Abstract
Colorectal cancer (CRC) is one of most common cancers and a leading cause of cancer-related death around the world. Identification of reliable biomarkers contributes to facilitate disease detection of this malignancy. This study aimed to explore the serum miR-106b expression in CRC and its potential clinical significance. Quantitative real-time polymerase chain reaction (qRT-PCR) was performed to measure the miR-106b expression levels in the serum from 122 CRC patients, 40 advanced adenomas and 50 healthy individuals. Serum miR-106b levels were significantly increased in CRC patients compared to healthy controls. Elevated serum miR-106b expression occurred more frequently in CRC patients with lymph node metastasis and distant metastasis. Moreover, receiver operating characteristic (ROC) curve analysis revealed that serum miR-106b could well discriminate CRC patients from healthy controls. In addition, miR-106b levels were greatly reduced in post-operative samples from CRC cases with early clinical stage. Furthermore, increased miR-106b expression was positively correlated with aggressive clinical variables and poor prognosis. Finally, serum miR-106b was identified as an independent prognostic predictor for CRC. Collectively, our findings suggest serum miR-106b might potentially serve as a promising biomarker in the diagnosis and prognosis of CRC.
Keywords: Colorectal cancer, MiR-106b, serum, prognosis, biomarker
Introduction
Colorectal cancer (CRC), which represents the fifth leading cause of malignant tumor-related deaths in China, is one of the most common cancers worldwide [1,2]. Although various treatment strategies including surgical resection, chemotherapy and radiotherapy have been developed over the past decades, the clinical outcome and prognosis of advanced CRC remains unfavorable [3,4]. Unfortunately, most patients with CRC are diagnosed at late stages, leading to dismal five-year overall survival [5]. Hence, identification of reliable markers for improving the prognosis of this disease are urgently required.
MiRNAs (miRNAs) are small non-coding RNAs RNA molecules (19~24 nt) that negatively regulate the expression of target messenger RNA (mRNA), resulting in either mRNA degradation or inhibition of its translation [6,7]. Previous studies have revealed that miRNAs are aberrantly expressed in diverse cancers and involved in the process of cell growth, invasion, and apoptosis [8,9]. Since miRNAs are highly stable in the plasma or serum, many serum miRNAs, such as miR-200c [10], miR-218 [11] or miR-21 [12], are used as biomarkers for CRC patients. Advanced adenoma (AD) is considered a high risk factor for CRC. It is defined as an adenoma (at least 10 mm) with significant villous components or high-grade dysplasia, representing the small subpopulation of adenomas that may ultimately progress to invasive cancer [13,14].
MiR-106b, located at Chr7, was found to be dysregulated in various cancers. For example, miR-106b was upregulated in CRC [15-17], cervical cancer [18], hepatocellular carcinoma [19-21], laryngeal carcinoma [22,23], melanoma [24] and pituitary adenoma [25], while downregulated in ovarian carcinoma [28] and gastric cancer [29]. However, no previous study had reported the role of serum miR-106b as an indicator for CRC detection. In this study, we sought to assess the clinical significance of serum miR-106b as a potential biomarker for CRC and AD diagnosis.
Materials and methods
Study population
This study was approved by the Ethics Committee of the Second Hospital of Shandong University. Written informed consent was provided by all participants. All specimens were handled and made anonymous according to the ethical and legal standards.
A total of 122 CRC patients, 40 AD patients and 50 healthy individuals as controls were included in this study. All 122 CRC cases had a clear histologic diagnosis, and none of them had received any treatment prior to surgery. Cancer staging was performed based on the fifth edition of the American Joint Commission on Cancer TNM criteria. The detailed information was obtained from their clinical and pathologic records and list in Table 1. Overall survival (OS) was defined as the time from diagnosis to the date of death or last follow up. Recurrence free survival (RFS) was defined as the time from diagnosis to the date of recurrence or death or last follow up.
Table 1.
Clinical characteristics of 122 subjects and serum miR-106b expression in CRC
| Variables | Total case | MiR-106b | P value | |
|---|---|---|---|---|
|
| ||||
| Low (n=57) | High (n=65) | |||
| Sex | 0.257 | |||
| Male | 75 | 32 (42.7%) | 43 (57.3%) | |
| Female | 47 | 25 (53.2%) | 22 (46.8%) | |
| Age (years) | 0.912 | |||
| <59 | 67 | 31 (46.3%) | 36 (53.7%) | |
| ≥59 | 55 | 26 (47.3%) | 29 (52.7%) | |
| Distant metastasis | 0.005 | |||
| Yes | 24 | 5 (20.8%) | 19 (79.2%) | |
| No | 98 | 52 (53.1%) | 46 (46.9%) | |
| Lymph node invasion | 0.013 | |||
| Yes | 53 | 18 (34.0%) | 35 (66.0%) | |
| No | 69 | 39 (56.5%) | 30 (43.5%) | |
| Tumor size | 0.255 | |||
| <6 cm | 77 | 39 (50.6%) | 38 (49.4%) | |
| ≥6 cm | 45 | 18 (40.0%) | 27 (60.0%) | |
| Tumor location | 0.243 | |||
| Colon | 51 | 27 (52.9%) | 24 (47.1%) | |
| Rectum | 71 | 30 (42.3%) | 41 (57.7%) | |
| Differentiation | 0.130 | |||
| Well/Moderate | 93 | 47 (50.5%) | 46 (49.5%) | |
| Poor | 29 | 10 (34.5%) | 19 (65.5%) | |
| Tumor invasion | 0.062 | |||
| T1/T2 | 43 | 25 (58.1%) | 18 (41.9%) | |
| T3/T4 | 79 | 32 (40.5%) | 47 (59.5%) | |
| TNM stage | <0.001 | |||
| I/II | 70 | 46 (65.7%) | 24 (34.3%) | |
| III/IV | 52 | 11 (21.2%) | 41 (78.8%) | |
Blood samples and RNA extraction
Peripheral blood was withdrawn from each participant and collected in EDTA-containing tubes. Blood samples were centrifuged at 2000 g for 10 min within an hour, and the supernatant was collected and centrifuged at 12000 g for 10 min, and stored at -80°C for RNA extraction. Total RNA was isolated from serum using Trizol reagent (Invitrogen, Carlsbad, CA, US) according to the manufacturer’s instructions, and RNA purity and concentrations were determined by a NanoDrop ND-1000 spectrophotometer (Wilmington, DE, US) at 260 and 280 nm.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Real-time PCR was carried out using a Taqman MicroRNA Assay Kit (Applied Biosystems, Foster City, CA, USA) on a 7500-fast real-time system (Applied Biosystems, Foster City, CA, USA). Cer-miR-39 was used as an internal control, and the relative amount of miR-106b to cer-miR-39 was calculated using the 2-ΔΔCt method. The primers were as follows: miR-106b primer, 5’-GACGCTAAAGTGCTGACAGT-3’ (forward), 5’-GTGCAGGGTCCGAGGT-3’ (reverse); cer-miR-39 primer, 5’-CAGAGTCACCGGGTGTAAAT-3’ (forward), 5’-CCAGTGCGTGTCGTGGAGTC-3’ (reverse). All qRT-PCRs were performed in triplicate.
Statistical analysis
All analyses were conducted using IBM SPSS 16.0 software (SPSS Inc, Chicago, IL, USA). The significance in serum miR-106b levels between groups was compared with the Mann-Whitney U test or Kruskal-Wallis test. Chi-square test was used to evaluate the relationship between miR-106b levels and clinicopathological characteristics. Receiver-operator characteristic (ROC) curve and the area under the ROC curve (AUC) was performed to assess the CRC detection potential of miR-106b. Cumulative survival rates were calculated by Kaplan-Meier method with log-rank tests. Multivariate Cox proportional hazards regression analysis was performed to assess the association between hazard ratio (HR) and survival. P<0.05 was considered statistically significant.
Results
Serum miR-106b expression is upregulated in CRC samples
MiR-106b levels in all samples were measured using qRT-PCR. The levels of miR-106b were significantly elevated in CRC patients and AD cases compared with the controls (Figure 1, both P<0.05). Moreover, CRC patients with distant metastasis exhibited higher miR-106b expression levels than those without (Figure 2A, both P<0.01). Likewise, a significantly increased miR-106b expression was observed in CRC patients with lymph node metastasis compared to those without (Figure 2B, both P<0.01).
Figure 1.
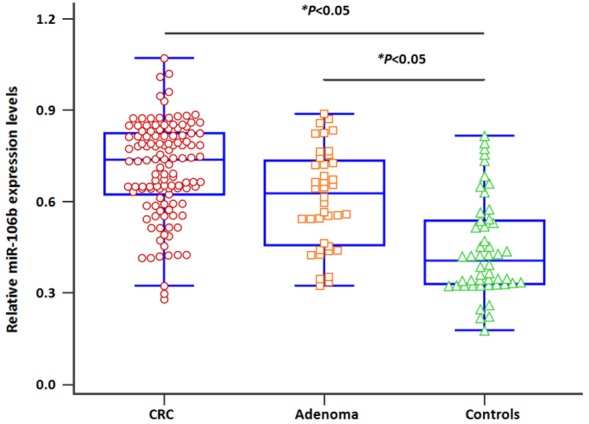
Serum miR-106b levels in CRC and advanced adenoma (AD) cases were higher than those of controls.
Figure 2.
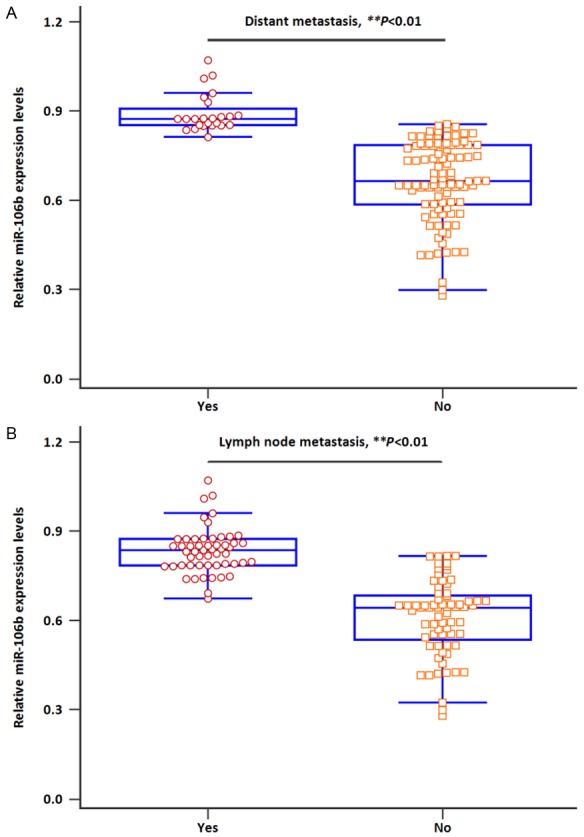
A. Serum miR-106b expression was higher in patients with distant metastasis. B. Serum miR-106b expression was higher in patients with lymph node metastasis.
The diagnostic value of miR-106b for CRC and AD
Next, ROC curve analysis was performed to validate the diagnostic potential of miR-106b as a non-invasive biomarker. As depicted in Figure 3A, serum miR-106b could successfully discriminate CRC patients from controls with an AUC of 0.869, and the diagnostic sensitivity and specificity were 85.2% and 78.0%, respectively. In addition, Figure 3B showed that serum miR-106b could be considered as a promising indicator for discriminating AD cases from healthy individuals, with a sensitivity and specificity of 72.5% and 76.0%, respectively (AUC: 0.776).
Figure 3.
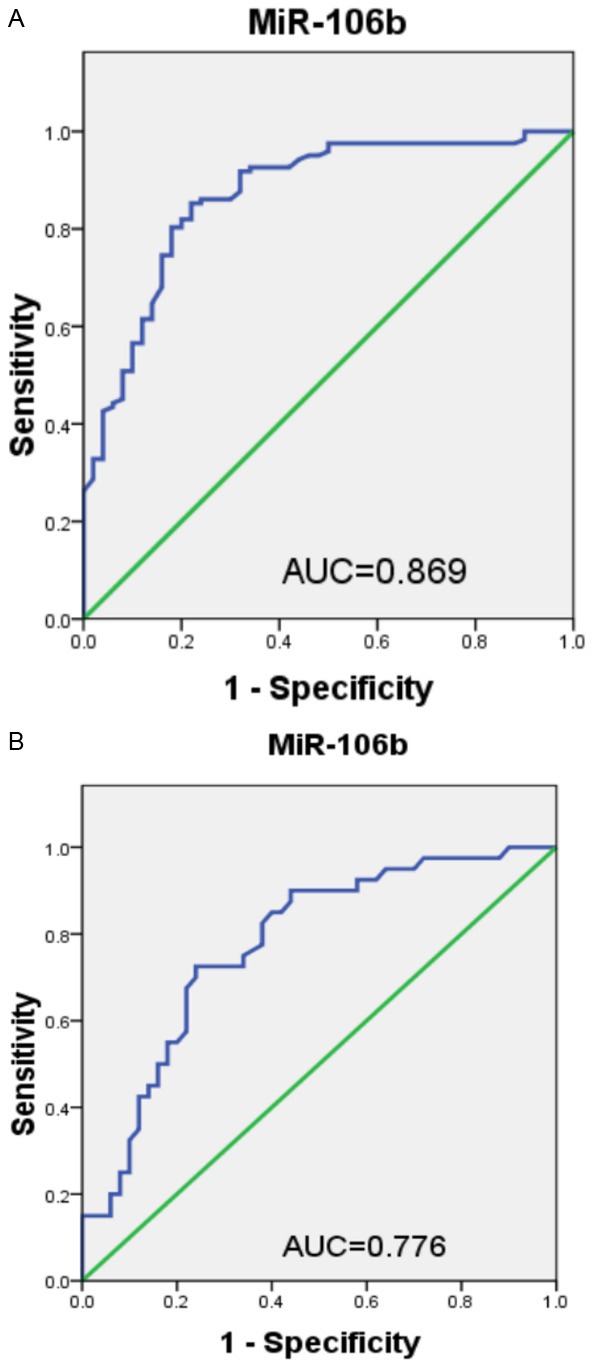
A. Diagnostic potential of serum miR-106b for CRC. B. Diagnostic potential of serum miR-106b for advanced adenoma (AD).
Relationship between serum miR-106b expression and clinical characteristics
Subsequently, the pre-operative serum was classified into high miR-106b and low miR-106b expression groups by using the median miR-106b value, and was shown in combination with the clinical features of CRC subjects in Table 1. Results revealed that serum miR-106b expression levels were closely correlated with TNM stage, distant metastasis and lymph node metastasis (all P<0.05). However, no significant difference was noted between serum miR-106b expression and sex, age, tumor size, tumor location, differentiation, as well as tumor invasion (all P>0.05).
Relationship of serum miR-106b expression with treatment response
The changes of serum miR-106b levels in seventy CRC patients with stage I/II were detected before and 1-3 months after treatment. Interestingly, a statistically significant reduction in miR-106b level was observed (Figure 4, P<0.05).
Figure 4.
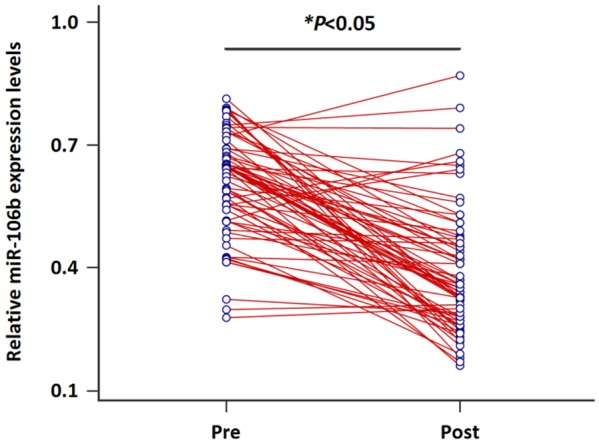
Changes of serum miR-106b levels in paired blood samples of early stage CRC patients.
Serum miR-106b expression and prognosis
To evaluate the prognostic significance of miR-106b, Kaplan-Meier survival analysis and multivariate analysis were carried out. The data demonstrated that CRC patients in the high miR-106b group had a significantly shorter OS (P=0.018, Figure 5A) and RFS (P=0.014, Figure 5B) than those in the low miR-106b group. By multivariate analysis, serum miR-106b (OS: HR=3.43, 95% CI=1.91-5.24, P=0.005; RFS: HR=3.87, 95% CI=2.15-5.71, P=0.002), distant metastasis (OS: HR=2.33, 95% CI=1.42-3.35, P=0.018; RFS: HR=3.15, 95% CI=1.76-4.78, P=0.007), lymph node metastasis (OS: HR=2.56, 95% CI=1.51-3.63, P=0.015; RFS: HR=3.04, 95% CI=1.72-4.52, P=0.008) and TNM stage (OS: HR=4.12, 95% CI=2.23-6.26, P=0.001; RFS: HR=5.23, 95% CI=2.56-8.17, P<0.001) were confirmed to have significant correlations with OS/RFS, suggesting all of these variables were potential risk factors for OS/RFS in CRC (Table 2).
Figure 5.
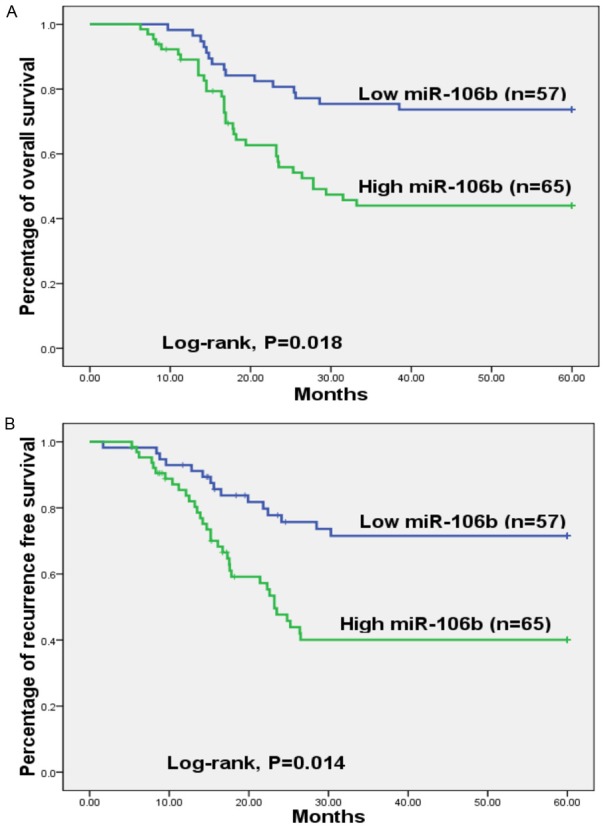
A. Patients with low miR-106b expression had a longer overall survival. B. Patients with low miR-106b expression had a longer recurrence-free survival.
Table 2.
Multivariate analysis for OS/RFS of CRC patients
| Clinical characteristic | Multivariate analysis | |
|---|---|---|
|
| ||
| HR (95% CI) | P value | |
| Overall survival | ||
| Serum miR-106b, high vs. low | 3.43 (1.91-5.24) | 0.005 |
| Distant metastasis, yes vs. no | 2.33 (1.42-3.35) | 0.018 |
| Lymph node invasion, yes vs. no | 2.56 (1.51-3.63) | 0.015 |
| TNM stage, III/IV vs. I/II | 4.12 (2.23-6.26) | 0.001 |
| Recurrence-free survival | ||
| Serum miR-106b, high vs. low | 3.87 (2.15-5.71) | 0.002 |
| Distant metastasis, yes vs. no | 3.15 (1.76-4.78) | 0.007 |
| Lymph node invasion, yes vs. no | 3.04 (1.72-4.52) | 0.008 |
| TNM stage, III/IV vs. I/II | 5.23 (2.56-8.17) | <0.001 |
HR, hazard ratio; CI, confidence interval.
Discussion
The current study investigated the potential clinical utility of miR-106b to serve as a diagnostic and prognostic marker in CRC patients. We found that serum miR-106b levels were highest in CRC cases, and significantly lower in healthy controls. Serum miR-106b expression was frequently and highly expressed in CRC patients with lymph node metastasis and distant metastasis. Moreover, ROC analysis showed serum miR-106b levels had high sensitivity and specificity in CRC diagnosis. Next, the serum concentration of miR-106b was dramatically reduced in the blood samples from I/II stage CRC patients after surgery. Also, high serum miR-106b expression was associated with aggressive clinical variables and worse OS/RFS, and emerged as an independent prognostic indicator in CRC patients.
Several previous reports suggested that miR-106b played an oncogenic role in CRC. Zhang et al reported miR-106b overexpression was observed in both metastatic CRC tissues and cell lines, and strongly correlated with worse clinical variables and shorter survival. Furthermore, high miR-106b expression stimulated cancer cell migration and invasion, and DLC1 was identified as its downstream target gene [15]. Wang et al performed a miRNA microarray and showed miR-106b was higher in colonic cancer tissues without lymph node metastasis than that in the para-cancerous tissues [16]. In vitroand in vivo analysis revealed CRC cell lines that express high miR-106b levels were more resistant to ionizing radiation, while miR-106b knockdown produced an opposite effect. Moreover, miR-106b upregulation increased the tumor-initiating cell ability under normal and ionizing radiation conditions. Both PTEN and p21 were confirmed to be the target genes [17]. Our data are consistent with the findings of these reports.
Besides CRC, accumulating evidence had suggested that miR-106b exerted oncogenic properties in various human cancers. Cheng et al demonstrated that miR-106b was highly expressed in tissues and cell lines of cervical cancer. Overexpressed MiR-106b, downregulating the expression of DAB2, greatly promoted the migration ability of cancer cell lines, and vice versa [18]. In hepatocellular carcinoma (HCC), Yen et al reported miR-106b expression was dramatically elevated in hepatitis B virus-associated HCC tissues, and its expression was inversely associated with shorter survival and poor differentiation of patients [19]. Likewise, increased miR-106b expression was found in HCC tissues, and highly correlated with reduced overall survival and aggressive clinical parameters [20]. Another report by Yau et al reported miR-106b was upregulated in HCC tissues. High miR-106b expression not only occurred more frequently in HCC patients with higher tumor grade, but also enhanced cell motility ability in vitro and metastatic ability in vivo [21]. In laryngeal carcinoma, miR-106b levels were dramatically lower in low stage carcinoma tissues than those with high stage, and repression of miR-106b significantly reduced cancer cell proliferation and stimulated cell cycle G0/G1 arrest via targeting RB [22], or inhibited tumorigenesis through targeted regulation of RUNX3 [23]. In melanoma, Prasad et al verified that an inverse association between miR-106b and the levels of p21/WAF1/Cip1 protein in cancer cells. Deregulated miR-106b expression repressed cell viability and proliferation, as well as resulted in cell cycle G0/G1 arrest in cells [24]. Zheng and colleagues found significantly higher miR-106b expression in pituitary adenoma tissues than that in normal samples. MiR-106b silencing greatly suppressed tumor cell proliferation, migration, invasion and anchorage-independent growth by directly regulating PTEN via PI3K/Akt pathway [25]. Similarly, Li et al reported miR-106b directly targeted PTEN, affected the activation of PI3K/Akt signal pathway, and enhanced tumorigenic properties of breast cancer (BC) cells in vitro and in vivo. Furthermore, miR-106b was overexpressed both in BC tissues and cell lines [26]. However, Ni et al demonstrated that miR-106b expression was negatively correlated with the expression of MMP2, which was closely related to bone metastasis. In addition, in vitro evidence showed ectopic miR-106b expression markedly suppressed the carcinogenesis of BC cells [27].
Additionally, miR-106b was reported to act as a tumor suppressor in some cancer types. In ovarian carcinoma, reduced miR-106b expression was found in cancerous tissues and inversely associated with worse cancer phenotype. Forced miR-106b expression significantly decreased cell migration in vitro and attenuated tumor growth in nude mice model [28]. The expression of serum miR-106b was greatly downregulated in patients with gastric cancer and benign gastric diseases compared to healthy volunteers, and a significant correlation was found between reduced miR-106b expression and elderly patients [29]. Based on these controversial findings, we suggest that the diverse function of miR-106b might depend on different tumor types.
In the current study, we demonstrate that serum miR-106b not only shows a high specificity and sensitivity for CRC diagnosis, but also has important clinical significance for the prognosis of CRC patients. Thus, serum miR-106b may serve as a non-invasive and promising indicator for patients with CRC.
Acknowledgements
This study was supported partly by the Digestive Department of The Second Hospital of Shandong University.
Disclosure of conflict of interest
None.
References
- 1.Liu S, Zheng R, Zhang M, Zhang S, Sun X, Chen W. Incidence and mortality of colorectal cancer in China, 2011. Chin J Cancer Res. 2015;27:22–28. doi: 10.3978/j.issn.1000-9604.2015.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 3.Migliore L, Migheli F, Spisni R, Coppedè F. Genetics, cytogenetics, and epigenetics of colorectal cancer. J Biomed Biotechnol. 2011;2011:792362. doi: 10.1155/2011/792362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chibaudel B, Tournigand C, André T, de Gramont A. Therapeutic strategy in unresectable metastatic colorectal cancer. Ther Adv Med Oncol. 2012;4:75–89. doi: 10.1177/1758834011431592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kraus S, Nabiochtchikov I, Shapira S, Arber N. Recent advances in personalized colorectal cancer research. Cancer Lett. 2014;347:15–21. doi: 10.1016/j.canlet.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 8.Zheng K, Li H, Huang H, Qiu M. MicroRNAs and glial cell development. Neuroscientist. 2012;18:114–118. doi: 10.1177/1073858411398322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 10.Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-200c is a novel prognostic and metastasis-predictive biomarker in patients with colorectal cancer. Ann Surg. 2014;259:735–743. doi: 10.1097/SLA.0b013e3182a6909d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu H, Gao G, Jiang L, Guo L, Lin M, Jiao X, Jia W, Huang J. Decreased expression of miR-218 is associated with poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2013;6:2904–2911. [PMC free article] [PubMed] [Google Scholar]
- 12.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y, Kusunoki M, Boland CR, Goel A. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105:849–859. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Butterly LF, Pohl H. CT colonography versus colonoscopy for the detection of advanced neoplasia. N Engl J Med. 2008;358:89. [PubMed] [Google Scholar]
- 14.El-Maraghi RH, Kielar AZ. CT colonography versus optical colonoscopy for screening asymptomatic patients for colorectal cancer a patient, intervention, comparison, outcome (PICO) analysis. Acad Radiol. 2009;16:564–571. doi: 10.1016/j.acra.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 15.Zhang GJ, Li JS, Zhou H, Xiao HX, Li Y, Zhou T. MicroRNA-106b promotes colorectal cancer cell migration and invasion by directly targeting DLC1. J Exp Clin Cancer Res. 2015;34:73. doi: 10.1186/s13046-015-0189-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YX, Zhang XY, Zhang BF, Yang CQ, Chen XM, Gao HJ. Initial study of microRNA expression profiles of colonic cancer without lymph node metastasis. J Dig Dis. 2010;11:50–54. doi: 10.1111/j.1751-2980.2009.00413.x. [DOI] [PubMed] [Google Scholar]
- 17.Zheng L, Zhang Y, Liu Y, Zhou M, Lu Y, Yuan L, Zhang C, Hong M, Wang S, Li X. MiR-106b induces cell radioresistance via the PTEN/PI3K/AKT pathways and p21 in colorectal cancer. J Transl Med. 2015;13:252. doi: 10.1186/s12967-015-0592-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng Y, Guo Y, Zhang Y, You K, Li Z, Geng L. MicroRNA-106b is involved in transforming growth factor β1-induced cell migration by targeting disabled homolog 2 in cervical carcinoma. J Exp Clin Cancer Res. 2016;35:11. doi: 10.1186/s13046-016-0290-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yen CS, Su ZR, Lee YP, Liu IT, Yen CJ. MiR-106b promotes cancer progression in hepatitis B virus-associated hepatocellular carcinoma. World J Gastroenterol. 2016;22:5183–5192. doi: 10.3748/wjg.v22.i22.5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li BK, Huang PZ, Qiu JL, Liao YD, Hong J, Yuan YF. Upregulation of microRNA-106b is associated with poor prognosis in hepatocellular carcinoma. Diagn Pathol. 2014;9:226. doi: 10.1186/s13000-014-0226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yau WL, Lam CS, Ng L, Chow AK, Chan ST, Chan JY, Wo JY, Ng KT, Man K, Poon RT, Pang RW. Over-expression of miR-106b promotes cell migration and metastasis in hepatocellular carcinoma by activating epithelial-mesenchymal transition process. PLoS One. 2013;8:e57882. doi: 10.1371/journal.pone.0057882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cai K, Wang Y, Bao X. MiR-106b promotes cell proliferation via targeting RB in laryngeal carcinoma. J Exp Clin Cancer Res. 2011;30:73. doi: 10.1186/1756-9966-30-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu Y, Wang K, Gao W, Zhang C, Huang F, Wen S, Wang B. MicroRNA-106b regulates the tumor suppressor RUNX3 in laryngeal carcinoma cells. FEBS Lett. 2013;587:3166–3174. doi: 10.1016/j.febslet.2013.05.069. [DOI] [PubMed] [Google Scholar]
- 24.Prasad R, Katiyar SK. Down-regulation of miRNA-106b inhibits growth of melanoma cells by promoting G1-phase cell cycle arrest and reactivation of p21/WAF1/Cip1 protein. Oncotarget. 2014;5:10636–10649. doi: 10.18632/oncotarget.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Z, Zhang Y, Zhang Z, Yang Y, Song T. Effect of miR-106b on invasiveness of pituitary adenoma via PTEN-PI3K/AKT. Med Sci Monit. 2017;23:1277–1285. doi: 10.12659/MSM.900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, Miao Y, Shan Y, Liu B, Li Y, Zhao L, Jia L. MiR-106b and miR-93 regulate cell progression by suppression of PTEN via PI3K/Akt pathway in breast cancer. Cell Death Dis. 2017;8:e2796. doi: 10.1038/cddis.2017.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ni X, Xia T, Zhao Y, Zhou W, Wu N, Liu X, Ding Q, Zha X, Sha J, Wang S. Downregulation of miR-106b induced breast cancer cell invasion and motility in association with overexpression of matrix metalloproteinase 2. Cancer Sci. 2014;105:18–25. doi: 10.1111/cas.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen S, Chen X, Xiu YL, Sun KX, Zhao Y. Inhibition of ovarian epithelial carcinoma tumorigenesis and progression by microRNA 106b mediated through the RhoC pathway. PLoS One. 2015;10:e0125714. doi: 10.1371/journal.pone.0125714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Q, Jin C, Chen W, Xia F, Wang Q, Fan F, Du J, Guo Y, Lin C, Yang K, Li J, Peng X, Li X, Cao K. Downregulation of serum miR-17 and miR-106b levels in gastric cancer and benign gastric diseases. Chin J Cancer Res. 2014;26:711–716. doi: 10.3978/j.issn.1000-9604.2014.12.03. [DOI] [PMC free article] [PubMed] [Google Scholar]


