Abstract
Background: Aberrant DNA methylation plays an important role in cancer and has been recognized to contribute to the activity of oncogenes and inactivity of tumor suppressor genes. RAS association domain family (RASSF) members have been shown to be epigenetically silenced by promoter methylation in cancers, including hepatoblastoma. Methods: We assessed the methylation patterns in the gene of RASSF5 from hepatoblastoma tissue samples harvested from patients using high-throughput mass spectrometry on a matrix-assisted laser desorption/ionization time-of-flight mass array. Results: Hypermethylation was found in the RASSF5 gene transcribed regionand was correlated with downregulation of RASSF5 RNA expression levels in the hepatoblastoma samples. Conclusions: The results indicate that aberrant methylation of RASSF5 may contribute to its downregulated mRNA expression in hepatoblastoma.
Keywords: RASSF5, methylation, hepatoblastoma
Introduction
Hepatoblastoma (HB) is the most common liver tumor in children with 0.5-1.5 cases per million children per year and an increasing incidence [1,2]. Previous studies suggested an increases risk in patients with familial adenomatous polyposis coli, both low and high birth weights, maternal tobacco exposure, and constitutional trisomy [3-6]. However, the etiology of HB remains obscure. Currently, despite recent advances in the treatment of HB, the mortality rate is 35-50% in high-risk patients [7], and alpha-fetoprotein level, histological analysis, tumor resectability, and metastasis are the only prognosis factors for HB. To better understand the underlying pathophysiology and treatment of this disease, novel targets for early detection and improved therapies and prognosis are required.
DNA methylation is one of the key mechanisms of epigenetic alteration and the identification of DNA methylation is important for understanding cancer pathogenesis. Hypermethylation of DNA in promoters is recognized to silence genes, and many studies have focused on promoter methylation patterns [8]. Recently, researchers have shown that aberrant DNA methylation in transcribed regions of genes is also correlated with gene expression [9-11].
The RAS association domain family (RASSF), including 10 members (RASSF1-10), was recognized to be frequently inactivated by promoter hypermethylation in cancers including HB [12-14]. Previously, we performed a genome-wide analysis of DNA methylation in HB tissues to identify novel targets for further study of HB, and found distinctly higher levels of methylation in HB tissues compared with non-tumor tissue [15], including in regions around the RASSF5 gene. In this study, we used high-throughput mass spectrometry on a matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass array to detected methylation changes in RASSF5 in HB tissues and investigated the possibility of RASSF5 becoming a therapeutic target in HB.
Material and methods
Sample collection and clinical information
Human HB and non-tumor liver tissues were obtained from patients who underwent resection of HB in the Children’s Hospital, Fudan University, Shanghai, China. Informed consent was obtained from the legal guardians of the patients. Clinical and pathologic data for these patients are listed in Table 1. The use of these human samples was approved by the Ethics Committee of the Children’s Hospital of Fudan University. HB tumors and non-tumor tissue sections were stored at -80°C immediately after surgery until the time of analysis.
Table 1.
Clinical and pathological characteristics of the study subjects
| Case | Age (months) | Sex | Diagnosis type | Alpha-fetoprotein (ng/ml) |
|---|---|---|---|---|
| 1 | 7 | Male | Mixed embryonal/fetal subtype | 68490 |
| 2 | 23 | Male | Mixed embryonal/fetal subtype | >121,000 |
| 3 | 11 | Female | Mixed embryonal/fetal subtype | >121,000 |
| 4 | 10 | Female | Mixed embryonal/fetal subtype | >121,000 |
| 5 | 20 | Female | Mixed embryonal/fetal subtype | >121,000 |
| 6 | 7 | Male | Mixed embryonal/fetal subtype | >121,000 |
| 7 | 30 | Male | Epithelial type | >121,000 |
| 8 | 19 | Male | Epithelial type | >121,000 |
| 9 | 7 | Female | Epithelial type | >121,000 |
DNA/RNA extraction and quantitative reverse transcription-PCR (qRT-PCR)
DNA was extracted from nine HB primary tumors and adjacent non-tumor tissues and stored until subsequent use for mass spectrometry analysis. Total RNA was isolated from HB and normal tissues using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s instructions and then reverse transcribed using a PrimeScript RT reagent kit (Perfect Real Time) with Gdna Eraser (Takara Biotechnology Co., Ltd., Dalian, China). qRT-PCRs were conducted using qPCR with a SYBRGreen PCR kit. Gene expression was normalized to the GAPDH expression level and represented as fold-change by the 2-ΔΔCt method and statistically analyzed [16]. The RASSF5 primers used for q-PCR were as follows: Forward, 5’-TGCTTGATCTCCTGCAGTGT-3’ and reverse, 5’-TCTCCAGAAAGCACCCTCAC-3’ (length, 20 bp).
Mass spectrometry
RASSF5 gene primers were designed to cover the regions with the most CpG sites. The genome DNA was treated with bisulfite, and a T7-promoter tag was attached to the reverse primer for the subsequent PCR amplification. The DNA samples were treated with shrimp alkaline phosphatase (SAP) in vitro transcription and uracil-specific cleavage, then robotically dispensed onto silicon matrix preloaded chips (SpectroCHIP; Sequenom, San Diego, CA, USA). Mass spectra were collected with a MassARRAY Compact MALDI-TOF system (Sequenom), and the methylation ratios of the spectra were generated using EpiTYPER software v1.0 (Sequenom).
Statistical methods
All data are presented as mean ± standard error of the mean and comparisons between groups were evaluated using a two-tailed Student’s t-test unless otherwise specified. Statistical analyses and graphical depiction of data were generated using GraphPad Prism 6.0 (GraphPad Software, Inc., La Jolla, CA, USA) and SPSS 19.0 software (IBM SPSS, Armonk, NY, USA) for Windows. P<0.05 was considering to be statistically significant.
Results
RASSF5 gene transcribed region (body) methylation is elevated in HB
We collected nine pairs of HB and adjacent normal liver tissue samples and detected the methylation patterns of RASSF5 in 9 HB in these tissues. The primers covered most of the regions with the most CpG sites. The CpGs located in the promoter regions could not be detected, but the CpGs located in the gene body were typically methylated. Five sites in the RASSF5 body regions were suitable for analysis. Using two-way hierarchical cluster analysis, we found that all five CpG sites had a significantly higher degree of methylation in the HB tissues compared with the same sites in the non-tumor tissues. In addition, these sites had a significantly higher level of methylation of the RASSF5 gene between HB tissues and their non-tumor counterparts (0.6391±0.02630 vs. 0.4688±0.02728, respectively; P<0.01; Figure 1).
Figure 1.
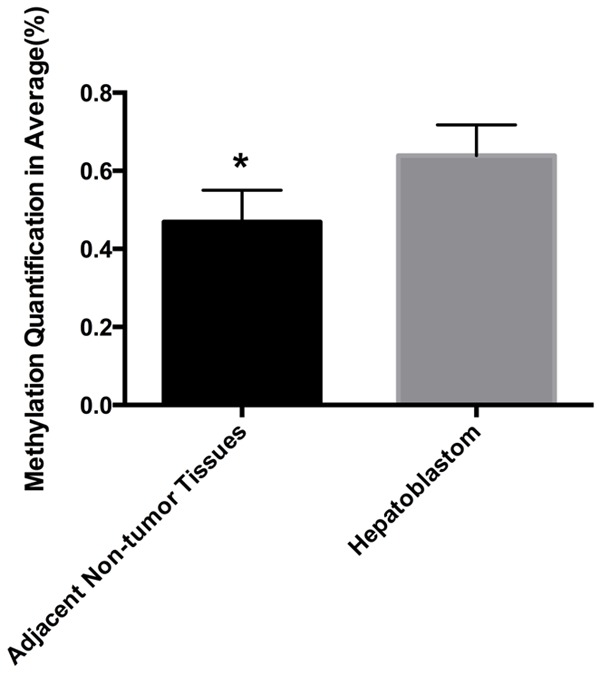
Methylation levels of RASSF5 body region were significantly elevated in the hepatoblastoma tissue samples (n=9) compared with adjacent non-tumor tissues (*P<0.01).
RASSF5 mRNA expression levels are reduced in HB
To determine whether RASSF5 is involved in tumorigenesis, we detected RASSF5 mRNA expression levels using qPCR. We found the expression level of RASSF5 was significantly downregulated in the HB tissues compared with the matched non-tumor liver tissues (0.0051±0.0013 vs. 0.0261±0.0020, respectively; P<0.01; Figure 2).
Figure 2.
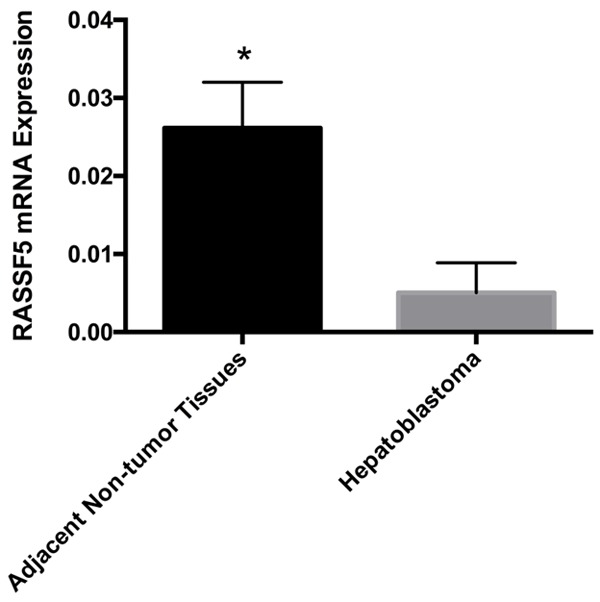
RASSF5 mRNA expression levels were significantly reduced in the HB tissue samples compared with the adjacent non-tumor tissues (*P<0.01).
Correlation between RASSF5 mRNA expression and percentage of DNA methylation
Using linear Pearson’s R correlation, we analyzed the correlations between RASSF5 mRNA expression and DNA methylation status of the CpG sites in the nine pairs of samples. Our data showed that RASSF5 mRNA expression was negatively correlated with its level of methylation (r=-0.5644; P=0.0147; Figure 3).
Figure 3.
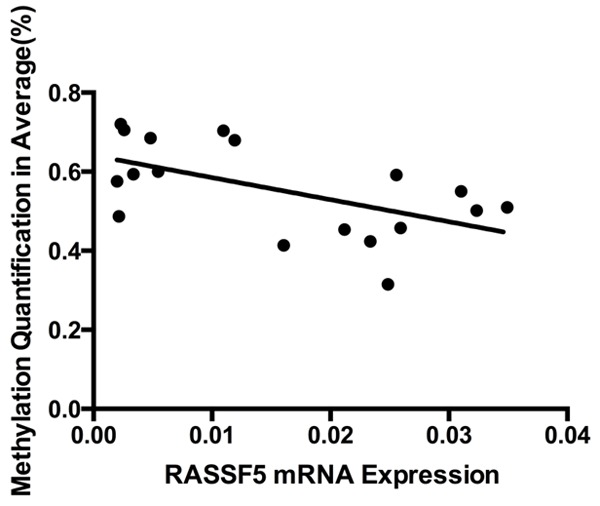
Correlation analysis of RASSF5 mRNA expression and the percentage of DNA methylation in nine hepatoblastoma primary tissues (two-tailed Pearson correlation: r=-0.05644; P=0.0147).
Discussion
DNA methylation and its effect on gene expression have been studied extensively for the last two decades [17]. Generally, methylation changes in CpG islands, CpG shores and CpG shelves [18], regulates several biological processes, including X chromosome inactivation, genomic imprinting, gene transcription and chomatin modification [19-21]. Most of these studies focused on gene promoter methylation [22,23]. Hypermethylation or hypomethylation of gene promoter regions can result in transcriptional silencing or activation [17]. It is generally recognized that hypermethylation of suppressor gene promoters or hypomethylation of oncogene promoters contribute to tumorigenesis. Although several studies have investigated how gene body DNA methylation impacts gene expression [10,24,25], the function of gene body DNA methylation still remains obscure. It has been suggested that gene body methylation may increase transcriptional activity by affecting the initiation of intragenic promoters or the activities of repetitive DNAs with the transcriptional unit [25]. Gene body DNA demethylation may lead to nucleosome destabilization in transcribed regions and reduced efficiencies of transcription elongation or slicing [11].
In our previous genome-wide analysis of DNA methylation in normal and HB liver tissues, which we performed using an Infinium Human Methylation 450 Beadchip, the data showed distinctively less methylation in positions near the RASSF5 gene [15]. To confirm the association between HB and RASSF5 methylation, in the present study, we performed a MALDI-TOF MS analysis of nine pair of tumor and adjacent normal liver tissues to specifically detect the RASSF5 methylation status.
RASSF5, also known as NORE1 (Novel Ras Effector 1), is the most commonly studied methylated gene in cancer so far. It is localized at 1q32.1 and has been reported as methylated and silenced in neuroblastoma, Wilms tumor, hepatocellular carcinoma and other cancer [26-30]. We detected five CpG sites in the gene body region of RASSF5 from each sample, and the degree of methylation of all five sites was significantly higher in the HB samples than in the normal liver tissues. In an independent cohort of nine adjacent HB-non-tumor tissue pairs, we investigated whether there was a correlation between RASSF5 methylation and mRNA expression. The RASSF5 mRNA expression levels in the nine paired samples was detected by qPCR, and the results showed that the RASSF5 mRNA expression levels in normal liver tissues were significantly higher than in HB tissues. Furthermore, the correlation analysis suggested that the methylation status of RASSF5 was significantly negatively correlated with its mRNA expression levels, which is in agreement with previously published data suggesting that RASSF5 may be an oncogene.
To our knowledge, this is the first report of hypermethylation of RASSF5 in HB. Although we failed to detect CpGs sites in the promoter regions of the RASSF5 gene, we found that DNA hypermethylation occurred in the RASSF5 gene body. Previous studies have suggested that gene body DNA methylation may be an intriguing additional target for therapy [11], so the results of the present study may help in understanding the role of DNA methylation in HB.
In conclusion, the current study indicated that aberrant body methylation of RASSF5 may contribute to its upregulated mRNA expression in HB. However, because HB is an uncommon disease of children, our sample size was limited. Therefore, further studies are required to fully understand whether aberrant methylation of RASSF5 is a consequence of gene expression or it can decrease gene expression levels.
Acknowledgements
This work was supported by grants from the China-US (NFSC-NIH) Program for Biomedical Collaborative Research (Grant No.812111102), the National Natural Science Foundation of China (Grant No.81171986), Research Grant from the Ministry of Public Health (Grant No.20110110001), the Basic and Advanced Technology Research Foundation from Science and Technology Department of Henan Province (Grant No.122300410155), Creative Research Team of Higher Education of Henan Province and the Innovation Team of the First Affiliated Hospital of Zhengzhou University, Shanghai Hospital Development Center (SHDC12014106), Shanghai Key Disciplines (no.2017ZZ02022).
Disclosure of conflict of interest
None.
References
- 1.McLaughlin CC, Baptiste MS, Schymura MJ, Nasca PC, Zdeb MS. Maternal and infant birth characteristics and hepatoblastoma. Am J Epidemiol. 2006;163:818–828. doi: 10.1093/aje/kwj104. [DOI] [PubMed] [Google Scholar]
- 2.Linabery AM, Ross JA. Trends in childhood cancer incidence in the U.S. (1992-2004) Cancer. 2008;112:416–432. doi: 10.1002/cncr.23169. [DOI] [PubMed] [Google Scholar]
- 3.Hirschman BA, Pollock BH, Tomlinson GE. The spectrum of APC mutations in children with hepatoblastoma from familial adenomatous polyposis kindreds. J Pediatr. 2005;147:263–266. doi: 10.1016/j.jpeds.2005.04.019. [DOI] [PubMed] [Google Scholar]
- 4.Spector LG, Feusner JH, Ross JA. Hepatoblastoma and low birth weight. Pediatr Blood Cancer. 2004;43:706. doi: 10.1002/pbc.20122. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KJ, Williams KS, Ross JA, Krailo MD, Tomlinson GE, Malogolowkin MH, Feusner JH, Spector LG. Parental tobacco and alcohol use and risk of hepatoblastoma in offspring: a report from the children’s oncology group. Cancer Epidemiol Biomarkers Prev. 2013;22:1837–1843. doi: 10.1158/1055-9965.EPI-13-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valentin LI, Perez L, Masand P. Hepatoblastoma associated with trisomy 18. J Pediatr Genet. 2015;4:204–206. doi: 10.1055/s-0035-1565265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.von Schweinitz D. Hepatoblastoma: recent developments in research and treatment. Semin Pediatr Surg. 2012;21:21–30. doi: 10.1053/j.sempedsurg.2011.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Pandiyan K, You JS, Yang X, Dai C, Zhou XJ, Baylin SB, Jones PA, Liang G. Functional DNA demethylation is accompanied by chromatin accessibility. Nucleic Acids Res. 2013;41:3973–3985. doi: 10.1093/nar/gkt077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pei L, Choi JH, Liu J, Lee EJ, McCarthy B, Wilson JM, Speir E, Awan F, Tae H, Arthur G, Schnabel JL, Taylor KH, Wang X, Xu D, Ding HF, Munn DH, Caldwell C, Shi H. Genome-wide DNA methylation analysis reveals novel epigenetic changes in chronic lymphocytic leukemia. Epigenetics. 2012;7:567–578. doi: 10.4161/epi.20237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Varley KE, Gertz J, Bowling KM, Parker SL, Reddy TE, Pauli-Behn F, Cross MK, Williams BA, Stamatoyannopoulos JA, Crawford GE, Absher DM, Wold BJ, Myers RM. Dynamic DNA methylation across diverse human cell lines and tissues. Genome Res. 2013;23:555–567. doi: 10.1101/gr.147942.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang X, Han H, De Carvalho DD, Lay FD, Jones PA, Liang G. Gene body methylation can alter gene expression and is a therapeutic target in cancer. Cancer Cell. 2014;26:577–590. doi: 10.1016/j.ccr.2014.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Richter AM, Pfeifer GP, Dammann RH. The RASSF proteins in cancer; from epigenetic silencing to functional characterization. Biochim Biophys Acta. 2009;1796:114–128. doi: 10.1016/j.bbcan.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 13.Oliveira C, Velho S, Domingo E, Preto A, Hofstra RM, Hamelin R, Yamamoto H, Seruca R, Schwartz S Jr. Concomitant RASSF1A hypermethylation and KRAS/BRAF mutations occur preferentially in MSI sporadic colorectal cancer. Oncogene. 2005;24:7630–7634. doi: 10.1038/sj.onc.1208906. [DOI] [PubMed] [Google Scholar]
- 14.Honda S, Miyagi H, Suzuki H, Minato M, Haruta M, Kaneko Y, Hatanaka KC, Hiyama E, Kamijo T, Okada T, Taketomi A. RASSF1A methylation indicates a poor prognosis in hepatoblastoma patients. Pediatr Surg Int. 2013;29:1147–1152. doi: 10.1007/s00383-013-3371-z. [DOI] [PubMed] [Google Scholar]
- 15.Cui X, Liu B, Zheng S, Dong K, Dong R. Genome-wide analysis of DNA methylation in hepatoblastoma tissues. Oncol Lett. 2016;12:1529–1534. doi: 10.3892/ol.2016.4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 17.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat Rev Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 18.Bibikova M, Barnes B, Tsan C, Ho V, Klotzle B, Le JM, Delano D, Zhang L, Schroth GP, Gunderson KL, Fan JB, Shen R. High density DNA methylation array with single CpG site resolution. Genomics. 2001;98:288–295. doi: 10.1016/j.ygeno.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Geiman TM, Robertson KD. Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem. 2002;87:117–125. doi: 10.1002/jcb.10286. [DOI] [PubMed] [Google Scholar]
- 20.Chow J, Heard E. X inactivation and the complexities of silencing a sex chromosome. Curr Opin Cell Biol. 2009;21:359–366. doi: 10.1016/j.ceb.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 21.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 22.Anwar SL, Krech T, Hasemeier B, Schipper E, Schweitzer N, Vogel A, Kreipe H, Buurman R, Skawran B, Lehmann U. Hsa-mir-183 is frequently methylated and related to poor survival in human hepatocellular carcinoma. World J Gastroenterol. 2017;23:1568–1575. doi: 10.3748/wjg.v23.i9.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo L, Huang C, Ji QJ. Aberrant promoter hypermethylation of p16, survivin, and retinoblastoma in gastric cancer. Bratisl Lek Listy. 2017;118:164–168. doi: 10.4149/BLL_2017_033. [DOI] [PubMed] [Google Scholar]
- 24.Kulis M, Heath S, Bibikova M, Queirós AC, Navarro A, Clot G, Martínez-Trillos A, Castellano G, Brun-Heath I, Pinyol M, Barberán-Soler S, Papasaikas P, Jares P, Beà S, Rico D, Ecker S, Rubio M, Royo R, Ho V, Klotzle B, Hernández L, Conde L, López-Guerra M, Colomer D, Villamor N, Aymerich M, Rozman M, Bayes M, Gut M, Gelpí JL, Orozco M, Fan JB, Quesada V, Puente XS, Pisano DG, Valencia A, López-Guillermo A, Gut I, López-Otín C, Campo E, Martín-Subero JI. Epigenomic analysis detects widespread gene-body DNA hypomethylation in chronic lymphocytic leukemia. Nat Genet. 2012;44:1236–1242. doi: 10.1038/ng.2443. [DOI] [PubMed] [Google Scholar]
- 25.Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D’Souza C, Fouse SD, Johnson BE, Hong C, Nielsen C, Zhao Y, Turecki G, Delaney A, Varhol R, Thiessen N, Shchors K, Heine VM, Rowitch DH, Xing X, Fiore C, Schillebeeckx M, Jones SJ, Haussler D, Marra MA, Hirst M, Wang T, Costello JF. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010;466:253–257. doi: 10.1038/nature09165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lazcoz P, Munoz J, Nistal M, Pestana A, Encio I, Castresana JS. Frequent promoter hypermethylation of RASSF1A and CASP8 in neuroblastoma. BMC Cancer. 2006;6:254. doi: 10.1186/1471-2407-6-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geli J, Kogner P, Lanner F, Natalishvili N, Juhlin C, Kiss N, Clark GJ, Ekström TJ, Farnebo F, Larsson C. Assessment of NORE1A as a putative tumor suppressor in human neuroblastoma. Int J Cancer. 2008;123:389–394. doi: 10.1002/ijc.23533. [DOI] [PubMed] [Google Scholar]
- 28.Morris MR, Hesson LB, Wagner KJ, Morgan NV, Astuti D, Lees RD, Cooper WN, Lee J, Gentle D, Macdonald F, Kishida T, Grundy R, Yao M, Latif F, Maher ER. Multigene methylation analysis of Wilms’ tumour and adult renal cell carcinoma. Oncogene. 2003;22:6794–6801. doi: 10.1038/sj.onc.1206914. [DOI] [PubMed] [Google Scholar]
- 29.Macheiner D, Gauglhofer C, Rodgarkia-Dara C, Grusch M, Brachner A, Bichler C, Kandioler D, Sutterlüty H, Mikulits W, Schulte-Hermann R, Grasl-Kraupp B. NORE1B is a putative tumor suppressor in hepatocarcinogenesis and may act via RASSF1A. Cancer Res. 2009;69:235–242. doi: 10.1158/0008-5472.CAN-08-2144. [DOI] [PubMed] [Google Scholar]
- 30.Hesson L, Dallol A, Minna JD, Maher ER, Latif F. NORE1A, a homologue of RASSF1A tumour suppressor gene is inactivated in human cancers. Oncogene. 2003;22:947–954. doi: 10.1038/sj.onc.1206191. [DOI] [PubMed] [Google Scholar]


