Abstract
Premature ovarian failure (POF) is the pathological aging of ovarian tissue. We have previously established a cyclophosphamide-induced mouse POF model and found that cyclophosphamide caused significant damage and apoptosis of mouse ovarian granulosa cells (mOGCs). To systematically explore the molecular biologic evidence of cyclophosphamide-induced mOGC damage at the gene transcription level, RNA-Seqwas used to analyse the differences in mOGC transcriptomes between POF and control (PBS) mice. The sequencing results showed that there were 18765 differential transcription genes between the two groups, of which 192 were significantly up-regulated (log2 [POF/PBS] > 2.0) and 116 were significantly down-regulated (log2 [POF/PBS] < -4.0). Kyoto Encyclopedia of Genes and Genomes analysis found that the neuroactive ligand-receptor interaction pathway was significantly up-regulated and metabolic pathways were significantly down-regulated in the POF group. Gene Ontology analysis showed that the expression of plasma membrane, regulation of transcription and ion binding functions were significantly up-regulated in the POF group, while the expression of cell and cell parts, catalytic activity and single-organism process functions were significantly down-regulated. Finally, protein interaction analysis reveals that in the ovarian steroidogenesis pathway, three Cytochrome P450 family proteins-Cyp1a1, Cyp11a1 and Cyp2u1-interact with Fdx1 to form an interactive network. These three proteins were down-regulated in POF cells, suggesting that they are likely direct regulatory targets of cyclophosphamide. RNA-Seq high-throughput screening analysis demonstrated that cyclophosphamide damage to mOGCs was achieved through its impacts on multiple pathways and on the transcription activities of multiple target genes. Among them, the protein network consisting of the cytochrome P450 family Fdx1, Cyp17a1, Cyp11a1 and Cyp2u1 is a potential new biomarker of mOGC damage in POF in mice.
Keywords: Premature ovarian failure, ovarian granulosa cells, cyclophosphamide, RNA sequencing, transcriptomic differences
Introduction
Premature ovarian failure (POF) is a common gynecological disease that causes female infertility [1-4]. The pathological features are amenorrhea, anovulation, absence of mature follicles,significantly increased gonadotropin levels,and significantly decreased estrogen levels in women before the age of 40 [1-4]. The mechanisms of POF are complex and diverse, and genetic factors, endocrine factors, psychological factors, and autoimmune factors can all lead to its occurrence [1-5]. In addition, there is still no efficacious treatment or medicine for POF [1-4,6]. Our previous study found that the injection of cyclophosphamide could cause POF in female mice [4,7,8] and cyclophosphamide could significantly damage mouse ovarian granulosa cells (mOGCs) [4,7,8]. Although we also revealed some mechanisms at the epigenetic level, the regulatory mechanism at the level of the entire genome is not yet clear.
RNA sequencing (RNA-Seq), also known as whole transcriptome shotgun sequencing (WTSS), uses next-generation sequencing to reveal changes in RNA transcription levels in biologic samples at a particular time point [9-13]. RNA-Seq is often used to analyse changes in cellular transcripts [9-13]. In particular, it focuses onalternative splicing and transcription of genes, modification at the post-transcription level, gene fusion, and transcription differences related to single-nucleotide polymorphisms (SNPs) and mutations [9-13]. Moreover, RNA-Seq can also be used to define the boundaries of exons and introns of a gene as well as the boundaries of the previously annotated 5’ and 3’ ends [9-13]. At present, RNA-Seq has been widely used in the field of genomic regulation in embryonic development, disease mechanisms and screening of drug resistance genes [9-13].
Ferredoxin 1 (Fdx1), an iron-sulfur protein, is a mono-oxygenase that promotes cytochrome P450 enzymatic reactions. The gene encodes a protein that resides in the mitochondrial matrix, and ferredoxin reductase transfers electrons to mitochondrial cytochrome P450. There are multiple Fdx1transcripts encoding different subtypes due to alternative splicing. Fdx1 is highly expressed in adult adrenal glands and ovaries [14-19]. Cyp2u1, which encodes polypeptide 1 of subfamily u in the cytochrome P450 family 2, Cyp11a1, which encodes polypeptide 1 of subfamily a in the cytochrome P450 family 11, and Cyp17a1, which encodes polypeptide 1 of subfamily a in the cytochrome P450 family 17, all belong to the cytochrome P450 family [20]. Tissue distribution of these three genes shows a significant preference, mostly in the ovary, testis, and adrenal gland [20]. Cytochrome P450 (Cyp) represents a large family of self-oxidizing heme proteinsand is a class of mono-oxygenases, named for its specific absorption at 450 nm [20,21]. It participates in the metabolism of endogenous substances and exogenous substances, including drugs and environmental compounds. According to the degree of homology of the amino acid sequence, its members are divided into the three levels of enzymes: family, subfamily and individual [20,21]. In cells, Cyp is mainly distributed in the endoplasmic reticulum and the mitochondrial inner membrane, and it acts as a terminal oxygenase to participate in the synthesis of steroid hormones in the body [20,21]. However, the relationship between the members of the cytochrome P450 enzyme family and the development of POF is still unclear.
To systematically explore the molecular biology evidence of cyclophosphamide-inducedmOGC damage at the transcriptome level and to exploit the related biomarkers, we used RNA-Seq technology to analyse the differences inmOGC transcriptomesbetween mice in the POF group and control group (PBS).
Material and methods
Establishment of a mouse model of POF
Briefly [8], 10-week-old female C57BL/6 mice (n = 6) were purchased from the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine, China. Mice were randomized into two groups,with three mice in each group. POF mice were first injected intraperitoneally with cyclophosphamide at 70 mg/kg (Sigma-Aldrich, St Louis, USA), followed by intraperitoneal injection of cyclophosphamide at 30 mg/kg once every 2 days for 3 consecutive weeks, to construct the POF mouse model. In addition, the control group mice were injected intraperitoneally with the same amount of PBS once every 2 days for 3 consecutive weeks. The study was approved by the Ethics Committee at the Shanghai Institute of Geriatrics (SHAGESYDW2017008). All experiments are in line with China National Science and Technology Commission animal laboratory regulations.
Isolation and culture of OGCs and establishment of the in vitro injury model
Briefly [8], 10-week-old female C57BL/6 mice (n = 10) were purchased from the Experimental Animal Center of Shanghai University of Traditional Chinese Medicine. Mice were euthanized by cervical dislocation, and ovarian tissues were isolated in sterile conditions and placed in PBS at 4°C. The ovarian tissues were shredded and digested with 2.0 ml of hyaluronidase (0.1%, Sigma-Aldrich, St Louis, MO, USA) for 1 minute at 37°C. The tissue suspension was gently pipetted,addedto 200 μl of fetal bovine serum (Gibco, Gaithersburg, MD, USA) to terminate the digestion, and then filtered through a 200-mesh cell sieve. The filtrate was added to 5.0 ml of PBS and mixed, then centrifuged at 1500 r/min for 5 min at 10°C. The supernatant was discarded, and the pellet was resuspended in 5.0 ml of PBS and centrifuged at 1500 r/min for 5 min at 10°C. The supernatant was discarded, and the cell pellet was resuspended in DMEM:F12 (1:1) medium containing 15% fetal bovine serum, 10 ng/ml basic fibroblast growth factor (bFGF), 10 ng/ml epidermal growth factor (EGF), 2 mM L-glutamine, 10 ng/ml growth hormone (Gh) and 15 ng/mlestradiol (E2) (all reagents were purchased from Gibco, Gaithersburg, MD, USA). Cells were seeded in 6-well-plates and incubated at 37°C with 5% CO2 until 80% confluent. MOGCs were divided into two groups, with 3 parallel controls in each group. The cells in each POF group were treated with cyclophosphamide (IC50 concentration: 38.721 μM) for 24 hours. The PBS (control) group cells were incubated with an equal volume of PBS for 24 hours.
Hematoxylin-eosin staining
Briefly [3], all fresh tissue was soaked in 4% paraformaldehyde (Sigma-Aldrich, St. Louis, USA) for 30 minutes of fixation at room temperature, followed by ethanol gradient dehydration, paraffin embedment, sectioning (6 μm in thickness), and deparaffinization in xylene. Tissue sections were stained with haematoxylin-eosin (H & E, Sigma- Aldrich, St. Louis, USA), clarified in xylene (Sigma-Aldrich, St. Louis, USA) and mounted in neutral resin (Sigma- Aldrich, St. Louis, USA).
Western blot
Briefly [7], total protein from each group of samples was used for 12% SDS-PAGE (Bio-Rad Laboratories, Inc., California, USA); upon completion, the proteinwas transferred to a PVDF membrane (Millipore, Bedford, MA, USA). After blocking and washing the membrane, incubation with a primary antibody was carried out at 37°C for 45 min. After sufficiently washing the membrane, incubation with a secondary antibody was carried out at 37°C for 45 min. The membrane was washed 4 times, with 14 minutes per wash, with TBST (Bio-Rad Laboratories, Inc., California, USA) at room temperature. The membrane was then developed by ECL enhanced chemiluminescence (Bio-Rad Laboratories, Inc., California, USA) and exposed to Kodak XAR-5 films (Sigma-Aldrich Chemical).
Flow cytometry-PI staining analysis of the cell cycle
Briefly [8], 5 × 105/mlcells were collected and fixed in 1 ml of 70% ice-cold ethanol for 48 hours. After centrifugation at 1500 r/min for 5 min at 4°C, cell pellets were harvested and stained withPI staining solution (Sigma-Aldrich, St. Louis, USA) in the darkat 4°C for 30 min. Flow cytometry (Quanta SC, Beckman Coulter INC) was then used to analyse the cell cycle distribution of each group of cells (a total of 20,000), and data analysis was conducted using CellQuest software.
Co-IP
Briefly [22], 1 × 108/ml cells were lysed using western and IP cell lysate (Beyotime Biotechnology). A total of 800 µl of total protein sample was taken, the protein concentration was adjusted to 1 mg/ml, and 1 µg of IgG and 20 µl of fully resuspended protein a agarose (Beyotime Biotechnology, HangZhou, China) were added to the samplesand shaken slowly at 4°C for 60 minutes, followed by centrifugation at 2500 r/min for 5 minutes. The centrifuged protein supernatant was collected, to which 1 µg of primary antibody was added,and the sample was then shaken slowly at 4°C for 12 hours, followed by the addition of another 20 µl of fully resuspended protein a agarose andshaking slowly at 4°C for 3 hours. After centrifugation at 2500 r/min for 5 minutes, the supernatant was discarded, and protein a agarose was washed with ice-cold PBS three times,with 15 minutes per wash. After centrifugation, protein a agarose was added to 100 μl of western and IP cell lysate (Beyotime Biotechnology, Hang Zhou, China), incubated in a 100°C water bath for 15 minutes and centrifuged at 12000 r/min for 10 minutes. The supernatant was collected and stored at -80°C.
RNA extraction and quantitative analysis
Total RNA was extracted from each group of cells according to theTRIzolmanual (Invitrogen). Subsequently, to each RNA sample, 10 U of DNase I (Sigma) was added,and the samples were incubated at 37°C for 30 min to remove residual DNA. The mRNA in the total RNA samples was isolated and purified using anOligotex mRNA Midi Kit (Qiagen). Quantification of the RNA concentration and integrity was determined using an Agilent 2100 Bioanalyzer and an Agilent RNA 6000 Nano Kit.
Establishment of cDNA sequencing libraries and high-throughput RNA-Seq
The following analysis was conducted by Shanghai FengHe InfoTech Ltd (Shanghai, China). According to their experimental procedures, a random fragment sequencing library was constructed using aSOLiD Whole Transcriptome Analysis Kit (Life technologies). Nucleic acid cleaving reagents were added, and the mRNA was randomly disrupted into short segments in a shaking incubator. First-strand cDNA was reverse transcribed using the fragmented mRNA as the template. Second-strand cDNA was synthesized using a second-strand DNAsynthesis reaction system consisting of DNA polymerase I, dNTPs and RNase H (Sigma). The synthesized DNA was purified using a DNA purification kit and recovered. Thebase ‘A’ was added to the 3’ end of the cDNA, followed by ligation to the adapter, to complete the blunt end repair reaction. Subsequently, DNA fragment size selection was performed. Finally, the cDNA was used for PCR amplification to obtain a sequencing library. The constructed library was qualified using an Agilent 2100 Bioanalyzer and the ABI StepOnePlus Real-Time PCR System and was subjected to high-throughput sequencing using an Illumina HiSeq™ 2000 Sequencer after passing quality control.
Statistical analysis
Each experiment was performed as least three times, and data are shown as the mean ± SE where applicable, and differences were evaluated using Student’s t-tests. P < 0.05 was considered significant.
Results
Cyclophosphamide significantly enhances OGC injury and apoptosis
Histopathological analysis of HE stained samples showed that ovarian tissues of mice in the POF group had severe atrophy, reduced volume, and dense interstitial areas (Figure 1). The proportion of atretic follicles out of the total number of follicles was significantly increased (49.51% ± 10.79%), while the proportion of normal follicles out of the total number of follicles was significantly decreased (50.49% ± 10.79%) (Figure 1). In contrast, many normal follicles (74.36% ± 4.72%) were found in the ovaries of mice in the PBS group (control group), whereas atretic follicles were rare (25.64% ± 4.72%) (Figure 1). In addition, the cell cycle analysis of OGCs by flow cytometry indicated that the number of S phase OGCs in the POF group was significantly decreased, while the number of OGCs in the G2/M phase was significantly increased. Due to the large number of cells arrested in the G2/M phase, the rate of cell cycle progression declined (Figure 1). mOGCs of WT adult C57 mice were isolated in vitro and were also purified and cultured. ThemOGCin vitro injury model was prepared using cyclophosphamide (POF group). Western blot results showed that mOGCs in the POF group expressed significantly higher levels of activated Caspase 3 fragment (ΔCaspase 3) and phosphorylated H2A.X (pho-H2A.X) proteinsthan did mOGCs in the PBS group, suggesting that cells entered the aging and apoptosis stage (Figure 1). However, mOGCs in the POF group showed significantly lower expression of AMH and Inhibin B proteins than did mOGCs in the PBS group, indicating a decrease in cellular health quality (Figure 1). The results suggest that cyclophosphamide significantly promotesOGC injury and apoptosis.
Figure 1.
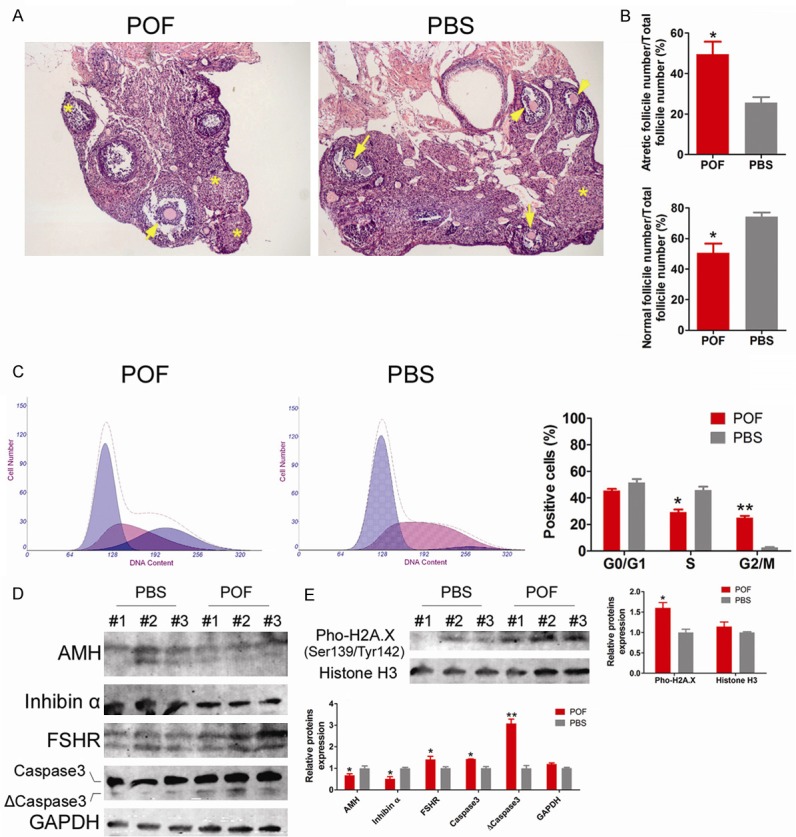
Cyclophosphamide significantly induced POFoccurrence and OGC damage and apoptosis. A. Histopathologic analysis of H & E staining showed that the POF group mice had severe ovarian atrophy, significantly increased atretic follicles (indicated by *), significantly reduced numbers of normal follicles in various stages (indicated by the black arrow), reduced ovarian volume, and dense interstitial area. Magnification is 200 ×. B. The proportion of ovarian atretic follicles in the POF group was significantly higher than that in the PBS group, and the proportion of normal follicles in the POF group was significantly lower than that in the PBS group. *P < 0.05 vs. PBS group, n = 3. C. Flow cytometry cell cycle analysis showed that the number of OGCs in the S phase of the cell cycle was significantly decreased in the POF group, while the number in the G2/M phase was significantly increased in the POF group. **P < 0.01 vs. PBS group, *P < 0.05 vs. PBS group, n = 3. D. Western blot results showed that the mOGCs of the POF group expressed significantly higher levels of activated Caspase 3 fragment (ΔCaspase 3) protein than did the mOGCs of the PBS group, while expressing significantly lower levels of AMH and Inhibin α proteins. **P < 0.01 vs. PBS group, *P < 0.05 vs. PBS group, n = 3. E. Western blot results showed that the mOGCs of the POF group expressed significantly higher levels of phosphorylated H2A.X (pho-H2A.X) protein than did the mOGCs of the PBS group, while expressing significantly lower levels of AMH and Inhibin α proteins. *P < 0.05 vs. PBS group, n = 3.
Cyclophosphamide causes transcription changes and functional disruptions of multiple mOGCs genes
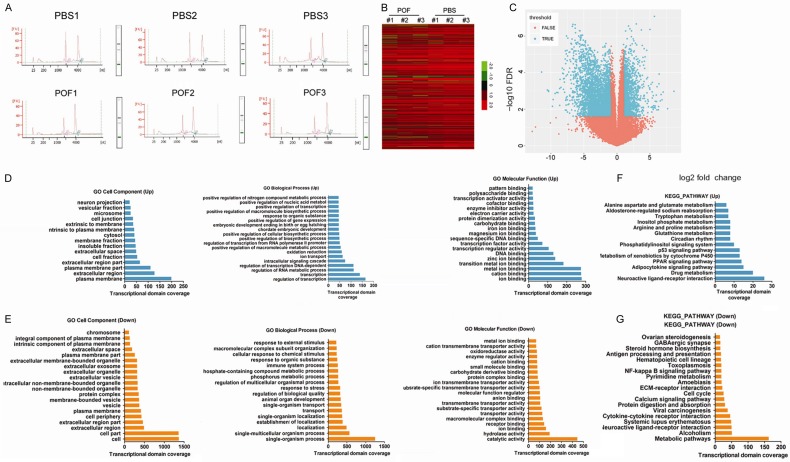
We first used chromatographic analysis to confirm that the RNA derived from each group of mOGCs had high purity (1.8 < OD260/OD280 < 2.0) and met the concentration requirementsfor RNA-Seq (Figure 2). Subsequently, RNA-Seq techniques were used to analyze the differences in mOGCtranscriptomes between POF and control (PBS) mice. After obtaining the RNA-Seq sequencing results, the raw data were statistically analyzed and calibrated. First, we removedreads for barcode sequences and adapter sequences; removedreads with > 5% N content; removed consecutive bases atthe 5’ and 3’ ends with quality less than 10; removed low quality reads (where the number of bases with quality < 20 was greater than 20% of read length); and removed reads less than 30 bases in length. Through the above processes, we obtained clean data from six samples, and follow-up statistical induction and in-depth data mining analysis were conducted using these clean reads. We first used TopHat software (v2.0.8) and aligned the clean reads to the mouse reference genome GRCm38 using the default parameters. We then used theStringtie tool (V1.2.2) and obtained the raw reads information of each mouse gene alignment according to mouse gene annotation information provided by Gencode. The Limmapackage method of the R language was used to screen for genes with significantly different expression between the sample and control. Finally, corrected RNA-Seq results showed that there were 18,765 differentially transcribed genes between the two groups (Figure 2; Table S1), of which 192 were significantly up-regulated (log2 [POF/PBS] > 2.0) and 116 were significantlydown-regulated (log2 [POF/PBS] < -4.0). Subsequently, differentially transcribed genes were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis using the DAVID online tool [23]. GO analysis suggested that transcription activities of genes related to the plasma membrane, extracellular region, cation binding, regulation of transcription, regulation of RNA metabolic process and ion binding functions were significantly increased in the POF group (Figure 2; Table S2), while transcription activities of genes related to the cell, cell parts, extracellular region, catalytic activity, single-organism process functions and single-multicellular organism process were significantly down-regulated (Figure 2; Table S2). KEGG analysis found that in the POF group, the transcription activities of genes belonging to the neuroactive ligand-receptor interaction and drug metabolism categories were significantly up-regulated, but genes belonging to the metabolic pathways category were significantly down-regulated (Figure 2; Table S3). The experimental results suggest that cyclophosphamide led to significanttranscription changes and functional disruptions in multiple mOGC genes.
Figure 2.
Cyclophosphamide led to transcription changes and functional disorders in multiple mOGC genes. A. Quantification of the purity and concentration of RNA of mOGCs from each group (1.8 < OD260/OD280 < 2.0). B. RNA-Seq data are graphically displayed using the clustering index, with green representing genes with down-regulated transcription levels and red representing genes with up-regulated transcription levels. C. The relationship between differential analysis tests based on negative binomial distributions (P value, FPR) and differential transcripts. The differential transcript region in blue is the reliable region; the transcript region of the red portion is a non-feasible region. D. Results of Gene Ontology (GO) analysis of differential transcripts with up-regulated expression. E. Results of GO analysis of differential transcripts with down-regulated expression. F. Results of Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differential transcripts with up-regulated expression. G. Results of KEGG enrichment analysis of differential transcripts with down-regulated expression.
Cyclophosphamide down-regulates the transcription activity of genes involved in mOGC ovarian steroid metabolism in a targeted fashion
Through KEGG pathway analysis, we identified a group of genes in POF mOGCs belonging to the ovarian steroidogenesis function withsignificantly reduced transcription activities compared to the control group (P < 0.01). Of the 122 genes, we screened 20 genes with log2 [POF/PBS] < -3.0 (Figure 3). Using the String V10.5 (https://string-db.org/) online analysis tool [24,25], wepredicted whether there were some relationshipsamongthe proteins encoded by these genes. The results of the analysis showed that the protein products of 10 genes (Hynu, Kmo, Hsd3d5, Hsd3b2, Cyp2u1, Cyp12a1, Cyp11a1, Hsd17d1, Fdx1, and Nos2) interacted with each other (Figure 3). We also found that the Fdx1 protein is an important common node that can simultaneously interact with proteins such as Cyp2u1, Cyp12a1 and Cyp11a1 and bind, catalyse and activate their activities (Figure 3). To verify the above results, co-immunoprecipitation (IP)western blot was used to detect protein interactions. The experimental results showed that if theanti-Fdx1 antibody (α Fdx1 ab) was used for IP, then the expression signals for the remaining proteins such as Cyp2u1, Cyp12a1 and Cyp11a1 could be detected both in the POF group and in the PBS group (Figure 3). However, in the POF group, the expression levels of Fdx1, Cyp2u1, Cyp12a1, and Cyp11a1 were significantly lower than those in the PBS group (Figure 3). Our results reveal that cyclophosphamide significantly down-regulates the transcription activity of genes involved in the ovarian steroid metabolic function in a targeted fashion in mOGCs as well as the expression level of protein-protein interaction networks.
Figure 3.
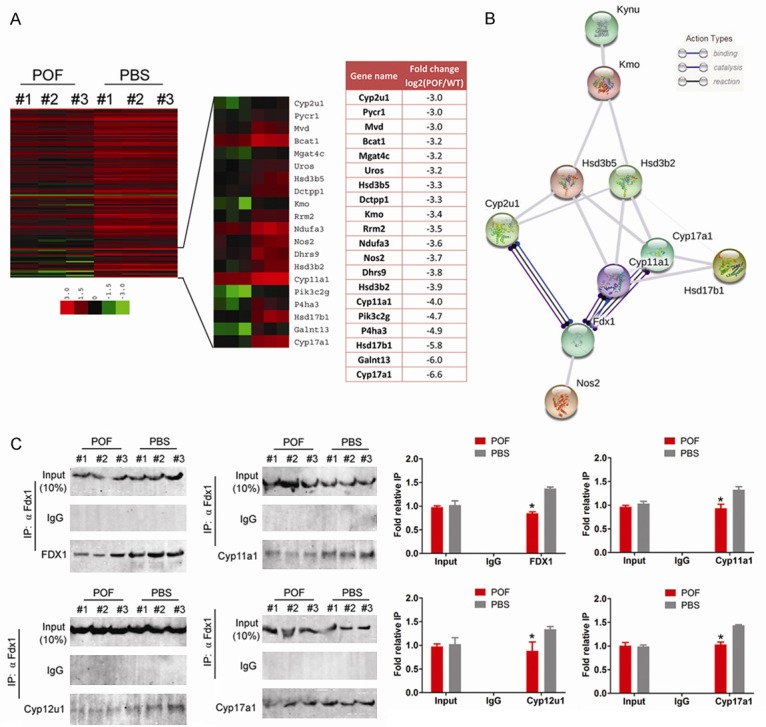
Cyclophosphamide down-regulated transcription activities of cytochrome P450 family genes involved in ovarian steroid metabolism in a targeted fashion in mOGCs. A. Clustered index results of differentially expressed genes involved in ovarian steroid genesis. KEGG pathway analysis identified a group of genes belonging to the functional group of ovarian steroid genesis, and its transcription activity was significantly lower in POF mOGCs than in the control group. Of the 122 genes, we screened 20 genes with log2 [POF/PBS] < -3.0. B. Prediction results using the String online tool for the differential transcripts suggest that there is interaction between the 10 proteins of the ovarian steroid genesis functional group. C. Co-IP western blot results show that the expression levels of Fdx1, Cyp2u1, Cyp12a1, and Cyp11a1 in the POF group are significantly lower than those in the PBS group, *P < 0.05 vs. PBS group, n = 3.
Discussion
POF is a disease with serious consequences for the reproductive health of women [1,2,26]. The pathogenesis of POF is diverse, involving genetic abnormalities, induction by environmental toxins and chemotherapy drugs, immune system abnormalities, and endocrine disorders caused by mental stress [1,2,4,26-28].However, its exact cause is still unclear. In previous studies, we first established a model of cyclophosphamide-induced POF in mice and a model of immunosuppressant tripterygium glycosides-induced POF in rats [3,4,22]. The pathologic features of these two models, such as ovarian atrophy, apoptosis or necrosis of OGCs, increased atretic follicles, and severe decline in peripheral blood estrogen levels and significantly elevated FSH levels are consistent with human POF [1,2]. This similarity suggests that the above two models are ideal and practical animal models to investigate drug-induced ovarian insufficiency. Although we have established animal models of POF, the mechanism of drug-induced POF has not yet been clearly established. In addition, genes with altered expression during the development of POF have not been clearly identified. OGCs play a very important role in oocyte maturation and ovarian function, as well as sex hormone release and endocrine maintenance, and our previous study has clarified that the apoptosis and necrosis of OGCs are important causes ofPOF [8,22]. Therefore, in this study, we decided to use OGCs as the source material. We focused on the differential gene expression profiles in OGCs after cyclophosphamide treatment to obtain complete gene expression profile information as the basis for further study of biological effects. Therefore, we chose RNA-Seqas a means tostudy the gene transcription profile of OGCs. RNA-Seq uses high-throughput sequencing technology to analyse the sequences of cDNA derived from RNA reverse transcription and PCR amplification. With this technology, almost all transcripts in a specific organ or tissue of a particular species in a particular state can be obtained rapidly and comprehensively at the single-nucleotide level [10,29,30]. Compared with traditional subtractive hybridization, suppression subtractive hybridization, and cDNA microarray, RNA-Seqhas the following advantages: compared with the traditional cDNA microarray hybridization screening technology, RNA-Seq does not need nucleic acid probes, and it is not necessary to know in advance the nucleic acid sequence of the gene to be sequenced. Therefore, RNA-Seq can use the sequencing method to conductfull transcriptome analysis of species with unknown genomesand to obtain differential information of gene transcripts. In addition, there are no issuesrelated to cross-reaction and background noise caused by the fluorescence analogue signal of traditional microarray hybridization, which greatly improves the resolution [10,29,30]. In addition, RNA-Seq has the obvious advantages of high throughput, low cost, and high sensitivity, and it enables us to obtain information about genes with low-expression levels [10,29,30]. We isolated OGCs from wild-type C57 mice and prepared the in vitro model of cyclophosphamide-induced apoptosis in OGCs. The RNA-Seq results showed that there were 18,765 differentially transcribed genes between the cyclophosphamide group and control group, of which 192 were significantly up-regulated and 116 were significantly down-regulated. In-depth analysis showed that the transcription activities of genes involved in the plasma membrane and extracellular region, regulation of transcription, RNA and drug metabolism and ion binding functions were significantly increased, while transcription activities of genes involved in the cells, catalytic activity, organism process and metabolic pathway functions were significantly down-regulated. Experimental data suggest that cyclophosphamide leads to significant transcription changes and functional disordersin multiple mOGCs genes.
In cells, cytochrome P450 is mainly distributed in the endoplasmic reticulum and the mitochondrial inner membrane, and it acts as a terminal oxygenase in the synthesis of steroid hormones in the body [20,21]. Some studies have shown that cytochrome P450 is a key enzyme in the metabolism of drugs and has a significant impact on cytokines and thermoregulation [20,21]. In the body, Cyp containing iron ions binds to drug molecules and accepts an electron delivered from NADPH-P450 reductase, which converts iron ions to divalent ferrous ions [20,21]. Subsequently, it binds with one molecule of oxygen, one proton, and the second electron to form the Fe2+OOH·DH complex, which binds to another proton to produce water and iron oxide complex (FeO)3+·DH [20,21]. (FeO)3+·DH extracts a hydrogen atom from ·DH to form a pair of transient free radicals. The oxidized drug is released from the complex, and the P450 enzyme is regenerated [20,21]. Dasari et al. found that mouse mitochondrial CYP1A1 is an outer membrane protein that exhibits high affinity for FDX1 and mediates the N-terminal demethylation of a wide range of tricyclic drugs such as anti-depressant drugs, analgesics, and anti-psychotic drugsby binding to FDXR [31]. This study confirms that thebinding ability between CYP1A1 and FDX1 is strong [31]. Roumaud et al. reported that in the mouse testicular stromal MA-10cells, the transcription factors SF1 and cJUNcould act together at a specific site of the Fdx1 promoter and activate its transcription [32]. Subsequently, the Fdx1 protein supports steroid biosynthesis in cells through electron transfer to the rate-limiting enzyme CYP11A1. CYP11A1 catalyses the conversion of cholesterol into pregnenolone in the mitochondria via sidechain cleavage [32]. The above study confirms that CYP1A1 and FDX1 are involved in steroid hormone synthesis [32]. In the present study, high-throughput RNA-Seq analysis showed that the transcription activities of the Cyp2u1, Cyp11a1, Cyp17a1 and Fdx1 genes were all significantly decreased after cyclophosphamide treatment of OGCs. Although the degree of decline was not the same, the trend was the same. In addition, coupled with the results of the protein-protein interaction network and several previous studies, we have reason to believe that the protein products of these four genes interact with each other. After cyclophosphamide treatment in mice, pathologic features of POF appeared randomly, and one of the most important phenomena was the significant decrease of AMH and E2 in OGCs, which marked the diminished ability of OGCs to synthesize hormones. It is likely that cyclophosphamide, by inhibiting the transcription activity of these four genes, eventually leads to a decrease in the ability of OGCs to synthesize and release hormones.
RNA-Seq high-throughput screening analysis demonstrated that the damage tomOGCsby cyclophosphamide was achieved through its impacts on multiple pathways and the transcription activities of multiple target genes. Among them, the protein network consisting of the cytochrome P450 family membersFdx1, Cyp17a1, Cyp11a1, and Cyp2u1 is a new potential biomarker of OGC damage in POF in mice.
Acknowledgements
This work was supported by grant from Shanghai Natural Science Foundation (No 16ZR1434000). And, grant from the projects sponsored by the development fund for Shanghai talents (2017054). And, grant from the projects sponsored by the fund for Xinglin talents of Shanghai University of TCM (201707081). And, grant from the graduate student innovation training project of Shanghai University of Traditional Chinese Medicine (Y201860). We declared no potential conflicts of interest.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Beck-Peccoz P, Persani L. Premature ovarian failure. Orphanet J Rare Dis. 2006;1:9. doi: 10.1186/1750-1172-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vujovic S, Ivovic M, Tancic-Gajic M, Marina L, Barac M, Arizanovic Z, Nenezic A, Ivanisevic M, Micic J, Sajic S, Micic D. Premature ovarian failure. Srp Arh Celok Lek. 2012;140:806–811. [PubMed] [Google Scholar]
- 3.Liu T, Huang Y, Zhang J, Qin W, Chi H, Chen J, Yu Z, Chen C. Transplantation of human menstrual blood stem cells to treat premature ovarian failure in mouse model. Stem Cells Dev. 2014;23:1548–1557. doi: 10.1089/scd.2013.0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu T, Wang S, Li Q, Huang Y, Chen C, Zheng J. Telocytes as potential targets in a cyclophosphamide-induced animal model of premature ovarian failure. Mol Med Rep. 2016;14:2415–2422. doi: 10.3892/mmr.2016.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Liu TE, Wang S, Zhang L, Guo L, Yu Z, Chen C, Zheng J. Growth hormone treatment of premature ovarian failure in a mouse model via stimulation of the Notch-1 signaling pathway. Exp Ther Med. 2016;12:215–221. doi: 10.3892/etm.2016.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu T, Huang Y, Guo L, Cheng W, Zou G. CD44+/CD105+ human amniotic fluid mesenchymal stem cells survive and proliferate in the ovary long-term in a mouse model of chemotherapy-induced premature ovarian failure. Int J Med Sci. 2012;9:592–602. doi: 10.7150/ijms.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu T, Qin W, Huang Y, Zhao Y, Wang J. Induction of estrogen-sensitive epithelial cells derived from human-induced pluripotent stem cells to repair ovarian function in a chemotherapy-induced mouse model of premature ovarian failure. DNA Cell Biol. 2013;32:685–698. doi: 10.1089/dna.2013.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xiong Y, Liu T, Wang S, Chi H, Chen C, Zheng J. Cyclophosphamide promotes the proliferation inhibition of mouse ovarian granulosa cells and premature ovarian failure by activating the lncRNA-Meg3-p53-p66Shc pathway. Gene. 2017;596:1–8. doi: 10.1016/j.gene.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 9.Chu Y, Corey DR. RNA sequencing: platform selection, experimental design, and data interpretation. Nucleic Acid Ther. 2012;22:271–274. doi: 10.1089/nat.2012.0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, Sam L, Barrette T, Palanisamy N, Chinnaiyan AM. Transcriptome sequencing to detect gene fusions in cancer. Nature. 2009;458:97–101. doi: 10.1038/nature07638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat Protoc. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kukurba KR, Montgomery SB. RNA Sequencing and analysis. Cold Spring Harb Protoc. 2015;2015:951–969. doi: 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, Patterson N, Lander ES, Mann M. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- 15.Dickinson ME, Flenniken AM, Ji X, Teboul L, Wong MD, White JK, Meehan TF, Weninger WJ, Westerberg H, Adissu H, Baker CN, Bower L, Brown JM, Caddle LB, Chiani F, Clary D, Cleak J, Daly MJ, Denegre JM, Doe B, Dolan ME, Edie SM, Fuchs H, Gailus-Durner V, Galli A, Gambadoro A, Gallegos J, Guo S, Horner NR, Hsu CW, Johnson SJ, Kalaga S, Keith LC, Lanoue L, Lawson TN, Lek M, Mark M, Marschall S, Mason J, McElwee ML, Newbigging S, Nutter LM, Peterson KA, Ramirez-Solis R, Rowland DJ, Ryder E, Samocha KE, Seavitt JR, Selloum M, Szoke-Kovacs Z, Tamura M, Trainor AG, Tudose I, Wakana S, Warren J, Wendling O, West DB, Wong L, Yoshiki A International Mouse Phenotyping Consortium; Jackson Laboratory; Infrastructure Nationale PHENOMIN, Institut Clinique de la Souris (ICS); Charles River Laboratories; MRC Harwell; Toronto Centre for Phenogenomics; Wellcome Trust Sanger Institute; RIKEN BioResource Center. MacArthur DG, Tocchini-Valentini GP, Gao X, Flicek P, Bradley A, Skarnes WC, Justice MJ, Parkinson HE, Moore M, Wells S, Braun RE, Svenson KL, de Angelis MH, Herault Y, Mohun T, Mallon AM, Henkelman RM, Brown SD, Adams DJ, Lloyd KC, McKerlie C, Beaudet AL, Bucan M, Murray SA. High-throughput discovery of novel developmental phenotypes. Nature. 2016;537:508–514. doi: 10.1038/nature19356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hansen J, Floss T, Van Sloun P, Fuchtbauer EM, Vauti F, Arnold HH, Schnutgen F, Wurst W, von Melchner H, Ruiz P. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A. A conditional knockout resource for the genome-wide study of mouse gene function. Nature. 2011;474:337–342. doi: 10.1038/nature10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diez-Roux G, Banfi S, Sultan M, Geffers L, Anand S, Rozado D, Magen A, Canidio E, Pagani M, Peluso I, Lin-Marq N, Koch M, Bilio M, Cantiello I, Verde R, De Masi C, Bianchi SA, Cicchini J, Perroud E, Mehmeti S, Dagand E, Schrinner S, Nurnberger A, Schmidt K, Metz K, Zwingmann C, Brieske N, Springer C, Hernandez AM, Herzog S, Grabbe F, Sieverding C, Fischer B, Schrader K, Brockmeyer M, Dettmer S, Helbig C, Alunni V, Battaini MA, Mura C, Henrichsen CN, Garcia-Lopez R, Echevarria D, Puelles E, Garcia-Calero E, Kruse S, Uhr M, Kauck C, Feng G, Milyaev N, Ong CK, Kumar L, Lam M, Semple CA, Gyenesei A, Mundlos S, Radelof U, Lehrach H, Sarmientos P, Reymond A, Davidson DR, Dolle P, Antonarakis SE, Yaspo ML, Martinez S, Baldock RA, Eichele G, Ballabio A. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011;9:e1000582. doi: 10.1371/journal.pbio.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choudhary D, Jansson I, Stoilov I, Sarfarazi M, Schenkman JB. Expression patterns of mouse and human CYP orthologs (families 1-4) during development and in different adult tissues. Arch Biochem Biophys. 2005;436:50–61. doi: 10.1016/j.abb.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 21.Meunier B, de Visser SP, Shaik S. Mechanism of oxidation reactions catalyzed by cytochrome p450 enzymes. Chem Rev. 2004;104:3947–3980. doi: 10.1021/cr020443g. [DOI] [PubMed] [Google Scholar]
- 22.Liu TE, Zhang L, Wang S, Chen C, Zheng J. Tripterygium glycosides induce premature ovarian failure in rats by promoting p53 phosphorylation and activating the serine/threonine kinase 11-p53-p21 signaling pathway. Exp Ther Med. 2015;10:12–18. doi: 10.3892/etm.2015.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. David: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 24.Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, Lin J, Minguez P, Bork P, von Mering C, Jensen LJ. STRING v9.1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–815. doi: 10.1093/nar/gks1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Correction to: Cytogenetic analysis of 179 Iranian women with premature ovarian failure. Gynecol Endocrinol. 2013;29:727. doi: 10.3109/09513590.2013.788625. [DOI] [PubMed] [Google Scholar]
- 27.Bandyopadhyay S, Chakrabarti J, Banerjee S, Pal AK, Goswami SK, Chakravarty BN, Kabir SN. Galactose toxicity in the rat as a model for premature ovarian failure: an experimental approach readdressed. Hum Reprod. 2003;18:2031–2038. doi: 10.1093/humrep/deg414. [DOI] [PubMed] [Google Scholar]
- 28.Qin Y, Sun M, You L, Wei D, Sun J, Liang X, Zhang B, Jiang H, Xu J, Chen ZJ. ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet J Rare Dis. 2012;7:5. doi: 10.1186/1750-1172-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tariq MA, Kim HJ, Jejelowo O, Pourmand N. Whole-transcriptome RNAseq analysis from minute amount of total RNA. Nucleic Acids Res. 2011;39:e120. doi: 10.1093/nar/gkr547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Richard H, Schulz MH, Sultan M, Nurnberger A, Schrinner S, Balzereit D, Dagand E, Rasche A, Lehrach H, Vingron M, Haas SA, Yaspo ML. Prediction of alternative isoforms from exon expression levels in RNA-Seq experiments. Nucleic Acids Res. 2010;38:e112. doi: 10.1093/nar/gkq041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dasari VR, Anandatheerthavarada HK, Robin MA, Boopathi E, Biswas G, Fang JK, Nebert DW, Avadhani NG. Role of protein kinase C-mediated protein phosphorylation in mitochondrial translocation of mouse CYP1A1, which contains a non-canonical targeting signal. J Biol Chem. 2006;281:30834–30847. doi: 10.1074/jbc.M510725200. [DOI] [PubMed] [Google Scholar]
- 32.Roumaud P, Rwigemera A, Martin LJ. Transcription factors SF1 and cJUN cooperate to activate the Fdx1 promoter in MA-10 Leydig cells. J Steroid Biochem Mol Biol. 2017;171:121–132. doi: 10.1016/j.jsbmb.2017.03.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.