Abstract
Since the initial discovery of ribozymes in the early 1980s, catalytic nucleic acids have been used in different areas. Compared with protein enzymes, catalytic nucleic acids are programmable in structure, easy to modify, and more stable especially for DNA. We take a historic view to summarize a few main interdisciplinary areas of research on nucleic acid enzymes that may have broader impacts. Early efforts on ribozymes in the 1980s have broken the notion that all enzymes are proteins, supplying new evidence for the RNA world hypothesis. In 1994, the first catalytic DNA (DNAzyme) was reported. Since 2000, the biosensor applications of DNAzymes have emerged and DNAzymes are particularly useful for detecting metal ions, a challenging task for enzymes and antibodies. Combined with nanotechnology, DNAzymes are key building elements for switches allowing dynamic control of materials assembly. The search for new DNAzymes and ribozymes is facilitated by developments in DNA sequencing and computational algorithms, further broadening our fundamental understanding of their biochemistry.
Subject Areas: Chemistry, Biochemistry, Biochemistry Applications, Enzymology, Biomolecules, Nanotechnology
Graphical Abstract
Chemistry; Biochemistry; Biochemistry Applications; Enzymology; Biomolecules; Nanotechnology
Introduction
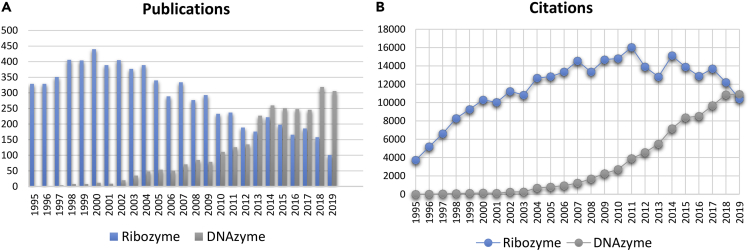
The notion that all enzymes are proteins was changed with the discovery of ribozymes in the early 1980s (Guerrier-Takada et al., 1983, Kruger et al., 1982). Since then, researchers have been motivated to search for new ribozymes in nature, in test tubes, and in silico. Although not yet found in nature, the first DNAzyme (also called deoxyribozyme or catalytic DNA) was isolated using in vitro selection in 1994 (Breaker and Joyce, 1994). For applications outside cells, DNA is more attractive than RNA for stability and cost considerations. A search in the ISI Web of Knowledge database revealed interesting trends (Figure 1). Publications on ribozymes peaked in the early 2000s, when the DNAzyme field just started to take off. Recently, the papers and citations on DNAzymes have outnumbered that on ribozymes, which was mainly driven by research on biosensors, intracellular RNA cleavage, and bionanotechnology.
Figure 1.
Publication and Citation Trends
Comparisons of the number of publications (A) and citations (B) on “ribozyme” and “DNAzyme” or “deoxyribozyme” as keywords from 1995 to 2019.
With nearly four decades of development, nucleic acid enzymes have impacted many fields from biosensing and anti-virus to materials science. Many reviews on ribozymes and DNAzymes have been published (Hwang et al., 2016, Liu et al., 2019, Liu et al., 2017b, Silverman, 2016, Sun et al., 2000, Weinberg et al., 2019, Zhou et al., 2017b), focusing these aspects. Herein, we wish to have a broad scope summary of some key areas of the field. This may set a basis for further development and attracting attention of researchers from other fields to use these interesting molecules. Limited by space, we cannot be comprehensive and only representative examples will be discussed to make the content broadly interesting and inspiring.
Ribozymes in the Early Days
RNA for a long time was only known for mediating the production of proteins. In the early 1980s, the initial discovery of ribozymes was related to the RNA cleavage and self-splicing reactions, leading to the award of the Nobel Prize to Cech and Altman (Guerrier-Takada et al., 1983, Kruger et al., 1982). Since then, ribozymes for many important biological reactions were discovered in nature, such as the ribosomal RNA catalyzing the formation of peptide bonds (Doudna and Cech, 2002, Müller et al., 2016). The ribosome in cells is composed of both protein and RNA, whereas the peptidyl transferase activity was performed by the RNA component (Figure 2A). Given that RNA possesses both genotype and enzymatic functions, the RNA world hypothesis was proposed, insisting that RNA dominated the early life before proteins (Gesteland et al., 1999, Joyce, 2002). In addition, many coenzymes or cofactors in protein enzymes, such as coenzyme A (CoA) and nicotinamide adenine dinucleotide (NAD), contain the basic structure of ribonucleotides, also supporting this hypothesis.
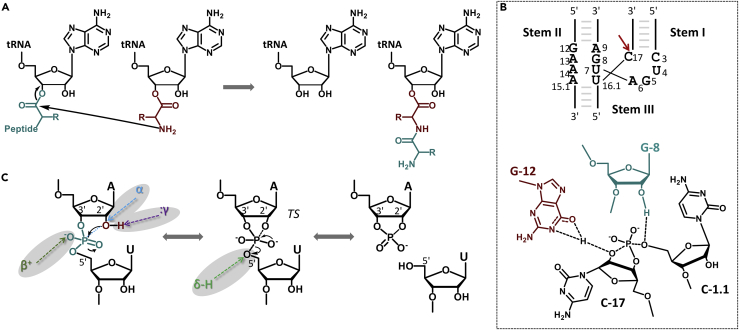
Figure 2.
Reactions and Mechanisms of Representative Ribozymes
(A) The peptidyl transferase activity of the ribosomal RNA during peptide bond formation.
(B) The minimal secondary structure of the hammerhead ribozyme containing 13 conserved nucleotides (top). The active site in the transition state where G-12 functions as a general base and G-8 as a general acid (bottom). Redrawn from Martick and Scott (2006).
(C) The cleavage of a 3′, 5′-phosphodiester bond catalyzed by self-cleaving ribozymes. The nucleophilic attack of the 2′-OH to the nearest phosphate causes the leaving of the 5′-O and produces a 2′, 3′-cyclic phosphate and a 5′-OH. The α, β, γ, and δ represent four catalytic mechanisms for the reaction.
Over the past 30 years, self-cleaving ribozymes catalyzing the cleavage of phosphodiester bonds have been continuously discovered. The hammerhead ribozyme (Figure 2B) was found in the tobacco ringspot virus satellite RNA. Other ribozymes, including the hairpin, hepatitis delta virus (HDV), varakud satellite, and twister ribozymes (Roth et al., 2013), were identified in various organisms. These enzymes catalyze the site-specific cleavage of 3′, 5′-phosphodiester bonds with up to 106-fold rate enhancement. Many structural and biochemical studies were performed to provide mechanistic understanding of ribozyme catalysis (Jimenez et al., 2015, Ren et al., 2017). Generally, the RNA cleavage reaction can be accelerated through four pathways (Figure 2C): arranging the in-line alignment between the 2′-O nucleophile, scissile phosphorus, and 5′-leaving oxygen (α factor); facilitating the deprotonation of 2′-OH in the nucleophilic attack (β factor); neutralizing the negative charge on the non-bridging phosphoryl oxygens in the transition state (γ factor); and stabilizing the negative charge on the 5′-leaving oxygen (δ factor) (Cochrane and Strobel, 2008). For a given ribozyme, critical nucleobases and metal ions in the catalytic core can be determined once the crystal structure becomes available. In the active site of the hammerhead ribozyme, a conserved guanine, G-12, acts as a general base by hydrogen bonding with the 2′-O nucleophile, whereas the 2′-OH of the other guanine G-8 stabilizes the leaving oxygen as a general acid (Figure 2B) (Martick and Scott, 2006). The HDV ribozyme utilizes a Mg2+-bound water molecule as a general base and a conserved cytosine (C75) as a general acid (Ferré-D'Amaré et al., 1998). In the case of the glms ribozyme, a small molecule, glucosamine-6-phosphate (GlcN6P), is required as a cofactor for its catalysis instead of divalent metal ions (Winkler et al., 2004).
Apart from these naturally occurring ribozymes, more were discovered in the laboratory through in vitro selection (Wilson and Szostak, 1999). In vitro selection resembles the process of natural evolution, in which a large RNA library is subject to a selection pressure and those sequences with a particular activity can survive and get amplified. Benefited from this accelerated process, ribozymes catalyzing a great variety of chemical reactions, such as RNA ligation, phosphorylation, self-alkylation, and Diels-Alder reaction, have been obtained over the past decades (Wilson and Szostak, 1999). Recently, interesting efforts were made for producing polymerizing ribozymes and self-replicating systems (Lincoln and Joyce, 2009, Robertson and Joyce, 2014). A self-replicating ribozyme represents an RNA ligase catalyzing the amplification of itself with an exponential growth. For instance, an optimized ribozyme from in vitro selection showed an exponential growth rate of 0.14 min−1, which is close to the limiting rate of product release. Continuous efforts will be made to broaden the scope of ribozyme catalysis. Meanwhile, with the development of sequencing technology and computational searching algorithm, more natural ribozymes might be discovered in the future (Hammann and Westhof, 2007, Perreault et al., 2011).
Ribozymes for Therapeutic and Chemical Biology Applications
After the discovery of self-cleaving ribozymes, their therapeutic potential in inhibiting gene expression was tested (Sullenger and Gilboa, 2002). For this purpose, several self-cleaving ribozymes (i.e., the hammerhead and hairpin ribozymes) were engineered to catalyze RNA cleavage in the trans-cleaving manner (Figure 3A). By altering the binding sequence to be complementary to target mRNA, trans-cleaving ribozymes can selectively cleave essentially any mRNA target, such as HIV (Rossi, 2000) and cancer-related genes (Pavco et al., 2000). However, many challenges remain in applying ribozymes in vivo, such as the short lifetime of ribozymes in vivo, ineffective delivery of ribozymes to target cells, and poor enzymatic activity. To improve the intracellular activity of the hammerhead ribozyme, in vivo selection of trans-cleaving hammerhead mutants was recently performed in E. coli cells (Huang et al., 2019). The selected variants showed an enhanced ability for intracellular gene silencing toward various RNA targets. For example, intracellularly expressed ribozymes were designed to cleave a reporter gene, spinach, and the mutant suppressed fluorescence by disrupting the binding of the DFHBI dye twice more than the original ribozyme (Figure 3B).
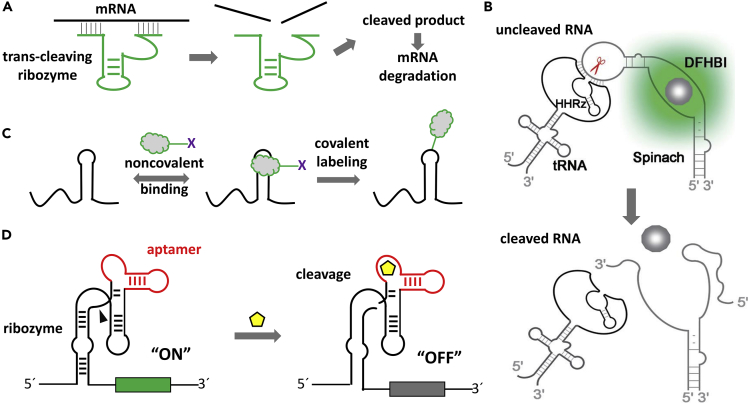
Figure 3.
Chemical Biology Applications of Ribozymes
(A) Trans-cleaving ribozymes for gene silencing.
(B) Intracellularly selected hammerhead ribozymes with enhanced cleavage activity in living cells. Spinach is a report RNA that binds with a small molecule, DFHBI, and emits fluorescence. Reprinted with permission from Huang et al. (2019). Copyright © 2019 Oxford University Press.
(C) Covalent labeling of a fluorophore on RNA by a self-alkylating ribozyme. Redrawn from Sharma et al. (2014).
(D) Rational design of an aptazyme for ligand-responsive gene expression.
Apart from RNA cleavage, another major direction is to develop ribozymes for chemical biology applications such as biomolecular synthesis and modification (Balke et al., 2014, Baskerville and Bartel, 2002). In a recent example, self-alkylating ribozymes were selected to achieve covalent labeling of a fluorophore on RNA (Figure 3C) (Sharma et al., 2014). Another branch of application is to combine aptamers and ribozymes to achieve ligand-responsive gene regulation. In nature, riboswitches are natural RNA aptamers that can regulate gene expression upon binding with small molecules. The glms ribozyme was identified in Gram-positive bacteria to cleave certain messenger RNA using GlcN6P as a cofactor (Winkler et al., 2004). Inspired by these, aptazymes or allosteric ribozymes have been rationally designed using existing aptamers and ribozymes, or selected from in vitro evolution (Famulok et al., 2007). A rationally designed aptazyme is composed of an aptamer domain (e.g., for ATP, theophylline [Wieland and Hartig, 2008]) and a ribozyme domain (e.g., hammerhead, hairpin) (Penchovsky and Breaker, 2005). Ligand binding induces conformational changes that subsequently influence the ribozyme activity and turns off (Figure 3D) or on the gene expression. Recently, hammerhead-based aptazymes have been embedded in the guide RNAs of the CRISPR-Cas9 system to perform ligand-responsive genome editing (Tang et al., 2017).
DNAzymes for Intracellular RNA Cleavage
Given the chemical similarity between DNA and RNA, a natural question is whether DNA can also perform catalysis. Since most DNA molecules in nature are double stranded, they are unlikely to be catalytically active. Single-stranded DNA, on the other hand, can form tertiary structures, allowing molecular recognition and catalysis. In the early 1990s, single-stranded DNAs with specific binding activity (aptamers) have been obtained through in vitro selection. Similar methods can also be used for the selection of DNAzymes, and the first DNAzyme was reported by Breaker and Joyce in 1994. This DNAzyme, named GR5 (Figure 5A), was selected for RNA cleavage in the presence of Pb2+ (Breaker and Joyce, 1994). GR5 is short with only 15 nucleotides in the catalytic loop, and its substrate contains a single RNA linkage serving as the cleavage site. In the original paper, the catalytic activity reached a turnover rate of 1 min−1 in the presence of 1 mM Pb2+. Later, after optimization, the activity easily reached >10 min−1 with 10 μM Pb2+ (Saran and Liu, 2016a). The initial choice of Pb2+ for the selection was probably motived by Pb2+-catalyzed RNA cleavage as observed in the leadzyme (Pan and Uhlenbeck, 1992).
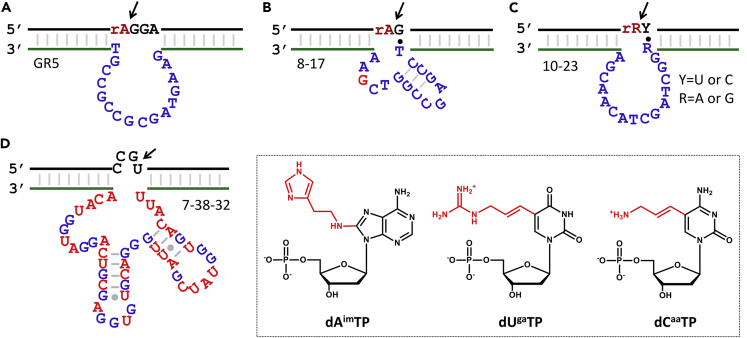
Figure 5.
DNAzymes and Modified DNAzymes for RNA Cleavage
(A–C) The secondary structures of the (A) GR5, (B) 8-17, and (C) 10-23 DNAzymes.
(D) The 7-38-32 DNAzyme containing three modified nucleotides: 8-histaminyl-deoxyadenosine (dAimTP), 5-guanidinoallyl-deoxyuridine (dUgaTP), and 5-aminoallyl-deoxycytidine (dCaaTP).
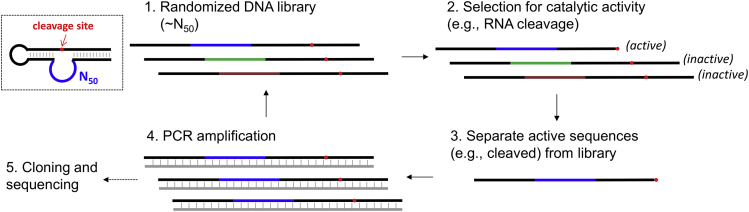
A general procedure of in vitro selection for RNA-cleaving DNAzymes is illustrated in Figure 4. For a typical selection experiment, a single-stranded DNA library with a random region is synthesized. The library is often designed to fold into a secondary structure to position the cleavage site close to the randomized region (the N50 region). A selection pressure is applied to the DNA library aiming for specific catalytic activity such as RNA cleavage. Sequences with catalytic activities are separated taking advantage that the reaction can decrease (e.g., cleavage reaction) the length of the library. After PCR amplification, the library is enriched with active sequences and can be used for the next round of selection. This method of test tube evolution is powerful and can be employed under both physiological and non-physiological conditions. In vitro selection of DNAzyme is easier than selection of ribozymes since no transcription/reverse transcription steps are needed.
Figure 4.
A Scheme Showing the Key Steps of In Vitro Selection for RNA-Cleaving DNAzymes Starting from a Random DNA Library
The boxed region shows the secondary structure of a typical random library with a single RNA linkage embedded (in red). After step 4, the PCR step, the resulting duplex product needs to be separated and only one of the strands is useful.
One of the first motivations to develop RNA-cleaving DNAzymes was to cleave viral RNA. GR5 is apparently not appropriate for this purpose since it cannot cleave all-RNA substrates (it only cleaves the substrate with a single RNA linkage), and it requires toxic Pb2+ (no activity in the presence of Mg2+ alone). Subsequently, Santoro and Joyce reported two DNAzymes named 8-17 and 10-23 (Figures 5B and 5C), both of which can cleave full-RNA substrates with Mg2+ alone (Santoro and Joyce, 1997). The catalytic rate reached ∼0.1 min−1 with 2 mM Mg2+, and the catalytic efficiency (kcat/Km) reached ∼109 M−1·min−1 for the 10-23 DNAzyme, which is comparable with the most efficient protein enzyme. The requirement of substrate sequence is very simple. Further studies revealed that all the 16 junctions can be cleaved at different efficiencies (Schlosser et al., 2008). The specificity of cleavage is defined by the two substrate binding arms. With each arm containing eight or more base pairs, these two DNAzymes can in principle target any specific RNA sequence in cells.
This discovery has excited the field for pursuing anti-viral and later anti-cancer applications (Huanhuan et al., 2017, Zhou et al., 2017c). However, to achieve a high cleavage activity, the required Mg2+ concentration is quite high and intracellular free Mg2+ can hardly meet the requirement. It has been argued that the observed intracellular inhibition of gene expression using DNAzymes could simply be due to the anti-sense effect (Young et al., 2010). To solve this problem, using modified nucleotides to mimic the chemical functionalities of RNase A has been attempted and metal-free cleavage was achieved in some cases. An example is the DNAzyme 7-38-32 (Figure 5D), which cleaves an RNA substrate with a catalytic rate of 1.06 min−1 in the presence of only 0.5 mM Mg2+ (Wang et al., 2018). Other efforts such as the selection of divalent metal-free DNAzymes have also been made, some of which required only Na+ for activity (Geyer and Sen, 1997, Ma and Liu, 2019, Torabi et al., 2015, Zhou et al., 2017a).
RNA-Cleaving DNAzymes as Biosensors
Sensing Metal Ions
Compared with biomedical applications, the work on using DNAzymes as biosensors has advanced more (Gong et al., 2015, Zhang et al., 2011, Zhou et al., 2017b). Lu and coworkers selected the 17E DNAzyme in the presence of Zn2+ (Li et al., 2000), and it is very similar to the 8-17. Both DNAzymes are much more active with Pb2+. Li and Lu first demonstrated using the 17E DNAzyme for Pb2+ detection (Li and Lu, 2000). Since then, the 17E has been used as a model system to develop different signaling mechanisms from fluorescence, color, electrochemistry to Raman spectroscopy (Liu et al., 2009), although later GR5 was found to have much better specificity for Pb2+ (Lan et al., 2010).
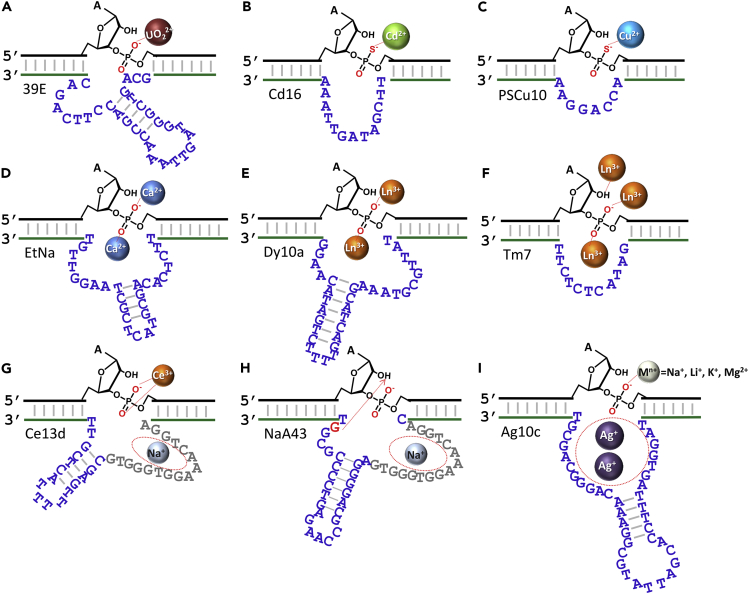
Many subsequent selections have been intentionally carried out in the presence of target metal ions to achieve specificity. The 39E DNAzyme was isolated in the presence of UO22+ and it has over 1-million-fold higher selectivity for UO22+ over other metal ions (Figure 6A) (Liu et al., 2007). The EtNa DNAzyme is highly specific for Ca2+ in water (Zhou et al., 2017a) but selective for Na+ in ethanol (Zhou et al., 2016b). EtNa requires a cooperative action of two Ca2+ ions, which may explain its excellent specificity (Figure 6D). For the Ce13d DNAzyme, all trivalent lanthanide ions exhibit similar activity by neutralizing the negative charge on the scissile phosphate (Figure 6G) (Huang et al., 2014). Two more lanthanide-dependent DNAzymes were found to require multiple lanthanides (Figures 6E and 6F) (Huang et al., 2015, Huang et al., 2016). By introducing a phosphorothioate modification at the cleavage site, Cd2+ (Figure 6B) (Huang and Liu, 2015), and Cu2+ (Figure 6C) specific DNAzymes were selected (Huang and Liu, 2016). The sulfur modification is critical for recognition of these softer metal ions, which in turn indicated that the binding of the scissile phosphate group is their main catalytic role.
Figure 6.
Metal-Dependent RNA-Cleaving DNAzymes
The secondary structures and metal binding sites of the (A) 39E, (B) Cd16, (C) PSCu10, (D) EtNa, (E) Dy10a, (F) Tm7, (G) Ce13d, (H) NaA43, and (I) Ag10c DNAzymes and their corresponding target meta ions. Some DNAzymes require multiple metal ions.
Normally, a well-defined metal binding pocket (e.g., an aptamer motif) cannot be identified in such metal-specific DNAzymes. The metal ions perform their catalytic function by transiently associating with the scissile phosphate. Interestingly, a few new DNAzymes were reported to contain aptamer motifs. The NaA43 DNAzyme (Figure 6H) was selected in the presence of Na+, and it can reach a rate of 0.1 min−1 with 400 mM Na+ alone (Torabi et al., 2015). It shares sequence similarity with the Ce13d DNAzyme, and the common loop is the main part of a Na+ aptamer (Zhou et al., 2016c). This aptamer can also bind K+, but this binding leads to misfolding and loss of catalytic activity (He et al., 2018). This is an interesting example in which catalysis affords better selectivity than simple binding exemplified on the same molecule. The Ag10c DNAzyme is another example (Saran and Liu, 2016b), and its aptamer loop binds two Ag+ ions (Figure 6I). The catalytic role of interacting with the scissile phosphate is fulfilled by the salt in buffer (e.g., Na+, Li+, K+, Mg2+) (Saran et al., 2017).
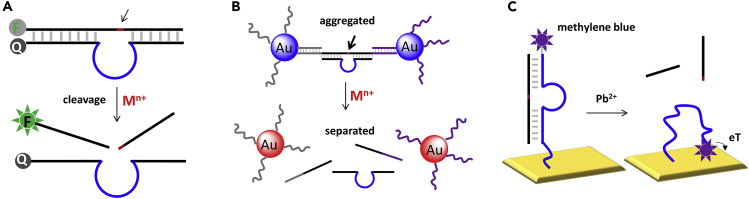
Since all these DNAzymes share a similar secondary structure, the same signaling strategies can in general be applied to all of them (Zhou et al., 2017b). For example, a fluorophore/quencher pair can be labeled on the two ends of the DNAzymes (Figure 7A), leading to an initially quenched state. After cleavage, the cleaved fragment bearing the fluorophore is released resulting in enhanced fluorescence. This fluorescence beacon design is commonly used for proof-of-concept sensing applications. An additional quencher can be attached to the other end of the substrate to further suppress the background signal (Torabi et al., 2015). Apart from fluorescence, cleavage-induced colorimetric signal can be achieved when utilizing DNA-functionalized gold nanoparticles (AuNPs). As illustrated in Figure 7B, a DNAzyme was used to assemble the AuNPs forming blue aggregates. Upon cleavage, the AuNPs were disassembled giving a blue-to-red color change (Liu and Lu, 2005). DNAzyme-based sensing can also be performed through electrochemical signals. For example, the methylene blue-labeled enzyme (8-17) can more efficiently transfer electrons to electrode surface after the Pb2+-induced cleavage (Xiao et al., 2007) (Figure 7C). Only a few representative examples are reviewed here, and more designs can be found in other reviews (Gong et al., 2015, Liu et al., 2009).
Figure 7.
Three Representative Signaling Strategies for RNA-cleaving DNAzymes
(A) Cleavage-induced fluorescent signal enhancement in a fluorescence beacon.
(B) The disassembly of AuNPs after the cleavage reaction.
(C) The conformational change of the 8-17 DNAzyme upon cleavage generates electrochemical signals for Pb2+ sensing.
Sensing of Non-metal Targets
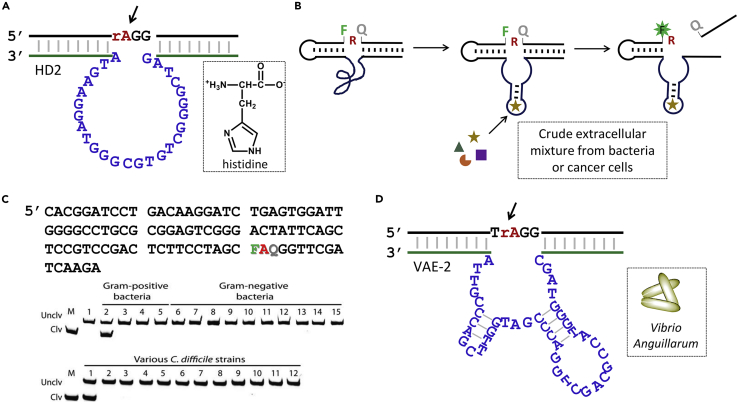
Aside from metal ions, RNA-cleaving DNAzymes have also been selected for other targets. An early example is the L-histidine DNAzyme selected by the Breaker group (Figure 8A) (Roth and Breaker, 1998). This sequence exhibits a stereospecific binding to histidine and relies on the general base catalyst of the imidazole group in the amino acid. Li and coworkers developed a new selection strategy by modifying a fluorophore and a quencher next to the cleavage site (Figure 8B) (Mei et al., 2003). An impressive aspect of it is to recognize a large number of bacterial and cancer cells. In these selections, the targets were the crude extracellular mixture, although the exact target molecule has yet to be identified in most cases. In one case, the RFD-CD1 shows very high specificity for a pathogenic strain of Clostridium difficile, and the target was identified to be a unique transcription factor (Figure 8C) (Shen et al., 2016). The fluorophore/quencher not only provides signal, but also likely directly participates in the molecular recognition/catalysis since the activity is often lost after removing them. Using the same method, Gu et al. recently selected an unmodified DNAzyme against an aquatic bacterium, Vibrio anguillarum (Figure 8D), and the target is also likely to be a protein (Gu et al., 2019). Most of these DNAzymes still require metal ions for catalysis, but they also need another molecule at the same time.
Figure 8.
Representative RNA-Cleaving DNAzymes Selected with Non-metal Targets
(A) The L-histidine DNAzyme.
(B) An in vitro selection strategy to identify DNAzymes using the crude extracellular mixture from cells. The two nucleotides near the cleavage site are labeled with a fluorophore and a quencher, respectively.
(C) The sequence of the RFD-CD1 DNAzyme and its cleavage activity with respect to various species of bacteria (top) and various strains of C. difficile (bottom). Reprinted with permission from Shen et al. (2016). Copyright © 2015 Wiley-VCH.
(D) The VAE-2 DNAzyme responsive to the crude extracellular mixture of Vibrio anguillarum.
G-Quadruplex Containing DNAzymes
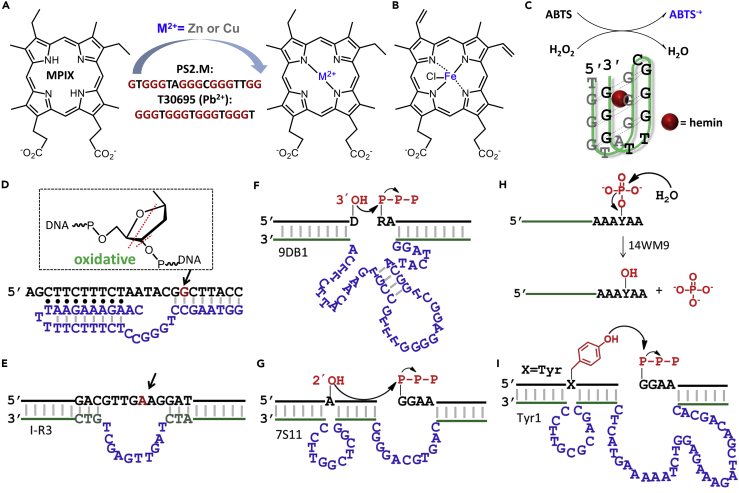
In 1996, Li and Sen reported another DNAzyme-catalyzed reaction called porphyrin metalation (Figure 9A) (Li and Sen, 1996). This reaction was also popular for developing catalytic antibodies and rich knowledge in terms of transition state analogs has been accumulated (Cochran and Schultz, 1990). In the same year, a ribozyme for the same reaction was reported by the Schultz group (Conn et al., 1996). The selected DNAzyme named PS5.ST1 has a G-quadruplex (G4) structure, yet it still showed low-micromolar binding affinity toward the anionic porphyrin mesoporphyrin IX (MPIX). This DNAzyme catalyzes the insertion of Cu2+ into MPIX with a catalytic efficiency of 79 min−1 M−1, ∼1400-fold higher than the uncatalyzed reaction.
Figure 9.
DNAzymes Catalyzing Other Reactions
(A) The two DNAzymes can accelerate the porphyrin metalation reaction (T30695 requires Pb2+).
(B) The structure of iron (III)-protoporphyrin IX (hemin).
(C) A G4 DNAzyme/hemin complex catalyzing the oxidation of ABTS in the presence of H2O2 into ABTS·+.
(D) Oxidative DNA cleavage catalyzed by DNA with the potential cleavage sites marked by dashed lines.
(E–G) (E) Hydrolytic DNA cleavage catalyzed by the I-R3 DNAzyme. RNA ligation reaction catalyzed by DNAzymes resulting in either (F) a linear 3′-5′ linkage or (G) a 2′,5′-branched linkage.
(H) Phosphatase activity of the 14WM9 DNAzyme.
(I) DNA-catalyzed nucleopeptide bond formation between a Tyr side chain and a 5′-triphosphorylated RNA.
Moreover, this G4 DNAzyme was found to exhibit peroxidase-mimicking activity upon binding to hemin (iron (III)-protoporphyrin IX, Figure 9B), and it can replace horseradish peroxidase (HRP) in many bioanalytical assays (Travascio et al., 1998). For example, the DNAzyme PS2.M-hemin complex can oxidize ABTS in the presence of H2O2 and produce colored ABTS·+ with ∼250-fold rate enhancement (Figure 9C). In fact, most G-rich sequences can fold into various G4 structures (e.g., parallel, antiparallel, or mixed), presenting different binding affinities to hemin and peroxidase-mimicking activities. The DNAzymes have good stability and can be conveniently linked to other DNA structures, gaining extensive bioanalytical applications. However, the catalytic activity of the DNAzymes is still relatively low compared with HRP. A recent effort was made to improve the catalytic rate of G4 DNAzyme by adding an adjacent 3′-adenine, which serves as a general acid-base catalyst to better mimic HRP (Xu et al., 2016).
The G4/hemin systems have been widely used for signal amplification (Kosman and Juskowiak, 2011). An example is to use rolling circle amplification (RCA) to generate multiple G4 sequences upon binding to a target DNA, allowing the detection of as low as picomolar DNA. The peroxidase-mimicking activity of G4 DNAzymes has also been applied to kill bacterial cells (Xing et al., 2018) and degrade organic pollutants (Kurapati and Bianco, 2018).
Recently, efforts have been made to select non-G4 metalation DNAzymes (Yang et al., 2019), and the effect of metal ions has been examined in more detail. A recent effort obtained a DNA aptamer with a high binding affinity toward MPIX but without a predictable G4 structure. The metalation rate of MPIX with Cu2+ enhanced by ∼3-fold in the presence of the aptamer. A guanine-rich sequence called T30695 (Figure 9A) was found to significantly accelerate the insertion of Cu2+ and Zn2+ into the MPIX in the presence of Pb2+ (Peng et al., 2019). With only 0.6 μM Pb2+, the rate constant improved by ∼4.3-fold.
New DNAzymes for New Reactions
Extensive efforts have also been made in searching for DNA sequences for catalyzing other reactions, and only a few representative examples are listed here. An early example is the oxidative DNA cleavage reaction by a DNAzyme with a triplex structures, requiring both Cu2+ and ascorbate (Figure 9D) (Carmi et al., 1998). Later, a Zn2+-dependent DNAzyme was selected, which catalyzes the DNA cleavage through hydrolysis with a rate constant of ∼1 min−1 (Figure 9E) (Gu et al., 2013).
A suite of RNA-ligating DNAzymes was reported by the Silverman group, achieving 3′-5′, 2′-5′, and lariat RNA products. When a 2′, 3′-cyclic phosphate and a 5′-hydroxyl group were used, the non-native 2′-5′ linkage was often favored in the DNA-catalyzed ligation (Flynn-Charlebois et al., 2003). In a selection using 2′, 3′-diol and 5′-triphosphate, the 9DB1 DNAzyme was obtained to produce the native 3′-5′ linkage with a kobs of ∼0.04 min−1 (Figure 9F) (Purtha et al., 2005). The high yield and sequence generality make it promising for practical applications. The crystal structure of the 9DB1 DNAzyme in the post-catalytic state was later solved by revealing a pseudoknot structure (Ponce-Salvatierra et al., 2016). A recent simulation effort further predicted the binding of two Mg2+ ions at the active site, which is commonly observed in proteins and ribozymes catalyzing this reaction. In another example, an internal 2′-hydroxyl group was ligated to a 5′-triphosphate by the 7S11 DNAzyme (Coppins and Silverman, 2004), leading to a 2′, 5′-branched RNA (Figure 9G). This DNA-catalyzed RNA ligation resembles the first step of RNA splicing in cells.
In biology, phosphorylation and dephosphorylation play important roles in many cellular processes, and they are mainly catalyzed by protein-based enzymes. The phosphatase activity was also explored in catalytic DNA. The 14WM9 DNAzyme catalyzes the Zn2+-dependent dephosphorylation of an internal phosphotyrosine (Figure 9H) with a single-turnover kobs of up to ∼0.65 min−1 (Chandrasekar and Silverman, 2013). Its phosphatase activity was demonstrated to be retained in the presence of human cell lysate and BSA. On the other hand, a self-phosphorylating DNAzyme was reported by Li and coworkers (Wang et al., 2002), requiring ATP/GTP as a phosphoryl donor and divalent metal ions (e.g., Cu2+, Mn2+) as cofactors.
Apart from all-nucleic-acid substrates, DNAzymes with activity toward peptide-containing substrates have been discovered (Silverman, 2015). For example, the Tyr1 DNAzyme (Figure 9I) catalyzes the formation of a nucleopeptide bond between a tyrosine (Tyr) side chain and a 5′-triphosphorylated RNA substrate with a kobs of 0.06 min−1 in 20 mM Mn2+ (Pradeepkumar et al., 2008). Covalent modifications on the side chain of amino acids can also be achieved by catalytic DNA. For instance, a sequence named 15MZ36 catalyzes the Tyr modification of a free tripeptide substrate with a kobs of 0.50 h−1 (Wong et al., 2011). Furthermore, the catalysis was extended to the cleavage of peptide bonds. Several DNAzymes were demonstrated to cleave the aromatic amide bond through hydrolysis (Brandsen et al., 2013).
Applications in Signal Amplification, Smart Materials Assembly, and Chemical Biology
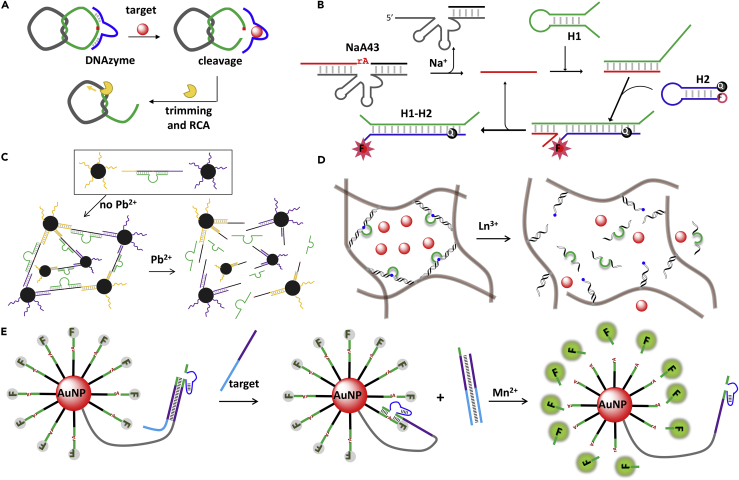
DNA is a highly programmable molecule, and many very sophisticated nanoscale structures have been designed using DNA. Compared with protein and RNA, DNA has advantages such as high stability, cost-effectiveness, and ease of modification. DNAzymes can be coupled with DNA amplification techniques for boosting sensitivity. Some common methods include protein enzyme-assisted (e.g., RCA and exonuclease III-based amplification) and enzyme-free mechanisms (e.g., hybridization chain reaction and catalytic hairpin assembly [CHA]) (Peng et al., 2018). For example, Li and coworkers coupled an E. coli-specific DNAzyme with RCA to achieve E. coli detection (Figure 10A) (Liu et al., 2016). The design of two interlocked single-stranded DNA rings prevents the template strand (in gray) from being used for RCA. The binding of the target cleaves the substrate, which then serves as a primer to initiate the RCA reaction. The RCA product was detected by a duplex-binding dye (i.e., 3, 3′-diethylthiadicarbocyanine), exhibiting a detection limit as low as 10 cells/mL. In another example, Wu et al. utilized the CHA reaction to enhance the sensitivity of the Na+-dependent DNAzyme (NaA43) (Wu et al., 2017). In the presence of Na+, the cleavage substrate fragment (red) initiates the CHA reaction by opening the hairpin DNA, H1 (Figure 10B). The opened H1 was designed to further unlock H2 and recover the fluorescence. Since the duplex formed between H1 and H2 is stronger, the cleavage substrate fragment is displaced and released. As a result, multiple CHA reactions can be induced with each cleavage product, resulting in an increased signal at low Na+ concentrations. With an excellent detection limit of 14 μM Na+, the CHA-amplified sensor was applied to image the endogenous Na+ in living cells.
Figure 10.
Biosensors Based on DNAzymes
(A) Coupling DNAzymes with RCA for sensitive detection of E. coli. Redrawn from Liu et al. (2016).
(B) A CHA-amplified DNAzyme sensor for imaging endogenous Na+. Redrawn from Wu et al. (2017).
(C) DNAzyme-conjugated magnetic nanoparticles for Pb2+ detection based on MRI signal. Redrawn from Xu et al. (2013).
(D) The target-responsive DNAzyme hydrogel for colorimetric detection of Ln3+. Redrawn from Huang et al. (2017).
(E) A microRNA-initiated DNAzyme motor constructed on AuNPs for intracellular imaging. Redrawn from Peng et al. (2017).
Nanoparticles have been functionalized with DNAzymes since they can quench fluorescence, absorb/conjugate DNA, and induce various signals (e.g., colorimetric, magnetic, surface-enhanced Raman scattering). The example in Figure 10C assembled the 8-17 DNAzyme and carboxyl-group modified magnetic beads to construct a magnetic resonance imaging (MRI) biosensor for Pb2+ (Xu et al., 2013). In the presence of Pb2+, the cleavage of the substrate increases the distance between the magnetic nanoparticles and decreases the relaxation time. Immobilization of DNAzymes in hydrogels is an interesting idea for improving metal sensing. For example, Huang et al. combined DNAzyme-assisted hydrogels and AuNPs for colorimetric detection of lanthanide ions (Figure 10D) (Huang et al., 2017). The polyacrylamide chains were grafted with short DNA strands that are complementary to two ends of the substrate strand. The addition of Ln3+ cleaves the substrate and breaks the hydrogel cross-links, releasing the AuNPs to the supernatant.
Intracellular sensing is an attractive application for functional nucleic acids (Hwang et al., 2019). However, like other nucleic acids, DNAzymes suffer from poor cellular uptake. To address this issue, nanomaterials (e.g., ZnO nanoparticles, AuNPs) were employed as transfection agents for DNAzyme-based intracellular biosensing (Wu et al., 2013). An interesting example is a DNAzyme motor constructed on AuNPs for microRNA detection (Figure 10E) (Peng et al., 2017). The AuNP was densely conjugated with FAM-labeled substrates along with a few DNAzyme strands that were silenced by the locking strands. The AuNP conjugate can be easily internalized by cells. In the presence of the target microRNA, the DNAzyme was activated and it can cleave multiple substrates on the AuNPs step by step allowing signal amplification. In addition, various strategies have been developed to avoid cleavage before the sensors entering cells such as caging DNAzymes (Hwang et al., 2014, Lin et al., 2019).
Biochemical Aspect to Understand the Reaction Mechanism
Biochemical insights into the function of catalytic nucleic acids is crucial for expanding our view of their mechanism. In this regard, the best studied examples are for RNA cleavage owing to their small size, high catalytic efficiency, and large quantity. The general mechanism of RNA cleavage is described in Figure 2C, and various factors can contribute to catalysis. The most direct insights are often from structural biology data such as NMR and X-ray crystallography. Many RNA-cleavage ribozymes have crystal structures, which contributed significantly to their fundamental understanding (Ward et al., 2014). In most cases, a guanine acts as a general base to help deprotonate the 2′-OH nucleophile. So far, only two DNAzymes have crystal structures (Liu et al., 2017a, Ponce-Salvatierra et al., 2016): one for RNA cleavage and the other for RNA ligation. These structures were obtained after more than 20 years of the report of the first DNAzyme. The challenges of crystallization suggest floppy and dynamic behavior of DNAzymes and coexistence of multiple structures, which also posed challenges for NMR. NMR has an additional barrier of isotope labeling for solving DNAzyme structures.
There was an interesting story in the structural study of the hammerhead ribozyme. The initial crystal structure was solved in 1994 on the minimal hammerhead ribozyme with only 13 nucleotides in the catalytic loop. However, this minimal structure failed to explain critical roles of invariant nucleotides (such as G-5, G-8, and G-12), since they appear far away from the active site (Figure 2B, top). In 2006, Martick et al. obtained crystal structures of a full-length hammerhead ribozyme composed of 63 nucleotides, revealing specific interactions between invariant nucleotides and the cleavage site. For example, the N1 position of G-12 is within hydrogen binding distance of the 2′-OH, serving as a general base during the catalysis (Figure 2B, bottom). Therefore, it is important to measure these enzymes as close to the native conditions as possible to get a more precise picture. Another aspect is the diluted in vitro assay conditions in contrast to the crowded intracellular environment. By adding crowding polymers, such as polyethylene glycol, the folding and activity of some ribozymes was promoted (Nakano and Sugimoto, 2017, Paudel et al., 2018). It was also reported that diphosphate can boost the activity of Mg2+ for both ribozymes and DNAzymes (Yamagami et al., 2019).
The only crystal structure of an RNA-cleaving DNAzyme (the 8-17) revealed a V-shaped folding. Upon binding to Pb2+, it contains two helical substrate-binding arms and a 15-nt pseudoknot catalytic core (Figure 11A) (Liu et al., 2017a). The 8-17 DNAzyme catalyzes the RNA cleavage via the general acid-base mechanism. At the active site, a Pb2+ ion activates a water molecule and stabilizes the 5′-leaving oxygen (general acid), whereas a conserved guanine serves as a general base to facilitate the deprotonation of 2′-OH group (Figure 11B). A recent dynamical simulation further revealed the supporting role of Na+ during the catalysis (Ekesan and York, 2019). Interestingly, the mechanism of 8-17 is similar to some RNA-cleaving ribozymes, especially the hammerhead ribozyme. Indeed, since RNA and DNA are chemically close, the abundant structural and biochemical observations of self-cleaving ribozymes help us to understand DNAzymes when a crystal structure is not available.
Figure 11.
Biochemical Characterization of DNAzymes and Ribozymes
(A) The secondary (left) and crystal structure (right) of the RNA-cleaving DNAzyme, 8-17, revealing a V-shaped overall folding (PDB: 5XM8).
(B) A proposed mechanism of 8-17 catalysis including a Pb2+-bound water molecule as a general acid and a conserved guanine (G13) as a general base. Reprinted with permission from Liu et al. (2017a). Copyright © 2017 Springer Nature.
(C) Phosphorothioate modification on the scissile phosphate.
(D) Na+-induced folding enhances the fluorescence of a 2AP-modification at the cleavage site of the Ce13d DNAzyme.
Owing to limited high-resolution structural information, so far, mechanistic studies of DNAzymes were achieved mainly by biochemical assays. Since metal binding is often important for DNAzyme catalysis, extensive efforts have been made in identifying metal-binding site in RNA-cleaving DNAzymes (Hwang et al., 2016, Ward et al., 2014, Zhou and Liu, 2018). Phosphorothioate modification on the cleavage site is a useful way to probe metal binding on the scissile phosphate (Figure 11C). In a typical case, a non-thiophilic metal ion such as Mg2+ directly interacts with one of the non-bridging oxygens. A PS modification significantly inhibits (often >100-fold) the cleavage activity compared with the original PO-substrate. The activity loss can normally be rescued by the addition of more thiophilic metal ions (e.g., Mn2+ or Cd2+). A PS modification makes the phosphorus a chiral center. Most metal ions bind to the pro-Rp oxygen in ribozymes and DNAzymes. This observation, however, has exceptions. For example, in the Dy10a DNAzyme (Figure 6E), a significant inhibition (∼1000-fold) was observed for both the Rp and Sp isomers, indicating that both the non-bridging oxygens are interacting with metal ions. This DNAzyme turned out to require two cooperative metals (Vazin et al., 2015).
When nucleobases are involved in metal binding, DNA footprinting can provide valuable information. For example, dimethyl sulfate (DMS) footprinting was used to probe guanines and showed the presence of a Na+ aptamer in the Ce13d DNAzyme (Zhou et al., 2016c) and an Ag+ aptamer in the Ag10c DNAzyme (Saran et al., 2017). Nevertheless, it needs to be noted that most DNAzymes and ribozymes do not have such well-defined metal-binding aptamers. Metals only need to transiently bind to the enzymes to exert their catalytic functions. Actually, for the above-mentioned Na+ and Ag+, they do not directly participate in catalysis. For Ce13d, a lanthanide is responsible for catalysis, whereas for Ag10c, a metal in buffer such as Na+ or Mg2+ is used. Biochemical insights can also be obtained from mutations including replacing a certain nucleotide with others or deletion. For example, in the NaA43 DNAzyme, mutating one to two nucleobases near the cleavage site resulted in a shifted pH-rate profile implying the general acid-base mechanism (Ma et al., 2019). Synthetic nucleobase analogs can be used to fine probe the interactions. Reselection can also be performed by partially randomizing enzymes to find out conserved and mutable regions. Such information is complementary to structural biology data and can often offer interesting insights too.
Spectroscopy is another type of characterization method. UV-vis, NMR (not for solving structures), EPR, and even XAFS are just a few examples to obtain metal binding information. The most popular method is fluorescence spectroscopy owing to its high sensitivity, rich molecular information, and simplicity in instrumentation. For example, the global or local folding of many RNA-cleaving DNAzymes have been explored by fluorescence techniques such as 2-aminopurine (2AP) modification (Zhou et al., 2016a), fluorescence resonance energy transfer (FRET) (Kim et al., 2007), and sensitized Tb3+ luminescence (Kim et al., 2008). An interesting example is the Na+-binding loop seen in the NaA43 and Ce13d DNAzymes. By introducing a 2AP-modification to the cleavage site, the Na+-induced folding enhances the fluorescence indicating a relaxed stacking environment near the 2AP base (Figure 11D) (Zhou et al., 2016a). This method also indicates that K+ can misfold and inactivate the DNAzyme, thus explaining the very high selectivity for Na+ (He et al., 2018).
Conclusions and Future Perspectives
In this review, we described catalytic nucleic acids from classic examples to recent developments. Nucleic acid catalysts have now been used in so many areas of research from inside cells to between nanomaterials, from bioanalytical chemistry to environmental monitoring. At the same time, fundamental studies have also provided better understanding of catalytic mechanism and structure. There are still many questions to be addressed, providing future research opportunities in this field.
In vitro selection is a main source of novel catalytic nucleic acids. New selection methods could bring in new properties, such as the use of circular DNA libraries for selecting DNA-cleaving DNAzymes (Gu et al., 2013). Recently, such library was also used for aptamer selection (Liu et al., 2019). The use of modified nucleotides has been tested since over 20 years ago. Although many interesting DNAzymes have been obtained, the modified dNTP complicates PCR. In addition, the selected enzymes often contain multiple such modifications, and such enzymes are usually not commercially available. We proposed to introduce a single modification at a critical site (Huang and Liu, 2015). For the RNA cleavage reaction, the scissile phosphate site can be modified for this purpose, whereas the rest of the nucleotides are natural. This has been successful for fluorophore- and quencher-based ligands as well (Shen et al., 2016). Modified nucleotides can also harness new functions such as sensitivity to external stimuli (Hwang et al., 2016, Xiao et al., 2019). For ribozymes, efforts will continue on the searching for more natural ribozymes. At the same time, bioinformatics has been widely applied yielding new ribozymes (e.g., the twister ribozyme) in recent years (Weinberg et al., 2015, Weinberg et al., 2019).
On the application side, going from proof-of-concept experiments to impactful real-world examples is needed for the field. Ribozymes will be developed for intracellular applications and to explore the limit of RNA catalysis. Compared with DNAzymes, the biggest advantage of ribozymes is that they do not need to be delivered but can be transcribed in cells via a plasmid. Some early ideas of using nucleic acid enzymes for inhibition of mRNA have not been proven to be clinically useful. With the development of siRNA, and some recent approval of anti-sense oligonucleotides for inhibition of gene expression, the use of ribozymes and DNAzymes for this purpose seems to be lagging. Intracellular RNA cleavage would require some highly efficient DNAzymes that can work inside cells. Some recent developments were seen to combine DNAzymes with nanotechnology, where a nanomaterial, such as MnO2 (Fan et al., 2015) and ZnO (Wang et al., 2019), is used to deliver DNAzymes and at the same time they can provide a source of metal ions for catalysis. More rigorous tests on these DNAzymes, especially under in vivo conditions, would be needed to fully validate this approach. Some RNA-cleaving ribozymes are often used by virus for processing viral RNA. Therefore, using ribozymes as a chemical biology tool for probing biological systems might be more productive. For DNAzymes, we expect to see more examples of application in metal detection, catalyzing new chemical and biochemical transformations, and controlling nanoscale devices.
On the fundamental biochemical aspect, since metal ions are indispensable for the catalysis of nucleic acids, more structural biology data are needed to capture the role of metal ions in catalysis. Techniques such as NMR and X-ray crystallography can provide valuable information in this aspect. Finally, the interest on the origin of life will continue to be an important part of the field.
Acknowledgments
The work from the Liu lab described in this review was mainly funded by the Natural Sciences and Engineering Research Council of Canada (NSERC).
Author Contributions
J.L. outlined the manuscript and supervised the writing. L.M. wrote the manuscript and prepared the figures.
References
- Balke D., Wichert C., Appel B., Müller S. Generation and selection of ribozyme variants with potential application in protein engineering and synthetic biology. Appl. Microbiol. Biotechnol. 2014;98:3389–3399. doi: 10.1007/s00253-014-5528-7. [DOI] [PubMed] [Google Scholar]
- Baskerville S., Bartel D.P. A ribozyme that ligates RNA to protein. Proc. Natl. Acad. Sci. U S A. 2002;99:9154–9159. doi: 10.1073/pnas.142153799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandsen B.M., Hesser A.R., Castner M.A., Chandra M., Silverman S.K. DNA-catalyzed hydrolysis of esters and aromatic amides. J. Am. Chem. Soc. 2013;135:16014–16017. doi: 10.1021/ja4077233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breaker R.R., Joyce G.F. A DNA enzyme that cleaves RNA. Chem. Biol. 1994;1:223–229. doi: 10.1016/1074-5521(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Carmi N., Balkhi H.R., Breaker R.R. Cleaving DNA with DNA. Proc. Natl. Acad. Sci. U S A. 1998;95:2233–2237. doi: 10.1073/pnas.95.5.2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrasekar J., Silverman S.K. Catalytic DNA with phosphatase activity. Proc. Natl. Acad. Sci. U S A. 2013;110:5315–5320. doi: 10.1073/pnas.1221946110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran A.G., Schultz P.G. Antibody-catalyzed porphyrin metalation. Science. 1990;249:781–783. doi: 10.1126/science.2389144. [DOI] [PubMed] [Google Scholar]
- Cochrane J.C., Strobel S.A. Catalytic strategies of self-cleaving ribozymes. Acc. Chem. Res. 2008;41:1027–1035. doi: 10.1021/ar800050c. [DOI] [PubMed] [Google Scholar]
- Conn M.M., Prudent J.R., Schultz P.G. Porphyrin metalation catalyzed by a small RNA molecule. J. Am. Chem. Soc. 1996;118:7012–7013. [Google Scholar]
- Coppins R.L., Silverman S.K. A DNA enzyme that mimics the first step of RNA splicing. Nat. Struct. Mol. Biol. 2004;11:270–274. doi: 10.1038/nsmb727. [DOI] [PubMed] [Google Scholar]
- Doudna J.A., Cech T.R. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- Ekesan Ş., York D.M. Dynamical ensemble of the active state and transition state mimic for the RNA-cleaving 8–17 DNAzyme in solution. Nucleic Acids Res. 2019;47:10282–10295. doi: 10.1093/nar/gkz773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Famulok M., Hartig J.S., Mayer G. Functional aptamers and aptazymes in biotechnology, diagnostics, and therapy. Chem. Rev. 2007;107:3715–3743. doi: 10.1021/cr0306743. [DOI] [PubMed] [Google Scholar]
- Fan H., Zhao Z., Yan G., Zhang X., Yang C., Meng H., Chen Z., Liu H., Tan W. A smart DNAzyme–MnO2 nanosystem for efficient gene silencing. Angew. Chem. Int. Ed. 2015;54:4801–4805. doi: 10.1002/anie.201411417. [DOI] [PubMed] [Google Scholar]
- Ferré-D'Amaré A.R., Zhou K., Doudna J.A. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- Flynn-Charlebois A., Wang Y., Prior T.K., Rashid I., Hoadley K.A., Coppins R.L., Wolf A.C., Silverman S.K. Deoxyribozymes with 2'-5' RNA ligase activity. J. Am. Chem. Soc. 2003;125:2444–2454. doi: 10.1021/ja028774y. [DOI] [PubMed] [Google Scholar]
- Gesteland R.F., Cech T.R., Atkins J.F. Second Edition. Cold Spring Harbor Laboratory Press; 1999. The RNA World. [Google Scholar]
- Geyer C.R., Sen D. Evidence for the metal-cofactor independence of an RNA phosphodiester-cleaving DNA enzyme. Chem. Biol. 1997;4:579–593. doi: 10.1016/s1074-5521(97)90244-1. [DOI] [PubMed] [Google Scholar]
- Gong L., Zhao Z., Lv Y.-F., Huan S.-Y., Fu T., Zhang X.-B., Shen G.-L., Yu R.-Q. DNAzyme-based biosensors and nanodevices. Chem. Commun. (Camb.) 2015;51:979–995. doi: 10.1039/c4cc06855f. [DOI] [PubMed] [Google Scholar]
- Gu H., Furukawa K., Weinberg Z., Berenson D.F., Breaker R.R. Small, highly active DNAs that hydrolyze DNA. J. Am. Chem. Soc. 2013;135:9121–9129. doi: 10.1021/ja403585e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L., Yan W., Wu H., Fan S., Ren W., Wang S., Lyu M., Liu J. Selection of DNAzymes for sensing aquatic bacteria: Vibrio anguillarum. Anal. Chem. 2019;91:7887–7893. doi: 10.1021/acs.analchem.9b01707. [DOI] [PubMed] [Google Scholar]
- Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- Hammann C., Westhof E. Searching genomes for ribozymes and riboswitches. Genome Biol. 2007;8:210. doi: 10.1186/gb-2007-8-4-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Chen D., Huang P.-J.J., Zhou Y., Ma L., Xu K., Yang R., Liu J. Misfolding of a DNAzyme for ultrahigh sodium selectivity over potassium. Nucleic Acids Res. 2018;46:10262–10271. doi: 10.1093/nar/gky807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-J.J., Lin J., Cao J., Vazin M., Liu J. Ultrasensitive DNAzyme beacon for lanthanides and metal speciation. Anal. Chem. 2014;86:1816–1821. doi: 10.1021/ac403762s. [DOI] [PubMed] [Google Scholar]
- Huang P.-J.J., Liu J. Rational evolution of Cd2+-specific DNAzymes with phosphorothioate modified cleavage junction and Cd2+ sensing. Nucleic Acids Res. 2015;43:6125–6133. doi: 10.1093/nar/gkv519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P.-J.J., Liu J. An ultrasensitive light-up Cu2+ biosensor using a new DNAzyme cleaving a phosphorothioate-modified substrate. Anal. Chem. 2016;88:3341–3347. doi: 10.1021/acs.analchem.5b04904. [DOI] [PubMed] [Google Scholar]
- Huang P.-J.J., Vazin M., Liu J. In vitro selection of a DNAzyme cooperatively binding two lanthanide ions for RNA cleavage. Biochemistry. 2016;55:2518–2525. doi: 10.1021/acs.biochem.6b00132. [DOI] [PubMed] [Google Scholar]
- Huang P.-J.J., Vazin M., Matuszek Ż., Liu J. A new heavy lanthanide-dependent DNAzyme displaying strong metal cooperativity and unrescuable phosphorothioate effect. Nucleic Acids Res. 2015;43:461–469. doi: 10.1093/nar/gku1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Zhao Y., Pu Q., Liu G., Peng Y., Wang F., Chen G., Sun M., Du F., Dong J. Intracellular selection of trans-cleaving hammerhead ribozymes. Nucleic Acids Res. 2019;47:2514–2522. doi: 10.1093/nar/gkz018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y.S., Wu X.M., Tian T., Zhu Z., Lin H., Yang C.Y. Target-responsive DNAzyme hydrogel for portable colorimetric detection of lanthanide(III) ions. Sci. China Chem. 2017;60:293–298. [Google Scholar]
- Huanhuan F., Xiaobing Z., Yi L. Recent advances in DNAzyme-based gene silencing. Sci. China Chem. 2017;60:591–601. [Google Scholar]
- Hwang K., Hosseinzadeh P., Lu Y. Biochemical and biophysical understanding of metal ion selectivity of DNAzymes. Inorg. Chim. Acta. 2016;452:12–24. doi: 10.1016/j.ica.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K., Mou Q., Lake R.J., Xiong M., Holland B., Lu Y. Metal-dependent DNAzymes for the quantitative detection of metal ions in living cells: recent progress, current challenges, and latest results on FRET ratiometric sensors. Inorg. Chem. 2019;58:13696–13708. doi: 10.1021/acs.inorgchem.9b01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang K., Wu P., Kim T., Lei L., Tian S., Wang Y., Lu Y. Photocaged DNAzymes as a general method for sensing metal ions in living cells. Angew. Chem. Int. Ed. 2014;53:13798–13802. doi: 10.1002/anie.201408333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez R.M., Polanco J.A., Lupták A. Chemistry and biology of self-cleaving ribozymes. Trends Biochem. Sci. 2015;40:648–661. doi: 10.1016/j.tibs.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce G.F. The antiquity of RNA-based evolution. Nature. 2002;418:214–221. doi: 10.1038/418214a. [DOI] [PubMed] [Google Scholar]
- Kim H.-K., Li J., Nagraj N., Lu Y. Probing metal binding in the 8–17 DNAzyme by Tb(III) luminescence spectroscopy. Chem. Eur. J. 2008;14:8696–8703. doi: 10.1002/chem.200701789. [DOI] [PubMed] [Google Scholar]
- Kim H.K., Rasnik I., Liu J.W., Ha T.J., Lu Y. Dissecting metal ion-dependent folding and catalysis of a single DNAzyme. Nat. Chem. Biol. 2007;3:762–768. doi: 10.1038/nchembio.2007.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosman J., Juskowiak B. Peroxidase-mimicking DNAzymes for biosensing applications: a review. Anal. Chim. Acta. 2011;707:7–17. doi: 10.1016/j.aca.2011.08.050. [DOI] [PubMed] [Google Scholar]
- Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kurapati R., Bianco A. Peroxidase mimicking DNAzymes degrade graphene oxide. Nanoscale. 2018;10:19316–19321. doi: 10.1039/c8nr06535g. [DOI] [PubMed] [Google Scholar]
- Lan T., Furuya K., Lu Y. A highly selective lead sensor based on a classic lead DNAzyme. Chem. Commun. (Camb.) 2010;46:3896–3898. doi: 10.1039/b926910j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Lu Y. A highly sensitive and selective catalytic DNA biosensor for lead ions. J. Am. Chem. Soc. 2000;122:10466–10467. [Google Scholar]
- Li J., Zheng W., Kwon A.H., Lu Y. In vitro selection and characterization of a highly efficient Zn(II)-dependent RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2000;28:481–488. doi: 10.1093/nar/28.2.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Sen D. A catalytic DNA for porphyrin metalation. Nat. Struct. Biol. 1996;3:743–747. doi: 10.1038/nsb0996-743. [DOI] [PubMed] [Google Scholar]
- Lin Y., Yang Z., Lake R.J., Zheng C., Lu Y. Enzyme-mediated endogenous and bioorthogonal control of a DNAzyme fluorescent sensor for imaging metal ions in living cells. Angew. Chem. Int. Ed. 2019;58:17061–17067. doi: 10.1002/anie.201910343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln T.A., Joyce G.F. Self-sustained replication of an RNA enzyme. Science. 2009;323:1229–1232. doi: 10.1126/science.1167856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Brown A.K., Meng X., Cropek D.M., Istok J.D., Watson D.B., Lu Y. A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity. Proc. Natl. Acad. Sci. U S A. 2007;104:2056–2061. doi: 10.1073/pnas.0607875104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Cao Z., Lu Y. Functional nucleic acid sensors. Chem. Rev. 2009;109:1948–1998. doi: 10.1021/cr030183i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H.H., Yu X., Chen Y.Q., Zhang J., Wu B.X., Zheng L.N., Haruehanroengra P., Wang R., Li S.H., Lin J.Z. Crystal structure of an RNA-cleaving DNAzyme. Nat. Commun. 2017;8:2006. doi: 10.1038/s41467-017-02203-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Chang D., Li Y. Discovery and biosensing applications of diverse RNA-cleaving DNAzymes. Acc. Chem. Res. 2017;50:2273–2283. doi: 10.1021/acs.accounts.7b00262. [DOI] [PubMed] [Google Scholar]
- Liu J., Lu Y. Stimuli-responsive disassembly of nanoparticle aggregates for light-up colorimetric sensing. J. Am. Chem. Soc. 2005;127:12677–12683. doi: 10.1021/ja053567u. [DOI] [PubMed] [Google Scholar]
- Liu M., Yin Q., Chang Y., Zhang Q., Brennan J.D., Li Y. In vitro selection of circular DNA aptamers for biosensing applications. Angew. Chem. Int. Ed. 2019;58:8013–8017. doi: 10.1002/anie.201901192. [DOI] [PubMed] [Google Scholar]
- Liu M., Zhang Q., Li Z., Gu J., Brennan J.D., Li Y. Programming a topologically constrained DNA nanostructure into a sensor. Nat. Commun. 2016;7:12074. doi: 10.1038/ncomms12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Kartik S., Liu B., Liu J. From general base to general acid catalysis in a sodium-specific DNAzyme by a guanine-to-adenine mutation. Nucleic Acids Res. 2019;47:8154–8162. doi: 10.1093/nar/gkz578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Liu J. An in vitro selected DNAzyme mutant highly specific for Na+ in slightly acidic conditions. ChemBioChem. 2019;20:537–542. doi: 10.1002/cbic.201800322. [DOI] [PubMed] [Google Scholar]
- Martick M., Scott W.G. Tertiary contacts distant from the active site prime a ribozyme for catalysis. Cell. 2006;126:309–320. doi: 10.1016/j.cell.2006.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S.H.J., Liu Z., Brennan J.D., Li Y. An efficient RNA-cleaving DNA enzyme that synchronizes catalysis with fluorescence signaling. J. Am. Chem. Soc. 2003;125:412–420. doi: 10.1021/ja0281232. [DOI] [PubMed] [Google Scholar]
- Müller S., Appel B., Balke D., Hieronymus R., Nübel C. Thirty-five years of research into ribozymes and nucleic acid catalysis: where do we stand today? F1000Res. 2016;5:1511. doi: 10.12688/f1000research.8601.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano S.-i., Sugimoto N. Model studies of the effects of intracellular crowding on nucleic acid interactions. Mol. Biosyst. 2017;13:32–41. doi: 10.1039/c6mb00654j. [DOI] [PubMed] [Google Scholar]
- Pan T., Uhlenbeck O.C. In vitro selection of RNAs that undergo autolytic cleavage with Pb(II) Biochemistry. 1992;31:3887–3895. doi: 10.1021/bi00131a001. [DOI] [PubMed] [Google Scholar]
- Paudel B.P., Fiorini E., Börner R., Sigel R.K.O., Rueda D.S. Optimal molecular crowding accelerates group II intron folding and maximizes catalysis. Proc. Natl. Acad. Sci. U S A. 2018;115:11917–11922. doi: 10.1073/pnas.1806685115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavco P.A., Bouhana K.S., Gallegos A.M., Agrawal A., Blanchard K.S., Grimm S.L., Jensen K.L., Andrews L.E., Wincott F.E., Pitot P.A. Antitumor and antimetastatic activity of ribozymes targeting the messenger RNA of vascular endothelial growth factor receptors. Clin. Cancer Res. 2000;6:2094–2103. [PubMed] [Google Scholar]
- Penchovsky R., Breaker R.R. Computational design and experimental validation of oligonucleotide-sensing allosteric ribozymes. Nat. Biotechnol. 2005;23:1424–1433. doi: 10.1038/nbt1155. [DOI] [PubMed] [Google Scholar]
- Peng D., Li Y., Huang Z., Liang R.-P., Qiu J.-D., Liu J. Efficient DNA-catalyzed porphyrin metalation for fluorescent ratiometric Pb2+ detection. Anal. Chem. 2019;91:11403–11408. doi: 10.1021/acs.analchem.9b02759. [DOI] [PubMed] [Google Scholar]
- Peng H., Li X.-F., Zhang H., Le X.C. A microRNA-initiated DNAzyme motor operating in living cells. Nat. Commun. 2017;8:14378. doi: 10.1038/ncomms14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng H., Newbigging A.M., Wang Z., Tao J., Deng W., Le X.C., Zhang H. DNAzyme-mediated assays for amplified detection of nucleic acids and proteins. Anal. Chem. 2018;90:190–207. doi: 10.1021/acs.analchem.7b04926. [DOI] [PubMed] [Google Scholar]
- Perreault J., Weinberg Z., Roth A., Popescu O., Chartrand P., Ferbeyre G., Breaker R.R. Identification of hammerhead ribozymes in all domains of life reveals novel structural variations. PLoS Comput. Biol. 2011;7:e1002031. doi: 10.1371/journal.pcbi.1002031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponce-Salvatierra A., Wawrzyniak-Turek K., Steuerwald U., Höbartner C., Pena V. Crystal structure of a DNA catalyst. Nature. 2016;529:231–234. doi: 10.1038/nature16471. [DOI] [PubMed] [Google Scholar]
- Pradeepkumar P.I., Höbartner C., Baum D.A., Silverman S.K. DNA-catalyzed formation of nucleopeptide linkages. Angew. Chem. Int. Ed. 2008;47:1753–1757. doi: 10.1002/anie.200703676. [DOI] [PubMed] [Google Scholar]
- Purtha W.E., Coppins R.L., Smalley M.K., Silverman S.K. General deoxyribozyme-catalyzed synthesis of native 3'-5' RNA linkages. J. Am. Chem. Soc. 2005;127:13124. doi: 10.1021/ja0533702. [DOI] [PubMed] [Google Scholar]
- Ren A., Micura R., Patel D.J. Structure-based mechanistic insights into catalysis by small self-cleaving ribozymes. Curr. Opin. Chem. Biol. 2017;41:71–83. doi: 10.1016/j.cbpa.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson M.P., Joyce G.F. Highly efficient self-replicating RNA enzymes. Chem. Biol. 2014;21:238–245. doi: 10.1016/j.chembiol.2013.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J.J. Ribozyme therapy for HIV infection. Adv. Drug Deliv. Rev. 2000;44:71–78. doi: 10.1016/s0169-409x(00)00085-5. [DOI] [PubMed] [Google Scholar]
- Roth A., Breaker R.R. An amino acid as a cofactor for a catalytic polynucleotide. Proc. Natl. Acad. Sci. U S A. 1998;95:6027–6031. doi: 10.1073/pnas.95.11.6027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A., Weinberg Z., Chen A.G.Y., Kim P.B., Ames T.D., Breaker R.R. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 2013;10:56. doi: 10.1038/nchembio.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro S.W., Joyce G.F. A general purpose RNA-cleaving DNA enzyme. Proc. Natl. Acad. Sci. U S A. 1997;94:4262–4266. doi: 10.1073/pnas.94.9.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saran R., Kleinke K., Zhou W., Yu T., Liu J. A silver-specific DNAzyme with a new silver aptamer and salt-promoted activity. Biochemistry. 2017;56:1955–1962. doi: 10.1021/acs.biochem.6b01131. [DOI] [PubMed] [Google Scholar]
- Saran R., Liu J. A comparison of two classic Pb2+-dependent RNA-cleaving DNAzymes. Inorg. Chem. Front. 2016;3:494–501. [Google Scholar]
- Saran R., Liu J. A silver DNAzyme. Anal. Chem. 2016;88:4014–4020. doi: 10.1021/acs.analchem.6b00327. [DOI] [PubMed] [Google Scholar]
- Schlosser K., Gu J., Sule L., Li Y.F. Sequence-function relationships provide new insight into the cleavage site selectivity of the 8-17 RNA-cleaving deoxyribozyme. Nucleic Acids Res. 2008;36:1472–1481. doi: 10.1093/nar/gkm1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A.K., Plant J.J., Rangel A.E., Meek K.N., Anamisis A.J., Hollien J., Heemstra J.M. Fluorescent RNA labeling using self-alkylating ribozymes. ACS Chem. Biol. 2014;9:1680–1684. doi: 10.1021/cb5002119. [DOI] [PubMed] [Google Scholar]
- Shen Z., Wu Z., Chang D., Zhang W., Tram K., Lee C., Kim P., Salena B.J., Li Y. A catalytic DNA activated by a specific strain of bacterial pathogen. Angew. Chem. Int. Ed. 2016;55:2431–2434. doi: 10.1002/anie.201510125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S.K. Pursuing DNA catalysts for protein modification. Acc. Chem. Res. 2015;48:1369–1379. doi: 10.1021/acs.accounts.5b00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S.K. Catalytic DNA: scope, applications, and biochemistry of deoxyribozymes. Trends Biochem. Sci. 2016;41:595–609. doi: 10.1016/j.tibs.2016.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullenger B.A., Gilboa E. Emerging clinical applications of RNA. Nature. 2002;418:252–258. doi: 10.1038/418252a. [DOI] [PubMed] [Google Scholar]
- Sun L.Q., Cairns M.J., Saravolac E.G., Baker A., Gerlach W.L. Catalytic nucleic acids: from lab to applications. Pharmacol. Rev. 2000;52:325–347. [PubMed] [Google Scholar]
- Tang W., Hu J.H., Liu D.R. Aptazyme-embedded guide RNAs enable ligand-responsive genome editing and transcriptional activation. Nat. Commun. 2017;8:15939. doi: 10.1038/ncomms15939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torabi S.-F., Wu P., McGhee C.E., Chen L., Hwang K., Zheng N., Cheng J., Lu Y. In vitro selection of a sodium-specific DNAzyme and its application in intracellular sensing. Proc. Natl. Acad. Sci. U S A. 2015;112:5903–5908. doi: 10.1073/pnas.1420361112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travascio P., Li Y., Sen D. DNA-enhanced peroxidase activity of a DNA aptamer-hemin complex. Chem. Biol. 1998;5:505–517. doi: 10.1016/s1074-5521(98)90006-0. [DOI] [PubMed] [Google Scholar]
- Vazin M., Huang P.-J.J., Matuszek Ż., Liu J. Biochemical characterization of a lanthanide-dependent DNAzyme with normal and phosphorothioate-modified substrates. Biochemistry. 2015;54:6132–6138. doi: 10.1021/acs.biochem.5b00691. [DOI] [PubMed] [Google Scholar]
- Wang J., Wang H., Wang H., He S., Li R., Deng Z., Liu X., Wang F. Nonviolent self-catabolic DNAzyme nanosponges for smart anticancer drug delivery. ACS Nano. 2019;13:5852–5863. doi: 10.1021/acsnano.9b01589. [DOI] [PubMed] [Google Scholar]
- Wang W., Billen L.P., Li Y. Sequence diversity, metal specificity, and catalytic proficiency of metal-dependent phosphorylating DNA enzymes. Chem. Biol. 2002;9:507–517. doi: 10.1016/s1074-5521(02)00127-8. [DOI] [PubMed] [Google Scholar]
- Wang Y.J., Liu E.K., Lam C.H., Perrin D.M. A densely modified M2+-independent DNAzyme that cleaves RNA efficiently with multiple catalytic turnover. Chem. Sci. 2018;9:1813–1821. doi: 10.1039/c7sc04491g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward W.L., Plakos K., DeRose V.J. Nucleic acid catalysis: metals, nucleobases, and other cofactors. Chem. Rev. 2014;114:4318–4342. doi: 10.1021/cr400476k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg C.E., Weinberg Z., Hammann C. Novel ribozymes: discovery, catalytic mechanisms, and the quest to understand biological function. Nucleic Acids Res. 2019;47:9480–9494. doi: 10.1093/nar/gkz737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z., Kim P.B., Chen T.H., Li S., Harris K.A., Lünse C.E., Breaker R.R. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat. Chem. Biol. 2015;11:606. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieland M., Hartig J.S. Improved aptazyme design and in vivo screening enable riboswitching in bacteria. Angew. Chem. Int. Ed. 2008;47:2604–2607. doi: 10.1002/anie.200703700. [DOI] [PubMed] [Google Scholar]
- Wilson D.S., Szostak J.W. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- Winkler W.C., Nahvi A., Roth A., Collins J.A., Breaker R.R. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- Wong O.Y., Pradeepkumar P.I., Silverman S.K. DNA-catalyzed covalent modification of amino acid side chains in tethered and free peptide substrates. Biochemistry. 2011;50:4741–4749. doi: 10.1021/bi200585n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P., Hwang K., Lan T., Lu Y. A DNAzyme-gold nanoparticle probe for uranyl ion in living cells. J. Am. Chem. Soc. 2013;135:5254–5257. doi: 10.1021/ja400150v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.K., Fan H.H., Satyavolu N.S.R., Wang W.J., Lake R., Jiang J.H., Lu Y. Imaging endogenous metal ions in living cells using a DNAzyme-catalytic hairpin assembly probe. Angew. Chem. Int. Ed. 2017;56:8721–8725. doi: 10.1002/anie.201703540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Gu C.M., Xiang Y. Orthogonal activation of RNA-cleaving DNAzymes in live cells by reactive oxygen species. Angew. Chem. Int. Ed. 2019;58:14167–14172. doi: 10.1002/anie.201908105. [DOI] [PubMed] [Google Scholar]
- Xiao Y., Rowe A.A., Plaxco K.W. Electrochemical detection of parts-per-billion lead via an electrode-bound DNAzyme assembly. J. Am. Chem. Soc. 2007;129:262. doi: 10.1021/ja067278x. [DOI] [PubMed] [Google Scholar]
- Xing Y., Liu X., Pu Q., Wu M., Zhao J.X. Biocompatible G-quadruplex/hemin for enhancing antibacterial activity of H2O2. ACS Appl. Bio. Mater. 2018;1:1019–1027. doi: 10.1021/acsabm.8b00211. [DOI] [PubMed] [Google Scholar]
- Xu F., Yi H., Yao S., Li W., Li Y., Liu Z., Nie Z., Lin B. Insight into G-quadruplex-hemin DNAzyme/RNAzyme: adjacent adenine as the intramolecular species for remarkable enhancement of enzymatic activity. Nucleic Acids Res. 2016;44:7373–7384. doi: 10.1093/nar/gkw634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Yin H., Ma W., Wang L., Kuang H., Xu C. MRI biosensor for lead detection based on the DNAzyme-induced catalytic reaction. J. Phys. Chem. B. 2013;117:14367–14371. doi: 10.1021/jp4087656. [DOI] [PubMed] [Google Scholar]
- Yamagami R., Huang R., Bevilacqua P.C. Cellular concentrations of nucleotide diphosphate-chelated magnesium ions accelerate catalysis by RNA and DNA enzymes. Biochemistry. 2019;58:3971–3979. doi: 10.1021/acs.biochem.9b00578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L., Ding P., Luo Y., Wang J., Lv H., Li W., Cao Y., Pei R. Exploration of catalytic nucleic acids on porphyrin metalation and peroxidase activity by in vitro selection of aptamers for N-methyl mesoporphyrin IX. ACS Comb. Sci. 2019;21:83–89. doi: 10.1021/acscombsci.8b00129. [DOI] [PubMed] [Google Scholar]
- Young D.D., Lively M.O., Deiters A. Activation and deactivation of DNAzyme and antisense function with light for the photochemical regulation of gene expression in mammalian cells. J. Am. Chem. Soc. 2010;132:6183–6193. doi: 10.1021/ja100710j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.-B., Kong R.-M., Lu Y. Metal ion sensors based on DNAzymes and related DNA molecules. Annu. Rev. Anal. Chem. 2011;4:105–128. doi: 10.1146/annurev.anchem.111808.073617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Ding J., Liu J. A highly specific sodium aptamer probed by 2-aminopurine for robust Na+ sensing. Nucleic Acids Res. 2016;44:10377–10385. doi: 10.1093/nar/gkw845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Saran R., Chen Q., Ding J., Liu J. A new Na+-dependent RNA-cleaving DNAzyme with over 1000-fold rate acceleration by ethanol. ChemBioChem. 2016;17:159–163. doi: 10.1002/cbic.201500603. [DOI] [PubMed] [Google Scholar]
- Zhou W., Zhang Y., Huang P.-J.J., Ding J., Liu J. A DNAzyme requiring two different metal ions at two distinct sites. Nucleic Acids Res. 2016;44:354–363. doi: 10.1093/nar/gkv1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W., Liu J. Multi-metal-dependent nucleic acid enzymes. Metallomics. 2018;10:30–48. doi: 10.1039/c7mt00268h. [DOI] [PubMed] [Google Scholar]
- Zhou W., Saran R., Huang P.-J.J., Ding J., Liu J. An exceptionally selective DNA cooperatively binding two Ca2+ ions. ChemBioChem. 2017;18:518–522. doi: 10.1002/cbic.201600708. [DOI] [PubMed] [Google Scholar]
- Zhou W., Saran R., Liu J. Metal sensing by DNA. Chem. Rev. 2017;117:8272–8325. doi: 10.1021/acs.chemrev.7b00063. [DOI] [PubMed] [Google Scholar]
- Zhou W.H., Ding J.S., Liu J.W. Theranostic DNAzymes. Theranostics. 2017;7:1010–1025. doi: 10.7150/thno.17736. [DOI] [PMC free article] [PubMed] [Google Scholar]