Abstract
Increasing evidence points to oxidative stress as a chief mediator of Parkinson’s disease (PD) characterized by progressive loss of dopamine neurons in the pars compacta of the substantia nigra. At present, microRNAs (miRNAs) have been recognized as important regulators in oxidative stress. Furthermore, miRNAs were also involved in the neuropathology of neurodegenerative disorders, including PD. In this study, we aimed to explore the influences of miR-153 and Nrf2 in oxidative stress during the development of PD. It was found that the expression of miR-13 and Nrf2 detected by qRT-PCR were significantly increased and decreased, respectively, in serum of PD patients and MPP+-induced SH-SY5Y cells. The target relationship between miR-153 and Nrf2 was determined by dual-luciferase assay. Moreover, after transfecting with miR-153 mimics and inhibitor, the expressions of Nrf2 in mRNA and protein were down-regulated and up-regulated, respectively. The indexes of oxidative stress were examined by biochemical methods. The data revealed that miR-153 could facilitate oxidative stress by negatively regulating Nrf2 in MPP+-treated SH-SY5Y cells. Finally, it was observed that miR-153 could suppress the Nrf2/HO-1 signaling pathway in MPP+-treated SH-SY5Y cells. Therefore, these findings indicated that overexpression of miR-153 could promote oxidative stress in PD by targeting the Nrf2/HO-1 signaling pathway, possibly providing a new way to treat PD.
Keywords: Parkinson’s disease (PD), microRNA-153 (miR-153), nuclear factor-E2-related factor 2 (Nrf2), oxidative stress
Introduction
Parkinson’s disease (PD) is a neurodegenerative disorder characterized clinically by tremors, rigidity (muscle stiffness), akinesia (loss or impairment of voluntary movements), bradykinesia (slowness of movement), poor balance, and difficulty in walking (Parkinsonian gait) [1]. In addition to the above-described motor symptoms, non-motor symptoms that include olfactory dysfunction, rapid eye movement disorder (IRBD), behavior disorder, depression, as well as cognitive impairments, are pressing issues [2]. Moreover, the most typical pathologic features of PD are degeneration of structure or function of dopaminergic neurons mainly located in the pars compacta of the substantia nigra, which eventually causes the biochemical abnormality of low levels of dopamine and the generation of intraneuronal protein aggregates called Lewy bodies [3,4]. Epidemiological survey revealed that approximately 1.0% of the population over the age of 60 suffered from PD around the world; meanwhile the number of new cases diagnosed is also gradually increasing every year [5]. However, there is still no effective cure for PD [6]. Previous experimental observations from postmortem brain analyses suggested that excessive production of reactive oxygen species (ROS) or reactive nitrogen species (RNS), which subsequently activated oxidative damage and mitochondrial dysfunction, play a central role in the neuropathology of PD [7,8]. Furthermore, in addition to PD, several other neurodegenerative disorders, including Alzheimer’s disease (AD), Huntington’s disease (HD), and amyotrophic lateral sclerosis (ALS) are closely associated with oxidative stress although there were a variety of distinct pathologic and clinical characteristics, implying that oxidative stress might be a dominant mechanism contributing to neuronal degeneration [9,10]. Therefore, neuroprotective antioxidant compounds were considered as a therapeutic strategy for inhibiting free radicals and oxidative damage in the development of PD [6].
microRNAs (miRNAs) are a class of 21~24 nucleotide long, single-stranded non-coding RNAs that regulate gene expression at the posttranscriptional level in a sequence-specific manner [11,12]. Recent data reported that miRNAs were involved in the development of the central nervous system and in neurodegeneration [13,14]. Therefore, it comes as no surprise that aberrant expression of miRNAs contributes to neurodevelopmental disorders and neurodegenerative diseases, including PD [15]. For example, miR-133b presented a decreased expression pattern in PD patients and physiologically targeted a transcription factor Pitx3 to regulate the development of dopaminergic neurons which were the main affected neurons in PD [16]. Decreased levels of miR-34b and miR-34c were observed in the frontal cortex, cerebellum and substantia nigra of PD brains, and they could indirectly reduce the expressions of both Parkin and DJ-1, as well as increase the rate of cell death [17,18]. Thus, to further investigate the possible role of miRNAs in pathogenesis of PD might provide novel diagnostic, prognostic and therapeutic strategies.
Currently, miR-153 has been demonstrated as highly expressed in mouse brain tissue [19]. However, its role in brain tissue is still unknown. The nuclear factor-E2-related factor 2 (Nrf2), a transcription factor with a high sensitivity to oxidative stress, binds to antioxidant response element (ARE) in the nucleus and promotes the transcription of a wide variety of antioxidant genes [20]. Moreover, it has been revealed that oxidative stress involving changes in Nrf2 and endoplasmic reticulum (ER) stress may constitute early events in AD pathogenesis, which provides a new strategy to study AD [21]. A prior study reported that miR-153 could negatively regulate Nrf2 in breast carcinogenesis [22]. Meanwhile, it also uncovered that the inhibition of miR-153 protected neurons against oxygen-glucose deprivation and reoxygenation (OGD/R)-induced injury by regulating Nrf2/heme oxygenase-1 (HO-1) signaling, which indicated that miR-153-mediated Nrf2/HO-1 pathway might be a therapeutic target for cerebral ischemia/reperfusion injury [23]. Nevertheless, the effects of miR-153 and Nrf2 on development of PD remain unclear. Hence, in this study, we aimed to investigate this problem.
Materials and methods
Ethical statement and tissue samples
The present study was approved by the Ethical Committee of The Seventh Affiliated Hospital of Sun Yat-sen University (Guangzhou, China). All patients provided written consent and were informed of the purposes of the study. About 5 ml of fresh whole-blood specimen was obtained from 5 PD patients who have been made a definite diagnosis by 3 clinicians at least and 1 health person between July 2016 and January 2017. Subsequently, the serum was immediately segregated by 3000 rmp centrifugation for 10 min and stored at -80°C for following quantitative reverse transcription-polymerase chain reaction (qRT-PCR) examination.
Cell culture and reagents
Human dopaminergic neuroblastoma SH-SY5Y cells purchased from American Type Culture Collection (ATCC) cultured in complete culture medium containing RPMI-1640 (Gibco, USA) containing 10% fetal bovine serum (FBS; Gibco, USA) plus 2 mmol/L L-glutamine, 100 U/mL penicillin and 100 µg/mL streptomycin. The cells were incubated at 37°C in a humidified 95% air and 5% CO2 atmosphere and passaged when the cells were grown to approximately 80% confluent.
293T cells supplied from the Cell Bank Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China) were cultivated in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, USA). The medium was supplemented with 10% FBS. Penicillin/streptomycin was added to the medium at a concentration of 100 IU/mL and applied to the cells, followed by incubation in a humidified incubator set at 37°C with 5% CO2.
RNA isolation and qRT-PCR for miR-153 and Nrf2 analysis
The total RNA enrichment procedure from clinical specimens and cell lines samples was firstly extracted with a TRIzol Reagent (TIANGEN, China) according to the manufacturer’s guidelines. The concentration and purity of the isolated RNA are assessed by optical density at 260 nm and 280 nm, respectively, using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA). The isolated RNA was cleared of contaminating genomic DNA by DNase treatment (Thermo Fisher Scientific, USA) and then converted to complementary DNA (cDNA) with a RevertAid First Strand cDNA Synthesis kit (Thermo Fisher Scientific, USA). qPCR was performed to determine the expression of miR-153 and Nrf2 on an ABI PRISM® 7500 Sequence Detection System (Applied Biosystems, USA) with their corresponding primers and the amplifications were achieved using a SYBR Green qPCR SuperMix Kit (Thermo Fisher Scientific, USA). The reactions were incubated in a 96-well optical plate at 95°C for 2 min, followed by 40 cycles of 15 s at 95°C and 32 s at 60°C and dissociation at 95°C for 60 s, 55°C for 30 s and 95°C for 30 s.
U6 snRNA and GAPDH were served as a normalization control for miR-153 and Nrf2, respectively, and the 2-ΔΔCt method was used to evaluate relative expression. The primers used in the qRT-PCR experiments were as follows: miR-153 forward, 5’-ACACTCCAGCTGGGTTGCATAGTCACAA-3’ and reverse, 5’-CTCAACTGGTGTCGTGGA-3’; U6 forward, 5’-CTCGCTTCGGCAGCACA-3’ and reverse, 5’-AACGCTTCACGAATTTGCGT-3’; Nrf2 forward, 5’-GTTGGAGCTGTTGATCTGTTG-3’ and reverse, 5’-GTTTTTTCTGTTTTTCCAGCTC-3’; GAPDH forward, 5’-GGGAAACTGTGGCGTGAT-3’ and reverse, 5’-GAGTGGGTGTCGCTGTTGA-3’.
MTT examination
Cell viability was usually evaluated using an MTT assay (Merck KGaA, Germany). In brief, SH-SY5Y cells were seeded into a 96-well tissue culture plate at a density of 5×103 cells/well. Once cells reached 80~90% confluence, they were incubated with different concentrations of MMP+ (i.e., 0 mM, 0.5 mM, 1 mM, 2 mM and 4 mM). Following incubation for 0 h, 6 h, 12 h, 18 h and 24 h at 37°C, cell proliferation was determined using fresh medium containing 0.5 mg/ml MTT at 37°C for another 4 h. Then, dimethyl sulfoxide (DMSO; Sigma, USA) was added to each well to dissolve the blue formazan products and the optical density (OD) was measured at an absorbance wavelength of 450 nm on a scanning microplate spectrophotometer (Thermo Fisher, USA). The experiments were performed in triplicate.
Dual-luciferase reporter assays
Wild-type (WT) or mutant (Mut) Nrf2 3’-UTR were amplified by PCR and cloned into a psiCHECK-2 plasmid (Promega, USA) with firefly luciferase. A total of 5×104 293T cells treated with control, miR-153 mimics, miR-153 inhibitors negative control (NC) plasmid, or NC inhibitor were transfected with WT or mutant Nrf2 3’-UTR luciferase reporters together with Renilla plasmid. After approximately 48 h posttransfection, the cells were lysed and luciferase activities were determined by Dual Luminescence Assay Kits (Promega, USA) according to the manufacturer’s protocols. The firefly luciferase activities were normalized to Renilla luciferase activities. The experiments in each group were repeated three times.
Bio-chemistry methods
ROS level, MDA content, SOD activity and GSH activity were the indexes of oxidative stress. These related indexes all used their corresponding commercial kits purchased from Nanjing Jiancheng Bioengineering Institute, China. The supernatants of all samples from different groups were utilized to examine the above indexes according to these respective instructions.
WB detection
Three independent samples from each group were harvested and homogenized, and total protein was extracted using ice-cold radioimmunoprecipitation assay (RIPA) lysis buffer (Beyotime, China) containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 100 mM phenylmethanesulfonyl fluoride (PMSF) and 10 μl protease inhibitor cocktail. After centrifugation at 12000 rpm for 10 min at 4°C, the insoluble material was removed, and the total protein concentration in supernatant was measured with a BCA protein assay kit (Beyotime, China). Equal amounts of protein (30 μg) were separated on an 8~15% discontinuous SDS-polyacrylamide gels for electrophoresis (SDS-PAGE) and electrophoretically transferred to polyvinylidene difluoride (PVDF) membranes at a constant current of 200 mA for 60 min. Following blocking the membrane with Tris-buffered saline containing 0.05% Tween-20 (TBST) containing 5% non-fat dry milk at room temperature for 1 h, the membranes were incubated overnight at 4°C with primary antibodies against Nrf2 (1:1000 diluted; Abcam, USA), NQO1 (1:1500 diluted; Abcam, USA), GCLC (1:2000 diluted; Abcam, USA), GCLM (1:1000 diluted; Abcam, USA), HO-1 (1:2000 diluted; Abcam, USA) and GAPDH (as an internal control, 1:5000 diluted; Abcam, USA). Subsequently, membranes were washed with TBST extensively three times and probed with the corresponding anti-mouse or anti-rabbit horseradish peroxidase-conjugated secondary antibodies (1:12000 diluted; Abmart, USA) for 1 hour at 37°C. Finally, band signal strength was determined by enhanced chemiluminescence reagents (Beyotime, China) and the densitometric analysis was conducted with ImageJ software version 1.8 (National Institutes of Health, USA).
Statistical analysis
All statistical calculations were performed using SPSS 18.0 software (IBM SPSS, USA). The values were expressed as the mean ± standard deviation (SD) in the figures and a value of P < 0.05 or 0.01 was considered statistically significant. Each experiment was repeated ≥ 3 times. Differences in measured values between two groups were analyzed using the Chi-square test and among multiple groups were analyzed by analysis of variance with Bonferroni’s multiple comparison correction.
Results
Clinical significance of miR-153 and Nrf2 expressions in PD
We first examined the levels of miR-153 in serum samples of PD patients. The results showed that the miR-153 expression was significantly increased in PD patients as compared to healthy people (Figure 1A). Subsequently, the Nrf2 expression was also detected by qRT-PCR which exhibited that the expression level of Nrf2 was notably decreased in PD patients when compared with health people (Figure 1B). Therefore, these results suggested that the changes of miR-153 and Nrf2 expression have an important clinical significance in PD patients.
Figure 1.

qRT-PCR was used to examine the expression of miR-153 and Nrf2 in serum specimens of PD patients. A. miR-153 expression level was increased in PD patients. B. Nrf2 expression level was decreased in PD patients.
Assessment of MPP+ treated cellular model
MTT assay found that the OD values representing cell viability were gradually decreased over time in different concentrations of MMP+ treatment (Figure 2A). Furthermore, the OD values presented the lowest level in the 4 mM MPP+ treated group at 24 h, thus SH-SY5Y cells treated with MPP+ for 24 h was selected for following study. In addition, as illustrated in Figure 2B and 2C, it was revealed that after giving MPP+ treatment, the miR-153 and Nrf2 expressions were dramatically up-regulated and down-regulated, respectively.
Figure 2.

MPP+ treatment promotes cytotoxic effect and influences the expression of miR-153 and Nrf2 in SH-SY5Y cells. A. Neuronal viability was monitored by MTT assay in SH-SY5Y cells treated with various concentrations (0~4 mM) of MPP+ for five time points (0 h, 6 h, 12 h, 18 h and 24 h). *P < 0.05. B. Expression of miR-153 was identified by qRT-PCR in SH-SY5Y cells with MPP+ treatment. *P < 0.05. C. Expression of Akt3 under MPP+ treatment was determined using qRT-PCR in SH-SY5Y cells. *P < 0.05.
Nrf2 was a critical functional target of miR-153
Based on the inverse correlation between miR-153 and Nrf2 expressions, the potential target relationship of miR-153 and Nrf2 was validated. As expected, Nrf2 was predicted as one target of miR-153 determined by TargetScan and miRDB online software and the base pairing between miR-153 and Nrf2 was presented in Figure 3A. Moreover, the dual-luciferase assay pointed out that compared with NC group, co-transfection of miR-153 mimics and Nrf2-3’-UTR displayed remarkably decreased luciferase activity, whereas co-transfection of miR-153 mimics and Nrf2-Mutant-3’-UTR did not result in marked decline in luciferase activity. These data suggested that Nrf2 was a direct target of miR-153. Additionally, when transfecting with miR-153 mimics or inhibitor, the expressions of Nrf2 in mRNA and protein levels were sharply reduced and elevated, respectively (Figure 3B and 3C).
Figure 3.

Nrf2 was the direct target gene of miR-153. A. Schematic diagram of the presumed binding site of miR-153 and on the 3’-UTR of Nrf2 was presented on the left panel, and the data from dual-luciferase were showed on the right panel. *P < 0.05 and **P < 0.01. B. mRNA level of Nrf2 was reduced following transfection of miR-153 mimics, as detected by qRT-PCR. C. Protein level of Nrf2 were also declined by miR-153 overexpression, as measured using WB.
The effects of miR-153 and Nrf2 on the indexes of oxidative stress in MPP+-treated SH-SY5Y cells
As seen in Figure 4, overexpression of miR-153 and Nrf2 significantly promoted and repressed the relative ROS levels, respectively (Figure 4A). Furthermore, up-regulation of miR-153 and Nrf2 also notably enhanced and hindered the MDA content, respectively (Figure 4B). ROS and MDA both belong to oxidative indexes which represented the oxidative degree [24]. Additionally, forced miR-153 and Nrf2 expression conspicuously decelerated and accelerated the SOD activity (Figure 4C), respectively, as well as the GSH activity (Figure 4D). SOD and GSH are the antioxidative indices which are indirectly related to degree of oxidation [25]. Meanwhile, it was also observed that there were an opposite effect in miR-153 inhibitor group and si-Nrf2 group. Hence, these findings manifested that miR-153 could facilitate the degree of oxidative stress in MPP pressions of Nrf2, NQO1, GCLM, GCLC and HO-1 in mRNA and protein levels, while forced expression of Nrf2 markedly decreased the expressions of Nrf2, NQO1, GCLM, GCLC and HO-1 in mRNA and protein levels. Moreover, the knockdown of miR-153 and Nrf2 showed an inverse trend which corresponded to the miR-153 group and Nrf2 group, respectively (Figure 5A and 5B). Thus, these data supported that miR-153 might enhance the oxidative stress in MPP+-treated SH-SY5Y cells via targeting Nrf2.
Figure 4.

The indexes of oxidative stress were tested by biochemical methods in SH-SY5Y cells with different treatments. A. miR-153 promoted the relative ROS levels and Nrf2 suppressed the relative ROS levels when compared with MPP+ group. *P < 0.05. B. Transfection with miR-153 or Nrf2 enhanced and inhibited the MDA content, respectively, in compared with MPP+ group. *P < 0.05. C. Overexpression of miR-153 and Nrf2 could hinder and facilitate the SOD activity, respectively, as compared to MPP+ group. *P < 0.05. D. The relative GSH activity was higher and lower in miR-153 group and Nrf2 group, respectively, than that in the MPP+ group. *P < 0.05.
Figure 5.
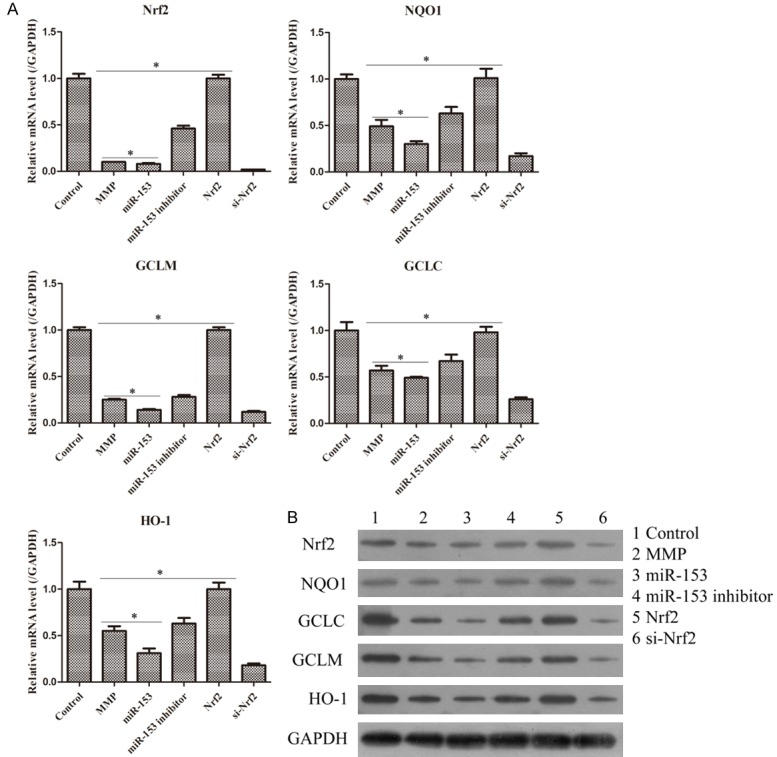
Effects of miR-153 and Nrf2 on the mRNA and protein expression alterations of oxidative stress-related genes in MPP+ treated SH-SY5Y cells. A. The mRNA levels of Nrf2, NQO1, GCLC, GCLM, and HO-1 were all relatively decreased in the miR-153 group and increased in the Nrf2 group as compared to the MPP+ group. *P < 0.05. B. The protein levels of Nrf2, NQO1, GCLC, GCLM, and HO-1 were all relatively down-regulated in miR-153 group and up-regulated in Nrf2 group as compared to MPP+ group.
Discussion
PD, a chronic neurodegenerative disease, is extremely common among elderly individuals [26]. Previous studies have reported that genetic and environmental factors have been implicated in the etiology of PD, but the pathogenesis is still largely unknown [27]. However, frequent aberrant miRNA expression has been detected in post-mortem human PD brain samples, thus miRNAs were considered as important regulators in progression of PD [28]. For example, miR-181c could promote cell viability and inhibit the apoptosis of PC12 cells treated with MPP+ as PD cellular model [29]. In our study, we first demonstrated that miR-153 expression was significantly increased in serum samples of PD patients and cellular samples of MPP+-treated SH-SY5Y cells, suggesting that abnormal miR-153 expression might exert an essential role in development of PD. Meanwhile, we also examined the expression of Nrf2 and the results showed that Nrf2 presented a conspicuous decline trend in vivo and in vitro. Nrf2, a leucine zipper/CNC protein widely expressed in organs with hyperoxia consumption, plays an important role in cellular resistance to exogenous toxic substances and oxidative stress [30]. Therefore, our results further indicated that oxidative stress might contribute to the progress of PD. Actually, it has been confirmed that oxidative stress is a common feature in many neurodegenerative disorders and it likely represents a crucial step in the neurodegenerative process and could therefore be a target for neuroprotective strategies [7]. Subsequently, based on the inverse expression of miR-153 and Nrf2 in vivo and in vitro, dual-luciferase assay was used to assess the target interaction between miR-153 and Nrf2. Our data revealed that Nrf2 was the direct target of miR-153.
In order to further investigate the effects of miR-153 and Nrf2 on the oxidative stress during the pathogenesis of PD, the indexes of oxidative stress were detected by biochemical methods. Oxidative stress is a state of oxidative damage in cells, tissues, or organs caused by the ROS or MDA, and is commonly linked to mitochondrial dysfunction [8]. Thus, the relative ROS level and MDA content were remarkably increased in miR-153 and si-Nrf2 groups as compared to the MPP+-treated group, while they were markedly decreased in miR-153 inhibitor and Nrf2 groups when compared with MPP+-treated group, which implied that miR-153 might activate oxidative stress by negatively regulating Nrf2. Nevertheless, SOD, as the natural enemy of oxygen free radicals, is a classic scavenger in the human body [9]. Additionally, GSH containing three amino acids mainly acts as an important antioxidant and free radical scavenger in the body. Hence, the two indexes both belong to antioxidants [24]. In the current study, the SOD activity and GSH activity did indeed show an opposite trend in each group as compared with the results of ROS or MDA. Moreover, accumulating evidence has verified that low levels of oxidative stress could induce Nrf2 activation which further regulated the antioxidant response in mixed neuron/astrocyte cultures and ultimately triggered neuroprotection to delay AD-like pathology in an AD mice model [31]. Therefore, we speculated whether the miR-153 could mediate the Nrf2-HO-1 signaling pathway participated in the development of PD. We chose some key molecules involved in Nrf2-HO-1 signaling pathway to test their expressions in mRNA and protein levels, such as NQO1, GCLM, GCLC. Finally, the data uncovered that Nrf2, NQO1, GCLM, GCLC and HO-1 mRNA and protein expressions were dramatically down-regulated in miR-153 and si-Nrf2 groups as compared to the MPP+-treated group, whereas they were sharply up-regulated in miR-153 inhibitor and Nrf2 groups when compared with the MPP+-treated group, pointing out that miR-153 might suppress the related molecules expressions by targeting Nrf2.
In conclusion, results obtained in this study provide evidence for the maladjustment of oxidative stress arising from aberrant miR-153 and Nrf2 expression. Furthermore, the Nrf2/HO-1 signaling pathway was the key factor to involve in the regulation of oxidative stress during the pathogenesis of PD via miR-153. Hence, miR-153 and Nrf2 might be novel therapeutic targets and the effective biomarkers for the diagnosis, evaluation, and treatment of PD.
Disclosure of conflict of interest
None.
References
- 1.Sheikh S, Safia , Haque E, Mir SS. Neurodegenerative diseases: multifactorial conformational diseases and their therapeutic interventions. J Neurodegener Dis. 2013;2013:563481. doi: 10.1155/2013/563481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Litvinenko IV, Krasakov IV, Bisaga GN, Skulyabin DI, Poltavsky ID. [Modern conception of the pathogenesis of neurodegenerative diseases and therapeutic strategy] . Zh Nevrol Psikhiatr Im S S Korsakova. 2017;117:3–10. doi: 10.17116/jnevro2017117623-10. [DOI] [PubMed] [Google Scholar]
- 3.Chen WW, Zhang X, Huang WJ. Role of neuroinflammation in neurodegenerative diseases (Review) Mol Med Rep. 2016;13:3391–3396. doi: 10.3892/mmr.2016.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weil RS, Lashley TL, Bras J, Schrag AE, Schott JM. Current concepts and controversies in the pathogenesis of Parkinson’s disease dementia and Dementia with Lewy Bodies. F1000Res. 2017;6:1604. doi: 10.12688/f1000research.11725.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller J, Myers J. Association between physical fitness, cardiovascular risk factors, and Parkinson’s disease. Eur J Prev Cardiol. 2018 doi: 10.1177/2047487318771168. 2047487318771168. [DOI] [PubMed] [Google Scholar]
- 6.Tarazi FI, Sahli ZT, Wolny M, Mousa SA. Emerging therapies for Parkinson’s disease: from bench to bedside. Pharmacol Ther. 2014;144:123–133. doi: 10.1016/j.pharmthera.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 7.Guo C, Sun L, Chen X, Zhang D. Oxidative stress, mitochondrial damage and neurodegenerative diseases. Neural Regen Res. 2013;8:2003–2014. doi: 10.3969/j.issn.1673-5374.2013.21.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Islam MT. Oxidative stress and mitochondrial dysfunction-linked neurodegenerative disorders. Neurol Res. 2017;39:73–82. doi: 10.1080/01616412.2016.1251711. [DOI] [PubMed] [Google Scholar]
- 9.Bernatoniene J, Kopustinskiene DM. The role of catechins in cellular responses to oxidative stress. Molecules. 2018:23. doi: 10.3390/molecules23040965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Z, Zhou T, Ziegler AC, Dimitrion P, Zuo L. Oxidative stress in neurodegenerative diseases: from molecular mechanisms to clinical applications. Oxid Med Cell Longev. 2017;2017:2525967. doi: 10.1155/2017/2525967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Achkar NP, Cambiagno DA, Manavella PA. miRNA biogenesis: a dynamic pathway. Trends Plant Sci. 2016;21:1034–1044. doi: 10.1016/j.tplants.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Huang Y, Shen XJ, Zou Q, Zhao QL. Biological functions of microRNAs. Bioorg Khim. 2010;36:747–752. doi: 10.1134/s1068162010060026. [DOI] [PubMed] [Google Scholar]
- 13.Maciotta S, Meregalli M, Torrente Y. The involvement of microRNAs in neurodegenerative diseases. Front Cell Neurosci. 2013;7:265. doi: 10.3389/fncel.2013.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goodall EF, Heath PR, Bandmann O, Kirby J, Shaw PJ. Neuronal dark matter: the emerging role of microRNAs in neurodegeneration. Front Cell Neurosci. 2013;7:178. doi: 10.3389/fncel.2013.00178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qiu L, Tan EK, Zeng L. microRNAs and neurodegenerative diseases. Adv Exp Med Biol. 2015;888:85–105. doi: 10.1007/978-3-319-22671-2_6. [DOI] [PubMed] [Google Scholar]
- 16.de Mena L, Coto E, Cardo LF, Diaz M, Blazquez M, Ribacoba R, Salvador C, Pastor P, Samaranch L, Moris G, Menendez M, Corao AI, Alvarez V. Analysis of the Micro-RNA-133 and PITX3 genes in Parkinson’s disease. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:1234–1239. doi: 10.1002/ajmg.b.31086. [DOI] [PubMed] [Google Scholar]
- 17.Kabaria S, Choi DC, Chaudhuri AD, Mouradian MM, Junn E. Inhibition of miR-34b and miR-34c enhances alpha-synuclein expression in Parkinson’s disease. FEBS Lett. 2015;589:319–325. doi: 10.1016/j.febslet.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Minones-Moyano E, Porta S, Escaramis G, Rabionet R, Iraola S, Kagerbauer B, Espinosa-Parrilla Y, Ferrer I, Estivill X, Marti E. Micro-RNA profiling of Parkinson’s disease brains identifies early downregulation of miR-34b/c which modulate mitochondrial function. Hum Mol Genet. 2011;20:3067–3078. doi: 10.1093/hmg/ddr210. [DOI] [PubMed] [Google Scholar]
- 19.Gui Y, Liu H, Zhang L, Lv W, Hu X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget. 2015;6:37043–37053. doi: 10.18632/oncotarget.6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bellezza I, Giambanco I, Minelli A, Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim Biophys Acta. 2018;1865:721–733. doi: 10.1016/j.bbamcr.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 21.Rojo AI, Pajares M, Rada P, Nunez A, Nevado-Holgado AJ, Killik R, Van Leuven F, Ribe E, Lovestone S, Yamamoto M, Cuadrado A. NRF2 deficiency replicates transcriptomic changes in Alzheimer’s patients and worsens APP and TAU pathology. Redox Biol. 2017;13:444–451. doi: 10.1016/j.redox.2017.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang B, Teng Y, Liu Q. MicroRNA-153 regulates NRF2 expression and is associated with breast carcinogenesis. Clin Lab. 2016;62:39–47. doi: 10.7754/clin.lab.2015.150518. [DOI] [PubMed] [Google Scholar]
- 23.Ji Q, Gao J, Zheng Y, Liu X, Zhou Q, Shi C, Yao M, Chen X. Inhibition of microRNA-153 protects neurons against ischemia/reperfusion injury in an oxygen-glucose deprivation and reoxygenation cellular model by regulating Nrf2/HO-1 signaling. J Biochem Mol Toxicol. 2017:31. doi: 10.1002/jbt.21905. [DOI] [PubMed] [Google Scholar]
- 24.Layali I, Shahriary A, Rahmani Talatappe N, Tahmasbpour E, Rostami H, Beigi Harchegani A. Sulfur mustard triggers oxidative stress through glutathione depletion and altered expression of glutathione-related enzymes in human airways. Immunopharmacol Immunotoxicol. 2018:1–7. doi: 10.1080/08923973.2018.1460754. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Wang M, Liu J, Ye J, Jiang H, Xu Y, Ye D, Wan J. Inhibition of TRPA1 attenuates doxorubicin-induced acute cardiotoxicity by suppressing oxidative stress, the inflammatory response, and endoplasmic reticulum stress. Oxid Med Cell Longev. 2018;2018:5179468. doi: 10.1155/2018/5179468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koppula S, Kumar H, More SV, Kim BW, Kim IS, Choi DK. Recent advances on the neuroprotective potential of antioxidants in experimental models of Parkinson’s disease. Int J Mol Sci. 2012;13:10608–10629. doi: 10.3390/ijms130810608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sarkar S, Raymick J, Imam S. Neuroprotective and therapeutic strategies against Parkinson’s disease: recent perspectives. Int J Mol Sci. 2016:17. doi: 10.3390/ijms17060904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Viswambharan V, Thanseem I, Vasu MM, Poovathinal SA, Anitha A. miRNAs as biomarkers of neurodegenerative disorders. Biomark Med. 2017;11:151–167. doi: 10.2217/bmm-2016-0242. [DOI] [PubMed] [Google Scholar]
- 29.Wei M, Cao LJ, Zheng JL, Xue LJ, Chen B, Xiao F, Xu CS. MicroRNA-181c functions as a protective factor in a 1-methyl-4-phenylpyridinium iodide-induced cellular Parkinson’s disease model via BCL2L11. Eur Rev Med Pharmacol Sci. 2017;21:3296–3304. [PubMed] [Google Scholar]
- 30.Chen B, Lu Y, Chen Y, Cheng J. The role of Nrf2 in oxidative stress-induced endothelial injuries. J Endocrinol. 2015;225:R83–99. doi: 10.1530/JOE-14-0662. [DOI] [PubMed] [Google Scholar]
- 31.Mota SI, Costa RO, Ferreira IL, Santana I, Caldeira GL, Padovano C, Fonseca AC, Baldeiras I, Cunha C, Letra L, Oliveira CR, Pereira CM, Rego AC. Oxidative stress involving changes in Nrf2 and ER stress in early stages of Alzheimer’s disease. Biochim Biophys Acta. 2015;1852:1428–1441. doi: 10.1016/j.bbadis.2015.03.015. [DOI] [PubMed] [Google Scholar]


