Abstract
Clear cell renal cell carcinoma (ccRCC) is the most common subtype of renal cell carcinoma. The chromo-helicase-DNA binding proteins (CHDs), containing nine members named CHD1-9, act as regulators of the chromatin remodeling process and gene expression. To determine the correlation between CHD9 expression and ccRCC, we performed an immunohistochemical staining in a tissue microarray (TMAs) containing tissue samples from 88 ccRCC patients. The results showed that cytoplasm CHD9 expression was statistically decreased in tumor tissues compared to adjacent tissues (8.54±2.90 vs 12.61±2.05, P=0.000), while nuclear CHD9 expression was upregulated in the tumor tissues (1.47±2.93 vs 0.29±1.24, P=0.000). A univariate analysis found that cytoplasm CHD9 expression in cancer tissues was correlated with the patients’ pathological grading (P=0.002, r=0.330), the clinical stages (P=0.02, r=0.250) and the T grading (P=0.024, r=0.241) significantly. In addition, cytoplasm CHD9 expression in non-tumor tissues was correlated with the ccRCC patients’ pathological grading (P=0.031, r=-0.231) significantly. Patients with high cytoplasm CHD9 expression had a significantly worse prognosis than those with low cytoplasm CHD9 expression levels (59.7% vs 85.7%, P=0. 042). In conclusion, our study indicated the important role of CHD9 in ccRCC and suggested CHD9 may be a potential biomarker for prognostic prediction and a new target for therapy.
Keywords: CHD9, clear cell renal cell carcinoma (ccRCC), tissue microarray, immunohistochemistry, survival time, chemotherapy
Introduction
Renal cell carcinoma (RCC), which comprises a heterogeneous group of four epithelial neoplasms with diverse biologic behaviors and variable clinical outcomes, is the most lethal of the common urological cancers. The most common subtype is the clear cell renal cell carcinoma, which makes up 75-80% of all RCC cases. The other three subtypes include papillary carcinoma (PRCC, 10% of RCC), chromophobic carcinoma (ChRCC, 5% of RCC), and collecting duct carcinoma (2% of RCC) makes up the other 20%-25% of RCC cases [1,2]. Although a number of targeted drugs have emerged in recent years, the overall survival times of patients with metastatic kidney cancer remain short [3].
The chromo-helicase-DNA binding proteins (CHDs) act as regulators of the chromatin remodeling process and gene expression. The CHD proteins contain two basal tandem chromo domains and different additional domains, according to which the CHDs are divided into three sub-groups: CHD1-2, CHD3-5 and CHD6-9 [4,5]. Chromatin remodeling plays a critical role in regulating gene expression throughout ontogenesis, and the CHDs associated with many human diseases and cancers include lymphoma, liver cancer, renal cancer, colorectal cancer, and stomach cancer [6-8]. For example, CHD1 deficiency may contribute to prostate cancer metastasis [9]. CHD4 and CHD3 bind ZGPAT (zinc finger, CCCH-type with G patch domain) could inhibit tumor development [10]. CHD5 expression could serve as an independent prognostic marker of survival for patients with ovarian cancer, neuroblastoma, glioma. and gallbladder carcinoma [8,11]. Respectively, the third sub-family CHD enzymes are orthologs of the drosophila kismet enzyme and are characterized by the brahma and kismet domains at C termini. The mutant of CHD7, 8 could lead to the distinct disease states of CHARGE syndrome and Kallman Syndrome [12,13]. There few studies on the relationship of CHD9 and human disease, especially on renal disease [14,15].
Materials and methods
Clinical material
Tumor samples from 88 clear cell renal cell carcinoma patients and their paired adjacent carcinoma tissues were assembled in a tissue microarray (HKidE180Su03, Shanghai Outdo Biotech Co., Ltd). All the typical pathological sites on the HE slices were labeled by two pathologists, and then drilled on the blank recipient paraffin (diameter was 1.5 mm) using a tissue microarray instrument. The patients’ clinical information, including pathological information, such as their age, gender, tumor size, tumor differentiation, stage, TNM stage, distant metastasis and clinical stage, was detailed in Table 1.
Table 1.
Details of the ccRCC patients’ clinical information
| Clinical index | N | Lost | Total N | |
|---|---|---|---|---|
| Gender | Male | 57 | 1 | 88 |
| Female | 30 | |||
| Age | ≤60 years | 49 | 2 | 88 |
| >60 years | 37 | |||
| Tumor size | ≤5 cm | 52 | 88 | |
| >5 cm | 36 | |||
| T staging | T1 | 57 | 88 | |
| T2 | 25 | |||
| T3 | 6 | |||
| N staging | N0 | 84 | 88 | |
| N1-3 | 2 | |||
| M staging | M0 | 86 | 88 | |
| M1 | 2 | |||
| Clinical staging | Stage I | 55 | 2 | 88 |
| Stage II | 23 | |||
| Stage III | 6 | |||
| Stage IV | 2 | |||
| Pathological grading | I-II | 34 | 88 | |
| III-IV | 24 |
The patients’ operations took place from October 2006 to February 2008. The final follow-up date was in August 2015, so follow-up times ranged from 90 to 106 months. During this follow-up time, 30 of the 88 ccRCC patients died of ccRCC (the overall survival time ranged from 2 to 79 months), and the other 58 survived. All patients were clinicopathologically diagnosed as having ccRCC and received no additional treatment before surgery.
Immunohistochemistry
As previously described, two-step Immunohistochemical staining was performed on a tissue microarray in this study [16,17]. Firstly, after they were treated with an EDTA buffer to retrieve the antigen, the tissue sections were blocked with goat serum. Then the tissue sections were incubated with a primary antibody (Proteintech, 13402-1-AP, 1:1500) at 4°C overnight.
Next, the tissue sections were incubated with a secondary antibody, washed with PBS, and then, examined using a diaminobenzidine (DAB) system and hematoxylin re-dying, observed and analyzed under microscope. The CHD9 expression was scored and the tissues grouped according to their “positive staining rate score” multiplied by their “staining intensity score”. The positive staining rate was defined according to the proportion of positively stained cancer cells: “Negative” is 0, “1%-20%” for 1, “21%-40%” for 2, “41-60%” for 3, “61-80%” for 4. The staining intensity score was defined as follows: “Negative” is 0, “1+” for 1, “2+” for 2, “3+” for 3. Thus, patients were placed into the low expression group when the score was ≤6, while the patients with a score >6 were placed into the high expression group.
Statistical analysis
Firstly, we statistically analyzed the positive staining rate and the intensity of immunohistochemical staining. The different expression of CHD9 in cancer and in the adjacent tissues was evaluated by NPar tests. Spearman’s correlation analysis was used to calculate the CHD9 expression in cancer and adjacent tissues. The relationships between the clinical indicators of the ccRCC patients and CHD9 expression were also analyzed by Spearman’s correlation analysis. In this study, the Kaplan-Meier survival analysis and a log-rank statistical test were used to analyze the survival factors. Finally, statistically significant variables in univariate analysis were also included in a COX multivariate regression survival analysis. Consider P<0.05 as statistically significant.
Results
The expression of CHD9 in the cancer tissues and their adjacent cancer
The NPar Tests showed that the cytoplasm CHD9 expression was statistically decreased in the cancer tissues compared to their adjacent tissues (8.54±2.90 vs 12.61±2.05, P=0.000) while nuclear CHD9 expression in cancer tissues was upregulated (1.47±2.93 vs 0.29±1.24, P=0.000). The detailed images are shown in Figure 1.
Figure 1.
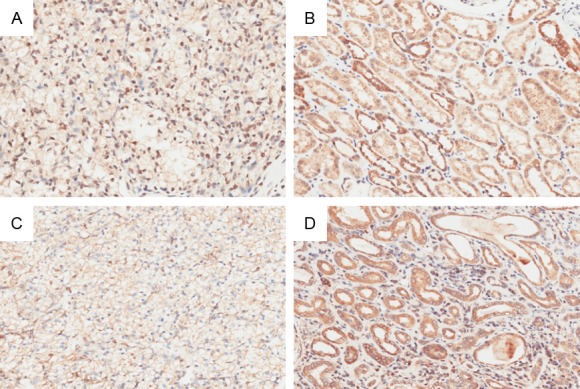
Representative immunohistochemistry images of CHD9 expression in ccRCC tissues and their para-carcinoma tissues: A. Nucleus CHD9 expression in tumor tissues; B. Nucleus CHD9 expression in adjacent tissues; C. Cytoplasm CHD9 expression in cancer tissues; D. Cytoplasm CHD9 expression in adjacent tissues (Magnification times: ×200).
The correlation of CHD9 expression between cancer tissues and their adjacent tissues
The Spearman’s correlation analysis showed that the relationships between cytoplasm CHD9 expression and the nuclear CHD9 expression in cancers or in non-tumors positively correlated with the nuclear CHD9 expression in adjacent tissues (P=0.002; r=0.330).
The relationship between the CHD9 expression and ccRCC patients’ clinical index
The Spearman’s correlation analysis was used to analyze the relationship between CHD9 expression and ccRCC patients’ clinical index including gender, age, tumor size, pathological grading, TNM stage. and clinical stages. The results showed that the cytoplasm CHD9 expression in cancer tissues correlated significantly with patients’ pathological grading (P=0.002, r=0.330), the clinical stages (P=0.02, r=0.250), and T grading (P=0.024, r=0.241). In addition, the cytoplasm expression of CHD9 in non-tumor tissues correlated significantly with ccRCC patients’ pathological grading (P=0.031, r=-231). The results are shown in Table 2.
Table 2.
The relationship between CHD9 expression and the ccRCC patients’ clinical index
| Gender | Age | Pathological grading | Tumor size | T | N | M | Clinical staging | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spearman’s rho | Cytoplasm CHD9 expression | Tumor | Correlation Coefficient | -0.084 | 0.027 | 0.330** | 0.057 | 0.241* | 0.029 | 0.026 | 0.250* |
| Sig. (2-tailed) | 0.437 | 0.805 | 0.002 | 0.596 | 0.024 | 0.792 | 0.808 | 0.020 | |||
| Adjacent | Correlation Coefficient | 0.024 | -0.138 | -0.231* | -0.188 | 0.017 | -0.028 | -0.119 | 0.008 | ||
| Sig. (2-tailed) | 0.826 | 0.207 | 0.031 | 0.081 | 0.879 | 0.802 | 0.274 | 0.945 | |||
| Nucleus CHD9 expression | Tumor | Correlation Coefficient | -0.004 | 0.027 | -0.108 | -0.056 | -0.046 | 0.111 | -0.092 | -.012 | |
| Sig. (2-tailed) | 0.969 | 0.804 | 0.318 | 0.606 | 00.672 | 0.307 | 0.395 | 0.913 | |||
| Adjacent | Correlation Coefficient | -0.045 | -0.069 | -0.005 | 0.021 | -0.081 | -0.039 | -0.038 | -0.091 | ||
| Sig. (2-tailed) | 0.682 | 0.530 | 0.961 | 0.850 | 0.455 | 0.723 | 0.728 | 0.412 | |||
Correlation is significant at the 0.01 level (2-tailed).
Correlation is significant at the 0.05 level (2-tailed).
The relationships between the overall survival time of ccRCC patients and the CHD9 expression
The relationship between CHD9 expression and the prognosis of ccRCC was defined according to the Kaplan-Meier method and the log-rank test. The results showed that patients with high cytoplasm CHD9 expression had a worse prognosis than those with a low cytoplasm CHD9 expression (59.7% vs 85.7%, P=0.042). The results are shown in Figure 2.
Figure 2.
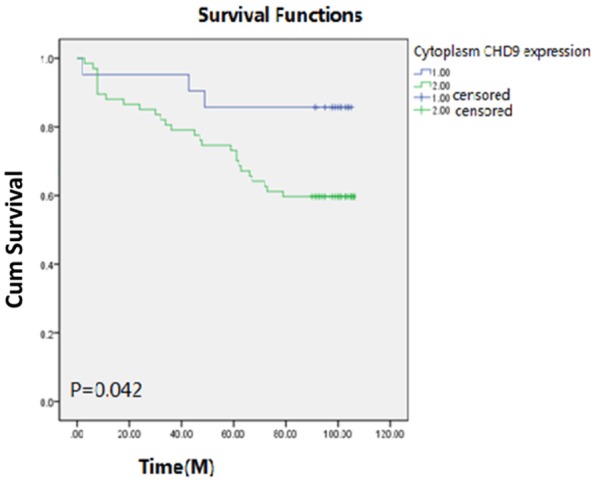
The relationships between the overall survival time of ccRCC patients and CHD9 expression: 1.00 represents the patients with low cytoplasm CHD9 expression; 2.00 represents the patients with high cytoplasm CHD9 expression.
Subsequently, the COX multi-factor survival analysis showed that cytoplasm CHD9 expression was not an independent predictive factor for ccRCC. The analysis results in detailed are shown in Table 3.
Table 3.
COX multivariate regression analysis of the independent predictors of CHD9 in ccRCC patients
| B | SE | Wald | Df | Sig | Exp (B) | 95.0% Exp (B) confidence interval | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Lower limit | Upper limit | |||||||
| Age | 1.294 | 0.427 | 9.195 | 1 | 0.002** | 3.649 | 1.581 | 8.425 |
| Pathological Grading | 0.613 | 0.277 | 4.917 | 1 | 0.027* | 1.847 | 1.074 | 3.176 |
| Tumor size | -0.068 | 0.462 | 0.021 | 1 | 0.884 | 0.935 | 0.378 | 2.312 |
| T | 0.675 | 0.369 | 3.343 | 1 | 0.067 | 1.964 | 0.953 | 4.050 |
| N | 1.508 | 0.860 | 3.074 | 1 | 0.080 | 4.519 | 0.837 | 24.392 |
| M | 1.283 | 0.941 | 1.858 | 1 | 0.173 | 3.608 | 0.570 | 22.839 |
| Clinical stages | . | 0 | . | |||||
| Cytoplasm CHD9 expression in cancer | 0.450 | 0.643 | 0.489 | 1 | 0.485* | 1.568 | 0.444 | 5.532 |
A. The degrees of freedom were reduced because of the constant or linear variation. B. Constant or linear covariance. Clinical stages = T + N + M.
Correlation is significant at the 0.01 level (2-tailed).
Correlation is significant at the 0.05 level (2-tailed).
Discussion
The Chromo Helicase DNA-binding proteins (CHDs) act as regulators of the chromatin remodeling process and gene expression in humans. It has been found that the CHD family contains nine members divided into three sub-groups according to the structural domains and features: CHD1-2, CHD3-5 and CHD6-9. The CHDs are associated with many human diseases and cancers including lymphoma, liver cancer, colorectal cancer, and stomach cancer [11]. Few studies have reported on the relationship between CHD9 and human renal disease or cancer.
We showed CHD9 expression in clear cell renal cell carcinoma through immunohistochemistry. The results show that the cytoplasm CHD9 expression was statistically significantly decreased, while nucleus CHD9 expression was upregulated in cancer tissues compared to their adjacent tissues. The nucleus CHD9 expression in cancer tissues were positively correlated with nucleus CHD9 expression in adjacent tissues. These alterations of the CHD9 expression in ccRCC indicate that CHD9 expression may be closely related to tumorigenesis [18]. Additionally, the cytoplasm CHD9 expression in tumor tissues was significantly correlated with ccRCC patients’ clinical index, including pathological grading, clinical grading and T stage. In line with previous studies, these findings indicate that cytoplasm CHD9 expression has a significant correlation with tumor cell progression, development, and invasiveness in ccRCC [19]. However, the cytoplasm expression of CHD9 in non-tumor tissue was correlated significantly negatively with the ccRCC patients’ pathological grading, indicating that CHD9 might have different regulatory effects in cancer and adjacent tissues which should be validated in the future work.
It is reported that the loss of CHD8 expression may serve as a novel indicator in gastric cancer [20], and CHD7 may be a novel biomarker in pancreatic cancer patients [21]. Thus, as the other member of CHD6-9, the CHD9 expression must have some predictive value with the prognosis of ccRCC, even though the COX multi-factor survival analysis showed that CHD9 expression was not an independent predictive factor. The statistical results showed that patients with high cytoplasm CHD9 expression in cancer tissues had an obviously worse prognosis than those with low cytoplasm CHD9 expression.
In summary, we found abnormal, upregulated expression of CHD9 to be associated with clinical index and overall survival in clear cell renal cell carcinoma. The findings are in line with CHD9 differential expression in the cytoplasm and nucleus, indicating that CHD9 expression correlates with survival and metastatic propensity in cancer. Though CHD9 expression could not serve as an independent prognostic marker, it still provides a reference for prognostic diagnosis. Our study indicated the important role of CHD9 in ccRCC and suggests that CHD9 is a potential biomarker for prognostic prediction and a new therapy target.
Disclosure of conflict of interest
None.
References
- 1.Wang Y, Wan F, Chang K, Lu X, Dai B, Ye D. NUDT expression is predictive of prognosis in patients with clear cell renal cell carcinoma. Oncol Lett. 2017;14:6121–6128. doi: 10.3892/ol.2017.6997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabjerg M, Bjerregaard H, Halekoh U, Jensen BL, Walter S, Marcussen N. Molecular characterization of clear cell renal cell carcinoma identifies CSNK2A1, SPP1 and DEFB1 as promising novel prognostic markers. APMIS. 2016;124:372–383. doi: 10.1111/apm.12519. [DOI] [PubMed] [Google Scholar]
- 3.Wang L, Yang G, Zhu X, Wang Z, Wang H, Bai Y, Sun P, Peng L, Wei W, Chen G, Li G, Zamyatnin AA Jr, Glybochko PV, Xu W. miR-93-3p inhibition suppresses clear cell renal cell carcinoma proliferation, metastasis and invasion. Oncotarget. 2017;8:82824–82834. doi: 10.18632/oncotarget.20458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Seldon CS, Colbert LE, Hall WA, Fisher SB, Yu DS, Landry JC. Chromodomain-helicase-DNA binding protein 5, 7 and pronecrotic mixed lineage kinase domain-like protein serve as potential prognostic biomarkers in patients with resected pancreatic adenocarcinomas. World J Gastrointest Oncol. 2016;8:358–365. doi: 10.4251/wjgo.v8.i4.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shur I, Benayahu D. Characterization and functional analysis of CReMM, a novel chromodomain helicase DNA-binding protein. J Mol Biol. 2005;352:646–655. doi: 10.1016/j.jmb.2005.06.049. [DOI] [PubMed] [Google Scholar]
- 6.Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, Wood WH 3rd, Zhang Y, Becker KG, Mattson MP, Pazin MJ. CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PLoS One. 2011;6:e24515. doi: 10.1371/journal.pone.0024515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin YW, Yan MD, Shih YL, Hsieh CB. The basal body gene, RPGRIP1L, is a candidate tumour suppressor gene in human hepatocellular carcinoma. Eur J Cancer. 2009;45:2041–2049. doi: 10.1016/j.ejca.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 8.Kim MS, Chung NG, Kang MR, Yoo NJ, Lee SH. Genetic and expressional alterations of CHD genes in gastric and colorectal cancers. Histopathology. 2011;58:660–668. doi: 10.1111/j.1365-2559.2011.03819.x. [DOI] [PubMed] [Google Scholar]
- 9.Kari V, Mansour WY, Raul SK, Baumgart SJ, Mund A, Grade M, Sirma H, Simon R, Will H, Dobbelstein M, Dikomey E, Johnsen SA. Loss of CHD1 causes DNA repair defects and enhances prostate cancer therapeutic responsiveness. EMBO Rep. 2016;17:1609–1623. doi: 10.15252/embr.201642352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D’Alesio C, Punzi S, Cicalese A, Fornasari L, Furia L, Riva L, Carugo A, Curigliano G, Criscitiello C, Pruneri G, Pelicci PG, Faretta M, Bossi D, Lanfrancone L. RNAi screens identify CHD4 as an essential gene in breast cancer growth. Oncotarget. 2016;7:80901–80915. doi: 10.18632/oncotarget.12646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li W, Mills AA. Architects of the genome: CHD dysfunction in cancer, developmental disorders and neurological syndromes. Epigenomics. 2014;6:381–395. doi: 10.2217/epi.14.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Batsukh T, Pieper L, Koszucka AM, von Velsen N, Hoyer-Fender S, Elbracht M, Bergman JE, Hoefsloot LH, Pauli S. CHD8 interacts with CHD7, a protein which is mutated in CHARGE syndrome. Hum Mol Genet. 2010;19:2858–2866. doi: 10.1093/hmg/ddq189. [DOI] [PubMed] [Google Scholar]
- 13.Xu C, Cassatella D, van der Sloot AM, Quinton R, Hauschild M, De Geyter C, Flück C, Feller K, Bartholdi D, Nemeth A, Halperin I, Pekic Djurdjevic S, Maeder P, Papadakis G, Dwyer AA, Marino L, Favre L, Pignatelli D, Niederländer NJ, Acierno J Jr, Pitteloud N. Evaluating CHARGE syndrome in congenital hypogonadotropic hypogonadism patients harboring CHD7 variants. Genet Med. 2017 doi: 10.1038/gim.2017.197. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 14.Shur I, Socher R, Benayahu D. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J Cell Physiol. 2006;207:374–378. doi: 10.1002/jcp.20586. [DOI] [PubMed] [Google Scholar]
- 15.Salomon-Kent R, Marom R, John S, Dundr M, Schiltz LR, Gutierrez J, Workman J, Benayahu D, Hager GL. New face for chromatin-related mesenchymal modulator: n-CHD9 localizes to nucleoli and interacts with ribosomal genes. J Cell Physiol. 2015;230:2270–2280. doi: 10.1002/jcp.24960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yamada H, Sano Y. The biotinylation of the rabbit serotonin antibody and its application to immunohistochemical studies using the two-step ABC method. Histochemistry. 1985;83:285–289. doi: 10.1007/BF00684372. [DOI] [PubMed] [Google Scholar]
- 17.Mazzoni A, Carrilho M, Papa V, Tjaderhane L, Gobbi P, Nucci C, Di Lenarda R, Mazzotti G, Tay FR, Pashley DH, Breschi L. MMP-2 assay within the hybrid layer created by a two-step etchand-rinse adhesive: biochemical and immunohistochemical analysis. J Dent. 2011;39:470–477. doi: 10.1016/j.jdent.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Wang G, Zeng Y, Chen S, Li D, Li W, Zhou Y, Singer RH, Gu W. Localization of TFPI-2 in the nucleus modulates MMP-2 gene expression in breast cancer cells. Sci Rep. 2017;7:13575. doi: 10.1038/s41598-017-14148-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helminen O, Huhta H, Leppanen J, Kauppila JH, Takala H, Lehenkari PP, Saarnio J, Karttunen TJ. Nuclear localization of Toll-like receptor 5 in Barrett’s esophagus and esophageal adenocarcinoma is associated with metastatic behavior. Virchows Arch. 2016;469:465–470. doi: 10.1007/s00428-016-1989-7. [DOI] [PubMed] [Google Scholar]
- 20.Sawada G, Ueo H, Matsumura T, Uchi R, Ishibashi M, Mima K, Kurashige J, Takahashi Y, Akiyoshi S, Sudo T, Sugimachi K, Doki Y, Mori M, Mimori K. CHD8 is an independent prognostic indicator that regulates Wnt/betacatenin signaling and the cell cycle in gastric cancer. Oncol Rep. 2013;30:1137–1142. doi: 10.3892/or.2013.2597. [DOI] [PubMed] [Google Scholar]
- 21.Colbert LE, Petrova AV, Fisher SB, Pantazides BG, Madden MZ, Hardy CW, Warren MD, Pan Y, Nagaraju GP, Liu EA, Saka B, Hall WA, Shelton JW, Gandhi K, Pauly R, Kowalski J, Kooby DA, El-Rayes BF, Staley CA 3rd, Adsay NV, Curran WJ Jr, Landry JC, Maithel SK, Yu DS. CHD7 expression predicts survival outcomes in patients with resected pancreatic cancer. Cancer Res. 2014;74:2677–2687. doi: 10.1158/0008-5472.CAN-13-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]


