Abstract
Background: Esophageal cancer is one of the most common malignant tumors threatening human health worldwide. Circular RNAs (circRNAs) are a large group of covalently closed continuous loops that are prevalently expressed in human cells and might be applied as novel esophageal cancer biomarkers. Purpose: To investigate the expression of a novel circular RNA, circFNDC3B, in esophageal cancer, as well as determine its function in the regulation of esophageal cell proliferation, apoptosis, migration, and invasion. Methods: Quantitative RT-PCR using circular RNA-specific primers was performed to analyze the existence and expressional change of circFNDC3B in esophageal cancer tissues. The esophageal cancer cell lines ECA109 and KYSE150 with inhibited circFNDC3B expression by gene silencing were subjected to proliferation analysis with the MTS method, FITC Annexin V apoptosis detection, and a proliferation and invasion evaluation using a transwell system. Results: circFNDC3B was specifically up-regulated in esophageal cancer tissues. Esophageal cancer cell lines ECA109 and KYSE150 with decreased circFNDC3B expression by gene silencing showed inhibited proliferation, increased apoptosis, and weakened migration and invasion abilities. Conclusion: The circFNDC3B encoded by two FNDC3B gene exons is an important regulator of esophageal cancer progression.
Keywords: CircFNDC3B, esophageal cancer, proliferation, apoptosis, migration, invasion
Introduction
Primary esophageal cancer is one of the most common cancers worldwide, with approximately 455,800 new cases and 400,200 deaths reported in 2012 [1,2]. Most cases occur in developing countries and approximately half of all these occur in China, bringing heavy economic and social burdens to these countries [2,3]. Esophageal cancer patients usually suffer severe symptoms, such as difficulty swallowing, a hoarse voice, and weight loss. Previous studies have shown that the causative factors for esophageal cancer include smoking, alcohol consumption, hot drinks, poor diet, and obesity. Esophageal cancers include esophageal adenocarcinoma (EAC) and esophageal squamous cell carcinoma (ESCC). Clinically, the diagnosis of esophageal cancer mainly depends on biopsy using an endoscope, and the most common treatments are still surgery, chemotherapy, and radiation, with consideration to the cancer stage and location [4]. The screening and identification of specific biomarkers for esophageal cancer would promote our understanding of the molecular mechanisms underlying disease progression, direct accurate diagnosis, precise treatment, and prognosis.
Circular RNAs (circRNAs) are a large group of endogenous RNAs characterized by the formation of covalently closed continuous loops, and prevalently expressed in multiple species including monkeys and humans [5,6]. Generally, circRNAs carry out their biological roles through multiple mechanisms, including interacting with miRNAs as sponges, modulating gene expression as regulators of RNA binding proteins (RBP), regulating gene transcription, and being translated into functional proteins or peptides [5-9]. Recent progress has shown that circRNAs might be applied as potential biomarkers for cancer. Due to the application of high-through RNA-seq technology and other investigations, an increasing number of circRNA molecules has been found to be differentially expressed in multiple cancers, including esophageal cancer [10,11], glioma [12], colorectal cancer [13], and hepatocellular carcinoma [14-16]. Specifically, one novel circRNA, hsa_circ_0067934, was shown to be remarkably over-expressed in esophageal squamous cell carcinoma tissues [11]. Furthermore, ESCC cell proliferation, migration, and cell cycle progression were significantly repressed when hsa_circ_0067934 was silenced in vitro by siRNA [11].
Fibronectin type III domain containing 3B (FNDC3B) is a newly identified oncogene-encoded protein that could activate multiple cancer pathways and promote cell migration and tumor metastasis in hepatocellular carcinoma and acute myeloid leukemia [17-19]. The existence of this circular RNA molecule encoded by the FNDC3B gene has been verified by several previous studies [20-23]. In order to further investigate the possible association of oncogene-related circRNAs with cancer progression and their possible application in cancer diagnosis and treatment, the expression of hsa_circ_0001361, a novel circRNA from the FNDC3B gene, termed circFNDC3B, was analyzed in this study, along with its function in regulating esophageal cell proliferation, apoptosis, migration, and tumor metastasis.
Material and methods
Ethical statement
This study was approved by the Ethical Committee of Guangdong General Hospital. A written informed consent was received from each patient, and all clinical data were collected after each surgery.
Tissue sample and cancer cells
The esophageal cancer tissues and adjacent control samples (n=23) were collected from patients that had undergone surgery at the Guangdong General Hospital, Guangzhou, China. After being confirmed by experienced clinical pathologists, the cancer and adjacent normal tissues were subjected to further analysis. The esophageal cancer cell lines ECA109 and KYSE150 used in this study were provided by the cell bank affiliated with the Chinese Academy of Sciences, Shanghai Branch.
RNA extraction and qRT-PCR assay
RNA extraction and qRT-PCR were performed as previously described with minor modifications [11]. Briefly, total RNA samples were extracted from cancer tissues or cultured cancer cells using Trizol reagent following the manufacturer’s instructions (Life Technologies, UK). The synthesis of cDNA was carried out by reverse transcription with 500 ng RNA and random primers using the Prime Script RT Master Mix kit (Takara, Cat: RR036A). The quantitative real-time polymerase chain reaction (qRT-PCR) was completed using the SYBR Select Master Mix kit (Applied Biosystems, Cat: 4472908) on an ABI7300 system (Applied Biosystems, USA) following the manufacturer’s instructions. The opposite-directed and divergent primers were used for the analysis of the linear and circular RNA, respectively. GAPDH was applied as the internal control for the quantitation of circFNDC3B expression. The following primers were used is this study: circFNDC3B divergent forward: 5’-CATCTCCATTCACCAAGTGGGG-3’; circFNDC3B divergent reverse, 5’-AGCAGGTTATTCTCGTTCAAG-3’; circFNDC3B opposite-directed forward: 5’-GACCGACCAAATCCCTCTGG-3’; circFNDC3B opposite-directed reverse, 5’-TTCACCAAGTGGGGCATCAT-3’; and GAPDH: forward, 5’-GAGTCAACGGATTTGGTCGT-3’, reverse, 5’-GACAAGCTTCCCGTTCTCAG-3’.
Esophageal cancer cell culture and siRNA transfection
The two esophageal cancer cell lines ECA109 and KYSE150 were purchased from the cell 112 bank of the Chinese Academy of Sciences, Shanghai. Esophageal cancer cell culture and gene silencing were carried out according to a previous description with minor modifications [11]. Briefly, the ECA109 and KYSE150 cells were cultured in DMEM (KeyGen, China) supplied with 10% FBS (Life Technologies, Australia) and penicillin-streptomycin at 37°C in a humidified atmosphere with 5% CO2. After being seeded in six-well plates, esophageal cancer cells were then transfected with siRNA or control virus for 24 h using the Lipofectamine RNAi MAX transfection kit following the manufacturer’s instructions (Invitrogen, USA). The primers used for gene silencing in this study were: circFNDC3B sense: 5’-AGUGCAUUCAAGGAAGCCA-3’; antisense: 5’-AGUGCAUUCAAGGAAGCCA-3’.
Cell proliferation analysis
The proliferation of esophageal cancer cells was measured using the MTS Cell Proliferation Assay Kit (Colorimetric) (197010; Abcam, UK) according to the manufacturer’s instructions. Briefly, esophageal cancer cells (104/well) were seeded in 96-well microtiter plates and cultured for 20-48 h depending on cell growth conditions. Then, 20 µL/well MTS reagent were added into each well and incubated for 2 hours at 37°C under standard culture conditions. After being shaken briefly on a shaker, the cell plates were measured using a plate reader at OD=490 nm. Three biological repeats were performed for the statistical analysis.
Cell apoptosis analysis
Esophageal cancer cell apoptosis was analyzed using the FITC Annexin V Apoptosis Detection Kit with 7-AAD (BioLegend, USA) according to the manufacturer’s instructions. Briefly, ECA109 and KYSE150 cells cultured in 6-well plates were transfected with siRNA for three days as described above. Esophageal cancer cells were then collected, washed three times with cold BioLegend’s Cell Staining Buffer, and were resuspended in Annexin V Binding Buffer at a concentration of 1 × 107 cells/ml. Next, 5 µl FITC Annexin V and 5 µl 7-AAD Viability Staining Solution was added to the cell suspension. After incubation at room temperature in the dark for 15 min with a gentle vortex, the cell suspension was mixed with 400 µl Annexin V Binding Buffer. Finally, esophageal cancer cell apoptosis was analyzed using FACScan flow cytometry (BD Biosciences, USA). Three biological repeats were performed for statistical analysis.
Cell migration and invasion assay
The esophageal cell migration was analyzed with the Boyden Transwell system in 12-well plates (BD Biosciences, USA) as described by previous literature with minor modifications [11]. Briefly, in order to analyze the cell migration, after being transfected with siRNA for 3 days, ECA109 and KYSE150 cells were harvested and resuspended in DMEM medium without serum and seeded into the upper chamber at a concentration of 105 cells per well. Then, 0.6 ml RPMI1640 medium with 10% fetal bovine serum was added into the bottom wells. After incubation for 24 h, the esophageal cancer cells that had migrated to the bottom wells were fixed in methanol and stained with a hematoxylin and eosin solution for cell number counting. For the invasion assay, 8 μm pore membrane (Corning Inc., Corning, NY) was pre-coated with Matrigel (BD Biosciences, USA) and solidified at 37°C. ECA109 and KYSE150 cells were kept in the top chamber with 500-μl serum-free DMEM after transfection, and the bottom chambers were filled with 750 μl of 10% FBS-DMEM. The cell staining and counting were performed after 48 h. Three biological replicates were performed for the statistical analysis.
Statistical analysis
All statistical analyses in this study were performed using the SPSS software package (version 18.0, SPSS). The expression level differences were analyzed using either the Student’s t test or the nonparametric Kruskal-Wallis test. Statistical significance was defined as P<0.05.
Results
Characterization of circFNDC3B in esophageal cancer tissues
According to the information provided by the circBase database, the circRNA circFNDC3B (circBase ID: hsa_circ_0001361) was generated from the FNDC3B gene located on the 7th chromosome (170013698-170015181) of the human genome (Figure 1A). CircFNDC3B is 215 bases long and is encoded and generated by the second and third exons of the FNDC3B gene by back splicing (Figure 1A). Total RNA and genomic DNA samples were extracted from esophageal cancer tissues and the expressions of linear and circular RNA were analyzed by PCR using opposite-directed and divergent primers, respectively. The RT-PCR assay showed that circFNDC3B was only detected in cDNA samples using circular RNA-specific primers, but not in the genomic DNA (Figure 1B). For further verification of circFNDC3B expression in esophageal cancer, total RNA samples were subjected to RNaseR digestion and RT-PCR analysis, showing that RNaseR treatment only slightly decreased the circFNDC3B level, compared with that of linear RNA, which is consistent with the properties of circular RNAs (Figure 1C). The splice junction of circFNDC3B was then confirmed by Sanger sequencing as shown in Figure 1D. Taken together, these results clearly demonstrate the existence of circFNDC3B in esophageal cancer tissues.
Figure 1.
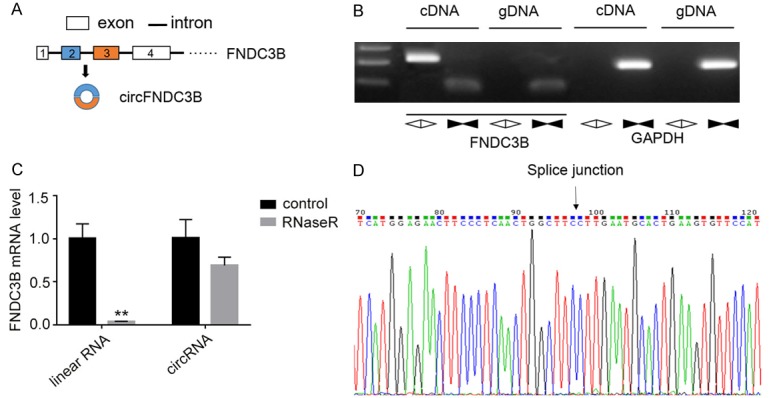
Detection of circFNDC3B in esophageal cancer tissues. A. Formation of circFNDC3B from two exons in the FNDC3B gene. B. Detection of circFNDC3B in esophageal cancer tissues using divergent primers. Expression of circFNDC3B was detected in cDNA samples, not in genomic DNA. C. Sensitivity of circFNDC3B to RNaseR digestion. The expression levels of linear and circular RNAs were measured by RT-PCR using specific primers after being incubated with RNaseR. D. Verification of splice junction by Sanger sequencing. The splice junction site of circFNDC3B was characterized by Sanger sequencing following amplification using circular RNA-specific primers.
Altered circFNDC3B expression level in esophageal cancer tissues
To address the expressional alteration of circFNDC3B in esophageal cancer, the esophageal cancer tissues and adjacent normal tissues from 23 esophageal cancer patients who underwent surgery were collected and analyzed with qRT-PCR. In the majority of the esophageal cancer samples, the expression level of circFNDC3B was greatly increased in the cancer tissues compared with the corresponding adjacent normal controls (Figure 2A). Statistical analysis also confirmed a significantly increased circFNDC3B expression level in esophageal cancer tissues (P=0.0414, Figure 2B).
Figure 2.
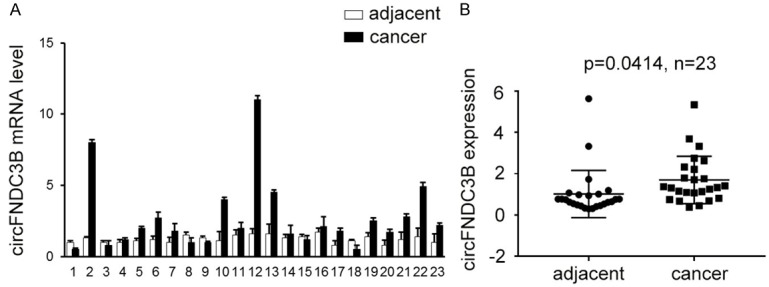
Increased circFNDC3B expression level in esophageal cancer tissues. A. Comparison of circFNDC3B expression between esophageal cancer tissues and adjacent normal tissues. The expression levels of circFNDC3B in 23 esophageal cancer tissues as well as the corresponding adjacent normal tissues were quantitatively measured by qRT-PCR. B. Statistically increased circFNDC3B level in esophageal cancer tissues. The expression of circFNDC3B between esophageal cancer and adjacent normal tissues of 23 patients was compared by statistical analysis. P=0.0414, n=23.
CircFNDC3B regulates esophageal cancer cell proliferation
To further investigate the potential implications of circFNDC3B in esophageal cancer development, we used gene silencing technology to repress the expression level of circFNDC3B in two cultured esophageal cancer cell lines, ECA109 and KYSE150, whereupon its role in esophageal cancer cell proliferation regulation was tested. The circFNDC3B expression levels in two esophageal cancer cells were successfully down-regulated by transfection with sh-circFNDC3B, compared with the negative controls (NC) (**P<0.01, Figure 3A). In order to quantitatively demonstrate the influence of circFNDC3B inhibition on esophageal cancer cell proliferation, the proliferations of these two esophageal cancer cell lines were measured using an MTS Cell Proliferation Assay Kit. We observed that the proliferation of esophageal cancer cells was significantly inhibited after the circFNDC3B expression was repressed for three days(**P<0.05, Figure 3B), clearly showing that circFNDC3B plays a role in the complexity of regulating the network responsible for esophageal cancer progression.
Figure 3.
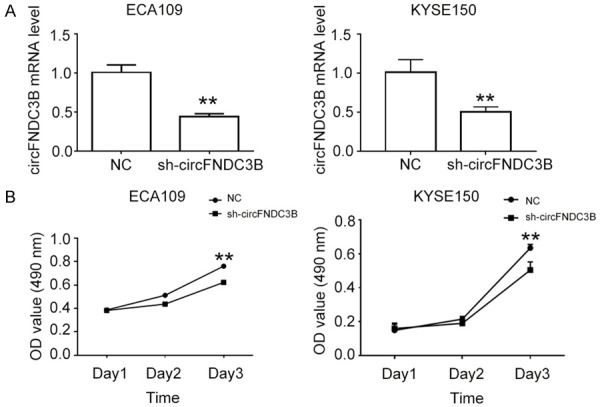
CircFNDC3B promotes esophageal cancer cell proliferation. A. Inhibition of circFNDC3B expression in esophageal cancer cells. The expressions of circFNDC3B in esophageal cancer cell lines ECA109 and KYSE150 by lentivirus-mediated gene silencing, and then qRT-PCR, were performed to measure the expression level. B. Proliferation of esophageal cancer cells with decreased circFNDC3B levels. The MTS Cell Proliferation Assay kit was used for the comparison of cell proliferation rates between esophageal cancer cells with normal and inhibited circFNDC3B expression. NC: negative control; sh-circFNDC3B: cells with inhibited circFNDC3B expression by lentivirus-mediated gene silencing. **P<0.01 vs NC.
CircFNDC3B regulates esophageal cancer cell apoptosis
For a comprehensive understanding of the role of circFNDC3B in esophageal cancer progression, cancer cell apoptosis was analyzed using the FITC Annexin V Apoptosis Detection Kit with 7-AAD (BioLegend, USA) according to the manufacturer’s instructions. Compared with the negative controls, the apoptotic esophageal cancer cell percentages of both ECA109 and KYSE150 were significantly increased in esophageal cancer cells transfected with siRNA (*P<0.05, **P<0.01, Figure 4A and 4B). These results convincingly suggest that circFNDC3B could regulate esophageal cancer progression through multiple cellular processes including cell proliferation and apoptosis regulation.
Figure 4.
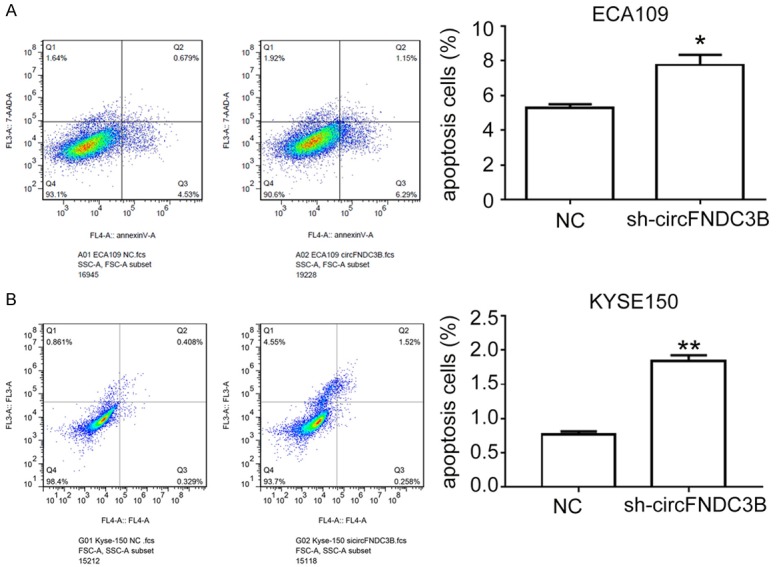
Decreased circFNDC3B expression represses esophageal cancer cell apoptosis. A. Apoptosis of esophageal cancer cell ECA109 with a decreased circFNDC3B level. The apoptosis of ECA109 cells transfected with siRNA, those treated with the FITC Annexin V Apoptosis Detection Kit with 7-AAD and then flow cytometry were performed to measure apoptotic cell percentages. B. Apoptosis of esophageal cancer cell KYSE150 with decreased circFNDC3B levels. The apoptosis of KYSE150 cells transfected with siRNA, treated with the FITC Annexin V Apoptosis Detection Kit with 7-AAD, and then flow cytometry were performed to measure apoptotic cell percentages. NC: negative control; sh-circFNDC3B: cells with inhibited circFNDC3B expression by lentivirus-mediated gene silencing. **P<0.01 vs NC.
CircFNDC3B regulates esophageal cancer cell migration and invasion
To evaluate the role of circFNDC3B in regulating esophageal cancer cell migration and invasion, the migration and invasion activities of ECA109 and KYSE150 cells were subjected to gene silencing and seeded into a transwell system for cell staining and counting. When the circFNDC3B expression level was down-regulated by gene silencing, the cell migration activity of ECA109 and KYSE150 cells was significantly decreased (**P<0.01, Figure 5A). Similarly, the invasion activity of ECA109 and KYSE150 cells was also shown to be greatly impaired in sh-circFNDC3B cells compared with the negative controls (**P<0.01, Figure 5B). The remarkably inhibited cell migration and invasion activities in sh-circFNDC3B cells indicate that circFNDC3B functions as an important regulator of cell migration and invasion during esophageal cancer progression.
Figure 5.
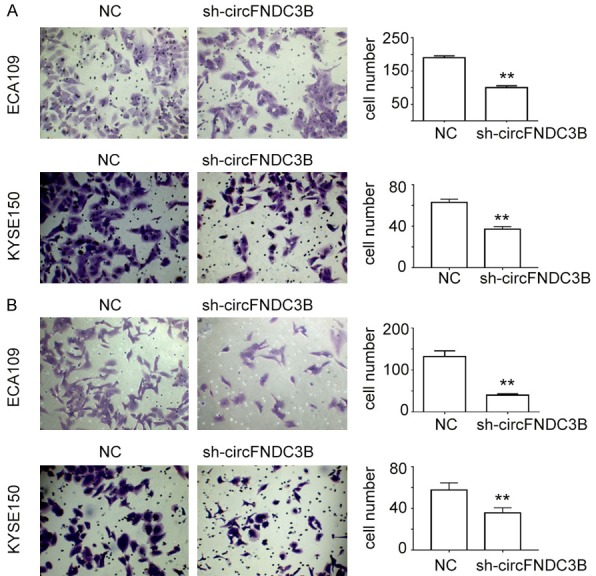
CircFNDC3B promotes esophageal cancer cell migration and invasion. A. Migration of esophageal cancer cell lines ECA109 and KYSE150 with silenced circFNDC3B expression. B. Invasion of esophageal cancer cells ECA109 and KYSE150 with silenced circFNDC3B expression. The migration and invasion of ECA109 and KYSE150 cells were evaluated using a transwell system. NC: negative control; sh-circFNDC3B: cells with inhibited circFNDC3B expression by gene silencing. **P<0.01 vs NC.
Discussion
The expression of circular RNAs in esophageal cancer tissues and cells has been demonstrated by a few previous studies [10,11], but available information about their expressional specificity and sensitivity as esophageal cancer biomarkers, as well as their functioning networks and mechanisms, is far from sufficient. In the present study, we investigated the expression of circFNDC3B in esophageal tissues and cell lines, clearly showing that this circular RNA encoded by the FNDC3B gene exons might be developed as a novel esophageal marker. A previous study showed that another circular RNA, has_circ_0067934, is also up-regulated in ESCC tumor tissues and cell lines [11]. The up-regulation of the two circular RNAs in both esophageal cancer tissue and cell lines indicates that enhanced circular RNA expression might be a prevalent molecular alteration during esophageal cancer progression. On the contrary, we also have reasons to speculate that the expression of certain circular RNAs would be inhibited in esophageal cancer, and the possibility of specifically down-regulated circular RNA molecules in esophageal cancer should also be investigated in future studies.
To investigation the potential roles of circFNDC3B, we repressed its expression by gene silencing in two esophageal cancer cell lines ECA109 and KYSE150, and the results proved that circFNDC3B is an important regulator of esophageal cancer cell proliferation, apoptosis, migration, and invasion, which are all significant cellular processes associated with cell malignant transformation. Similarly, has_circ_0067934 has also been shown to be involved in regulating these key cellular processes [11], indicating that circular RNAs, at least as a group, might function as important upstream components in the complex signaling networks responsible for the initiation and progression of esophageal cancer development. It has been widely accepted that early events during cancer initiation might possess a valuable application potential for early screening and diagnosis of malignant tumors [24]. The pleiotropic regulating roles of circular RNAs shown by recent reports indicate that circular RNAs specifically expressed in esophageal cancer tissues might be suitable early diagnostic markers for esophageal cancer.
The focus of this study was the expression and functions of circFNDC3B in esophageal cancer cell functions, and the underlying molecular mechanisms deserve further investigation. As revealed by recent progress in circular RNA function analysis, circRNAs can perform their biological roles by interacting with miRNAs as sponges and regulating RNA binding protein (RBP), thus modulating gene expression [7,9]. They have also been shown to regulate gene transcription and be translated into functional proteins or peptides [5,6,8,9]. These results show that circRNAs might mainly function as regulators of gene transcription and post-transcriptional modification, through which multiple genes associated with various biological processes might be influenced. This hypothesis was supported by our results and by has_circ_0067934 research showing the involvement of both circRNAs in regulating multiple key cellular processes including proliferation, apoptosis, migration, and invasion [11]. For instance, hsa_circ_001569 positively regulates cell proliferation and invasion of colorectal cancer (CRC) by targeting miR-145, which further regulates E2F5, BAG4, and FMNL2.1 gene expression [13]. We further speculate that FNDC3B might regulate the transcription of key components of cell proliferation and other cellular pathways to break the normal balance of cell proliferation, differentiation, and apoptosis, finally promoting the initiation and progression of esophageal cancer. Further studies of the specific signaling pathways regulated by circFNDC3B and other circular RNAs would shed light on the significant roles and functioning mechanisms mediating their effects on esophageal cancer progression.
Although the expression level of circFNDC3B was shown to be slightly decreased in several cases of esophageal cancer tissues which might be caused by sample collection or individual differences, these statistically significant results clearly demonstrate that circFNDC3B expression is greatly enhanced in clinical esophageal cancer samples, suggesting possible functions of this novel circular RNA in the progression of esophageal cancer.
In summary, we verified the up-regulated expression of one novel circular RNA, circFNDC3B, in esophageal cancer tissues and cell lines. By gene silencing, the important regulating functions of circFNDC3B for esophageal cancer cell proliferation, apoptosis, migration, and invasion were also revealed by in vitro assays in this study, providing the necessary scientific basis for the application of circular RNAs in cancer diagnosis and treatment.
Disclosure of conflict of interest
None.
References
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 2.Zeng H, Zheng R, Zhang S, Zuo T, Xia C, Zou X, Chen W. Esophageal cancer statistics in China, 2011: estimates based on 177 cancer registries. Thoracic Cancer. 2016;7:232–7. doi: 10.1111/1759-7714.12322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tran GD, Sun XD, Abnet CC, Fan JH, Dawsey SM, Dong ZW, Mark SD, Qiao YL, Taylor PR. Prospective study of risk factors for esophageal and gastric cancers in the Linxian general population trial cohort in China. Int J Cancer. 2005;113:456–463. doi: 10.1002/ijc.20616. [DOI] [PubMed] [Google Scholar]
- 4.Stahl M, Mariette C, Haustermans K, Cervantes A, Arnold D. Oesophageal cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;24:vi51–vi56. doi: 10.1093/annonc/mdt342. [DOI] [PubMed] [Google Scholar]
- 5.Dong Y, Dan H, Peng Z, Wei P, Shi W, Wang J, Li B, Zhang C, Duan C. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10:2. doi: 10.1186/s13045-016-0370-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Mo Y, Gong Z, Yang X, Yang M, Zhang S, Xiong F, Xiang B, Zhou M, Liao Q. Circular RNAs in human cancer. Mol Cancer. 2017;16:25. doi: 10.1186/s12943-017-0598-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–8. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 8.Matsumoto T, Urushido M, Ide H, Ishihara M, Hamada-Ode K, Shimamura Y, Ogata K, Inoue K, Taniguchi Y, Taguchi T, Horino T, Fujimoto S, Terada Y. Small heat shock protein Beta-1 (HSPB1) is upregulated and regulates autophagy and apoptosis of renal tubular cells in acute kidney injury. PLoS One. 2015;10:e0126229. doi: 10.1371/journal.pone.0126229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNAs. Biochim Biophys Acta. 2016;1859:1245–51. doi: 10.1016/j.bbagrm.2016.07.009. [DOI] [PubMed] [Google Scholar]
- 10.Su H, Lin F, Deng X, Shen L, Fang Y, Fei Z, Zhao L, Zhang X, Pan H, Xie D, Jin X, Xie C. Profiling and bioinformatics analyses reveal differential circular RNA expression in radioresistant esophageal cancer cells. J Transl Med. 2016;14:225. doi: 10.1186/s12967-016-0977-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia W, Qiu M, Chen R, Wang S, Leng X, Wang J, Xu Y, Hu J, Dong G, Xu PL. Circular RNA has_circ_0067934 is upregulated in esophageal squamous cell carcinoma and promoted proliferation. Sci Rep. 2016;6:35576. doi: 10.1038/srep35576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Song X, Zhang N, Han P, Byoung-San M, Lai RK, Wang K, Lu W. Circular RNA profile in gliomas revealed by identification tool UROBORUS. Nucleic Acids Res. 2016;44:e87. doi: 10.1093/nar/gkw075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qin M, Liu G, Huo X, Tao X, Sun X, Ge Z, Yang J, Fan J, Liu L, Qin W. Hsa_circ_0001649: a circular RNA and potential novel biomarker for hepatocellular carcinoma. Cancer Biomark. 2015;16:161–9. doi: 10.3233/CBM-150552. [DOI] [PubMed] [Google Scholar]
- 16.Shang X, Li G, Liu H, Li T, Liu J, Zhao Q, Wang C. Comprehensive circular RNA profiling reveals that hsa_circ_0005075, a new circular RNA biomarker, is involved in hepatocellular crcinoma development. Medicine. 2016;95:e3811. doi: 10.1097/MD.0000000000003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cai C, Rajaram M, Zhou X, Liu Q, Marchica J, Li J, Powers RS. Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle. 2012;11:1773–1781. doi: 10.4161/cc.20121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lin CH, Lin YW, Chen YC, Liao CC, Jou YS, Hsu MT, Chen CF. FNDC3Bpromotes cell migration and tumor metastasis in hepatocellular carcinoma. Oncotarget. 2016;7:49498–49508. doi: 10.18632/oncotarget.10374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang HY, Mcmahon C, Ali SM, Young LE, Yekezare S, Ross JS, Ball ED. Novel FNDC3B and MECOM fusion and WT1 L378fs* 7 frameshift mutation in an acute myeloid leukaemia patient with cytomorphological and immunophenotypic features reminiscent of acute promyelocytic leukaemia. Br J Haematol. 2015;172:987–990. doi: 10.1111/bjh.13552. [DOI] [PubMed] [Google Scholar]
- 20.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 21.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2012;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO. Cell-type specific features of circular RNA expression. PLoS Genet. 2013;9:e1003777. doi: 10.1371/journal.pgen.1003777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 24.Shah AK, Saunders NA, Barbour AP, Hill MM. Early diagnostic biomarkers for esophageal adenocarcinoma--the current state of play. Cancer Epidemiol Biomarkers Prev. 2013;22:1185–1209. doi: 10.1158/1055-9965.EPI-12-1415. [DOI] [PubMed] [Google Scholar]


