Abstract
Ontologically distinct populations of macrophages differentially contribute to organ fibrosis through unknown mechanisms.
We applied lineage tracing, single-cell RNA sequencing and single-molecule fluorescence in situ hybridisation to a spatially restricted model of asbestos-induced pulmonary fibrosis.
We demonstrate that tissue-resident alveolar macrophages, tissue-resident peribronchial and perivascular interstitial macrophages, and monocyte-derived alveolar macrophages are present in the fibrotic niche. Deletion of monocyte-derived alveolar macrophages but not tissue-resident alveolar macrophages ameliorated asbestos-induced lung fibrosis. Monocyte-derived alveolar macrophages were specifically localised to fibrotic regions in the proximity of fibroblasts where they expressed molecules known to drive fibroblast proliferation, including platelet-derived growth factor subunit A. Using single-cell RNA sequencing and spatial transcriptomics in both humans and mice, we identified macrophage colony-stimulating factor receptor (M-CSFR) signalling as one of the novel druggable targets controlling self-maintenance and persistence of these pathogenic monocyte-derived alveolar macrophages. Pharmacological blockade of M-CSFR signalling led to the disappearance of monocyte-derived alveolar macrophages and ameliorated fibrosis.
Our findings suggest that inhibition of M-CSFR signalling during fibrosis disrupts an essential fibrotic niche that includes monocyte-derived alveolar macrophages and fibroblasts during asbestos-induced fibrosis.
Short abstract
Monocyte-derived alveolar macrophages orchestrate the development of the fibrotic niche, causally related to fibrosis and maintained via M-CSF/M-CSFR signalling http://bit.ly/2nDjS20
Introduction
Pulmonary fibrosis is a complex process that is clinically characterised by a progressive increase in the number and size of spatially restricted areas of fibrosis [1]. Indeed, the three-dimensional distribution of these lesions on chest computed tomography combined with radiographic features of fibrotic regions is critical to the diagnosis and classification of pulmonary fibrosis. Single-cell RNA sequencing offers the opportunity to examine interactions between cell populations within these areas of fibrosis, but the process of tissue dissociation precludes understanding the spatial relationships between the cells [2]. We reasoned that a combination of genetic lineage tracing, single-cell RNA sequencing and single-molecule fluorescence in situ hybridisation could be combined with genetic or pharmacological interventions to identify conserved intracellular signalling events that might promote or sustain multicellular fibrotic niches in the lung. To test this hypothesis, we used a model of asbestos-induced lung fibrosis. Exposure to asbestos can induce the development of pulmonary fibrosis years after exposure has ceased [3], and historical and ongoing exposure to asbestos fibres remains an important occupational cause of pulmonary fibrosis. After inhalation, asbestos fibres remain lodged in small airways in rodents, creating spatially restricted regions of lung fibrosis [4–6].
Recently, investigators have identified several distinct macrophage populations in the lung. Tissue-resident alveolar macrophages originate from fetal monocytes, populate the alveolar niche soon after birth, are capable of self-renewal and, in mouse models, persist in the lung without appreciable input from myeloid cells over the lifespan of the animal [7–10]. Lung tissue-resident interstitial macrophages have been shown to include perivascular and peribronchial macrophages, which demonstrate distinct anatomical localisation and function [11–14]. In response to alveolar macrophage depletion and/or injury, monocytes are recruited to the lung where factors present in the microenvironment drive their differentiation into alveolar macrophages [15, 16]. Alveolar macrophages were recognised to play a critical role in asbestos-induced injury when Dostert et al. [17] reported that potassium currents in alveolar macrophages attempting to engulf asbestos fibres resulted in activation of the NLRP3 inflammasome, which is essential for the development of asbestos-mediated lung fibrosis [18]. Whether one or more of these macrophage populations is necessary for the development of fibrosis in response to asbestos remains unclear.
Here, we used a genetic lineage tracing system to show that tissue-resident alveolar macrophages and tissue-resident peribronchial and perivascular interstitial macrophages are present within the fibrotic niche, and that monocyte-derived alveolar macrophages do not originate from tissue-resident interstitial lung macrophages. Specific deletion of monocyte-derived alveolar macrophages reduced fibrosis severity. Single-molecule fluorescence in situ hybridisation demonstrated that monocyte-derived alveolar macrophages, expressing pro-fibrotic genes causally linked to fibrosis, were present within pathogenic multicellular niches that included injured epithelial cells and tissue fibroblasts. We then used single-cell transcriptomic analysis of lungs from mice treated with asbestos or bleomycin and patients with pulmonary fibrosis to look for intercellular interactions that sustain monocyte-derived alveolar macrophages. This analysis revealed macrophage colony-stimulating factor (M-CSF)/M-CSF receptor (M-CSFR) signalling as one of the key factors controlling monocyte-derived alveolar macrophages. Moreover, it provided unexpected evidence that expression of M-CSF in monocyte-derived alveolar macrophages could be involved in the maintenance of these cells in an autocrine manner. Interruption of M-CSF/M-CSFR signalling resulted in a selective loss of monocyte-derived alveolar macrophages within the fibrotic niche and an amelioration of lung fibrosis. These findings suggest that single-cell RNA sequencing data from multiple laboratories can be combined and validated to identify conserved intercellular interactions within multicellular fibrotic microdomains that might be targeted for the treatment of pulmonary fibrosis.
Materials and methods
Mice
All mouse procedures were approved by the Institutional Animal Care and Use Committee at Northwestern University (Chicago, IL, USA). All strains including wild-type mice were bred and housed at a barrier and specific pathogen-free facility at the Center for Comparative Medicine at Northwestern University. Mice aged 10–12 weeks were used for all experiments. C57BL/6J, Cx3cr1ER-Cre mice [19] and ZsGreen [20] mice were obtained from Jackson Laboratories (Farmington, CT, USA). Floxed Casp8fl/fl, CD11cCreCasp8fl/fl and CD11cCreCasp8fl/flRipk3−/− mice have been described previously [9].
Asbestos-induced lung fibrosis and drug administration
Mice aged 8–10 weeks were instilled intratracheally with control particles (TiO2; 100 µg in 50 µL PBS) or crocidolite asbestos fibres (100 µg in 50 µL PBS) to induce lung fibrosis as described previously [21]. For lineage tracing studies, anesthetised mice were administered tamoxifen by oral gavage (100 µL of 10 mg tamoxifen; Sigma, St Louis, MO, USA) dissolved in sterile corn oil (Sigma). Anti-CSF1 (M-CSF) antibody (0.5 mg, clone BE0204; BioXCell, West Lebanon, NH, USA) was administered intraperitoneally every 5 days and PLX3397 (40 mg·kg−1; MedKoo Biosciences, Morrisville, NC, USA) was administered orally every day as described previously [22, 23]. Lungs were harvested at indicated time-points for flow cytometry, single-cell RNA sequencing, histopathology, immunohistochemistry and immunofluorescence.
Tissue preparation and flow cytometry
Tissue preparation for flow cytometry analysis and cell sorting was performed as described previously [9], with modifications. Briefly, mice were euthanised and their lungs were perfused through the right ventricle with 10 mL Hanks’ balanced salt solution (HBSS). The lungs were removed and infiltrated with 2 mg·mL−1 collagenase D (Roche, Indianapolis, IN, USA) and 0.2 mg·mL−1 DNase I (Roche) dissolved in HBSS with Ca2+ and Mg2+, using a syringe with a 30G needle. Lungs were chopped with scissors, tissue was transferred into C-tubes (Miltenyi Biotech, Auburn, CA, USA) and processed in a GentleMACS dissociator (Miltenyi Biotech) using the m_lung_01 program, followed by incubation for 30 min at 37°C with gentle agitation, followed by the m_lung_02 program. The resulting single-cell suspension was filtered through a 40 µm nylon cell strainer mesh to obtain a single-cell suspension. The cells were incubated with anti-mouse CD45 microbeads (Miltenyi Biotech) and CD45+ cells were collected using a MultiMACS Cell24 Separator (Miltenyi Biotech) according to the manufacturer's protocol. Automated cell counting was performed (Cellometer K2; Nexcelom, Lawrence, MA, USA) with acridine orange/propidium iodide reagent. Cells were stained with fixable viability dye eFluor 506 (eBioscience, San Diego, CA, USA), incubated with FcBlock (BD Biosciences, San Jose, CA, USA) and stained with the following mixture of fluorochrome-conjugated antibodies (listed as antigen (clone, fluorochrome; manufacturer)): major histocompatibility complex II (2G9, BUV395; BD Biosciences), Ly6C (HK1.4, eFluor450; eBioscience), CD45 (30-F11, fluorescein isothiocyanate; eBioscience), CD64 (X54-5/7.1, phycoerythrin (PE); BioLegend, San Diego, CA, USA), Siglec F (E50-2440, PECF594; BD Biosciences), CD11c (HL3, PECy7; BD Biosciences), CD24 (M1/69, allophycocyanin; eBioscience), CD11b (M1/70, ACPCy7; BioLegend), Ly6G (1A8, Alexa 700; BD Biosciences) and NK1.1 (PK136, Alexa 700; BD Biosciences). Single-colour controls were prepared using BD CompBeads (BD Biosciences) and Arc beads (Invitrogen, Carlsbad, CA, USA). Flow cytometry was performed at the Northwestern University Robert H. Lurie Comprehensive Cancer Center Flow Cytometry Core Facility (Chicago, IL, USA). Data were acquired on a custom BD FACSymphony instrument using BD FACSDiva software (BD Biosciences). Compensation and analysis were performed using FlowJo software (TreeStar; www.flowjo.com). Each cell population was identified using a sequential gating strategy (supplementary figure S1a). The percentage of cells in the live/singlets gate was multiplied by the number of live cells using a Cellometer K2 image cytometer to obtain the cell count.
Single-cell RNA sequencing
Single-cell suspensions were prepared as described earlier with slight modification. Mice were euthanised with sodium pentobarbital overdose. The chest cavity was opened and lungs were perfused through the right ventricle with 10 mL HBSS. The lungs were removed and, using a 30G needle, infused with 1 mL dispase (Corning, Corning, NY, USA) with DNase I (Sigma) before incubation at room temperature with gentle agitation for 45 min, followed by gentle teasing using forceps into small (1–2 mm) fragments and incubation in digestion buffer for another 15 min. The resulting suspension was passed through a 70 µm cell strainer (Falcon; Corning), washed with DMEM (Corning) supplemented with 5% FBS (Corning), pelleted by centrifugation and erythrocytes were lysed using BD Pharm Lyse (BD Biosciences). The resulting single-cell suspension was kept in DMEM/FBS and passed through a 40 µm cell strainer (Falcon) two times. Cells were counted using a Cellometer K2 with nucleic acid binding dyes acridine orange to calculate total number of nucleated cells and propidium iodide to count dead cells; cell viability exceeded 85%. All manipulations were performed using wide-bore tips (Axygen; Corning). Single-cell 3′ RNA sequencing libraries were prepared using Chromium Single Cell v2 Reagent Kit and Controller (10X Genomics, Pleasanton, CA, USA). Libraries were assessed for quality (TapeStation 4200; Agilent, Santa Clara, CA, USA) and then sequenced on an HiSeq 4000 instrument (Illumina, San Diego, CA, USA). Initial data processing was performed using the Cell Ranger version 2.0 pipeline (10X Genomics); reads were mapped to the mm10 version of the mouse genome. Analysis was performed using the Seurat R toolkit version 2.3.4 [24] for the dataset from Xie et al. [25] and version 3.0.2 [26] with sctransform normalisation [27] for the asbestos dataset. Analysis of the putative interactions between cells was performed using a curated list of ligands and receptors from the FANTOM5 project [28]. Analysis of RNA velocity in single cells was performed using the velocyto pipeline [29]. All code used for analysis can be found at https://github.com/NUPulmonary/JoshiWatanabe2019.
Data availability
Single-cell RNA sequencing data from TiO2- and asbestos-exposed mice have been deposited with the Gene Expression Omnibus (GSE127803). We also used bulk RNA sequencing data on flow-sorted alveolar macrophages from GSE82158 [9], single-cell RNA sequencing data from patients with pulmonary fibrosis from GSE122960 [2] and mice exposed to bleomycin from GSE104154 [25]. Interactive plots from figures 4a, 4c and 7a are available for exploration at www.nupulmonary.org/resources.
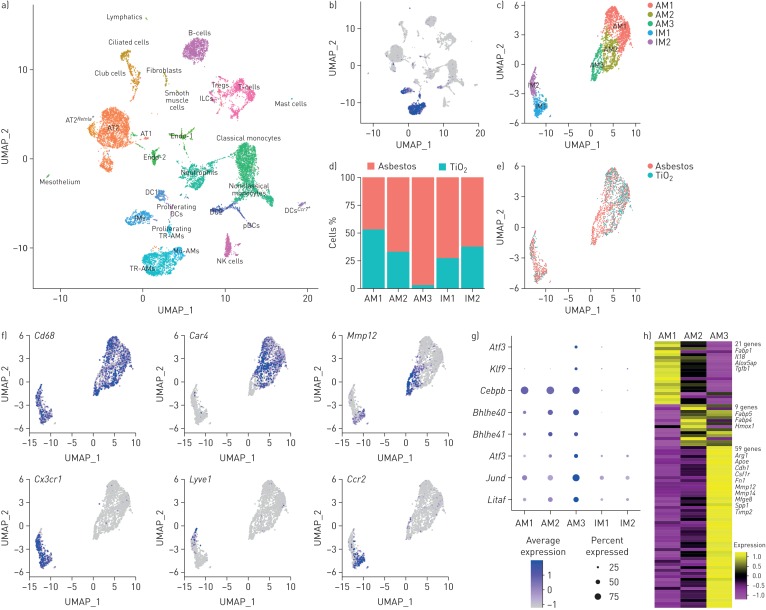
FIGURE 4.
Single-cell RNA sequencing reveals pro-fibrotic monocyte-derived alveolar macrophages (Mo-AMs) during asbestos-induced pulmonary fibrosis. UMAP: uniform manifold approximation and projection; Treg: regulatory T-cell; AT1: alveolar epithelial type I cell; AT2: alveolar epithelial type II cell; Endo: endothelial cell; DC: dendritic cell; IM: interstitial macrophage; pDC: plasmacytoid DC; TR-AM: tissue-resident AM. a) UMAP plot demonstrating 26 cell clusters from 15 452 cells identified by single-cell RNA sequencing 14 days after asbestos or TiO2 exposure (one mouse per condition). b) Macrophages were identified using canonical lineage-restricted markers, such as Mrc1, as shown on the UMAP plot. c) Clusters of cells expressing Mrc1 were subset from the main dataset and re-clustered, revealing two subclusters of tissue-resident lung IMs (IM1 and IM2) and three subclusters of AMs (AM1, AM2 and AM3). d) Bar plot and e) feature plot demonstrating the composition of macrophage subclusters in cells from asbestos- and TiO2-exposed animals. f) Feature plots demonstrating expression of cluster-specific genes: Cd68 as a pan-macrophage marker, Car4 as a marker of mature TR-AMs (AM1 and AM2) and Mmp12 as a marker of Mo-AMs (AM3). Tissue-resident IMs are characterised by expression of Cx3cr1, and can be further subdivided into perivascular (Lyve1) and peribronchial (Ccr2) IMs. g) Dot plot demonstrating the expression of transcription factors differentially expressed in Mo-AMs (AM3). h) Heatmap of 93 genes overlapping between AMs and pulmonary fibrosis-associated genes from the Comparative Toxicogenomic Database (947 genes as of February 2019). Selected genes characterising clusters are shown; see supplementary figure S4e for the full list of genes. Interactive plots are available for exploration at www.nupulmonary.org/resources.
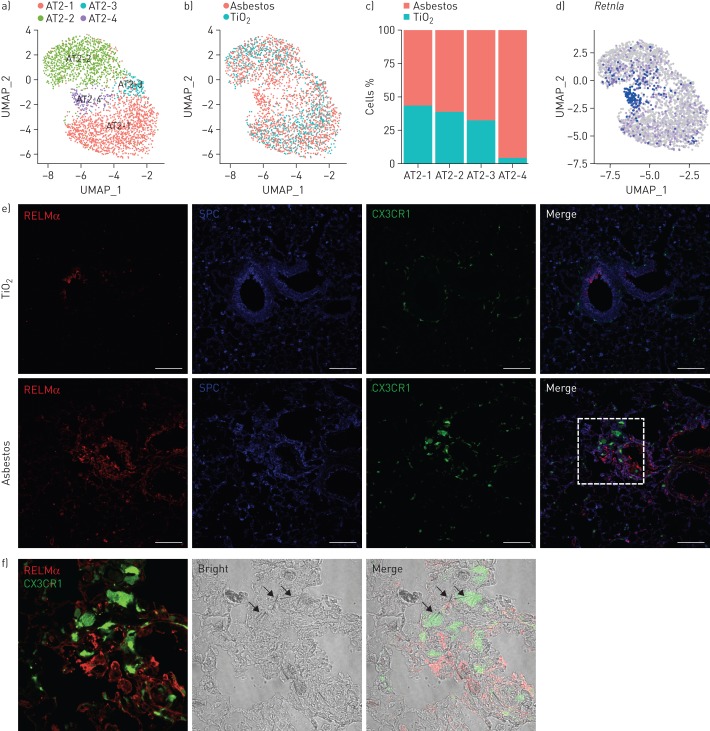
FIGURE 7.
Expression of resistin-like molecule-α (RELMα) is restricted to epithelial cells located in the areas of fibrosis. UMAP: uniform manifold approximation and projection; SPC: surfactant protein C. a) UMAP plot demonstrating subclusters of alveolar type II cells. b) UMAP plot and c) bar plot demonstrating composition of the alveolar type II cell subclusters. d) Feature plot demonstrating increased expression of Retnla in alveolar type II cells 14 days after asbestos exposure. e) Cx3cr1ER-Cre×ZsGreen mice were administered with asbestos intratracheally and treated with tamoxifen at days 14 and 15 after exposure; lungs were harvested for analysis at day 21. Representative fluorescent images showing expression of RELMα (red), SPC (blue) and CX3CR1–green fluorescent protein (green) in lungs from TiO2- or asbestos-treated animals at day 21 post-exposure. RELMα is detected in the airway epithelial cells and alveolar type II cells in the fibrotic regions in the asbestos model, but not in alveolar type II cells after TiO2 exposure. Scale bar: 100 µm. f) Enlargement of the box in (e): RELMα-positive epithelial cells (red) and monocyte-derived alveolar macrophages (green) are co-localised with asbestos fibres. Experimental design same as in (e).
Fluorescence in situ RNA hybridisation
Multiplex fluorescence in situ hybridisation was performed using RNAscope (Advanced Cell Diagnostics, Newark, CA, USA). Mouse lungs were inflated to 15 cmH2O and fixed with 4% paraformaldehyde (EMS, Hatfield, PA, USA) for 24 h. Lungs were paraffin embedded and 5 µm tissue sections were mounted on Superfrost Plus slides (Thermo Fisher Scientific, Waltham, MA, USA). Slides were baked for 1 h at 60°C, deparaffinised in xylene and dehydrated in 100% ethanol. Sections were treated with H2O2 (Advanced Cell Diagnostics) for 10 min at room temperature and then heated to mild boil (98–102°C) in 1× target retrieval reagent buffer (Advanced Cell Diagnostics) for 15 min. Protease Plus (Advanced Cell Diagnostics) was applied to sections for 30 min at 40°C in a HybEZ Oven (Advanced Cell Diagnostics). Hybridisation with target probes, pre-amplifier, amplifier, fluorescent labels and wash buffer (Advanced Cell Diagnostics) was done according to Advanced Cell Diagnostics’ instructions for Multiplex Fluorescent Reagent Kit version 2 and 4-Plex Ancillary Kit version 2 when needed. Parallel sections were incubated with Advanced Cell Diagnostics positive and negative control probes. Sections were covered with ProLong Diamond Antifade Mountant (Invitrogen). Probes used were: mouse Mrc1, mouse Pdgfa, mouse Pdgfra and mouse Csf1 (all Advanced Cell Diagnostics). Opal fluorophores (Opal 520, Opal 620 and Opal 690; Perkin Elmer, Shelton, CT, USA) were used at 1:1500 dilution in Multiplex TSA buffer (Advanced Cell Diagnostics). To validate colocalisation of Mrc1 and either Pdgfa or Csf1 probe, images were captured on a Nikon A1R confocal microscope (Tokyo, Japan) with ×20 and ×40 objectives, followed by uniform processing and pseudo-colouring in Fiji (https://imagej.net/Fiji). For quantification of Mrc1 and Pdgfa co-staining, images of whole mouse lung tissue were captured on a Nikon Ti2 wide-field microscope with a ×20 objective and 0.45 NA. Quantification was performed using Nikon NIS-Elements software version 5.20.0 using the General Analysis module. Images were pre-processed with rolling ball background subtraction and segmented with thresholding. Objects positive for 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) and Mrc1, DAPI and Pdgfa, and DAPI, Mrc1 and Pdgfa were normalised to total number of cells.
Histopathology, immunohistochemistry and immunofluorescence
For histopathology and immunohistochemistry, mouse lung tissue from the regions adjacent to the regions used for flow cytometry and single-cell RNA sequencing was fixed in 4% paraformaldehyde for 24 h, dehydrated and embedded in paraffin; 4 µm thick sections were prepared. Haematoxylin/eosin staining and Masson's trichrome staining were performed for the analysis of fibrosis scoring. Immunohistochemistry was performed at the Northwestern University Mouse Histology and Phenotyping Laboratory Core Facility (Chicago, IL, USA).
For immunofluorescence, mouse lung tissue was fixed in 4% paraformaldehyde for 6 h and transferred into 20% sucrose for overnight incubation. Tissue was embedded in Tissue-Tek OCT compound (Sakura, Torrance, CA, USA), flash frozen in liquid nitrogen and cut on a cryostat at 14 µm thickness. Sections were air-dried and stained with PE-conjugated anti-proto-oncogene tyrosine-protein kinase MER (MERTK) (BioLegend), Alexa Fluor 647-conjugated anti-Siglec F (BD Biosciences), rabbit anti-resistin-like molecule-α (RELMα) (Abcam, Cambridge, MA, USA) and goat anti-surfactant protein C (SPC) (Santa Cruz Biotechnology, Dallas, TX, USA). Appropriate secondary antibodies were used for unconjugated primary antibody, including donkey anti-rabbit Alexa Fluor 647 (Invitrogen) and donkey anti-goat Alexa Flour 568 (Invitrogen). DAPI was used for nuclear staining and sections were mounted with ProLong Diamond Antifade Mountant. Images were acquired on a Nikon A1R confocal microscope or Nikon Ti2 wide-field microscope at Northwestern University Nikon Cell Imaging Facility (Chicago, IL, USA), processed using Nikon Elements software.
Fibrosis scores and lung collagen determination
Fibrosis scores in mice were determined from haematoxylin/eosin- and Masson's trichrome-stained specimens in a blinded manner, in accordance with the code set by the Pathology Standards for Asbestosis as described previously [30]. Collagen levels were determined using the Sircol assay as described previously [21].
Statistical analysis
Statistical tests and tools for each analysis are explicitly described with the results or detailed in the figure legends.
Results
Recruitment of monocyte-derived alveolar macrophages distinguishes the response to asbestos from a nonfibrogenic particle (TiO2)
We quantified monocyte and macrophage populations in the mouse lung via flow cytometry 14 days after intratracheal administration of asbestos, which reproducibly induces lung fibrosis, or a standard control particle TiO2, which does not induce fibrosis (both 100 µg) (figure 1a; see supplementary figure S1a for gating strategy). We found that the numbers of monocytes and alveolar macrophages (CD64+Siglec F+) were similarly increased in mice administered either asbestos or TiO2 when compared with naive mice (supplementary figure S1b). While the increase in alveolar macrophages was largely attributable to expansion of a Siglec Fhigh population in TiO2-treated animals, the increase in alveolar macrophages in animals administered asbestos was attributable to both an expansion in Siglec Fhigh alveolar macrophages and the emergence of a population of Siglec Flow alveolar macrophages (∼15% of the total alveolar macrophage pool) (figure 1b and c, and supplementary figure S1b).
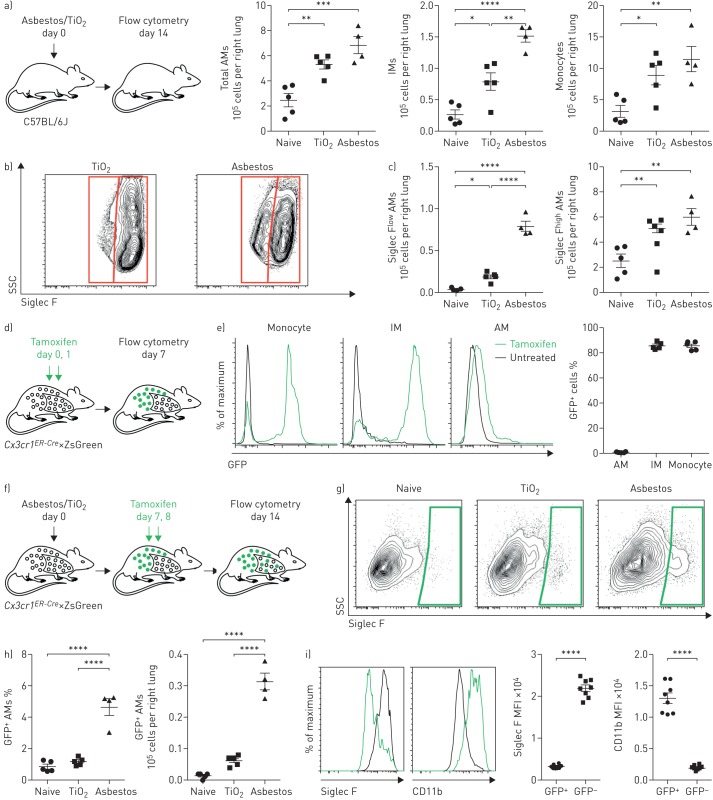
FIGURE 1.
Exposure to asbestos or TiO2 is distinguished by the recruitment of monocyte-derived alveolar macrophages (Mo-AMs) to the lung. IM: interstitial macrophage; SSC: side scatter; FSC: forward scatter; GFP: green fluorescent protein; MFI: median fluorescence intensity. a) Mice were administered crocidolite asbestos or TiO2 (both at 100 µg intratracheally), and monocyte and macrophage populations were quantified by flow cytometry 14 days later (see supplementary figure S1a and b for gating strategy and quantification of other myeloid cell populations). b) Representative contour plots gated on AMs (CD64+Siglec F+) from asbestos- or TiO2-treated animals. c) Quantification of Siglec Flow and Siglec Fhigh AMs from naive, TiO2- or asbestos-exposed animals according to gating in (b). d) Schematic of the experimental design for (e). e) Cx3cr1ER-Cre×ZsGreen mice were treated with tamoxifen, and the percentage of GFP+ classical monocytes, IMs and AMs was assessed by flow cytometry. Representative histograms showing GFP expression are shown. f) Schematic of the experimental design for (g) and (h): lineage tracing system to track the ontogeny of AMs after intratracheal administration of asbestos or TiO2. Cx3cr1ER-Cre×ZsGreen mice were treated with asbestos or TiO2 and tamoxifen was administered as two boluses at days 7 and 8. The number of GFP+ AMs was analysed 7 days later. g) Representative contour plots and h) quantification of GFP+ Mo-AMs after asbestos or TiO2 exposure. i) Representative histograms and MFI demonstrating expression of Siglec F and CD11b on Mo-AMs 14 days after exposure to asbestos. All data are presented as mean±sem. n=4–5 mice per group. One-way ANOVA with the Tukey–Kramer test for multiple comparisons. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001. Representative data from two independent experiments.
We used a genetic lineage tracing system to determine whether Siglec Flow alveolar macrophages that were expanded in asbestos-treated animals but absent in TiO2-treated animals were derived from the recruitment of monocytes or the expansion of tissue-resident alveolar macrophages. Circulating Ly6Chigh classical monocytes express high levels of Cx3cr1, while tissue-resident alveolar macrophages do not [11, 19]. Accordingly, we crossed Cx3cr1ER-Cre animals to ZsGreen reporter mice to track the fate of monocyte-derived cells in the lung during asbestos-mediated fibrosis (figure 1d). In this system, 7 days after tamoxifen administration 86.8% of circulating monocytes are green fluorescent protein (GFP)+ (figure 1e), but because they are short-lived, they disappear after tamoxifen pulse [19]. However, if GFP-labelled monocytes give rise to long-living macrophages, such as monocyte-derived alveolar macrophages, they will be permanently labelled with GFP, even after the expression of endogenous Cx3cr1 has ceased. As expected, long-lived tissue-resident alveolar macrophages, which do not express Cx3cr1, were not labelled in this system (figure 1e). In contrast, 86.7% of the tissue-resident peribronchial and perivascular interstitial macrophages, which also express Cx3cr1, became GFP+ after tamoxifen pulse (figure 1e).
To assess the contribution of monocyte-derived macrophages to the expansion of the alveolar macrophage pool, reporter mice were treated with tamoxifen via oral gavage 7 and 8 days after administration of asbestos or TiO2 and lungs were analysed by flow cytometry at day 14 (figure 1f). Administration of asbestos resulted in an influx of GFP+ alveolar macrophages, which were Siglec Flow and CD11bhigh (figure 1g–i). In total, 4.9% of monocyte-derived alveolar macrophages (CD64+Siglec Flow) were GFP+ 1 week after tamoxifen pulse in asbestos-exposed mice (figure 1h and i). In contrast, in mice treated with TiO2, only 1.4% of alveolar macrophages were GFP+, which was comparable to naive mice (figure 1h and i).
Monocyte-derived alveolar macrophages and not tissue-resident interstitial macrophages contribute to the macrophage pool in the fibrotic niche
We took advantage of the GFP label on monocyte-derived alveolar macrophages in our lineage tracing system to determine whether recruitment of these cells in response to asbestos was spatially restricted to areas near the fibres. Fibrosis was restricted to regions of bronchoalveolar duct junctions where asbestos fibres had lodged, but was absent in the distal lung (figure 2a). Immunofluorescent microscopy demonstrated that GFP+ cells were specifically found in these fibrotic areas, and both GFP+ and GFP– cells appeared to engulf asbestos fibres (figure 2b and c). These cells are located in the alveolar space and express MERTK and Siglec F, confirming their identity as monocyte-derived alveolar macrophages (supplementary figure S2).
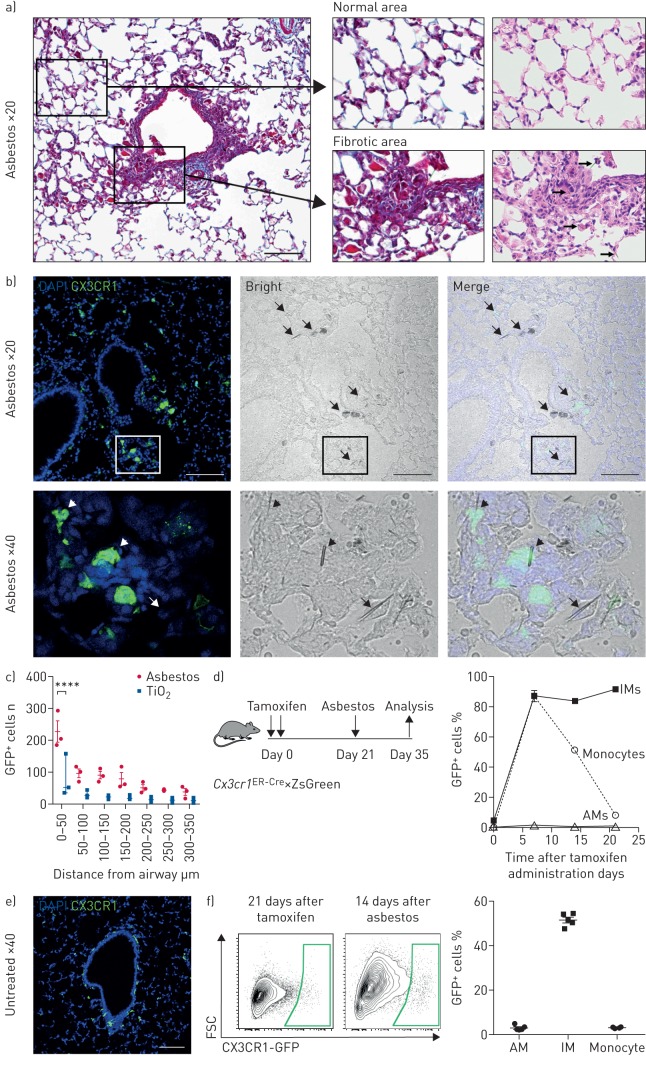
FIGURE 2.
Recruitment of monocyte-derived alveolar macrophages (Mo-AMs) is spatially restricted to areas near asbestos fibres. DAPI: 4′,6-diamidino-2-phenylindole; GFP: green fluorescent protein; IM: interstitial macrophage; FSC: forward scatter. a) The intratracheal administration of asbestos fibres induces fibrosis near bronchoalveolar duct junctions where asbestos fibres lodge. Left panel: low-power image of a medium-sized airway (Mason's trichrome). Scale bar: 100 µm. Right panels: high-power images (Mason's trichrome and haematoxylin/eosin, respectively). Areas of fibrosis develop adjacent to the airway in which asbestos fibres can be observed (arrows, bottom right panel). In contrast, alveolar structures in the distal lung parenchyma are relatively preserved. b) Top panels: representative lung histology from Cx3cr1ER-Cre×ZsGreen mice treated with tamoxifen on day 14 and 15 (10 mg via oral gavage) and harvested 21 days after asbestos exposure. Scale bar: 100 µm. Bottom row panels correspond to areas outlined in the boxes. Left panels: monocyte-derived cells are GFP+; nuclei stained with DAPI. Middle panels: phase contrast images; asbestos fibres are indicated by arrows. Right panels: merge. Bottom panels: asbestos fibres are surrounded by GFP+ cells (short arrows) and GFP− cells (arrow) with macrophage morphology. c) Quantification of GFP+ Mo-AMs in peribronchial regions in asbestos- and TiO2-treated mice. Two-way ANOVA with Tukey's multiple comparisons test. ****: p<0.0001. d) Schematic of the experimental design, and kinetics of GFP+ monocytes, tissue-resident IMs and tissue-resident AMs after tamoxifen pulse in naive Cx3cr1ER-Cre×ZsGreen mice. Percentage of GFP+ cells was assessed by flow cytometry. e) Representative fluorescent image showing GFP+ tissue-resident IMs 21 days after tamoxifen pulse in naive animals. f) Representative contour plots showing GFP expression gated on AMs from Cx3cr1ER-Cre×ZsGreen mice 21 days after tamoxifen and 14 days after asbestos instillation. Percentage of GFP+ classical monocytes, IMs and AMs was assessed by flow cytometry. All data are presented as mean±sem. n=3–5 mice per group or time-point.
Tissue-resident interstitial macrophages (CD64+Siglec F−) have recently been reported to contribute to the population of tumour-associated macrophages [12]. To exclude the possibility that these tissue-resident peribronchial or perivascular interstitial macrophages can migrate to the alveolar space in response to asbestos-induced epithelial injury and differentiate into alveolar macrophages, we treated Cx3cr1ER-Cre×ZsGreen reporter mice with tamoxifen to label tissue-resident interstitial macrophages (figure 2d). By 21 days after tamoxifen treatment, virtually all GFP-labelled monocytes disappeared from circulation, while 82% of tissue-resident interstitial macrophages remained GFP+ (figure 2d and e). These mice were treated with asbestos intratracheally and the lungs were analysed by flow cytometry 14 days later (figure 2d). The percentage of GFP+ alveolar macrophages in asbestos-exposed mice was not different from nonexposed mice, suggesting that monocytes, and not tissue-resident interstitial macrophages, drive the expansion of alveolar macrophages at the sites of asbestos-mediated injury and fibrosis (figure 2f).
Genetic deletion of monocyte-derived alveolar macrophages attenuates asbestos-induced pulmonary fibrosis
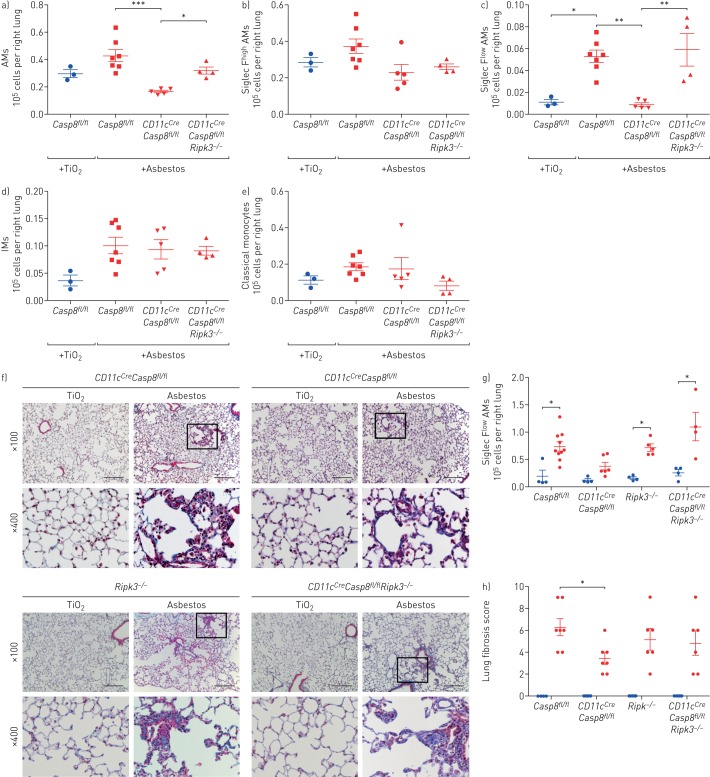
Previously, we have shown that a loss of caspase-8 in monocyte-derived alveolar macrophages results in their death via necroptosis during differentiation [9]. Accordingly, we used macrophage-specific deletion of Casp8 with or without prevention of necroptosis through deletion of Ripk3 to determine whether monocyte-derived alveolar macrophages are necessary for the development of asbestos-induced pulmonary fibrosis. Control mice (Casp8fl/fl) and mice lacking caspase-8 in alveolar macrophages (CD11cCreCasp8fl/fl) were administered either asbestos or TiO2 intratracheally. The number of monocyte-derived alveolar macrophages (CD64+Siglec Flow) was significantly reduced in CD11cCreCasp8fl/fl compared with Casp8fl/fl controls 28 days after the administration of asbestos (figure 3a–d). Other cell populations were unchanged in CD11cCreCasp8fl/fl mice compared with Casp8fl/fl controls with the exception of eosinophils, which were reduced in CD11cCreCasp8fl/fl mice compared with Casp8fl/fl controls (figure 3e and supplementary figure S3a). CD11cCreCasp8fl/fl mice showed less asbestos-induced fibrosis compared with Casp8fl/fl controls (figure 3f–h). To confirm that the protection from fibrosis in CD11cCreCasp8fl/fl mice resulted from necroptotic loss of monocyte-derived alveolar macrophages, we performed the same experiments in CD11cCreCasp8fl/flRipk3−/− mice and Ripk3−/− mice. While the global loss of Ripk3 did not affect the recruitment of monocyte-derived alveolar macrophages or the severity of fibrosis, the loss of Ripk3 in CD11cCreCasp8fl/flRipk3−/− mice restored monocyte-derived alveolar macrophages and rescued asbestos-induced fibrosis (figure 3a–h). To determine whether tissue-resident alveolar macrophages also contributed to the development of asbestos-induced fibrosis, we depleted tissue-resident alveolar macrophages by treating mice with intratracheal liposomal clodronate 24 h prior to the administration of intratracheal asbestos or TiO2. There was no difference in fibrosis 28 days later as determined by quantitative scoring of lung sections and measurements of lung soluble collagen using Picrosirius red precipitation, suggesting that tissue-resident alveolar macrophages are not required for the development of fibrosis (supplementary figure S3b). These data demonstrate that monocyte-derived alveolar macrophages are necessary to develop a spatially restricted fibrotic niche in response to asbestos.
FIGURE 3.
Deletion of monocyte-derived alveolar macrophages (AMs) attenuates asbestos-induced pulmonary fibrosis. IM: interstitial macrophage. Casp8fl/fl, CD11cCreCasp8fl/fl, CD11cCreCasp8fl/flRipk3−/−and Ripk3−/− mice were administered crocidolite asbestos or TiO2 (both at 100 µg intratracheally) and lungs were harvested 28 days later. a–e) Lungs were analysed using flow cytometry to quantify monocyte and macrophage populations. f) Representative histological images (Mason's trichrome). Scale bar: 100 µm. g) Quantification of soluble collagen in lung homogenates. h) Lung fibrosis score: blinded scoring of a single longitudinal section from each mouse. Blue circles: TiO2 treatment; red symbols: asbestos administration. All data are presented as mean±sem. n=3–7 mice per group. One-way ANOVA with the Tukey–Kramer test for multiple comparisons. *: p<0.05; **: p<0.01; ***: p<0.001.
Single-cell RNA sequencing distinguishes responses to fibrogenic asbestos fibres and control particles
We reasoned that single-cell RNA sequencing combined with spatial techniques, such as in situ RNA hybridisation, could be used to gain insights into cellular interactions by which monocyte-derived alveolar macrophages are maintained and signal within fibrotic niches in the asbestos-exposed lung. Accordingly, we performed single-cell RNA sequencing on unenriched single-cell suspensions from the lungs of wild-type mice 14 days after exposure to either asbestos or TiO2, when fibrosis is just beginning (one mouse per condition). Clustering was performed using the Seurat R toolkit and clusters were annotated based on the expression of cell-type-specific genes (figure 4a and supplementary table S1). After excluding doublets and low-quality cells, 15 288 cells from two libraries were used for subsequent analysis. We captured and resolved all major cell populations in the mouse lung, including interstitial and alveolar macrophages, alveolar epithelial type I and type II cells, dendritic cells, fibroblasts, mesothelial cells, smooth muscle cells, and subsets of endothelial cells. Each cluster included cells from both experimental groups (i.e. mice exposed to asbestos or TiO2) (supplementary figure S4a and b). In agreement with our lineage tracing studies indicating influx of monocyte-derived alveolar macrophages, we identified a subcluster of alveolar macrophages (figure 4f) that was disproportionately represented by the cells from the asbestos-exposed animal (supplementary figure S4b).
We identified macrophages based on the expression of canonical macrophage-associated genes (Cd68, Mrc1, Lyz2, Adgre1 and Axl) (figure 4b and supplementary figure S4c). Focused analysis of macrophage populations resolved two major populations identified as alveolar macrophages and tissue-resident interstitial macrophages. Tissue-resident interstitial macrophages could be further subdivided into peribronchial and perivascular macrophages, distinguished by expression of Ccr2, and F13a1 and Lyve1, respectively (figure 4c–f and supplementary figure S4c) [11]. While some markers were expressed in both alveolar and interstitial macrophages (Cd68, Mrc1, Lyz2, Adgre1 and Axl), others were restricted to alveolar macrophages (Siglecf, Marco and Il18) (supplementary figure S4c and supplementary table S2).
Alveolar macrophages contained three subclusters. Cluster AM1 was comprised of alveolar macrophages from mice exposed to either asbestos or TiO2 (figure 4d and e). In contrast, clusters AM2 and AM3 were predominantly represented by cells from asbestos-exposed animals. Alveolar macrophages from cluster AM1 were characterised by expression of genes associated with normal homeostatic function of alveolar macrophages (Ear1 and Fabp1) (supplementary figure S4d). Macrophages from cluster AM2 expressed genes involved in the inflammatory response, cytokine production and matrix metalloproteinase activation (Car4, Ctsk, Chil3, S100a1 and Wfdc21) (supplementary figure S4d). In agreement with fate mapping studies, macrophages from cluster AM3 exhibited a more immature alveolar macrophage phenotype, characterised by lower expression of Pparg, Car4, Ear1, Siglecf and Marco, increased expression of Itgam, Cd36 and Gpnmb (supplementary figure S4c and d), and increased expression of transcription factors involved in macrophage development and maintenance (Litaf, Jund, Bhlhe40, Bhlhe41 and Klf9) and the unfolded protein response (Atf3 and Atf4) (figure 4g). RNA velocity analysis [29] suggests that cluster AM3 represents a transcriptionally stable cell state, rather than an intermediate state between monocyte to tissue-resident macrophage differentiation (supplementary figure S4e and f).
We then queried the expression of 947 genes associated with pulmonary fibrosis using the Comparative Toxicogenomic Database [31] and found that 89 genes were detected in alveolar macrophages (figure 4h and supplementary figure S4g). Clusters AM1 and AM2 contained “generic” genes (21 and nine genes, correspondingly) associated with alveolar macrophage cellular identity and function (Fabp1, Fabp4, Fabp5 and Tgfb1) [32]. In contrast, cluster AM3 was enriched for pro-fibrotic genes, and contained 59 genes involved in matrix remodelling and cell–cell interactions (Cdh1, Fn1, Mmp12, Mmp14, Spp1 and Timp2) (figure 4h and supplementary figure S4g).
In a recent publication, Xie et al. [25] evaluated the response of fibroblasts in bleomycin-induced pulmonary fibrosis using single-cell RNA sequencing. While the authors focused their analysis on fibroblasts, their dataset contained a large number of nonmesenchymal cells, including alveolar macrophages (supplementary figure S4h and i, and supplementary table S3). Analysis of the macrophage population demonstrated the presence of three subclusters: AM1, AM2 and AM3 (supplementary figure S4j and supplementary table S4). Similar to our findings in asbestos, cluster AM1 was predominantly comprised of cells from PBS-treated control animals, while clusters AM2 and AM3 were comprised of cells from bleomycin-treated animals (supplementary figure S4k and l). Out of 947 genes associated with pulmonary fibrosis in the Comparative Toxicogenomic Database, we found 80 to be detected in alveolar macrophages from the bleomycin dataset (supplementary figure S4m). Cluster AM3 was enriched for expression of fibrosis-associated genes (53 genes), many of which overlapped with the AM3 cluster genes from our asbestos dataset (Gpnmb, Fn1, Mmp12, Mmp14, Spp1 and Timp2) (supplementary figure S4g and m).
We have previously reported the emergence of a novel subpopulation of alveolar macrophages in lung explants from patients with pulmonary fibrosis compared with biopsies from normal donor lung [2]. 20 genes, including SPP1, MMP14, TREM2 and GPNMB, overlapped between the genes characterising the cluster of human pro-fibrotic alveolar macrophages from patients with pulmonary fibrosis and genes observed in the AM3 clusters from asbestos-induced and bleomycin-induced pulmonary fibrosis (supplementary figure S4n and supplementary table S5). Thus, our analysis has demonstrated the emergence of homologous populations in different models of pulmonary fibrosis (asbestos and bleomycin) and in patients with pulmonary fibrosis.
Single-cell RNA sequencing identifies ligand–receptor interactions that emerge during pulmonary fibrosis
Our data causally implicate monocyte-derived alveolar macrophages in the development of pulmonary fibrosis. To understand the molecular basis by which they signal to promote pulmonary fibrosis, we analysed possible ligand–receptor interactions that were identified in single-cell data from asbestos-treated lungs but were absent in TiO2-treated lungs. Our initial analysis identified more than 50 000 possible interactions (supplementary figure S4o–q; see Data availability and online code for details) [28]. To narrow this list, we focused on the interactions between cell types located in physical proximity to areas of fibrosis (macrophages, fibroblasts and alveolar type II cells) and specifically focused on the two cell types that emerged exclusively during asbestos-induced pulmonary fibrosis: monocyte-derived alveolar macrophages and Retnla+ alveolar type II cells (supplementary figure S4r and supplementary table S6). We then went on to validate some of the identified interactions, specifically focusing on 1) factors that can promote recruitment of monocyte-derived alveolar macrophages by injured alveolar epithelial cells, 2) factors that can promote persistence of monocyte-derived alveolar macrophages in the fibrotic regions and 3) factors produced by monocyte-derived alveolar macrophages that can drive proliferation of fibroblasts.
M-CSF/M-CSFR signalling is necessary for the maintenance of monocyte-derived alveolar macrophages within fibrotic niches
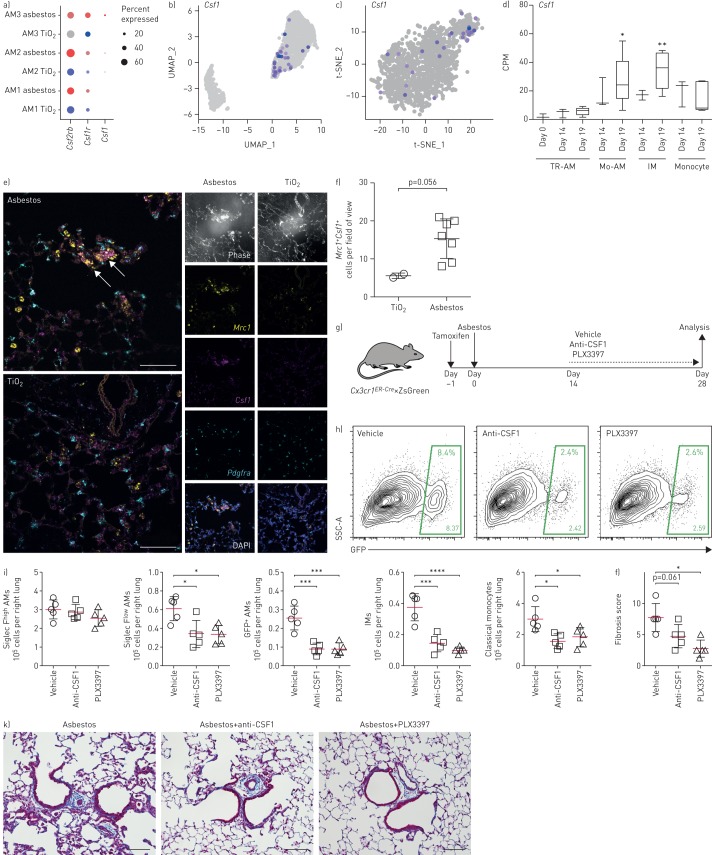
Tissue-resident alveolar macrophages rely on granulocyte−macrophage colony-stimulating factor (GM-CSF; Csf2) produced by alveolar epithelial cells for their homeostatic maintenance [33]. In contrast, data from M-CSF-deficient mice and pharmacological blockade of M-CSFR (Csf1r) suggest that M-CSF (Csf1) signalling is dispensable for homeostatic maintenance of alveolar macrophages [34–36]. We found that Csf1r was upregulated in the AM3 cluster both in asbestos- and bleomycin-treated mice (figure 5a, and supplementary figure S4g and m). We asked which cells express Csf1 and Il34 (ligands for M-CSFR), and particularly whether any new Csf1/Il34-producing cells emerge during fibrosis. In TiO2- and asbestos-exposed animals, Il34 was expressed in alveolar type II cells and club cells, while Csf1 was expressed in fibroblasts, mesothelium and endothelium (supplementary figure S5a and b). Interestingly, ∼10% of macrophages in cluster AM3 expressed Csf1 in both asbestos- and bleomycin-induced pulmonary fibrosis (figure 5a–c and supplementary table S6). Expression of Csf1 was also detected by bulk RNA sequencing in flow-sorted macrophages from mice with bleomycin-induced fibrosis (from our previously published dataset [9]) (figure 5d). In humans, expression of CSF1 was detected in alveolar macrophages from the lungs of normal donors and patients with pulmonary fibrosis (supplementary figure S5c), and expression of CSF1R was increased in patients with pulmonary fibrosis, specifically in the cluster containing pro-fibrotic alveolar macrophages (supplementary figure S5d and e). We then validated our findings using in situ RNA hybridisation. Double-positive Mrc1+Csf1+ alveolar macrophages were detected in the areas of fibrosis in proximity to Pdgfra+ fibroblasts 28 days after asbestos exposure (figure 5e and f, and supplementary figure S5f).
FIGURE 5.
Autocrine macrophage colony-stimulating factor (M-CSF) signalling is required for maintenance of monocyte-derived alveolar macrophages (Mo-AMs) within fibrotic niches. UMAP: uniform manifold approximation and projection; t-SNE: t-distributed stochastic neighbour embedding; TR-AM: tissue-resident AM; IM: interstitial macrophage; SSC: side scatter; GFP: green fluorescent protein. a) Dot plot showing expression of Csf2rb, Csf1r and Csf1 in subclusters of AMs after TiO2 and asbestos exposure. b, c) Feature plots showing expression of Csf1 in subcluster AM3 after b) asbestos and c) bleomycin exposure. d) Box and whisker plot shows expression of Csf1 in flow-sorted AMs during the course of bleomycin-induced pulmonary fibrosis (data from Misharin et al. [9]). Box plot centre lines are median, box limits are upper and lower quartiles, and whiskers are minimal and maximal values. One-way ANOVA with Bonferroni correction for multiple comparisons. e) In situ RNA hybridisation confirms expression of Csf1 in AMs during asbestos-induced pulmonary fibrosis. Analysis performed on day 28 after TiO2 or asbestos exposure. Macrophages were identified as Mrc1+ cells and fibroblasts were identified as Pdgfra+ cells. Arrows indicate Mrc1+Csf1+ AMs. Scale bar: 50 µm. f) The number of Mrc1+Csf1+ AMs is increased after asbestos exposure. Data are from one mouse per condition. Mean±sd. Mann–Whitney test. g) Schematic of experimental design. Cx3cr1ER-Cre×ZsGreen mice received a pulse of tamoxifen via oral gavage 1 day prior to administration of crocidolite asbestos (100 µg intratracheally). Starting at day 14 mice were treated with anti-CSF1 antibody (0.5 mg intraperitoneally, every 5 days) or PLX3397 (40 mg·kg−1 orally, every day) and harvested at day 28. Numbers of monocytes and macrophages were measured by flow cytometry and the fibrosis score was analysed by histology at day 28. h) Representative flow cytometry plots gated on CD64+Siglec F+ AMs. i) Number of TR-AMs, Mo-AMs, IMs and classical monocytes from asbestos-exposed animals. j) Fibrosis score 28 days after asbestos exposure. k) Representative histological findings. Mason's trichrome. Scale bar 100 µm. Data are presented as mean±sem. n=5 mice per group. One-way ANOVA with the Tukey–Kramer test for multiple comparisons. *: p<0.05; **: p<0.01; ***: p<0.001; ****: p<0.0001.
These data suggest that monocyte-derived alveolar macrophages are capable of generating an autocrine signal for their persistence. We tested this hypothesis using the Cx3cr1ER-Cre×ZsGreen reporter mice described earlier. At 1 day prior to induction of pulmonary fibrosis via intratracheal administration of asbestos, we treated these mice with a pulse of tamoxifen to transiently label circulating monocytes and monocyte-derived alveolar macrophages derived from these monocytes (figure 5g). An established population of monocyte-derived alveolar macrophages was present in the lung 14 days after induction of fibrosis (figure 1g and h). We then treated mice with neutralising anti-CSF1 antibody or with PLX3397 (a small molecule inhibitor of M-CSFR kinase signalling), and analysed pulmonary macrophage populations and pulmonary fibrosis severity 14 later (figure 5g). Pharmacological blockade of M-CSFR signalling decreased the number of monocyte-derived alveolar macrophages and decreased the severity of pulmonary fibrosis, while the number of tissue-resident alveolar macrophages was not affected and the level of circulating monocytes was only moderately reduced (figure 5h–k).
Monocyte-derived alveolar macrophages provide a link between epithelial injury and activation of resident fibroblasts within spatially restricted pro-fibrotic niches
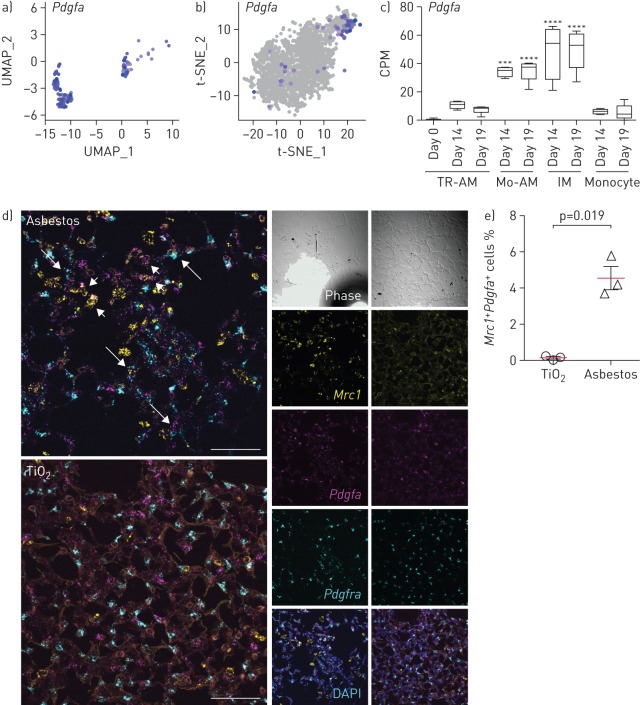
Next, we queried both our asbestos and published bleomycin [25] alveolar macrophage single-cell RNA sequencing datasets for the expression of ligands that have partnering receptors on fibroblasts and could be involved in their proliferation. Using a curated database of ligand–receptor pairs [28], we identified that macrophages from the AM3 cluster in both asbestos- and bleomycin-treated mice expressed several molecules that could drive fibroblast proliferation, including Pdgfa (figure 6a and b, and supplementary table S6). We confirmed increased expression of Pdgfa in monocyte-derived alveolar macrophages by querying our bulk RNA sequencing data from flow-sorted macrophages from the bleomycin model (figure 6c) [9]. Furthermore, we validated our findings by performing in situ RNA hybridisation for Mrc1, Pdgfa and Pdgfra after asbestos exposure, and detected double-positive Mrc1+Pdgfa+ alveolar macrophages in the areas of fibrosis in proximity to Pdgfra+ fibroblasts (figure 6d and e, and supplementary figure S6).
FIGURE 6.
Monocyte-derived alveolar macrophages (Mo-AMs) express Pdgfa which is required for fibroblast proliferation. UMAP: uniform manifold approximation and projection; t-SNE: t-distributed stochastic neighbour embedding; TR-AM: tissue-resident AM; IM: interstitial macrophage. a, b) UMAP and t-SNE plots showing expression of Pdgfa in AMs after a) asbestos and b) bleomycin exposure. c) Bar plot showing expression of Pdgfa in flow-sorted AMs during the course of bleomycin-induced pulmonary fibrosis. Data from Misharin et al. [9]. Box plot centre lines are median, box limits are upper and lower quartiles, and whiskers are minimal and maximal values. One-way ANOVA with Bonferroni correction for multiple comparisons. ***: p<0.001; ****: p<0.0001. d) In situ RNA hybridisation confirms expression of Pdgfa in AMs during pulmonary fibrosis. Analysis performed on day 28 after TiO2 or asbestos exposure. Macrophages were identified as Mrc1+ cells and fibroblasts were identified as Pdgfra+ cells. Short arrows indicate Mrc1+Pdgfa+ macrophages and long arrows indicate Pdgfra+ fibroblasts adjacent to Mrc1+Pdgfa+ macrophages. Scale bar: 50 µm. e) The percentage of Mrc1+Pdgfa+ AMs is increased after asbestos exposure. Data are from three mice per condition. Mean±sd. Mann–Whitney test.
Finally, to explore the possibility that monocyte-derived alveolar macrophages represent a link between asbestos-induced epithelial injury and fibroblast activation, we performed focused analysis of alveolar epithelial type II cells, which were well represented in our dataset. The analysis identified four subclusters. Subclusters AT2-1 and AT2-2 represented typical alveolar type II cells (figure 7a–c and supplementary figure S7). Subcluster AT2-3 contained cells from asbestos- and TiO2-treated animals, and was characterised by the expression of Krt8, Krt18, Ndnf and Lgals3 and matched recently described KRT8+ epithelial cells in the distal lung parenchyma (supplementary figure S7) [37]. Finally, cluster AT2-4 was comprised of cells found after asbestos exposure and expressed several genes linked to pulmonary fibrosis, including Retnla (RELMα), Il33 and Chia1 (supplementary figure S7d and supplementary table S7). Retnla is primarily expressed in epithelial cells and is thought to act as a chemoattractant for monocytes and monocyte-derived cells. Moreover, Retnla-deficient mice have been reported to be protected from experimental pulmonary fibrosis [38]. To see if the Retnla-expressing epithelial cells are restricted to areas of fibrosis, we treated Cx3cr1ER-Cre×ZsGreen mice with tamoxifen 14 and 15 days after administration of asbestos or TiO2 and analysed lungs using confocal immunofluorescent microscopy at day 21. Double-staining for RELMα and SPC confirmed that alveolar type II cells expressing RELMα were spatially restricted to areas of fibrosis, which also contained GFP+ monocyte-derived alveolar macrophages (figure 7e and f). Thus, our data suggest that monocyte-derived alveolar macrophages link spatially restricted epithelial injury and fibroblast proliferation.
Discussion
We performed an integrated analysis of single-cell RNA sequencing data from lung tissue of patients with pulmonary fibrosis and two murine models of lung fibrosis to identify putative mechanisms of intracellular communication within the fibrotic niche. From this analysis, we predicted that monocyte-derived alveolar macrophages drive fibrosis and are capable of self-maintenance via autocrine M-CSF/M-CSFR signalling. We used a genetic lineage tracing system and single-molecule in situ RNA hybridisation to show that Csf1- and Pdgfa-expressing monocyte-derived alveolar macrophages, Retnla-expressing epithelial cells, and Pdgfra-expressing fibroblasts are localised to fibrotic niches within the lung near asbestos fibres. We then used genetic and pharmacological strategies to causally link Csf1-expressing monocyte-derived alveolar macrophages to fibrosis. These findings demonstrate the power of a combined approach using single-cell RNA sequencing from humans and mice, spatial transcriptomics, genetic lineage tracing, and causal interventions in mouse models to unravel the complex intercellular interactions necessary for fibrosis.
Single-cell RNA sequencing is transforming our understanding of biology by revealing heterogeneity within cell populations that emerge during disease. We show that single-cell RNA sequencing can be used to combine data generated by different laboratories in different animal models of disease with homologous data from diseased patients to identify common mechanisms in disease pathogenesis [2, 25]. By using a genetic lineage tracing system that marks the ontogeny of recruited macrophages, we were able to spatially localise intercellular signals predicted from single-cell RNA sequencing data to areas of fibrosis surrounding asbestos fibres in the mouse lung. Computational consolidation of single-cell RNA sequencing datasets generated in other models of fibrosis is a promising approach to generate hypotheses with respect to fibrosis pathogenesis that can be tested with causal genetic or pharmacological interventions [39].
Our findings suggest the emergence of spatially restricted multicellular pro-fibrotic niches during pulmonary fibrosis. In these niches, injured epithelial cells drive recruitment of monocyte-derived alveolar macrophages, which, in turn, provide signals for fibroblast proliferation. Tissue-resident macrophages outside the lung rely on growth factors produced by other cells comprising the niche [40], particularly on M-CSF produced by fibroblasts [41]. In contrast, tissue-resident alveolar macrophages in the lung do not require M-CSF but do require GM-CSF for their maintenance, which is normally produced by alveolar type II cells [8, 33]. We found that monocyte-derived alveolar macrophages in mouse models of pulmonary fibrosis and (possibly monocyte-derived) pro-fibrotic alveolar macrophages in patients with pulmonary fibrosis were characterised by increased expression of Csf1r/CSF1R. Similar upregulation of CSF1R expression on fibrosis-associated macrophages can be found in the recent independent single-cell transcriptomic analysis of the lungs from patients with idiopathic pulmonary fibrosis, indicating high reproducibility of our findings [42]. Detection of Csf1/CSF1 expression in these alveolar macrophages suggests they can maintain their population in the fibrotic niche via autocrine production of M-CSF, thus becoming independent of signals from other resident lung cells for their survival as was recently postulated in an elegant computational model of fibrosis containing macrophages and fibroblasts [41, 43]. We confirmed this hypothesis by showing two pharmacological strategies to inhibit CSF1 signalling reduced the number of monocyte-derived alveolar macrophages in the niche and reduced the severity of asbestos-induced fibrosis. Autocrine M-CSF signalling may provide a mechanism by which macrophages are sustained at the “advancing front” of fibroblastic foci where normal signals from the alveolar epithelium and mesenchyme are lacking [44]. In this niche, pro-fibrotic macrophages secrete factors essential for fibroblast proliferation, including platelet-derived growth factor subunit A (PDGFA), which was also elevated in alveolar macrophages from patients with pulmonary fibrosis [45, 46]. Our results and those from another group studying radiation-induced lung fibrosis support therapeutic consideration of M-CSF/M-CSFR inhibition for the treatment of some forms of pulmonary fibrosis [47]. It is of interest that monocyte-derived alveolar macrophages recruited to the lung during bleomycin also express M-CSF and M-CSFR, but bleomycin-induced fibrosis resolves spontaneously. Combining our lineage tracing system with single-cell RNA sequencing data from resolving bleomycin-induced fibrosis and persistent asbestos-induced fibrosis might suggest how monocyte-derived alveolar macrophages change as fibrosis resolves.
Using previously validated genes as markers, we resolved three previously described transcriptionally and anatomically distinct populations of macrophages in the normal murine lung from single-cell RNA sequencing data: alveolar macrophages, peribronchial interstitial macrophages and perivascular interstitial macrophages [11, 13]. We used a genetic lineage tracing system to distinguish between tissue-resident interstitial macrophages, tissue-resident alveolar macrophages and monocyte-derived alveolar macrophages that we predicted would emerge during asbestos-induced fibrosis based on work from our group that has been confirmed by others [9, 48, 49]. All three populations were present within the fibrotic niche, but only monocyte-derived alveolar macrophages were uniquely found in areas of fibrosis. Lineage tracing studies showed these monocyte-derived alveolar macrophages do not arise from interstitial macrophages. Tissue-resident interstitial macrophages did not expand, become activated or differentiate into alveolar macrophages during fibrosis and depletion of monocyte-derived but not tissue-resident alveolar macrophages ameliorated fibrosis severity. We combined our single-cell RNA sequencing data of asbestos-induced fibrosis with a recently published single-cell RNA sequencing dataset from bleomycin-induced fibrosis [25] to demonstrate the existence of the homologous macrophage populations in both models. Moreover, we show that during the later stages of fibrosis monocyte-derived alveolar macrophages represent a transcriptionally homogenous population during fibrosis. In contrast, high-resolution profiling of the early stages of bleomycin-induced pulmonary fibrosis across multiple time-points demonstrated heterogeneity of monocyte-derived macrophages [37]. These effects are likely a combination of cell autonomous changes related to monocyte to macrophage differentiation and environmentally driven transcriptional programs related to the spatial localisation of monocyte-derived alveolar macrophages to areas of injury as we report here, and as we found in bleomycin-induced fibrosis using a distinct lineage tracing system [10].
We have previously shown that macrophage-specific deletion of Casp8 results in the death of monocytes as they differentiate into alveolar macrophages via necroptosis as evidenced by a rescue of fibrosis severity when Casp8 and Ripk3 are simultaneously deleted [9]. We took advantage of this system to genetically demonstrate a causal role for monocyte-derived alveolar macrophages in the development of asbestos-induced lung fibrosis. In contrast, deletion of tissue-resident alveolar macrophages through the administration of intratracheal liposomal clodronate prior to asbestos exposure had no effect on fibrosis severity. These findings suggest caution in interpreting the results of “alveolar macrophage-specific deletion” of genes during fibrosis. Monocyte to alveolar macrophage differentiation involves reshaping of >70% of the cellular transcriptome and requires multiple transcriptional pathways. Deletion of any gene necessary for monocyte to alveolar macrophage differentiation might therefore reduce or eliminate monocyte-derived alveolar macrophages and ameliorate fibrosis. These effects might be independent of the described function of the gene in mature alveolar macrophages [15]. For example, monocyte/alveolar macrophage deletion of Tgfb1 [50], Torc1 [51], Pparg [52], Casp8 [9] or Cflar [48] will prevent monocyte to macrophage differentiation, which likely in part explains their salutary effects on fibrosis or airway inflammation.
Our study has limitations. First, while we used a validated lineage tracing system to exclude the possibility that tissue-resident peribronchial or perivascular interstitial macrophages serve as a significant source of pro-fibrotic alveolar macrophages, it is possible that they regulate the recruitment or differentiation of monocytes in response to pro-fibrotic stimulus as has been recently described [14]. Second, while our single-cell RNA sequencing analysis was able to capture and resolve many of the canonical cell types identified in the mouse lung, several cell types, including alveolar type I cells, fibroblasts and smooth muscle cells/myofibroblasts, were under-represented in our dataset. Thus, we were not able to resolve heterogeneity within these populations during pulmonary fibrosis. Other techniques, such as enrichment for specific populations of interest or single-nucleus RNA sequencing, may address these limitations [25, 53, 54].
In summary, we found that recruitment of monocyte-derived alveolar macrophages is limited to spatially restricted fibrotic niches in pulmonary fibrosis. Genes expressed by injured epithelial cells, monocyte-derived macrophages and fibroblasts can be detected using single-cell RNA sequencing and then used to guide spatial assays (such as in situ RNA hybridisation) to reconstruct multicellular niches necessary for the development and maintenance of fibrosis. Genetically inducing deletion of monocyte-derived alveolar macrophages by necroptosis during their differentiation ameliorated lung fibrosis, causally linking them to disease pathogenesis. Single-cell RNA sequencing suggested monocyte-derived alveolar macrophages sustain themselves through M-CSF/M-CSFR signalling and the administration of antibodies to M-CSF or blockade of M-CSFR with small molecule inhibitors resulted in a loss of already established populations of monocyte-derived alveolar macrophages. While other populations, such as activated fibroblasts, could act as a source of M-CSF to sustain the population of monocyte-derived alveolar macrophages, single-cell RNA sequencing and fluorescence in situ RNA hybridisation data suggest that monocyte-derived alveolar macrophages express Csf1, raising an intriguing possibility that these cells can maintain their population via an autocrine M-CSF/M-CSFR loop. If true, this finding could be of high importance, suggesting that pathogenic monocyte-derived alveolar macrophages can sustain their population and drive fibroblast proliferation even in the absence of active epithelial injury [43]. Collectively, our results support the combination of lineage tracing, computational integration of single-cell RNA sequencing datasets from murine and human fibrosis, and in situ RNA hybridisation imaging as a powerful method to identify pathways that can be targeted for the treatment of different disease endotypes in patients with pulmonary fibrosis.
Supplementary material
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures ERJ-00646-2019_Figures.supplement (22.8MB, pdf)
Shareable PDF
Acknowledgements
This work was supported by the Office of the Assistant Secretary of Defense for Health Affairs, through the Peer Reviewed Medical Research Program under Award W81XWH-15-1-0215 to G.R.S. Budinger and A.V. Misharin. Opinions, interpretations, conclusions and recommendations are those of the authors and are not necessarily endorsed by the Dept of Defense. Next-generation sequencing on the Illumina HiSeq 4000 was performed by the NUSeq Core Facility, which is supported by the Northwestern University Center for Genetic Medicine, Feinberg School of Medicine, and Shared and Core Facilities of the University's Office for Research. Northwestern University Flow Cytometry Facility, Center for Advanced Microscopy and Pathology Core Facility are supported by NCI Cancer Center Support Grant P30 CA060553 awarded to the Robert H. Lurie Comprehensive Cancer Center. Multiphoton microscopy was performed on a Nikon A1R multiphoton microscope, acquired through the support of NIH 1S10OD010398-01. This research was supported in part through the computational resources and staff contributions provided by the Genomics Computing Cluster (Genomic Nodes on Quest), which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg's Dept of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology.
Footnotes
This article has supplementary material available from erj.ersjournals.com
Author contributions: N. Joshi: designed the study, performed experiments, analysed results, performed bioinformatics analysis, wrote the manuscript. S. Watanabe: designed the study, performed experiments, analysed results, wrote the manuscript. R. Verma: performed bioinformatics analysis, wrote the manuscript. R.P. Jablonski, C-I Chen, P. Cheresh: performed experiments, analysed results. P.A. Reyfman, N.S. Markov: performed bioinformatics analysis. A.C. McQuattie-Pimentel, Z. Lu, L. Sichizya, R. Piseaux-Aillon, A.S. Flozak: performed experiments, analysed results. D. Kirchenbuechler: performed analysis of the RNAscope experiments. C.J. Gottardi: analysed results, wrote the manuscript. C.M. Cuda, H. Perlman: developed and provided genetically modified animals, provided reagents and resources. M. Jain: provided reagents and resources, wrote the manuscript. D.W. Kamp: designed and supervised the study, provided reagents and resources, wrote the manuscript. G.R.S. Budinger and A.V. Misharin: designed and supervised the study, performed analysis, wrote the manuscript, provided funding for the project. All authors discussed the results and commented on the manuscript.
Conflict of interest: S. Watanabe has nothing to disclose.
Conflict of interest: R. Verma has nothing to disclose.
Conflict of interest: R.P. Jablonski has nothing to disclose.
Conflict of interest: C-I Chen has nothing to disclose.
Conflict of interest: P. Cheresh has nothing to disclose.
Conflict of interest: N.S. Markov has nothing to disclose.
Conflict of interest: P.A. Reyfman has nothing to disclose.
Conflict of interest: A.C. McQuattie-Pimentel has nothing to disclose.
Conflict of interest: L. Sichizya has nothing to disclose.
Conflict of interest: Z. Lu has nothing to disclose.
Conflict of interest: R. Piseaux-Aillon has nothing to disclose.
Conflict of interest: D. Kirchenbuechler has nothing to disclose.
Conflict of interest: A.S. Flozak has nothing to disclose.
Conflict of interest: C.J. Gottardi has nothing to disclose.
Conflict of interest: C.M. Cuda has nothing to disclose.
Conflict of interest: H. Perlman has nothing to disclose.
Conflict of interest: M. Jain has nothing to disclose.
Conflict of interest: D.W. Kamp has nothing to disclose.
Conflict of interest: G.R.S. Budinger has nothing to disclose.
Conflict of interest: A.V. Misharin has nothing to disclose.
Support statement: S. Watanabe is supported by MSD Life Science Foundation, Public Interest Incorporated Foundation, Japan, and David W. Cugell and Christina Enroth-Cugell Fellowship Program at Northwestern University. P.A. Reyfman is supported by Northwestern University's Lung Sciences Training Program 5T32HL076139-13 and 1F32HL136111-01A1. H. Perlman is supported by NIH grants AR064546, HL134375, AG049665 and UH2AR067687; and the United States–Israel Binational Science Foundation (2013247), the Rheumatology Research Foundation (Agmt 05/06/14), Mabel Greene Myers Professor of Medicine and generous donations to the Rheumatology Precision Medicine Fund. C.J. Gottardi is supported by NIH grant HL143800. M. Jain is supported by Veterans Administration grant BX000201. D.W. Kamp is supported by Veterans Affairs Merit Award 2IO1BX000786-05A2. G.R.S. Budinger is supported by NIH grants ES013995, HL071643 and AG049665, Veterans Administration grant BX000201 and Dept of Defense grant PR141319. A.V. Misharin is supported by NIH grants HL135124, AG049665 and AI135964 and US Dept of Defense grant PR141319. Funding information for this article has been deposited with the Crossref Funder Registry.
Conflict of interest: N. Joshi has nothing to disclose.
References
- 1.Raghu G, Remy-Jardin M, Myers JL, et al. . Diagnosis of idiopathic pulmonary fibrosis. An official ATS/ERS/JRS/ALAT clinical practice guideline. Am J Respir Crit Care Med 2018; 198: e44–e68. doi: 10.1164/rccm.201807-1255ST [DOI] [PubMed] [Google Scholar]
- 2.Reyfman PA, Walter JM, Joshi N, et al. . Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am J Respir Crit Care Med 2019; 199: 1517–1536. doi: 10.1164/rccm.201712-2410OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cugell DW, Kamp DW. Asbestos and the pleura: a review. Chest 2004; 125: 1103–1117. doi: 10.1378/chest.125.3.1103 [DOI] [PubMed] [Google Scholar]
- 4.Adamson IY, Bowden DH. Response of mouse lung to crocidolite asbestos. 2. Pulmonary fibrosis after long fibres. J Pathol 1987; 152: 109–117. doi: 10.1002/path.1711520207 [DOI] [PubMed] [Google Scholar]
- 5.Adamson IY, Bowden DH. Response of mouse lung to crocidolite asbestos. 1. Minimal fibrotic reaction to short fibres. J Pathol 1987; 152: 99–107. doi: 10.1002/path.1711520206 [DOI] [PubMed] [Google Scholar]
- 6.Roggli VL, George MH, Brody AR. Clearance and dimensional changes of crocidolite asbestos fibers isolated from lungs of rats following short-term exposure. Environ Res 1987; 42: 94–105. doi: 10.1016/S0013-9351(87)80010-5 [DOI] [PubMed] [Google Scholar]
- 7.Janssen WJ, Barthel L, Muldrow A, et al. . Fas determines differential fates of resident and recruited macrophages during resolution of acute lung injury. Am J Respir Crit Care Med 2011; 184: 547–560. doi: 10.1164/rccm.201011-1891OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guilliams M, De Kleer I, Henri S, et al. . Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 2013; 210: 1977–1992. doi: 10.1084/jem.20131199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Misharin AV, Morales-Nebreda L, Reyfman PA, et al. . Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med 2017; 214: 2387–2404. doi: 10.1084/jem.20162152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McQuattie-Pimentel AC, Ren Z, Joshi N, et al. . The aging microenvironment shapes alveolar macrophage identity in aging. bioRxiv 2019; preprint [ 10.1101/717033]. doi: 10.1101/717033 [DOI] [Google Scholar]
- 11.Gibbings SL, Thomas SM, Atif SM, et al. . Three unique interstitial macrophages in the murine lung at steady state. Am J Respir Cell Mol Biol 2017; 57: 66–76. doi: 10.1165/rcmb.2016-0361OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loyher PL, Hamon P, Laviron M, et al. . Macrophages of distinct origins contribute to tumor development in the lung. J Exp Med 2018; 215: 2536–2553. doi: 10.1084/jem.20180534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lim HY, Lim SY, Tan CK, et al. . Hyaluronan receptor LYVE-1-expressing macrophages maintain arterial tone through hyaluronan-mediated regulation of smooth muscle cell collagen. Immunity 2018; 49: 326–341. doi: 10.1016/j.immuni.2018.06.008 [DOI] [PubMed] [Google Scholar]
- 14.Chakarov S, Lim HY, Tan L, et al. . Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 2019; 363: eaau0964. doi: 10.1126/science.aau0964 [DOI] [PubMed] [Google Scholar]
- 15.Lavin Y, Winter D, Blecher-Gonen R, et al. . Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell 2014; 159: 1312–1326. doi: 10.1016/j.cell.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gibbings SL, Goyal R, Desch AN, et al. . Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood 2015; 126: 1357–1366. doi: 10.1182/blood-2015-01-624809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dostert C, Petrilli V, Van Bruggen R, et al. . Innate immune activation through Nalp3 inflammasome sensing of asbestos and silica. Science 2008; 320: 674–677. doi: 10.1126/science.1156995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.dos Santos G, Rogel MR, Baker MA, et al. . Vimentin regulates activation of the NLRP3 inflammasome. Nat Commun 2015; 6: 6574. doi: 10.1038/ncomms7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yona S, Kim KW, Wolf Y, et al. . Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013; 38: 79–91. doi: 10.1016/j.immuni.2012.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madisen L, Zwingman TA, Sunkin SM, et al. . A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 2010; 13: 133–140. doi: 10.1038/nn.2467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheresh P, Morales-Nebreda L, Kim SJ, et al. . Asbestos-induced pulmonary fibrosis is augmented in 8-oxoguanine DNA glycosylase knockout mice. Am J Respir Cell Mol Biol 2015; 52: 25–36. doi: 10.1165/rcmb.2014-0038OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruffell B, Chang-Strachan D, Chan V, et al. . Macrophage IL-10 blocks CD8+ T cell-dependent responses to chemotherapy by suppressing IL-12 expression in intratumoral dendritic cells. Cancer Cell 2014; 26: 623–637. doi: 10.1016/j.ccell.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stafford JH, Hirai T, Deng L, et al. . Colony stimulating factor 1 receptor inhibition delays recurrence of glioblastoma after radiation by altering myeloid cell recruitment and polarization. Neuro-oncology 2016; 18: 797–806. doi: 10.1093/neuonc/nov272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler A, Hoffman P, Smibert P, et al. . Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 2018; 36: 411–420. doi: 10.1038/nbt.4096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xie T, Wang Y, Deng N, et al. . Single-cell deconvolution of fibroblast heterogeneity in mouse pulmonary fibrosis. Cell Rep 2018; 22: 3625–3640. doi: 10.1016/j.celrep.2018.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stuart T, Butler A, Hoffman P, et al. . Comprehensive integration of single-cell data. Cell 2019; 177: 1888–1902. doi: 10.1016/j.cell.2019.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hafemeister C, Satija R. Normalization and variance stabilization of single-cell RNA sequencing data using regularized negative binomial regression. bioRxiv 2019; preprint [ 10.1101/576827]. doi: 10.1101/576827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramilowski JA, Goldberg T, Harshbarger J, et al. . A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun 2015; 6: 7866. doi: 10.1038/ncomms8866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.La Manno G, Soldatov R, Zeisel A, et al. . RNA velocity of single cells. Nature 2018; 560: 494–498. doi: 10.1038/s41586-018-0414-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kamp DW, Israbian VA, Yeldandi AV, et al. . Phytic acid, an iron chelator, attenuates pulmonary inflammation and fibrosis in rats after intratracheal instillation of asbestos. Toxicol Pathol 1995; 23: 689–695. doi: 10.1177/019262339502300606 [DOI] [PubMed] [Google Scholar]
- 31.Davis AP, Grondin CJ, Johnson RJ, et al. . The Comparative Toxicogenomics Database: update 2017. Nucleic Acids Res 2017; 45: D972–D978. doi: 10.1093/nar/gkw838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gautier EL, Shay T, Miller J, et al. . Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat Immunol 2012; 13: 1118–1128. doi: 10.1038/ni.2419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitsett JA, Alenghat T. Respiratory epithelial cells orchestrate pulmonary innate immunity. Nat Immunol 2015; 16: 27–35. doi: 10.1038/ni.3045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sauter KA, Pridans C, Sehgal A, et al. . Pleiotropic effects of extended blockade of CSF1R signaling in adult mice. J Leukoc Biol 2014; 96: 265–274. doi: 10.1189/jlb.2A0114-006R [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibata Y, Zsengeller Z, Otake K, et al. . Alveolar macrophage deficiency in osteopetrotic mice deficient in macrophage colony-stimulating factor is spontaneously corrected with age and associated with matrix metalloproteinase expression and emphysema. Blood 2001; 98: 2845–2852. doi: 10.1182/blood.V98.9.2845 [DOI] [PubMed] [Google Scholar]
- 36.Witmer-Pack MD, Hughes DA, Schuler G, et al. . Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J Cell Sci 1993; 104: 1021–1029. [DOI] [PubMed] [Google Scholar]
- 37.Strunz M, Simon LM, Ansari M, et al. . Longitudinal single cell transcriptomics reveals Krt8+ alveolar epithelial progenitors in lung regeneration. bioRxiv 2019; preprint [ 10.1101/705244]. doi: 10.1101/705244 [DOI] [Google Scholar]
- 38.Liu T, Yu H, Ullenbruch M, et al. . The in vivo fibrotic role of FIZZ1 in pulmonary fibrosis. PLoS One 2014; 9: e88362. doi: 10.1371/journal.pone.0088362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lotfollahi M, Wolf FA, Theis FJ. scGen predicts single-cell perturbation responses. Nat Methods 2019; 16: 715–721. doi: 10.1038/s41592-019-0494-8 [DOI] [PubMed] [Google Scholar]
- 40.Guilliams M, Scott CL. Does niche competition determine the origin of tissue-resident macrophages? Nat Rev Immunol 2017; 17: 451–460. doi: 10.1038/nri.2017.42 [DOI] [PubMed] [Google Scholar]
- 41.Zhou X, Franklin RA, Adler M, et al. . Circuit design features of a stable two-cell system. Cell 2018; 172: 744–757. doi: 10.1016/j.cell.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morse C, Tabib T, Sembrat J, et al. . Proliferating SPP1/MERTK-expressing macrophages in idiopathic pulmonary fibrosis. Eur Respir J 2019; 54: 1802441. doi: 10.1183/13993003.02441-2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adler M, Mayo A, Zhou X, et al. . Principles of cell circuits for tissue repair and fibrosis. bioRxiv 2019; preprint [ 10.1101/710012]. doi: 10.1101/710012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Herrera J, Forster C, Pengo T, et al. . Registration of the extracellular matrix components constituting the fibroblastic focus in idiopathic pulmonary fibrosis. JCI Insight 2019; 4: 125185. doi: 10.1172/jci.insight.125185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martinet Y, Rom WN, Grotendorst GR, et al. . Exaggerated spontaneous release of platelet-derived growth factor by alveolar macrophages from patients with idiopathic pulmonary fibrosis. N Engl J Med 1987; 317: 202–209. doi: 10.1056/NEJM198707233170404 [DOI] [PubMed] [Google Scholar]
- 46.Nagaoka I, Trapnell BC, Crystal RG. Upregulation of platelet-derived growth factor-A and -B gene expression in alveolar macrophages of individuals with idiopathic pulmonary fibrosis. J Clin Invest 1990; 85: 2023–2027. doi: 10.1172/JCI114669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meziani L, Mondini M, Petit B, et al. . CSF1R inhibition prevents radiation pulmonary fibrosis by depletion of interstitial macrophages. Eur Respir J 2018; 51: 1702120. doi: 10.1183/13993003.02120-2017 [DOI] [PubMed] [Google Scholar]
- 48.McCubbrey AL, Barthel L, Mohning MP, et al. . Deletion of c-FLIP from CD11bhi macrophages prevents development of bleomycin-induced lung fibrosis. Am J Respir Cell Mol Biol 2018; 58: 66–78. doi: 10.1165/rcmb.2017-0154OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aran D, Looney AP, Liu L, et al. . Reference-based analysis of lung single-cell sequencing reveals a transitional profibrotic macrophage. Nat Immunol 2019; 20: 163–172. doi: 10.1038/s41590-018-0276-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu X, Buttgereit A, Lelios I, et al. . The cytokine TGF-beta promotes the development and homeostasis of alveolar macrophages. Immunity 2017; 47: 903–912. doi: 10.1016/j.immuni.2017.10.007 [DOI] [PubMed] [Google Scholar]
- 51.Sinclair C, Bommakanti G, Gardinassi L, et al. . mTOR regulates metabolic adaptation of APCs in the lung and controls the outcome of allergic inflammation. Science 2017; 357: 1014–1021. doi: 10.1126/science.aaj2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider C, Nobs SP, Kurrer M, et al. . Induction of the nuclear receptor PPAR-gamma by the cytokine GM-CSF is critical for the differentiation of fetal monocytes into alveolar macrophages. Nat Immunol 2014; 15: 1026–1037. doi: 10.1038/ni.3005 [DOI] [PubMed] [Google Scholar]
- 53.Zepp JA, Zacharias WJ, Frank DB, et al. . Distinct mesenchymal lineages and niches promote epithelial self-renewal and myofibrogenesis in the lung. Cell 2017; 170: 1134–1148. doi: 10.1016/j.cell.2017.07.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu H, Kirita Y, Donnelly EL, et al. . Advantages of single-nucleus over single-cell RNA sequencing of adult kidney: rare cell types and novel cell states revealed in fibrosis. J Am Soc Nephrol 2019; 30: 23–32. doi: 10.1681/ASN.2018090912 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Please note: supplementary material is not edited by the Editorial Office, and is uploaded as it has been supplied by the author.
Supplementary figures ERJ-00646-2019_Figures.supplement (22.8MB, pdf)
This one-page PDF can be shared freely online.
Shareable PDF ERJ-00646-2019.Shareable (327.9KB, pdf)
Data Availability Statement
Single-cell RNA sequencing data from TiO2- and asbestos-exposed mice have been deposited with the Gene Expression Omnibus (GSE127803). We also used bulk RNA sequencing data on flow-sorted alveolar macrophages from GSE82158 [9], single-cell RNA sequencing data from patients with pulmonary fibrosis from GSE122960 [2] and mice exposed to bleomycin from GSE104154 [25]. Interactive plots from figures 4a, 4c and 7a are available for exploration at www.nupulmonary.org/resources.
FIGURE 4.
Single-cell RNA sequencing reveals pro-fibrotic monocyte-derived alveolar macrophages (Mo-AMs) during asbestos-induced pulmonary fibrosis. UMAP: uniform manifold approximation and projection; Treg: regulatory T-cell; AT1: alveolar epithelial type I cell; AT2: alveolar epithelial type II cell; Endo: endothelial cell; DC: dendritic cell; IM: interstitial macrophage; pDC: plasmacytoid DC; TR-AM: tissue-resident AM. a) UMAP plot demonstrating 26 cell clusters from 15 452 cells identified by single-cell RNA sequencing 14 days after asbestos or TiO2 exposure (one mouse per condition). b) Macrophages were identified using canonical lineage-restricted markers, such as Mrc1, as shown on the UMAP plot. c) Clusters of cells expressing Mrc1 were subset from the main dataset and re-clustered, revealing two subclusters of tissue-resident lung IMs (IM1 and IM2) and three subclusters of AMs (AM1, AM2 and AM3). d) Bar plot and e) feature plot demonstrating the composition of macrophage subclusters in cells from asbestos- and TiO2-exposed animals. f) Feature plots demonstrating expression of cluster-specific genes: Cd68 as a pan-macrophage marker, Car4 as a marker of mature TR-AMs (AM1 and AM2) and Mmp12 as a marker of Mo-AMs (AM3). Tissue-resident IMs are characterised by expression of Cx3cr1, and can be further subdivided into perivascular (Lyve1) and peribronchial (Ccr2) IMs. g) Dot plot demonstrating the expression of transcription factors differentially expressed in Mo-AMs (AM3). h) Heatmap of 93 genes overlapping between AMs and pulmonary fibrosis-associated genes from the Comparative Toxicogenomic Database (947 genes as of February 2019). Selected genes characterising clusters are shown; see supplementary figure S4e for the full list of genes. Interactive plots are available for exploration at www.nupulmonary.org/resources.
FIGURE 7.
Expression of resistin-like molecule-α (RELMα) is restricted to epithelial cells located in the areas of fibrosis. UMAP: uniform manifold approximation and projection; SPC: surfactant protein C. a) UMAP plot demonstrating subclusters of alveolar type II cells. b) UMAP plot and c) bar plot demonstrating composition of the alveolar type II cell subclusters. d) Feature plot demonstrating increased expression of Retnla in alveolar type II cells 14 days after asbestos exposure. e) Cx3cr1ER-Cre×ZsGreen mice were administered with asbestos intratracheally and treated with tamoxifen at days 14 and 15 after exposure; lungs were harvested for analysis at day 21. Representative fluorescent images showing expression of RELMα (red), SPC (blue) and CX3CR1–green fluorescent protein (green) in lungs from TiO2- or asbestos-treated animals at day 21 post-exposure. RELMα is detected in the airway epithelial cells and alveolar type II cells in the fibrotic regions in the asbestos model, but not in alveolar type II cells after TiO2 exposure. Scale bar: 100 µm. f) Enlargement of the box in (e): RELMα-positive epithelial cells (red) and monocyte-derived alveolar macrophages (green) are co-localised with asbestos fibres. Experimental design same as in (e).