Abstract
Objective: This study aims to observe changes in the expression of the hepatic endoplasmic reticulum stress PERK-eIF2α-ATF4 signaling pathway protein in the progression of CCl4-induced hepatic fibrosis in rats. Methods: Male Wistar rats were sacrificed at the end of the 2nd, 4th, 8th and 12th weeks, respectively. A specific test was performed to compare the pathological changes of hepatic tissues in the model and normal groups. Immunohistochemical staining and Western blot were carried out to detect the expression of p-PERK, p-eIF2α and ATF4 proteins in the hepatic tissue group. Furthermore, real-time PCR was used to detect changes in the expression of ATF4 mRNA in hepatic tissues. Results: In the eight-week and twelve-week hepatic fibrosis group, significant fibrosis hyperplasia was identified in the livers of rats, and pseudo-lobules were also formed in the livers of rats in the twelve-week hepatic fibrosis group. Immunohistochemical staining and Western blot results indicated that the expression levels of p-PERK, p-eIF2α and the ATF4 protein in the livers of rats were significantly increased from the 8th week compared with the normal group (P<0.05). Real-time PCR results revealed that the expression of ATF4 mRNA was significantly increased in hepatic tissues in the hepatic fibrosis group compared with the normal group (P<0.05), and this was gradually enhanced as hepatic fibrosis progressed. Conclusions: CCl4 can induce an increase in the expression of the PERK-eIF2α-ATF4 signaling protein in the development of hepatic fibrosis along with phosphorylation-mediated activation, indicating that the activation of the PERK-eIF2α-ATF4 signaling pathway may contribute to the onset and development of hepatic fibrosis by regulating downstream target genes.
Keywords: Rats, hepatic fibrosis, p-PERK, p-eIF2α, ATF4
Introduction
Hepatic fibrosis is a pathological process that appears when the liver fibrillar connective tissues present with abnormal hyperplasia, and the extracellular matrix (ECM) is overloaded in the liver. In severe cases, this can develop into hepatic cirrhosis and even liver cancer [1]. Recent studies have found that endoplasmic reticulum stress and mediated cell apoptosis are correlated to many liver diseases, such as viral hepatitis, alcoholic liver disease, non-alcoholic fatty liver disease, drug-induced liver disease, acute liver failure and liver cancer [2]. Endoplasmic reticulum stress (ERS) is a subcellular organelle pathological process that manifests imbalance in the cellular endoplasmic reticulum homeostasis or protein processing and transportation disorders and physiological function disorders, as part of the cell stress defensive reaction. The Protein kinase R-like ER kinase (PERK) signaling pathway is an unfolded protein response (UPR) signal transduction pa thway in ERS and it’s the most widely researched [3,4]. Previous studies indicated that significant ERS and hepatic cell apoptosis occurs in liver cells in the process of carbon tetrachloride (CCl4)-induced rat hepatic fibrosis, and that the expression of GRP78 and CHOP proteins in the liver of rats is obviously increased in the hepatic fibrosis group [11,12]. However, at present, there are no reports on how to regulate the protein expression of CHOP after the expression of GRP78 increases in the process of hepatic fibrosis.
Studies on ERS signal transduction reveal that when ERS occurs in cells, highly expressed ERS protein, glucose regulated protein 78 (GRP78), can phosphorylate, dimerize and even activate PERK signaling protein [5,6], and activated PERK signaling protein can specifically phosphorylate and activate the 51st serine of the eukaryotic translation initiator factor 2α (eIF2α). Phosphorylated eIF2α further induces the transcription of activating transcription factor 4 (ATF4) downstream, allowing the latter to activate the CCAAT/enhancer-binding protein homologous protein (CHOP); and the increased expression of these proteins can cause cell apoptosis [7,8]. In ERS endoplasmic reticulum stress, UPR can also induce the protein expression CHOP via other signaling pathways, but PERKeIF2α-ATF4 is necessary [9,10]. Therefore, this study observed changes in the expression of the ERS pathway PERK-eIF2α-ATF4 signaling proteins in the development of hepatic fibrosis caused by CCl4 and investigated its potential roles in the pathogenetic mechanism of hepatic fibrosis.
Materials and methods
Main reagents
CCl4 (pure; Chengdu Jinshan Chemical Reagents Co., Ltd., Chengdu, China); Phosphorylated protein kinase R-like ER kinase (PERK), eukaryotic translation initiator factor 2α (eIF2α), and activating transcription factor 4 (ATF4) antibody I (Beijing Biosynthesis Biotechnology Co., Ltd., Beijing, China); β-Actin antibody (Wuhan Boster Biological Technology Co., Ltd.); TRIzol kit (Invitrogen, USA); Reverse transcription kit (Fermentas, USA); 2× SYBR green I kits (Applied Biosystems, USA); the β-Actin and ATF4 primers were designed and synthesized by TaKaRa Bioengineering (Dalian) Co., Ltd. The primer sequence was as follows: internal reference β-actin: upstream primer 5’-TCCTCCTGAGCGCAAGTACTCT-3’, downstream primer 5’-GCTCAGTAACAGTCCGCCTAGA-3’; ATF4: upstream primer 5’-CCCAAACCTTATgACCCACCT-3’, downstream primer 5’-gCTgTCTTgTTTTgCTCCATCTT-3’.
Groupings and treatment
A total of 56 healthy male Wistar rats (180±20 g, provided by the Laboratory Animal Center of Guizhou Medical University) were included in this study. After one week of feeding, these animals were randomly divided into two groups: the normal group (n=28) and the hepatic fibrosis group (n=28). In the hepatic fibrosis group, rats were subcutaneously injected with newly prepared 40% CCl4 peanut oil solutions (3 l/kg) every three days for the convenience in establishing the model. In the normal control group, rats were subcutaneously injected with peanut oil solutions in equal doses. Seven rats from each group were sacrificed by femoral artery bleeding at the end of the 2nd, 4th, 8th and 12th weeks, respectively. Before the rats were sacrificed, all of the rats were weighed. Representative sections were collected from the left liver lobe of all rats, and the wet liver weight was measured. The left lobe of the liver collected from each rat was cut into 3-mm3 sized sections and fixed in 4% neutral formalin. Remaining liver tissues were stored at -80°C for western blot and real-time PCR detection.
Measurement of the hepatic index
Hepatic index (%) = liver weight/body weight ×100%.
Pathomorphological observation of hepatic tissues
Hepatic tissues were routinely embedded in paraffin, cut into slices, and stained with hematoxylin and eosin (H&E). Under a lighted microscope, liver pathological changes were observed; and the semi-quantitative analysis of the degree of collagen fiber hyperplasia was carried out with reference to the fibrosis staging method.
Detection of the protein expression of p-PERK, p-eIF2α and ATF4 by immunohistochemical staining
This procedure was performed using the streptavidin-biotin complex (SABC) method, according to the kit’s instructions. In the negative control group, antibody I was replaced by phosphate buffered saline (PBS) solution. Five samples of each type were extracted and observed under a high-power microscope to detect for cytoplasm stained into pale brown cells, which was described as positive. The number of positive cells and the total number of cells were counted, and the percentage (%) of positive cells was calculated.
Western blot
Total protein was extracted from hepatic tissues, and the concentration of the total protein was detected using the BCA method. Each well was coated with 120 µg of the samples and separated by 10% SDS-PAGE electrophoresis. The membrane was transferred and washed by tris-buffered saline (TBS), then antibody I was added and the membrane was sealed for incubation for one hour at room temperature (p-PERK: 1:200, p-eIF2α: 1:250, ATF4: 1:200, and β-actin: 1:100). Then, the membrane was shaken and placed in a refrigerator at 4°C overnight. The next day, the membrane was washed, antibody II marked by HRP (1:5,000) was added and them membrane was incubated for one hour at room temperature. Next, the membrane was developed using the ECL chemiluminescence method, and the X-ray film was exposed. The stripes obtained were scanned and analyzed by the gel image analysis system. The relative expression level of the target proteins = gray value of the target proteins/gray value of the β-Actin protein.
Real-time PCR detection and analysis
The TRIzol method was used to extract the total RNA of rat livers for quantification and reverse transcription, and to synthesize cDNA, as follows: The total volume of the reaction system was 25 µl (1 µg of total RNA, 4 µl of 5× PCR buffer, 2 µl of 10 mmol/L of dNTP, 1 µl of 10 µmol/L of Oligod (T), 1 µl of RNase inhibitor, and 1 µl of MMLV reverse transcriptase supplemented with DEPC H2O), and the reaction condition was: 25°C for 10 minutes, 48°C for 60 minutes, and 95°C for five minutes. The total volume of the real-time PCR reaction system was 25 μl (4 μl of the reverse transcription product, 1.0 μl of the 10 μmol/L upstream and downstream primers, and 12.5 μl of 2× SYBR Green I supplemented with DEPC H2O), and the reaction condition was: 50°C for two minutes, 95°C for 10 minutes, 95°C for 15 seconds, and 60°C for one minute, with the last two steps repeated 40 times. With β-actin as the internal reference, the 2-ΔCT method was used to calculate the relative mRNA expression of ATF4 in rat hepatic tissues in each group. All samples were coated in three wells, respectively.
Statistical analysis
All the data were analyzed using SPSS 19.0 statistical software. The results of the pathological semi-quantitative method were detected using the Wilcoxon rank sum test. The comparison of the mean among multiple groups was performed using one-way ANOVA (for equal variances, the least significant difference [LSD] method was used; for variance heterogeneity, Games Howell method was used), with an inspection level of α=0.05. A P-value <0.05 was considered statistically significant.
Results
Changes in the hepatic index of rats in each group
As shown in Table 1, the hepatic index of rats in the two-week hepatic fibrosis group was a little higher than that of the rats in the two-week normal control group, but the difference was not statistically significant (P>0.05). The hepatic index of rats in the four-week hepatic fibrosis group, eight-week hepatic fibrosis group, and twelve-week hepatic fibrosis group was higher than that in the normal group (P<0.01). Furthermore, the hepatic index of the rats in the eight-week hepatic fibrosis group increased more significantly than that of the rats in the four-week hepatic fibrosis group (P<0.05). Moreover, the hepatic index of the rats in the twelve-week hepatic fibrosis group slightly decreased compared with the rats in the eight-week hepatic fibrosis group. The difference was not statistically significant (P>0.05).
Table 1.
The level of liver index in the rats of each group (x̅±s)
| Group | Cases | Liver index (mg/g) | Degree of hepatic fibrosis | mean rank | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
|
|
||||||||||
| 0 | I | II | III | IV | V | VI | ||||
| 2W normal group | 7 | 3.85±0.385 | 5 | 0 | 0 | 0 | 0 | 0 | 0 | 8.0 |
| 2W HF group | 7 | 4.84±0.495 | 1 | 2 | 1 | 1 | 0 | 0 | 0 | 19.5Δ |
| 4W normal group | 7 | 3.81±0.175 | 4 | 1 | 0 | 0 | 0 | 0 | 0 | 10.3 |
| 4W HF group | 7 | 5.16±0.34Δ | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 32.3Δ |
| 8W normal group | 7 | 3.78±0.44 | 2 | 3 | 0 | 0 | 0 | 0 | 0 | 14.9 |
| 8W HF group | 7 | 5.98±0.69Δ,a,b,﹡ | 0 | 0 | 0 | 2 | 2 | 1 | 0 | 30.3Δ,a |
| 12W normal group | 7 | 3.44±0.20 | 3 | 2 | 0 | 0 | 0 | 0 | 0 | 12.6 |
| 12W HF group | 7 | 5.94±0.55Δ,a,b | 0 | 0 | 0 | 0 | 1 | 3 | 1 | 36.1Δ,a,b |
P<0.01 vs normal group;
P<0.01 vs two weeks hepatic fibrosis group;
P<0.01 vs four weeks hepatic fibrosis group;
P<0.05 vs four weeks hepatic fibrosis group.
Pathological changes in the liver of rats in each group
After the hepatic tissues of the rats in each group underwent H&E staining, it was found that the hepatic cells of the rats in the normal group exhibited a radial arrangement with the central veins as the center in a normal structure. There was no collagen proliferation in the mesenchyme without inflammatory cell infiltration. The hepatic cords of rats in the four-week hepatic fibrosis group were arranged chaotically. Hepatic cells exhibited significant fat-like changes. Fiber connective tissue proliferation was observed in the portal area and the hepatic lobules, and the hepatic cords of the rats in the twelve-week hepatic fibrosis group were arranged chaotically with massive fiber connective tissue proliferation. Different-sized round or oval pseudolobules formed around the hepatic tissue, as shown in Figure 1. According to Wang Baoen’s staging method, it was found by rank sum analysis that early hepatic fibrosis formed at the 4th week after the rats were subcutaneously injected with CCl4. The degree of fibrosis at the 12th week group after subcutaneous injection was significantly aggravated compared with the normal group, the four-week hepatic fibrosis group and the eight-week hepatic fibrosis group (P<0.01), as shown in Table 1.
Figure 1.
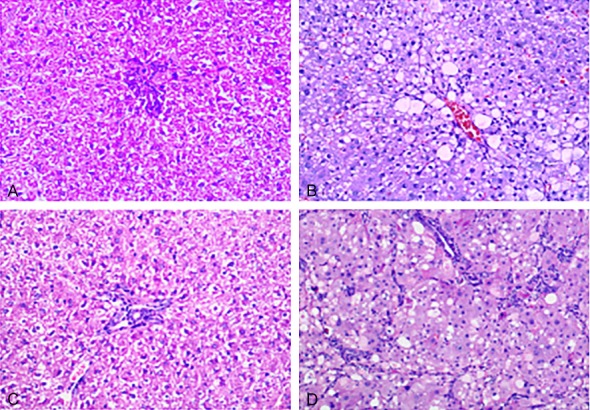
Liver tissue of the rats in the control and hepatic fibrosis groups (HE, ×200). A: 4-week snormal group; B: 4-weeks hepatic fibrosis group; C: 12-weeks normal group; D: 12-weeks hepatic fibrosis group.
The expression of p-PERK and p-eIF2α proteins in the liver of rats in each group
The hepatic tissues of rats in each group underwent immunohistochemical staining and were observed under a light microscope. Pale-brown stained particles were observed in the cytoplasms of the liver cells and were described as positive cells. The p-PERK and p-eIF2α proteins were positively expressed in the cytoplasm. When the semi-quantitative method was used to count the percentage of positive cells, it was found that there was almost no positive p-PERK and p-eIF2α protein expression in the hepatic tissues of rats in each normal control group. In the four-week hepatic fibrosis group, the positive protein expression of p-PERK and p-eIF2α increased in the liver cells of the rats and were mainly distributed around the fibrous septum. Compared with the rats in the four-week normal group, the increase in positive expression was statistically significant (P<0.05), as shown in Figure 2. In the eight-week hepatic fibrosis group, the protein expression of p-PERK and p-eIF2α in the cytoplasm of liver cells was more significant than that in the normal control group, two-week hepatic fibrosis group, and four-week hepatic fibrosis group. The difference was statistically significant (P<0.01), and most of these exhibited a strong positive expression. In the twelve-week hepatic fibrosis group, the p-PERK and p-eIF2α protein in the liver cells of the rats widely exhibited a strong positive expression. The difference in percentage of the positive expression among this group, the normal control groups and the other hepatic fibrosis groups was statistically significant (P<0.01).
Figure 2.
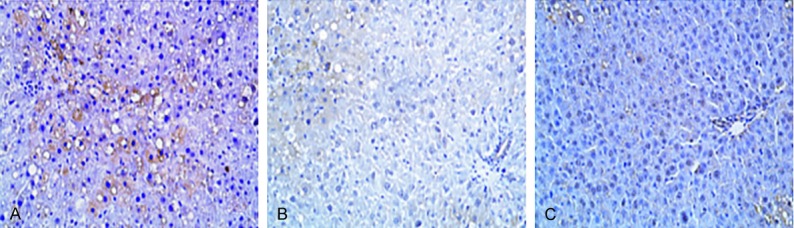
The expression of p-PERK, p-eIF2α, ATF4 protein in liver of rats (SABC, ×200).
The expression of ATF4 protein in the liver of rats in each group
The hepatic tissues of the rats in each group were subjected to immunohistochemical staining and observed under a lighted microscope. Pale-brown stained particles were observed in the cytoplasm of the liver cells, and they were described as positive cells. The positive protein expression of AFT4 was mainly distributed in the cytoplasm. The semi-quantitative method was used to count the percentage of positive cells. No positive AFT4 protein expression was found in hepatic tissues of rats in each corresponding normal group. In the two-week hepatic fibrosis group and the four-week hepatic fibrosis group, a weak positive expression of AFT4 was observed in the liver cells of the rats, but its difference from the corresponding normal group was not statistically significant (P>0.05), as shown in Figure 2. In the eight-week hepatic fibrosis group, a strong positive expression of p-eIF2α was found in the cytoplasm of liver cells, and the significance of this expression was more than that of the normal group, two-week hepatic fibrosis group, and four-week hepatic fibrosis group. The differences were statistically significant (P<0.01). The p-eIF2α protein in the liver cells of the rats in the twelve-week hepatic fibrosis group widely exhibited a strong positive expression, and the significance of this expression was more than that of the normal group, two-week hepatic fibrosis group, and four-week hepatic fibrosis group. The difference was statistically significant (P<0.01), but the differences from the eight-week hepatic fibrosis group were not statistically significant.
Analysis results on the expression of p-PERK, p-eIF2α and ATF4 proteins by western blot
The protein expression of p-PERK and ATF4 was detected in hepatic tissues of rats in each group, but there was almost no p-eIF2α expression detected in the hepatic tissues of the rats in the normal group. Since the 2nd week of hepatic fibrosis, the protein expression of p-PERK and p-eIF2α in the hepatic tissues of the rats in the hepatic fibrosis groups increased compared with that in the normal control group, and this significantly increased as the time to establish the model passed and the degree of hepatic fibrosis was aggravated. In the four-week hepatic fibrosis group, the protein expression of p-PERK and p-eIF2α in the liver of rats significantly increased compared with the corresponding normal control group. In the two-week and four-week hepatic fibrosis groups, the protein expression of ATF4 in the liver of the rats slightly increased compared with the normal group, but the differences were not statistically significant. Compared with the two-week hepatic fibrosis group and the four-week hepatic fibrosis group, the expression of p-PERK, p-eIF2α and ATF4 proteins in hepatic tissues significantly increased (P<0.05) (Figure 3).
Figure 3.
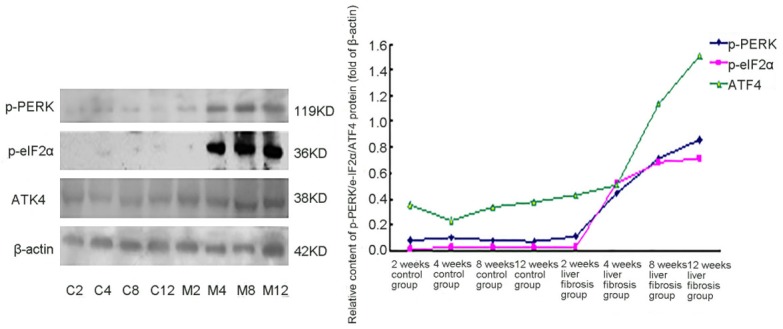
The expressions of p-PERK, p-eIF2α and ATF4 proteins in the livers of the rats. C2: 2-week normal group; C4: 4-week normal group; C8: 8-week normal group; C12: 12-week normal group; M2: 2-week hepatic fibrosis group; M4: 4-week hepatic fibrosis group; M8: 8-week hepatic fibrosis group; M12: 12-week hepatic fibrosis group.
Real-time PCR analysis results on ATF4 mRNA expression
The expression of ATF4 mRNA was detected in the livers of the rats in each group. There were no significant differences in the ATF4 mRNA expression in the livers of the rats between the two-week hepatic fibrosis group and the corresponding normal group (P>0.05). In the four-week, eight-week, and twelve-week hepatic fibrosis groups, the mRNA expression of ATF4 in the livers of the rats gradually increased as the time to establish the model passed. This was significantly higher than that of the control group (P<0.05 or P<0.01), and significantly higher than that of the two-week hepatic fibrosis group (P<0.01). In the twelve-week hepatic fibrosis group, the expression of ATF4 mRNA in the hepatic tissues of the rats was significantly higher than that of the rats in the 2nd, 4th (P<0.01), and 8th (P<0.05) weeks of the hepatic fibrosis model, as shown in Table 2.
Table 2.
The expression of hepatic fibrosis p-PERK, p-eIF2α, ATF4 protein and ATF4 mRNA in each group (mean ± s, n=7)
| Group | p-PERK (%) | p-eIF2α (%) | ATF4 (%) | ATF4 mRNA |
|---|---|---|---|---|
| 2W normal group | 7.48±3.38 | 1.02±0.56 | 35.04±9.75 | 86.85±16.68 |
| 2W HF group | 10.50±2.91 | 2.60±0.59 | 43.21±13.51 | 63.31±25.29 |
| 4W normal group | 9.63±6.51 | 2.49±1.11 | 22.76±6.77 | 79.03±34.55 |
| 4W HF group | 44.71±8.90Δ,* | 52.03±5.55Δ,* | 50.40±18.64 | 195.48±22.42ΔΔ,* |
| 8W normal group | 7.78±1.30 | 2.50±0.59 | 33.42±7.75 | 84.45±44.35 |
| 8W HF group | 71.13±11.96Δ,*,# | 68.03±6.08Δ,*,# | 113.14±30.28Δ,*,# | 291.43±83.49Δ,* |
| 12W normal group | 7.25±2.91 | 2.21±0.90 | 37.16±7.36 | 77.88±43.18 |
| 12W HF group | 85.488±9.84Δ,*,#,○ | 71.35±8.16Δ,*,# | 151.02±30.40Δ,*,#,○ | 461.54±139.37Δ,*,#,☆ |
P<0.01 vs normal group;
P<0.05 vs normal group;
P<0.01 vs two weeks hepatic fibrosis group;
P<0.01 vs four weeks hepatic fibrosis group;
P<0.01vs eight weeks hepatic fibrosis group;
P<0.05 vs eight weeks hepatic fibrosis group.
Discussion
To date, the mechanism of hepatic fibrosis remains unknown. As research progresses on the pathogenetic mechanism of hepatic fibrosis, wide attention has been given on the roles of subcellular levels, such as endoplasmic reticulum and mitochondrial injuries, in the pathogenetic mechanism of hepatic fibrosis. ERS is a series of reactions in a cell caused by endoplasmic reticulum homeostasis disorder, which is part of the cellular defensive response. These reactions can be induced by misfolded protein aggregation, Ca2+ exhaustion and lipid synthesis disorders through corresponding signaling pathways [13]. Our previous studies have revealed that the expression of ERS protein GRP78 in liver cells significantly increased when CCl4 was induced in the onset of hepatic fibrosis [11], and the expression of ERS-related CHOP and TRB3 proteins and genes also significantly increased. These changes were positively correlated to the apoptosis rate of liver cells in the study rats, which indicated that ERS may promote apoptosis in liver cells by regulating the expression of downstream signal CHOP and TRB3 proteins [12]. However, there are no reports on how to regulate the mechanism of CHOP and TRB3 through ERS protein GRP78 in liver cells when hepatic fibrosis occurs.
PERK is a type I trans-membrane protein kinase located in the endoplasmic reticulum membrane. As described in previous studies, when cells undergo ERS, the GRP78 in cells would be separated from the PERK proteins in cells and be transferred to unfolded proteins. Separated PERK proteins could be phosphorylated and activated, and phosphorylated PERK proteins are dimerized to further phosphorylate eukaryotic translation initiator factor 2α (eIF2α) protein and activate the gene and protein expression of ATF4, and this in turn activates the PERK-eIF2α-ATF4 signaling pathway and induces the gene and protein expression of CHOP [14]. Some studies have indicated that p-PERK/eIF2α/ATF4 is a signaling pathway necessary for regulating the protein expression of CHOP [15]. In vitro experiments have revealed that the silent PERK gene could significantly increase the vitality of liver cells under the ERS state and reduce their apoptosis rate, indicating that the GRP78/p-PERK/eIF2α/ATF4/CHOP signaling pathway is an important pathway for ERS to induce apoptosis in liver cells [16]. However, there are no reports on whether this signaling pathway is activated in the development of hepatic fibrosis, as well as the roles of its activation on target proteins in the downstream.
In this study, we found that as the tCCl4 subcutaneous injection extended, the degree of hepatic fibrosis continuously was aggravated, and the hepatic index and histopathological grade of hepatic fibrosis continuously increased. This indicates that 40% CCl4 subcutaneous injection may cause hepatic fibrosis in rats and is consistent with the results of previous studies. The expression of p-PERK and p-eIF2α proteins in the hepatic tissues of rats in the four-week hepatic fibrosis group significantly increased compared with the normal control group, and the expression of p-PERK and p-eIF2α proteins was obviously increased as the hepatic fibrosis was aggravated. The difference was statistically significant. This proves that the GRP78/p-PERK/eIF2 signaling pathway is activated in the development of hepatic fibrosis, and this signaling pathway is continuously activated in the overall development of hepatic fibrosis and is closely correlated to the progression of fibrosis. In addition, this study indicated that the time pattern for the increased expression of p-PERK and p-eIF2α proteins was consistent with that of the increased protein expression of GRP78 and CHOP in the liver of rats with hepatic fibrosis [12]. This indicates that in the hepatic fibrosis process caused by CCl4, early liver cells of hepatic fibrosis undergo ERS, and the GRP78 protein increased by ERS in liver cells rapidly phosphorylates type I transmembrane protein kinase PERK in the endoplasmic reticulum membrane and further phosphorylates eIF2α downstream, which activates the downstream gene transcription regulated by the GRP78-PERK-eIF2α signaling pathway.
It was found that by observing the protein and mRNA expression of ATF4 in the development of hepatic fibrosis, the ATF4 protein in the liver of rats increased in the four-week hepatic fibrosis group, but the difference was not statistically significant. In the eight-week hepatic fibrosis group, the protein expression of ATF4 in the liver of rats was significantly higher than that of rats in the normal group and four-week hepatic fibrosis group. As time passed, the expression of ATF4 protein in the liver of the rats in the hepatic fibrosis group significantly increased compared with that in the normal group and four-week hepatic fibrosis group. In the two-week hepatic fibrosis group, the mRNA expression of ATF4 in the liver of the rats was significantly higher than that of the rats in the normal control group; and the difference was statistically significant. As time passed, the mRNA expression of ATF4 in the liver of the rats in the hepatic fibrosis group was significantly higher than that of the rats in the control group and the two-week hepatic fibrosis group. In this study, phosphorylated eIF2α selectively activated ATF4 gene transcription and protein synthesis, inducing the protein expression of ATF4, to gradually increase in hepatic tissues of the rats with hepatic fibrosis as the time to establish the model was extended. Furthermore, the time point when the protein expression of ATF4 in the hepatic tissue of the rats with hepatic fibrosis was later than that for the increased mRNA expression of ATF4. Hence, the activation of the GRP78/p-PERK/eIF2α signaling pathway increases the expression of ATF4 mRNA, and subsequently increases the protein expression level of ATF4.
This study revealed that the increment in the expression of p-eIF2α in the liver of the rats in the twelve-week hepatic fibrosis group was not as significant as that of the rats in the eight-week hepatic fibrosis group (P>0.05). Some studies have found that phosphorylated eIF2α selectively increased the mRNA expression of ATF4 when ERS persistently existed, and ATF4 induced the expression of CHOP. The latter induced the expression of the growth arresting and DNA-damage-inducing protein 153 (GADD34). The expression of GADD34 is related to dephosphorylated eIF2α, which forms a negative feedback loop [16,17]. This may be one reason why the protein expression level of p-eIF2α in the liver of the rats in the twelve-week hepatic fibrosis group was not significantly increased compared with protein expression level of p-eIF2α in the eight-week hepatic fibrosis group.
In conclusion, this study indicates that ERS in the liver cells plays an important role in the pathogenetic mechanism of hepatic fibrosis. ERS in liver cells regulates the protein expression of CHOP molecules downstream by activating the PERK-eIF2α-ATF4 signaling pathway, promoting the apoptosis of liver cells, and participating in the onset and development of hepatic fibrosis.
Acknowledgements
We are particularly grateful to all the people who have given us help on our article. This work was supported by the National Natural Science Foundation of China (No: 81160064).
Disclosure of conflict of interest
None.
References
- 1.Wu FL, Liu WY, Van Poucke S, Braddock M, Jin WM, Xiao J, Li XK, Zheng HM. Targeting endoplasmic reticulum tress in liver disease. Expert Rev Gastroenterol Hepatol. 2016;10:1041–52. doi: 10.1080/17474124.2016.1179575. [DOI] [PubMed] [Google Scholar]
- 2.Malhi H, Kaufman RJ. Endoplasmic reticulum stress in liver disease. J Hepatol. 2011;54:795–809. doi: 10.1016/j.jhep.2010.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Chen L, Shen Y, Xu J. Rifampicin-induced injury in L02 cells is alleviated by 4-PBA via inhibition of the PERK-ATF4-CHOP pathway. Toxicol In Vitro. 2016;36:186–96. doi: 10.1016/j.tiv.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 4.Liu Z, Shi Q, Song X, Wang Y, Wang Y, Song E, Song Y. Activating transcription Factor 4 (ATF4)-ATF3-C/EBP homologous protein (CHOP) cascade shows an essential role in the ER stress-induced sensitization of tetrachlorobenzoquinone-challenged PC12 cells to ROS-mediated apoptosis via death receptor 5 (DR5) signaling. Chem Res Toxicol. 2016;29:1510–8. doi: 10.1021/acs.chemrestox.6b00181. [DOI] [PubMed] [Google Scholar]
- 5.Dally S, Monceau V, Corvazier E, Bredoux R, Raies A, Bobe R, del Monte F, Enouf J. Compartmentalized expression of three novel sarco/endoplasmic reticulum Ca2+ATPase 3 isoforms including the switch to ER stress, SERCA3f, in non-failing and failing human heart. Cell Calcium. 2009;45:283–90. doi: 10.1016/j.ceca.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Tardif KD, Waris G, Siddiqui A. Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 2005;13:1592–1620. doi: 10.1016/j.tim.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 7.Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. Translational resistance of late alphavirus mRNA to eIF2alpha phosphorylation: a strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev. 2006;20:87–100. doi: 10.1101/gad.357006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenwald IB, Pechet L, Han A, Lu L, Pihan G, Woda B, Chen JJ, Szymanski I. Expression of translation initiation factors elF-4E and elF-2alpha and a potential physiologic role of continuous protein synthesis in human platelets. Thromb Haemost. 2001;85:142–51. [PubMed] [Google Scholar]
- 9.Guo G, Meng Y, Tan W, Xia Y, Cheng C, Chen X, Gu Z. Induction of apoptosis coupled to endoplasmic reticulum stress through regulation of CHOP and JNK in bone marrow mesenchymal stem cells from patients with systemic lupus erythematosus. J Immunol Res. 2015;2015:183738. doi: 10.1155/2015/183738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang W, Chen L, Shen Y, Xu J. Rifampicin-induced injury in L02 cells is alleviated by 4-PBA via inhibition of the PERK-ATF4-CHOPpathway. Toxicol In Vitro. 2016;36:186–96. doi: 10.1016/j.tiv.2016.07.017. [DOI] [PubMed] [Google Scholar]
- 11.Wen JJ, Xie RJ, Han B, Yang T, Qian C, Yang Q. Morphology of endoplasmic reticulum and expression of GRP78 in rat fibrotic liver. Chinese Journal of Pathophysiology. 2011;27:2210–2213. [Google Scholar]
- 12.Du XC, Han B, Xie RJ, Zou H, Yang Q. Changes of endoplasmic reticulum stress-related molecule CHOP and TRB3 in rat fibrotic liver. Chinese Journal of Pathophysiology. 2013;29:906–912. [Google Scholar]
- 13.Mahdi AA, Rizvi SH, Parveen A. Role of endoplasmic reticulum stress and unfolded protein responses in health and diseases. Indian J Clin Biochem. 2016;31:127–37. doi: 10.1007/s12291-015-0502-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sidrauski C, Walter P. The transmembrane kinase Ire1 is a site-specific endonuclease that initiates mRNA splicing in the unfolded protein response. Cell. 1997;90:1031–9. doi: 10.1016/s0092-8674(00)80369-4. [DOI] [PubMed] [Google Scholar]
- 15.Cao J, Yang ZX, Shen W, Yao L. Construction of shRNA eukaryotic expression vectors targeting PERK gene and effect of PERK gene knockdown on apoptosis in endoplasmic reticulum stress-induced L02 hepatocytes. Chinese Journal of Pathophysiology. 2011;27:2376–2381. [Google Scholar]
- 16.Reid DW, Tay AS, Sundaram JR, Lee IC, Chen Q, George SE, Nicchitta CV, Shenolikar S. Complementary roles of GADD34- and CReP-Containing Eukaryotic initiation factor 2α phosphatases during the unfolded protein response. Mol Cell Biol. 2016;36:1868–80. doi: 10.1128/MCB.00190-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Akai R, Hosoda A, Yoshino M, Iwawaki T. Constitutive role of GADD34and CReP in cancellation of phospho-eIF2α-dependent translational attenuation and insulin biosynthesis in pancreatic cells. Genes Cells. 2015;20:871–86. doi: 10.1111/gtc.12279. [DOI] [PubMed] [Google Scholar]


