL Technologies) under conditions of 37°C, 95% air, and 5% CO2 in a humidified incubator as described previously (Schinzel et al., 2011). The WA01 and BJ-SIPS cell lines were cultured on Geltrex-coated (100 μL matrix/10 mL basal medium) plates in StemFlex medium (Gibco) under the same conditions.
hPSC Adaption and Maintenance in Spinner Flasks
hPSCs were cultured in 125-mL disposable spinner flasks (Corning, VWR) on a nine-position stir plate (Dura-Mag) at a speed of 65 rpm, in a 37°C incubator with 5% CO2, which is a higher speed than previously reported (Rigamonti et al., 2016). Before adaption to spinner flask culture, hPSCs were expanded in 10-cm dishes (Corning) until they reached confluence. Cells were dissociated using Accutase (Innovative Cell Technologies) for approximately 5 min at room temperature or until colonies detached from the plate. Cells were counted using a Bio-Rad automated cell counter. Forty million individual hPSCs were seeded into a spinner flask in 120 mL of StemFlex medium supplemented with 10 μM ROCK inhibitor Thiazovivin (STEMCELL Technologies). Spheres formed spontaneously, and, after 48 h, approximately half of the culture medium was replaced. The cells were maintained as undifferentiated pluripotent spheres in spin culture, with medium changes every other day until the spheres were approximately 500 μm (organoid area was measured using Nikon software in brightfield).
Differentiation into Midbrain DNs in Spin Culture
Differentiation was initiated through a full medium switch using differentiation medium d0. Subsequently medium was changed every other day by removing approximately 50% of the medium after spheres were allowed to settle by gravity. Differentiation medium to pattern toward midbrain has a changing composition outlined in experimental scheme in Figure 1 and contains DMEM-F12, N2, B27, 10 μM transforming growth factor β inhibitor SB431542 (R&D Systems) and 100 nM BMP inhibitor LDN193189 (Stemgent) (dual-SMAD inhibition), 1 μM CHIR99021 (Stemgent), 2 μM purmorphamine (Stemgent or STEMCELL Technologies), and 1 μM SAG (Cursi or Cayman Chemical). Terminal differentiation medium contains 10 ng/mL brain-derived neurotrophic factor (R&D), 10 ng/mL glial cell line-derived neurotrophic factor (R&D), 0.2 mM ascorbic acid (Sigma), 0.1 mM butyryl cAMP (Sigma or Biolog), and 10 μM N-[N-(3,5-difluorophenacetyl)-L-alanyl]-S-phenylglycine t-butyl ester(Tocris or Cayman Chemical) (BAGTC).
Data Access
The accession number for the RNA sequencing data generated in this paper is GEO: GSE140076. Table S2 lists all quantified proteins as well as associated TMT reporter ion intensities used for quantitative analysis.
Statistical Analysis
If not otherwise stated, experiments were carried out in clonal isogenic cell lines in at least three differentiation experiments that were separated by time in culture. The experiments were not randomized. In other experiments three independent clonal lines were used. The investigators were not blinded during experiments and outcome assessment. Immunofluorescence experiments were repeated independently 2 times and at least 20 cells were analyzed from a single experiment. When analyzing immunohistochemistry staining data of sectioned spheres, several spheres were analyzed using sections representing outer, inner, and middle layers. Error bars are mostly presented as the means ± SD unless otherwise specified. Statistical comparisons between pairs were made using Student's t test. Statistical significance across multiple samples for proteomics data was determined using one-way ANOVA (artificial within groups variance was set to 1 and correction for multiple hypothesis testing was done by permutation-based false discovery rate [FDR 1%] followed by a post hoc test [1% FDR Bonferroni-Holm]), Statistical analysis for gene expression was performed using standard normalization and multiple comparison methods of the edgeR package or denoted in the Supplemental Information.
Author Contributions
T.A. and L.L.R. conceived of the experiments, supervised the experimental work and wrote the final manuscript. T.A. carried out most of the experiments and data analysis presented. A.O., C.B., J.A.P. and J.W.H. collected the proteomics data and contributed to the related data analysis and manuscript writing. L.S., S.P., T.G. and G.M.P. collected cell biological data, assisted in the data analysis and manuscript writing. C.S. performed and interpreted the electrophysiological characterization of all cell lines. F.Y., T.U. and Y.K. assisted with the acquisition, analysis and interpretation of live cell imaging data. All authors discussed the results, provided critical feedback and contributed to the final manuscript.
Acknowledgments
The authors thank Geraldine Jowett and Aya Alame for technical help and discussions, Methodios Ximerakis for help with PARKIN antibody staining conditions, Rich Krolewski, Kathleen Pfaff, and Jane LaLonde for critical reading, Matt LaVoie for ideas and discussions, Claire Reardon, Abbie Groff, Chiara Gerhardinger, and John Rinn for their help in making RNA-sequencing libraries, generating the dataset, and alignments, as well as advice on the experimental design and analysis, Silvia Ionescu and Joyce LaVecchio from the HSCRB-HSCI Flow Cytometry Core for discussions and technical expertise sorting dissociated neurons, Christopher Bare from the Deans Flow Cytometry CoRE at Icahn School of Medicine, Mount Sinai for flow cytometry training, and Aaron Bell and Valeriy Borukhov from the Neuropathology Brain Bank at the Icahn School of Medicine at Mount Sinai for technical expertise in sectioning, staining, and imaging spheres. This work was supported by a generous gift to the Harvard Stem Cell Institute from the Elizabeth Miller fund, and grants from Nikon Corporation and Biogen. In addition, this work was supported by the Michael J. Fox Foundation, NIH grant R37NS083524, and the Ned Goodnow Fund to J.W.H., and by a postdoctoral fellowship from the Edward R. and Anne G. Lefler Center to A.O.
Published: January 2, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.stemcr.2019.12.005.
Contributor Information
Tim Ahfeldt, Email: tim.ahfeldt@mssm.edu.
Lee L. Rubin, Email: lee_rubin@harvard.edu.
Supplemental Information
Set 1, intersection of d35 proteins; set 2, intersection of pluripotent samples, all detected with confidence in two analyses that were bridged using WT TH spheres from three independent differentiation replicates. ANOVA significance indicated with asterisk in column ANOVA, stringent pairwise significance indicated in significance columns.
Summarized table of propagated gates, FlowJo Table editor (FlowJo, LLC), FSC-A/SSC-A, and pulse geometry gate FSC-W/SSCH. TH was quantified on a GFP-A/PE-Texas red-A gate.
References
- Aits S., Jaattela M. Lysosomal cell death at a glance. J. Cell Sci. 2013;126:1905–1912. doi: 10.1242/jcs.091181. [DOI] [PubMed] [Google Scholar]
- Amit M., Laevsky I., Miropolsky Y., Shariki K., Peri M., Itskovitz-Eldor J. Dynamic suspension culture for scalable expansion of undifferentiated human pluripotent stem cells. Nat. Protoc. 2011;6:572–579. doi: 10.1038/nprot.2011.325. [DOI] [PubMed] [Google Scholar]
- Blesa J., Trigo-Damas I., Quiroga-Varela A., Jackson-Lewis V.R. Oxidative stress and Parkinson's disease. Front Neuroanat. 2015;9:91. doi: 10.3389/fnana.2015.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodea G.O., Blaess S. Establishing diversity in the dopaminergic system. FEBS Lett. 2015;589:3773–3785. doi: 10.1016/j.febslet.2015.09.016. [DOI] [PubMed] [Google Scholar]
- Burbulla L.F., Song P., Mazzulli J.R., Zampese E., Wong Y.C., Jeon S., Santos D.P., Blanz J., Obermaier C.D., Strojny C. Dopamine oxidation mediates mitochondrial and lysosomal dysfunction in Parkinson's disease. Science. 2017;357:1255–1261. doi: 10.1126/science.aam9080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudle W.M., Bammler T.K., Lin Y., Pan S., Zhang J. Using 'omics' to define pathogenesis and biomarkers of Parkinson's disease. Expert Rev. Neurother. 2010;10:925–942. doi: 10.1586/ern.10.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C., Lim K.L. Genetic insights into sporadic Parkinson's disease pathogenesis. Curr. Genomics. 2013;14:486–501. doi: 10.2174/1389202914666131210195808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers S.M., Fasano C.A., Papapetrou E.P., Tomishima M., Sadelain M., Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper O., Seo H., Andrabi S., Guardia-Laguarta C., Graziotto J., Sundberg M., McLean J.R., Carrillo-Reid L., Xie Z., Osborn T. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci. Transl. Med. 2012;4:141ra190. doi: 10.1126/scitranslmed.3003985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowan C.A., Klimanskaya I., McMahon J., Atienza J., Witmyer J., Zucker J.P., Wang S., Morton C.C., McMahon A.P., Powers D. Derivation of embryonic stem-cell lines from human blastocysts. N. Engl. J. Med. 2004;350:1353–1356. doi: 10.1056/NEJMsr040330. [DOI] [PubMed] [Google Scholar]
- Csobonyeiova M., Danisovic L., Polak S. Induced pluripotent stem cells for modeling and cell therapy of Parkinson's disease. Neural Regen. Res. 2016;11:727–728. doi: 10.4103/1673-5374.182692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauer W., Przedborski S. Parkinson's disease: mechanisms and models. Neuron. 2003;39:889–909. doi: 10.1016/s0896-6273(03)00568-3. [DOI] [PubMed] [Google Scholar]
- Dias V., Junn E., Mouradian M.M. The role of oxidative stress in Parkinson's disease. J. Parkinsons Dis. 2013;3:461–491. doi: 10.3233/JPD-130230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampshire D.J., Roberts E., Crow Y., Bond J., Mubaidin A., Wriekat A.L., Al-Din A., Woods C.G. Kufor-Rakeb syndrome, pallido-pyramidal degeneration with supranuclear upgaze paresis and dementia, maps to 1p36. J. Med. Genet. 2001;38:680–682. doi: 10.1136/jmg.38.10.680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H., Ren Y., Yuen E.Y., Zhong P., Ghaedi M., Hu Z., Azabdaftari G., Nakaso K., Yan Z., Feng J. Parkin controls dopamine utilization in human midbrain dopaminergic neurons derived from induced pluripotent stem cells. Nat. Commun. 2012;3:668. doi: 10.1038/ncomms1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney P.M., Xie J., Capaldi R.A., Bennett J.P., Jr. Parkinson's disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. 2006;26:5256–5264. doi: 10.1523/JNEUROSCI.0984-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein C., Westenberger A. Genetics of Parkinson's disease. Cold Spring Harb. Perspect. Med. 2012;2:a008888. doi: 10.1101/cshperspect.a008888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S., Shim J.W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn K., Zhu X.R., Lubbert H., Stichel C.C. Parkin expression in the developing mouse. Brain Res. Dev. Brain Res. 2004;149:131–142. doi: 10.1016/j.devbrainres.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Marton R.M., Ioannidis J.P.A. A comprehensive analysis of protocols for deriving dopaminergic neurons from human pluripotent stem cells. Stem Cells Transl. Med. 2018;8:366–374. doi: 10.1002/sctm.18-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordureau A., Paulo J.A., Zhang W., Ahfeldt T., Zhang J., Cohn E.F., Hou Z., Heo J.M., Rubin L.L., Sidhu S.S. Dynamics of PARKIN-dependent mitochondrial Ubiquitylation in induced neurons and model systems revealed by Digital Snapshot proteomics. Mol. Cell. 2018;70:211–227.e8. doi: 10.1016/j.molcel.2018.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paisan-Ruiz C., Guevara R., Federoff M., Hanagasi H., Sina F., Elahi E., Schneider S.A., Schwingenschuh P., Bajaj N., Emre M. Early-onset L-dopa-responsive parkinsonism with pyramidal signs due to ATP13A2, PLA2G6, FBXO7 and spatacsin mutations. Mov. Disord. 2010;25:1791–1800. doi: 10.1002/mds.23221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plotegher N., Duchen M.R. Crosstalk between lysosomes and mitochondria in Parkinson's disease. Front. Cell Dev. Biol. 2017;5:110. doi: 10.3389/fcell.2017.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polymeropoulos M.H., Lavedan C., Leroy E., Ide S.E., Dehejia A., Dutra A., Pike B., Root H., Rubenstein J., Boyer R. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276:2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhardt P., Schmid B., Burbulla L.F., Schondorf D.C., Wagner L., Glatza M., Hoing S., Hargus G., Heck S.A., Dhingra A. Genetic correction of a LRRK2 mutation in human iPSCs links parkinsonian neurodegeneration to ERK-dependent changes in gene expression. Cell stem cell. 2013;12:354–367. doi: 10.1016/j.stem.2013.01.008. [DOI] [PubMed] [Google Scholar]
- Rigamonti A., Repetti G.G., Sun C., Price F.D., Reny D.C., Rapino F., Weisinger K., Benkler C., Peterson Q.P., Davidow L.S. Large-scale production of mature neurons from human pluripotent stem cells in a three-dimensional suspension culture system. Stem Cell Reports. 2016;6:993–1008. doi: 10.1016/j.stemcr.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan S.D., Dolatabadi N., Chan S.F., Zhang X., Akhtar M.W., Parker J., Soldner F., Sunico C.R., Nagar S., Talantova M. Isogenic human iPSC Parkinson's model shows nitrosative stress-induced dysfunction in MEF2-PGC1alpha transcription. Cell. 2013;155:1351–1364. doi: 10.1016/j.cell.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinzel R.T., Ahfeldt T., Lau F.H., Lee Y.K., Cowley A., Shen T., Peters D., Lum D.H., Cowan C.A. Efficient culturing and genetic manipulation of human pluripotent stem cells. PLoS One. 2011;6:e27495. doi: 10.1371/journal.pone.0027495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulte C., Gasser T. Genetic basis of Parkinson's disease: inheritance, penetrance, and expression. Appl. Clin. Genet. 2011;4:67–80. doi: 10.2147/TACG.S11639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaltouki A., Sivapatham R., Pei Y., Gerencser A.A., Momcilovic O., Rao M.S., Zeng X. Mitochondrial alterations by PARKIN in dopaminergic neurons using PARK2 patient-specific and PARK2 knockout isogenic iPSC lines. Stem Cell Reports. 2015;4:847–859. doi: 10.1016/j.stemcr.2015.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner N.C., Campbell R.E., Steinbach P.A., Giepmans B.N., Palmer A.E., Tsien R.Y. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- Shi W.X. Slow oscillatory firing: a major firing pattern of dopamine neurons in the ventral tegmental area. J. Neurophysiol. 2005;94:3516–3522. doi: 10.1152/jn.00317.2005. [DOI] [PubMed] [Google Scholar]
- Shubin A.V., Demidyuk I.V., Komissarov A.A., Rafieva L.M., Kostrov S.V. Cytoplasmic vacuolization in cell death and survival. Oncotarget. 2016;7:55863–55889. doi: 10.18632/oncotarget.10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmann O., Zou H., Gerber S.A., Gygi S.P., Kirschner M.W. Dual inhibition of sister chromatid separation at metaphase. Cell. 2001;107:715–726. doi: 10.1016/s0092-8674(01)00603-1. [DOI] [PubMed] [Google Scholar]
- Tabata Y., Imaizumi Y., Sugawara M., Andoh-Noda T., Banno S., Chai M., Sone T., Yamazaki K., Ito M., Tsukahara K. T-type calcium channels determine the vulnerability of dopaminergic neurons to mitochondrial stress in familial Parkinson disease. Stem Cell Reports. 2018;11:1171–1184. doi: 10.1016/j.stemcr.2018.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H.C., Schuman E.M. Ubiquitin, the proteasome and protein degradation in neuronal function and dysfunction. Nat. Rev. Neurosci. 2008;9:826–838. doi: 10.1038/nrn2499. [DOI] [PubMed] [Google Scholar]
- Thomas B., Beal M.F. Parkinson's disease. Hum. Mol. Genet. 2007;16 Spec No. 2:R183–R194. doi: 10.1093/hmg/ddm159. [DOI] [PubMed] [Google Scholar]
- Uhl G.R., Hedreen J.C., Price D.L. Parkinson's disease: loss of neurons from the ventral tegmental area contralateral to therapeutic surgical lesions. Neurology. 1985;35:1215–1218. doi: 10.1212/wnl.35.8.1215. [DOI] [PubMed] [Google Scholar]
- Xia N., Zhang P., Fang F., Wang Z., Rothstein M., Angulo B., Chiang R., Taylor J., Reijo Pera R.A. Transcriptional comparison of human induced and primary midbrain dopaminergic neurons. Sci. Rep. 2016;6:20270. doi: 10.1038/srep20270. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Section 1: WT-Well-1 10× magnification. Section 2: WT-Well-1 representative area zoomed in. Section 3: PARKIN−/−-Well-1 10× magnification. Section 4: PARKIN−/−-Well-1 representative area zoomed in. Section 5: WT-Well-2 10× magnification. Section 6: WT-Well-2 representative area zoomed in. Section 7: PARKIN−/−-Well-2 10× magnification. Section 8: PARKIN−/−-Well-2 representative area zoomed in.
Set 1, intersection of d35 proteins; set 2, intersection of pluripotent samples, all detected with confidence in two analyses that were bridged using WT TH spheres from three independent differentiation replicates. ANOVA significance indicated with asterisk in column ANOVA, stringent pairwise significance indicated in significance columns.
Summarized table of propagated gates, FlowJo Table editor (FlowJo, LLC), FSC-A/SSC-A, and pulse geometry gate FSC-W/SSCH. TH was quantified on a GFP-A/PE-Texas red-A gate.