Abstract
Aberrant expression of miR-206 has been repeatedly found and demonstrated to play crucial roles in cancers. However, the role of miR-206 in brain glioma remains unclear. To address this issue, we detected miR-206 expression of 60 gliomas and 18 normal peritumor tissues, and found that miR-206 is significantly down-regulated in gliomas. Further in silico analysis of 198 glioma samples from the Chinese Glioma Genome Atlas (CGGA) indicated that miR-206 is significantly down-regulated in high grade gliomas and that miR-206 predicts favorable patients’ prognosis. Notably, we found that miR-206 expression is negatively correlated with Ki-67 staining, indicating a proliferative inhibition of miR-206 in gliomas. To explore the crucial role of miR-206 in gliomas, we constructed miR-206 stably overexpressed LN229 glioma cell lines and found that the proliferation is significantly inhibited. Through flow cytometry (FCM) analyses, we found that the apoptotic rate is increased and the cell cycle is arrested in LN229 cells after overexpression of miR-206. Bioinformatic analysis, qPCR, western blot and luciferase assay indicated that the Forkhead Box Protein 1 (FOXP1) is a direct target of miR-206 in gliomas. Overexpression of FOXP1 could partially rescue the proliferative inhibition in the miR-206 stably overexpressed LN229 cells. In summary, our results suggest that miR-206 might function as a tumor suppressor of gliomas by inhibition of proliferation and could serve as a promising candidate for therapeutic applications in glioma by targeting FOXP1.
Keywords: miR-206, brain glioma, cell proliferation, FOXP1
Introduction
Brain gliomas, the most common type of primary tumors, account for about 40% of intracranial malignant tumors and remain a leading cause of morbidity and mortality worldwide [1]. In recent years, despite the introduction of modern therapeutic approaches including surgical resection, radiotherapy, and chemotherapy, a majority of glioma patients still succumb to this disease within 2 years after initial diagnosis [1]. Therefore, there is an urgent need to explore the molecular mechanisms underlying glioma initiation and progression, so as to design effective therapeutic strategies to prolong patients’ survival time and improve the quality of life.
miRNA is a non-coding RNAs with 18-22 nt length and suppresses gene expression by complementarily binding with the 3’ untranslated region (UTR) of the target mRNA [2]. Accumulating evidence has reported that miRNAs play a key role in tumor initiation, progression and drug resistance, including miR-206 [3]. miRNA-206 was earlier mentioned as a muscle-specific miRNA which plays important role in skeletal muscle development and functional disorders [4]. Recently, miR-206 has attracted intense attention because of the reporting of its tumor suppressor role in breast cancer by regulation of the estrogen receptor alpha gene [5]. In gliomas, miR-206 was first reported to be a biomarker that decreasing of miR-206 predicted a poor prognosis in patients with malignant astrocytoma [6]. Subsequently, miR-206 was found to regulate glioma proliferation and apoptosis by negatively regulating Otx2 and Bcl-2 [7,8]. These findings indicate that miR-206 might act as a tumor suppressor and its down-regulation might promote proliferation in glioma. Nonetheless, to our knowledge, few papers have elucidated the detailed mechanisms of miR-206 in gliomas.
Forkhead Box Protein 1 (FOXP1), a member of FOX family, has been reported to be related to a wide range of biological and pathological functions [9]. From previous reports, we know that FOXP1 has dual characteristics in tumors. In gastrointestinal and lung cancers, FOXP1 acted as a tumor suppressor, while in B-cell neoplasia [9] and multiple myeloma [10], FOXP1 functions as an oncogene. In gliomas, FOXP1 was mainly reported as an oncogene, which can promote glioma proliferation and enhance tumorigenicity [11]. Here, we show that miR-206 is down-regulated in gliomas, and this down-regulation correlates with glioma grade. Moreover, miR-206 can inhibit tumor growth by targeting FOXP1.
Materials and methods
Glioma tissues and patients
In a total of 61 cases of glioma tissues and 18 cases of peritumor tissues were recruited from Xinqiao Hospital (XQ cohort), Third Military Medical University (TMMU). Samples were collected during surgery and were immediately flash frozen and stored at -80°C for follow-up experiments. Pathologic diagnosis was evaluated according to the World Health Organization classification (2016) by two experienced pathologists. Written informed consent was obtained from all patients. Specimen collection was approved by the ethics committee of Xinqiao Hospital, TMMU. In addition, another glioma cohort from the Chinese Glioma Genome Atlas (CGGA) database (http://www.cgga.org.cn/) was investigated in silico.
Cell culture
Human glioma cell line LN229 was obtained from the Department of Pathology, Southwest Hospital, TMMU. The cell line was incubated at 37°C in a humidified incubator with 5% CO2 and 20% O2 in Dulbecco’s-modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum and 100 ug/ml penicillin/streptomycin. Cells were passaged at 80%-90% confluency. Culture medium was refreshed every 48 hours.
Lentivirus vector, plasmid construction and transduction
For target overexpression, the human FOXP1 cDNA were cloned into the pcDNA3.1 vector. A scramble control vector was also constructed. For lentivirus production, mature miR-206 sequence was obtained from the miRBase database and cloned into the GV217 vector (Ubi-EGFP-MCS) (Genechem, China). HEK-293T cells were seeded in 6-well plates and transfected with transfer and packaging vectors using Lipofectamine 2000 (Invitrogen, USA). Then, virus particles were harvested after 72 hours of transfection.
For construction of miR-206 overexpressed cell lines, LN229 cells were seeded into 6-well plates 1 day before transduction. When cells reached 50-80% confluency, lentivirus particles were added to the culture medium. After 96 hours of transduction, cells were harvested for following experiments.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from tissues or cells using TRIzol Reagent (Invitrogen, USA) and treated with DNase I to eliminate genomic DNA contamination. MiRNA/mRNA qPCR analysis was performed as previously described [12]. The FOXP1 sequences for qPCR detection are: Forward: 5’-TCCCGTGTCAGTGGCTATGAT-3’ and Reverse: 5’-CTCTTTAGGCTGTTTTCCAGCAT-3’.
Cell proliferation assay
Cell proliferation was measured by Cell Counting Kit-8 (Dojindo Laboratories, Japan) according to the manufacturer’s instructions. Cells were seeded into 96-well plate and the proliferation rate was detected at 0, 24, 48, 72 and 96 hours. Single cell proliferation was measured by Edu cell proliferation Kit with Alexa Fluor 647 (Beyotime Biotechnology, China) according to the manufacturer’s instructions.
Dual luciferase assay
Hek-293T cells were seeded in 24-well plates on day before transfection. 400 ng FOXP1 3’UTR plasmid (Genecopoeia, USA) was co-transfected with 400 ng miR-206 overexpression or negative control plasmids using Lipofectamine 2000 (Invitrogen, USA), and culture medium refreshed after 6 hours of transfection. After 48 hours of incubation, supernatant was harvested for dual luciferase assay using Secrete-Pair™ Dual Luminescence Assay Kit (Genecopoeia, USA).
Western blot analysis
LN229 cell extracts equivalent to 10 ug protein were subjected to 12% SDS-PAGE and PVDF membranes. The membranes were blocked with 5% nonfat milk in Tris-buffered saline containing 0.05% Tween 20, and incubated with rabbit antibodies against human FOXP1 and GAPDH (Abcam, USA), and Cyclin D1 and β-tublin (Cell Signaling Technology, USA). After that, the membranes were incubated with horseradish peroxidase-conjugated goat anti-rabbit IgG (Santa Cruz Biotechnology, USA) and visualized with BeyoECL Star chemiluminescence assay kits (Beyotime, China).
Apoptosis detection with annexin V-FITC and PI staining
Cells were digested, collected, and washed with cold PBS. Then, 1×105 cells were collected and re-suspended with 195 ul binding Buffer with 5 ul Annexin V-FITC and 5 ul PI Staining Solution (Beyotime, China). After incubation for 15 min in dark at room temperature, cells were analyzed by a flow cytometer (FACS Calibur; Becton-Dickinson). Subsequent analyses of flow cytometry data were performed using Cell Quest software.
Statistical analysis
Data analysis were conducted by SPSS for Windows version 13.0 (SPSS Inc, USA). P-values less than 0.05 were considered significant. Student’s t-test was used to analyze the difference between the means of treatment and control groups.
Results
MiR-206 is significantly down-regulated in gliomas and predicts favorable patients’ prognosis
We first detected miR-206 expression using 18 cases of peritumor tissues and 61 cases of glioma. We found that miR-206 expression is significantly down-regulated in glioma tissues, and this down-regulation tends to be correlated with tumor grade (Figure 1A). Subsequently, 18 pairs of gliomas and corresponding peritumor tissues were selected and we also obtained the same results (Figure 1B). Among these glioma tissues, 44 samples performed Ki-67 immunochemistry staining. Interestingly, Pearson correlation analysis shows that miR-206 expression is significantly negatively correlated with Ki-67 index in these samples (Figure 1C), indicating a proliferative inhibition of miR-206 in gliomas. We further validated these results via in silico analysis of 198 glioma samples from the Chinese Glioma Genome Atlas (CGGA). The results show that miR-206 is significantly down-regulated in high grade gliomas (WHO grade III and IV) when compared with low grade gliomas (WHO grade II) (Figure 1D). Moreover, we defined these glioma samples into low expression of miR-206 (N=119) and high expression of miR-206 (N=79) groups using online software “Cutoff Finder” (Figure 1E). Kaplan-Meier survival analysis shows that the survival time of the high miR-206 expression group is significantly longer than that of the low miR-206 expression group (Figure 1F). As summarized in Table 1, low miR-206 expression significantly correlates with an advanced patient age and advanced tumor grade in samples from CGGA, and shows similar trends in samples from XQ cohort. Together, these results show that miR-206 may function as a tumor suppressor in gliomas by inhibiting cell proliferation.
Figure 1.
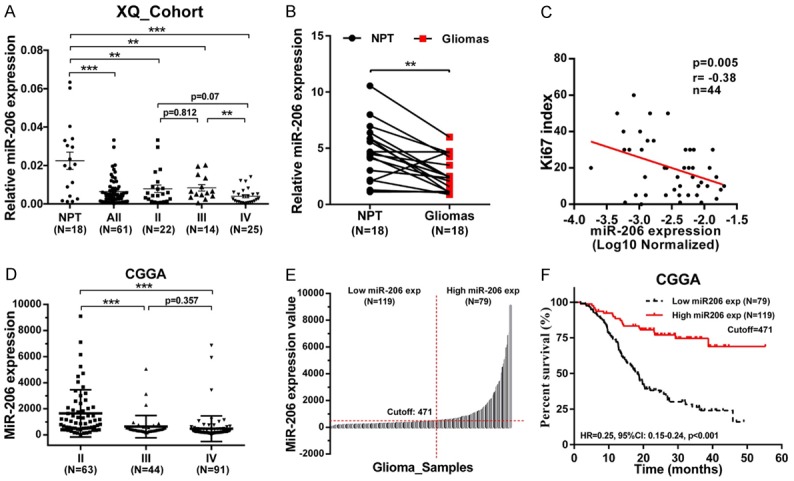
miR-206 is down-regulated and may function as a tumor suppressor in gliomas. A, B. miR-206 is down-regulated in glioma tissues when compared with peritumor tissues; C. miR-206 expression is negatively correlated with Ki-67 immunochemistry staining. D. miR-206 is down-regulated in high grade glioma tissues from CCGA; E, F. miR-206 predicts favorable prognosis of glioma patients. The cutoff value is 471 and determined by online software “Cutoff Finder”. (**: P<0.01; ***, P<0.001; exp: expression).
Table 1.
Characteristics of glioma samples with low/high expression of miR-206
| Characteristics | XQ_Cohort | CGGA | |||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Low miR-206 | High miR-206 | P Value | Low miR-206 | High miR-206 | P Value | ||
| No. of patients | 30 | 30 | 119 | 79 | |||
| Age at diagnosis | |||||||
| Mean | 50.1 | 41.6 | P=0.392 | 45.62 | 38.6 | P<0.001 | |
| Range | 13-76 | 18-69 | 13-71 | 18-61 | |||
| >50/≤50 | 13/17 | 8/22 | 73/46 | 13/66 | |||
| Gender | |||||||
| Male | 19 | 21 | P=0.548 | 74 | 49 | P=0.982 | |
| Female | 11 | 9 | 45 | 30 | |||
| Male-to-Female | 1.72 | 2.33 | 1.64 | 1.63 | |||
| WHO grade | |||||||
| Low grade | (Grade II) | 8 | 13 | P=0.176 | 15 | 48 | P<0.001 |
| High grade | (Grade III) | 7 | 8 | 29 | 15 | ||
| (Grade IV) | 15 | 9 | 75 | 16 | |||
MiR-206 significantly inhibits the proliferation of LN229 glioma cells
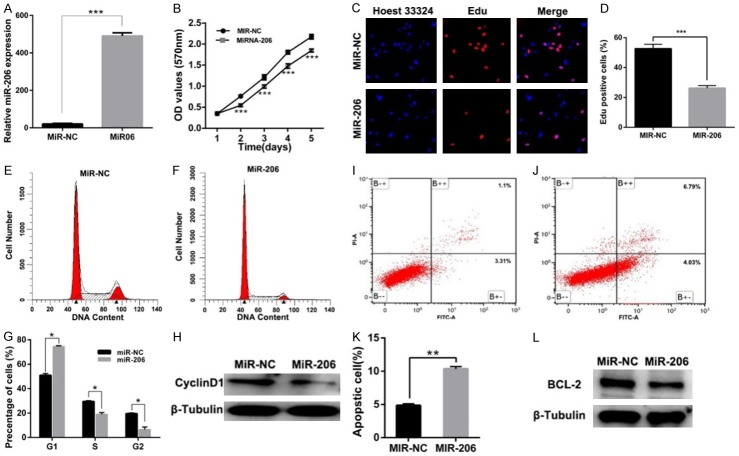
To investigate the role of miR-206 in cell proliferation, we established miR-206 stably overexpressed and negative control (miR-NC) LN229 glioma cell lines (Figure 2A). Through CCK-8 assay, we found that the proliferation of LN229 glioma cells was significantly inhibited over a 5-day period (Figure 2B). For further confirmation, Edu staining assay was performed, and we observed a lower Edu incorporation ratio in miR-206-overexpressed cells, when compared with the miR-NC group (Figure 2C, 2D). These results show us that miR-206 can indeed inhibit glioma cell proliferation.
Figure 2.
miR-206 can significantly inhibit glioma cell line LN229 proliferation by promoting apoptosis and inducing cell cycle arrest. A. qPCR validation of the establishment of miR-206 overexpressed LN229 glioma cells; B. CCK-8 assay shows that the proliferation of LN229 is significantly inhibited after miR-206 overexpression; C, D. Edu assay shows that miR-206 overexpression inhibits the Edu incorporation of LN229; E-G. miR-206 significantly inhibits cell cycle progression: LN229 cells in G1 phase of was significantly up-regulated while in S/G2 phase was significantly down-regulated. H.Western blot shows that Cyclin D1 was inhibited in miR-206 overexpressed LN229 cells; I-K. miR-206 significantly promotes cell apoptosis; L. Western blot shows that Cyclin D1 was inhibited in miR-206 overexpressed LN229 cells. (*, P<0.05; **, P<0.01; ***, P<0.001).
MiR-206 predominantly promotes apoptosis and induces cell cycle arrest of glioma cells
Since cell cycle and apoptosis play key roles in regulating cell proliferation, we performed flow cytometry assay to detect the alterations in these processes in miR-206 overexpressed cells. As shown in Figure 2E-G, after miR-206 overexpression, cell counts were significantly increased in the G1 phase while significantly inhibited in S/G2 phase. Because it is known that Cyclin D1 can promote cell cycle transition from the G1 to S phase, we performed western blot assay and found that Cyclin D1 was down-regulated in the miR-206 overexpressed group (Figure 2H). On the other hand, the percentage of apoptotic LN229 cells was also found to be significantly up-regulated after miR-206 overexpression (Figure 2I-K). Meanwhile, western blot results show that the known anti-apoptotic protein Bcl-2 is significantly down-regulated (Figure 2L). These results strongly indicate that miR-206 can inhibit glioma proliferation by inhibiting cell cycle progression and promoting cell apoptosis.
MiR-206 regulates glioma cell growth by down-regulating FOXP1
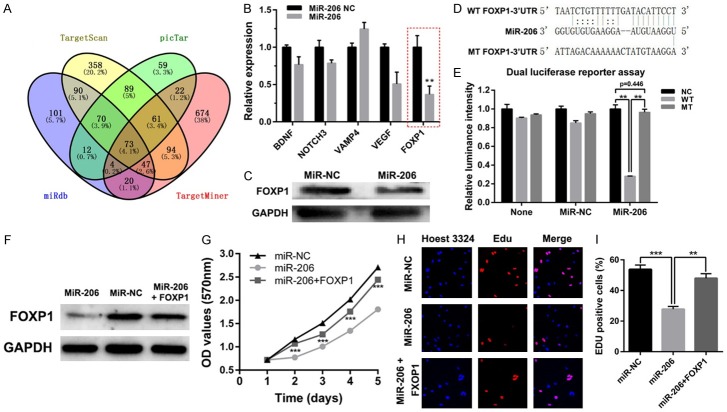
To identify the downstream target of miR-206 in gliomas, we firstly combined miR-206 target prediction lists from miRDB, TargetScan, TargetMiner and picTar, and got 73 targets predicted by the four databases (Figure 3A). To further narrow down the targets number, we referred to the published papers finding genes in the 73 targets that may play important roles in gliomas. By this way, we finally selected 5 targets: BDNF, NOTCH3, VAMP4, VEGF and FOXP1. We then performed qPCR analysis to detect the alterations of the 5 genes after miR-206 overexpression, and found that FOXP1 is the most significantly down-regulated gene (Figure 3B). As it was reported that FOXP1 can enhance glioblastoma tumorigenicity [11], we hypothesized that miR-206 may inhibit glioma proliferation by targeting FOXP1. Indeed, further western blot results show that FOXP1 expression was down-regulated by miR-206 overexpression (Figure 3C). Moreover, we identified two potential miR-206 binding sites in FOXP1 mRNA 3’UTR using the TargetScan database. As shown in Figure 3D, we mutated one of the binding sites, which was predicted with the highest miR-206 binding score, to perform dual luciferase assay. As shown in Figure 3E, the luciferase reporter activity was only significantly down-regulated in the wildtype FOXP1-3’UTR group, indicating that FOXP1 is a bona fide downstream target of miR-206.
Figure 3.
FOXP1 overexpression can partially rescue miR-206-mediated inhibition of glioma cell proliferation. A. Venn diagram shows that in total of 73 targets were commonly predicted by TargetScan, TargetMiner, miRDB and picTar databases. B. qPCR analysis shows that FOXP1 is the most down-regulated gene among the 5 selected miR-206 potential downstream targets. C. Western blot shows that FOXP1 is down-regulated by miR-206. D. Sequence alignment of the FOXP1 3’UTR binding site of miR-206. E. Dual luciferase reporter assay shows that miR-155 can only down-regulate the wildtype FOXP1 3’UTR-containing reporter activity. F. Western blot shows that FOXP1 protein was up-regulated after FOXP1 overexpression in miR-206 stably overexpressed LN229 cells. G. The proliferation of miR-206 stably overexpressed LN229 cells can be partially rescued by FOXP1 overexpression. H, I. Edu incorporation of miR-206 stably overexpressed LN229 cells can be partially rescued by FOXP1 overexpression. (*: P<0.05; **: P<0.01; ***, P<0.001).
Next, we wondered whether FOXP1 is involved in the proliferative inhibition of miR-206. To validate our speculation, we transfected the miR-206 stably overexpressed LN229 cells with pcDNA3.1-FOXP1 (Figure 3F). Subsequently, CCK-8 and Edu cell proliferation assays were performed. The results show that FOXP1 overexpression can partially reverse the proliferative inhibition induced by miR-206 overexpression (Figure 3G-I). Furthermore, the flow cytometry assay reveals that overexpression of FOXP1 can partially rescue the promotion of cell apoptosis and inhibition of cell cycle arrest effect in the miR-206 overexpressed group (Figure 4A-D, F-I). Consistent with these functional studies, the western blot results show that, after overexpression of FOXP1 in miR-206 overexpressed LN229 glioma cells, the anti-apoptotic protein Bcl-2 and cell cycle promoting protein Cyclin D1 can be partially rescued (Figure 4A, 4E). Together, these data indicate that FOXP1 is involved in miR-206-dependent negative regulation of glioma cell growth.
Figure 4.
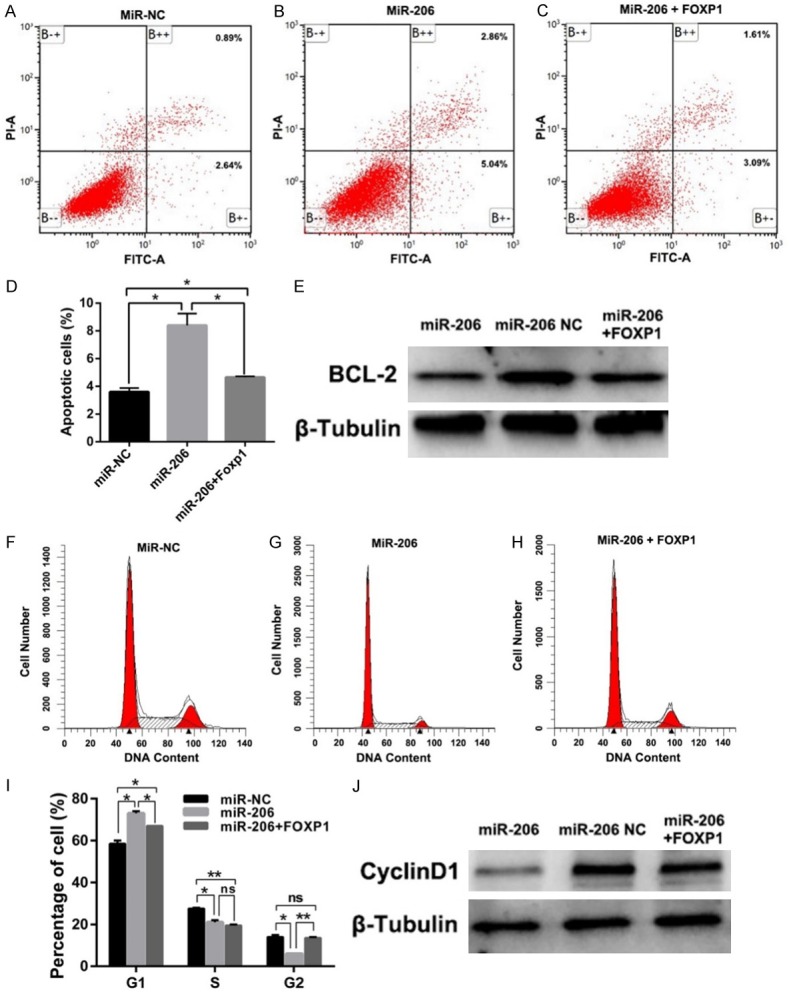
FOXP1 overexpression can partially rescue miR-206 mediated glioma cell cycle blockage and apoptosis promotion. A-D. The promotion of apoptosis of miR-206 stably overexpressed LN229 cells can be partially rescued by FOXP1 overexpression; E. Western blot shows that Bcl-2 was up-regulated after FOXP1 overexpression in miR- 206 stably overexpressed LN229 cells. F-I. The cell cycle blockage of miR-206 stably overexpressed LN229 cells can be partially rescued by FOXP1 overexpression; J. Western blot shows that Cyclin D1 was up-regulated after FOXP1 overexpression in miR-206 stably overexpressed LN229 cells.
Discussion
Over the past decade, the role of miRNAs in cancers has been extensively studied. Researchers have agreed that miRNAs play extremely important roles in tumor formation, development, and progression. By regulating the expression of different target genes, miRNAs can act as oncogenes or tumor suppressors in various human cancers. A large number of miRNAs also have been defined as potential diagnostic and prognostic factors in certain tumors. The aberrant expression and dysregulated function of miRNAs merit broad investigation, and a therapeutic strategy specifically aimed at targeting miRNA is also on the way.
Accumulating experimental evidences have pointed out that there is a close correlation between aberrant expression of miR-206 and tumors. Most related studies indicate a suppressive role of miR-206 on various human tumors. For example, miR-206 expression level was very low in non-small cell lung cancer (NSCLC) cell lines. Overexression of miR-206 suppressed c-Met and Bcl-2 expression and activated apoptosis, and inhibits tumor cell proliferation, migration and colony formation in NSCLC [13]. Similarly, miR-206 inhibited the growth of cervical cancer cell lines and hepatocellular carcinoma cells through targeting Bcl-2 [14] and CDK9 [15], respectively. Another study demonstrated that miR-206 suppressed several cancers’ growth by blocking Cyclin D1 and inducing a G1 arrest [16]. Furthermore, miR-206 has been proven to be a critical factor in epithelial-mesenchymal transition (EMT) and consequently invasion and metastasis of many cancers. A recent study showed that miR-206 regulates EMT in human lung adenocarcinoma cells and partly reduces cisplatin resistance [17]. miR-206-PAX3-MET signaling is also found to be critical to gastric cancer metastasis, and the activation of PAX3-MET pathways is due to miR-206 loss in clinical cancers [18]. In estrogen receptor positive breast cancers, miR-206 inhibits TGF-β transcription and autocrine production, as well as downstream target genes of EMT [19]. Moreover, miR-206 was found to inhibit epithelial-mesenchymal transition and angiogenesis via tar-geting classical c-Met/PI3k/Akt/mTOR pathway in non-small cell lung cancer [13,20]. miR-206 also inhibits tumor invasion and migration in colorectal cancer [21]. To date, miR-206 has been found to be an independent prognostic factor in cervical cancer, colorectal cancer.
However, few experiments have elucidated the role of miR206 in gliomas. Therefore, exploring the functions and clinical significance of miR-206 in glioma is very attractive. In the present study, we showed that miR-206 was significantly down-regulated in high grade gliomas and predicts favorable prognosis. One study by S. Wang, et al. detected the expression level of miR-206 in 108 malignant astrocytomas and 20 normal brain tissues. Consistent with our findings, that study reported decreased miR-206 expression in higher grade astrocytomas and miRNA-206 negatively correlated with poor clinical outcome [6]. In this study, we specifically selected 18 pairs of glioma--peritumor tissue to confirm the lower miRNA-206 expression in brain gliomas. Our findings are further confirmed by the data from CGGA.
The study of R. Wang et al. showed that miR-206 inhibits neural cells proliferation and promote apoptosis via regulating Otx2. MicroRNA-206 also inhibited the progression of glioblastoma through Bcl-2. Thus, we assumed miR-206 would inhibit glioma cell proliferation. Interestingly, we found that miR-206 expression was significantly negatively correlated with the proliferation maker Ki-67, indicating a potential proliferative inhibition of miR-206 in gliomas. Therefore, we up-regulated miR-206 in LN229 glioma cells, and observed the proliferative inhibition in these cells. Subsequently, the flow cytometry analysis indicates that the inhibition of proliferation may be caused by promotion of cell apoptosis and cell cycle arrest.
To explore how miR-206 regulates glioma, we employed four target gene predicting algorithms: miRDB, TargetScan, TargetMiner and picTar. After integrating public online data and performing validation experiment via miR-206 overexpression, we identified FOXP1 to be a potential downstream target of miR-206. We further elucidated the direct down-regulation of FOXP1 by miR-206 after qPCR analysis, western blot and luciferase assay. Moreover, our study found that FOXP1 was involved in miR-206-dependent negative regulation of glioma cell growth. The regulatory role of miR-206 on FOXP1 is partially evidenced by a research showing that upregulation of microRNA-206 induces apoptosis of vascular smooth muscle cells by modulating FOXP1 [22].
FOXP1 belongs to the winged helix or forkhead transcription factor family which has a wide range of functions. The role of FOXP1 in cancers is context-dependent. In lymphoma, up-regulation of FOXP1 was correlated with poor patient prognosis and was indicated as an oncogene [23]. Overexpression of FOXP1 in ovarian cancer cells increased spheroid formation, expression of stemness-related genes and epithelial to mesenchymal transition-related genes, cell migration, and resistance to paclitaxel or cisplatin treatment [24,25]. The role of FOXP1 in breast cancer is quite complicated and different or even controversial results are reported different research groups. As a downstream effector, FOXP1 takes on the tumorigenic effect of PI3K/Akt/p70S6K signaling pathway in breast cancer cells [26]. A few study also observed that FOXP1 regulates the transcriptional activity of a serial of protein-coding genes and enhances the proliferation, migration, drug-resistant of breast cancer cells [27,28]. On the contrary, in more common circumstances, unfavorable relapse-free survival (RFS) in breast cancer patients with decreased FOXP1 expression. Another study suggested that FOXP1 demonstrates different expression patterns in familial breast cancers than sporadic tumours, and expression of the FOXP1 is associated with estrogen receptor alpha, estrogen receptor beta and improved survival in familial breast cancers [29]. In addition, high expression of FOXP1 is found to be associated with improved survival in patients with non-small cell lung cancer [30].
However, the role of FOXP1 in the tumorigenesis and clinical prognosis of brain glioma remains poorly understood. An earlier report provided evidence that FOXP1 can assist the enhancement of glioblastoma tumorigenicity [11]. In the present study, we demonstrated that FOXP1 may function as an oncogene which reverses the tumorigenic inhibition of glioma cells induced by miR-206 overexpression. Overexpression of FOXP1 could partially rescue the proliferative inhibition in the miR-206 stably overexpressed LN229 cells. Similar to our study, FOXP1 is found to be regulated by other microRNAs in glioma. Cui et al. reported that miR-504 inhibits glioma cell proliferation and promotes apoptosis by down-regulating FOXP1 [31]. Glioblastoma tumorigenicity could be enhanced by FOXP1 after removing the inhibition effect exerted by miR-9 [11]. The above mentioned studies all indicated an oncogene role of FOXP1 in brain glioma. On the contrary, in the study by Xue, et al. FOXP1 was found to be down-regulated in clinical glioma samples its overexpression could inhibit proliferation, invasion and migration of human glioma U251 cells [32]. The reason for these controversial results is not clarified at the moment, and further investigation about the role of FOXP1 in malignant glioma is warranted.
In summary, our study shows that miR-206 is significantly down-regulated in gliomas. Overexpression of miR-206 in LN229 glioma cells can inhibit proliferation by promoting cell apoptosis and cell cycle arrest. FOXP1 is a direct downstream target of miR-206 which is involved in the inhibition of proliferation. Together, our results indicate that miR-206 can act as a tumor suppressor in gliomas and serve as a novel therapeutic target via FOXP1.
Acknowledgements
The authors would like to thank Professor Xiu-Wu Bian, Department of Pathology, Southwest Hospital, TMMU for providing LN229 glioma cell line, and would like to thank Mrs. Qing-Rui Li from Department of Pathology, Southwest Hospital, TMMU for collecting clinical data and technical supports. This research was supported by the National Natural Science Foundation of China (NSFC-81272783).
Disclosure of conflict of interest
None.
References
- 1.Vredenburgh JJ, Desjardins A, Herndon JE 2nd, Dowell JM, Reardon DA, Quinn JA, Rich JN, Sathornsumetee S, Gururangan S, Wagner M, Bigner DD, Friedman AH, Friedman HS. Phase II trial of bevacizumab and irinotecan in recurrent malignant glioma. Clin Cancer Res. 2007;13:1253–1259. doi: 10.1158/1078-0432.CCR-06-2309. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 3.Mitchelson KR, Qin WY. Roles of the canonical myomiRs miR-1, -133 and -206 in cell development and disease. World J Biol Chem. 2015;6:162–208. doi: 10.4331/wjbc.v6.i3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ma G, Wang Y, Li Y, Cui L, Zhao Y, Zhao B, Li K. MiR-206, a key modulator of skeletal muscle development and disease. Int J Biol Sci. 2015;11:345–352. doi: 10.7150/ijbs.10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kondo N, Toyama T, Sugiura H, Fujii Y, Yamashita H. miR-206 expression is down-regulated in estrogen receptor alpha-positive human breast cancer. Cancer Res. 2008;68:5004–5008. doi: 10.1158/0008-5472.CAN-08-0180. [DOI] [PubMed] [Google Scholar]
- 6.Wang S, Lu S, Geng S, Ma S, Liang Z, Jiao B. Decreased expression of microRNA-206 correlates with poor clinical outcome in patients with malignant astrocytomas. Pathol Oncol Res. 2014;20:343–348. doi: 10.1007/s12253-013-9701-6. [DOI] [PubMed] [Google Scholar]
- 7.Hao W, Luo W, Bai M, Li J, Bai X, Guo J, Wu J, Wang M. MicroRNA-206 inhibited the progression of glioblastoma through BCL-2. J Mol Neurosci. 2016;60:531–538. doi: 10.1007/s12031-016-0824-6. [DOI] [PubMed] [Google Scholar]
- 8.Wang R, Hu Y, Song G, Hao CJ, Cui Y, Xia HF, Ma X. MiR-206 regulates neural cells proliferation and apoptosis via Otx2. Cell Physiol Biochem. 2012;29:381–390. doi: 10.1159/000338493. [DOI] [PubMed] [Google Scholar]
- 9.Koon HB, Ippolito GC, Banham AH, Tucker PW. FOXP1: a potential therapeutic target in cancer. Expert Opin Ther Targets. 2007;11:955–965. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gu Y, Xiao X, Yang S. LncRNA MALAT1 acts as an oncogene in multiple myeloma through sponging miR-509-5p to modulate FOXP1 expression. Oncotarget. 2017;8:101984–101993. doi: 10.18632/oncotarget.21957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gomez GG, Volinia S, Croce CM, Zanca C, Li M, Emnett R, Gutmann DH, Brennan CW, Furnari FB, Cavenee WK. Suppression of microRNA-9 by mutant EGFR signaling upregulates FOXP1 to enhance glioblastoma tumorigenicity. Cancer Res. 2014;74:1429–1439. doi: 10.1158/0008-5472.CAN-13-2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu QF, Pan YW, Li LC, Zhou Z, Huang QL, Pang JC, Zhu XP, Ren Y, Yang H, Ohgaki H, Lv SQ. MiR-22 is frequently downregulated in medulloblastomas and inhibits cell proliferation via the novel target PAPST1. Brain Pathol. 2014;24:568–583. doi: 10.1111/bpa.12136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun C, Liu Z, Li S, Yang C, Xue R, Xi Y, Wang L, Wang S, He Q, Huang J, Xie S, Jiang W, Li D. Down-regulation of c-Met and Bcl2 by microRNA-206, activates apoptosis, and inhibits tumor cell proliferation, migration and colony formation. Oncotarget. 2015;6:25533–25574. doi: 10.18632/oncotarget.4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen AH, Qin YE, Tang WF, Tao J, Song HM, Zuo M. MiR-34a and miR-206 act as novel prognostic and therapy biomarkers in cervical cancer. Cancer Cell Int. 2017;17:63. doi: 10.1186/s12935-017-0431-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pang C, Huang G, Luo K, Dong Y, He F, Du G, Xiao M, Cai W. miR-206 inhibits the growth of hepatocellular carcinoma cells via targeting CDK9. Cancer Med. 2017;6:2398–2409. doi: 10.1002/cam4.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliman SJ, Howley BV, Mehta DS, Fearnhead HO, Kemp DM, Barkley LR. Selective repression of the oncogene cyclin D1 by the tumor suppressor miR-206 in cancers. Oncogenesis. 2014;3:e113. doi: 10.1038/oncsis.2014.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen QY, Jiao DM, Wang J, Hu H, Tang X, Chen J, Mou H, Lu W. miR-206 regulates cisplatin resistance and EMT in human lung adenocarcinoma cells partly by targeting MET. Oncotarget. 2016;7:24510–24526. doi: 10.18632/oncotarget.8229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ren J, Huang HJ, Gong Y, Yue S, Tang LM, Cheng SY. MicroRNA-206 suppresses gastric cancer cell growth and metastasis. Cell Biosci. 2014;4:26. doi: 10.1186/2045-3701-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin K, Yin W, Wang Y, Zhou L, Liu Y, Yang G, Wang J, Lu J. MiR-206 suppresses epithelial mesenchymal transition by targeting TGF-beta signaling in estrogen receptor positive breast cancer cells. Oncotarget. 2016;7:24537–24548. doi: 10.18632/oncotarget.8233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen QY, Jiao DM, Wu YQ, Chen J, Wang J, Tang XL, Mou H, Hu HZ, Song J, Yan J, Wu LJ, Chen J, Wang Z. MiR-206 inhibits HGF-induced epithelial-mesenchymal transition and angiogenesis in non-small cell lung cancer via c-Met /PI3k/Akt/mTOR pathway. Oncotarget. 2016;7:18247–18261. doi: 10.18632/oncotarget.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun P, Sun D, Wang X, Liu T, Ma Z, Duan L. miR-206 is an independent prognostic factor and inhibits tumor invasion and migration in colorectal cancer. Cancer Biomark. 2015;15:391–396. doi: 10.3233/CBM-150489. [DOI] [PubMed] [Google Scholar]
- 22.Xing T, Du L, Zhuang X, Zhang L, Hao J, Wang J. Upregulation of microRNA-206 induces apoptosis of vascular smooth muscle cells and decreases risk of atherosclerosis through modulating FOXP1. Exp Ther Med. 2017;14:4097–4103. doi: 10.3892/etm.2017.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood. 2004;104:2933–2935. doi: 10.1182/blood-2004-03-1209. [DOI] [PubMed] [Google Scholar]
- 24.Choi EJ, Seo EJ, Kim DK, Lee SI, Kwon YW, Jang IH, Kim KH, Suh DS, Kim JH. FOXP1 functions as an oncogene in promoting cancer stem cell-like characteristics in ovarian cancer cells. Oncotarget. 2016;7:3506–3519. doi: 10.18632/oncotarget.6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z, Zhu L, Gao J, Cai M, Tan M, Liu J, Lin B. Expression of FOXP1 in epithelial ovarian cancer (EOC) and its correlation with chemotherapy resistance and prognosis. Tumour Biol. 2015;36:7269–7275. doi: 10.1007/s13277-015-3383-5. [DOI] [PubMed] [Google Scholar]
- 26.Halacli SO, Dogan AL. FOXP1 regulation via the PI3K/Akt/p70S6K signaling pathway in breast cancer cells. Oncol Lett. 2015;9:1482–1488. doi: 10.3892/ol.2015.2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oskay Halacli S. FOXP1 enhances tumor cell migration by repression of NFAT1 transcriptional activity in MDA-MB-231 cells. Cell Biol Int. 2017;41:102–110. doi: 10.1002/cbin.10702. [DOI] [PubMed] [Google Scholar]
- 28.Shigekawa T, Ijichi N, Ikeda K, Horie-Inoue K, Shimizu C, Saji S, Aogi K, Tsuda H, Osaki A, Saeki T, Inoue S. FOXP1, an estrogen-inducible transcription factor, modulates cell proliferation in breast cancer cells and 5-year recurrence-free survival of patients with tamoxifen-treated breast cancer. Horm Cancer. 2011;2:286–297. doi: 10.1007/s12672-011-0082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rayoo M, Yan M, Takano EA, Bates GJ, Brown PJ, Banham AH, Fox SB. Expression of the forkhead box transcription factor FOXP1 is associated with oestrogen receptor alpha, oestrogen receptor beta and improved survival in familial breast cancers. J Clin Pathol. 2009;62:896–902. doi: 10.1136/jcp.2009.065169. [DOI] [PubMed] [Google Scholar]
- 30.Feng J, Zhang X, Zhu H, Wang X, Ni S, Huang J. High expression of FoxP1 is associated with improved survival in patients with non-small cell lung cancer. Am J Clin Pathol. 2012;138:230–235. doi: 10.1309/AJCPDHQFNYJZ01YG. [DOI] [PubMed] [Google Scholar]
- 31.Cui R, Guan Y, Sun C, Chen L, Bao Y, Li G, Qiu B, Meng X, Pang C, Wang Y. A tumor-suppressive microRNA, miR-504, inhibits cell proliferation and promotes apoptosis by targeting FOXP1 in human glioma. Cancer Lett. 2016;374:1–11. doi: 10.1016/j.canlet.2016.01.051. [DOI] [PubMed] [Google Scholar]
- 32.Xue L, Yue S, Zhang J. FOXP1 has a low expression in human gliomas and its overexpression inhibits proliferation, invasion and migration of human glioma U251 cells. Mol Med Rep. 2014;10:467–472. doi: 10.3892/mmr.2014.2197. [DOI] [PubMed] [Google Scholar]