Abstract
Previous studies have showed that bile acids (BAs) play essential roles in the progression of various human cancers, and the G-protein coupled bile acid receptor-1 (Gpbar-1, or TGR5), a receptor of BAs, has been reported to connect BAs with cancers. However, little is known about the prognostic role of TGR5 in pancreatic cancer. In this study, we found that the expression of TGR5 was significantly higher in the cancerous tissues than the adjacent normal tissues by immunohistochemical staining (81.6% vs. 36.8%). Meanwhile, TGR5 was positively correlated with lymph node metastasis (P=0.021) and advanced stage (P=0.011). Finally, univariate analysis showed that patients with high TGR5 expression (P<0.001), lymph node metastasis (P=0.002) and advanced tumor stage (P=0.008) had decreased overall survival, and Cox proportional hazards regression analysis confirmed that TGR5 expression was an independent predictor of the overall survival of patients with pancreatic cancer (P=0.019). Our findings suggested that TGR5 might serve as an important predictor of poor survival in pancreatic cancer.
Keywords: Pancreatic cancer, bile acids, TGR5, prognosis, lymph node metastasis
Introduction
Pancreatic cancer is currently one of leading causes of tumor-related death across the world [1]. Although the incidence of pancreatic cancer varies greatly across regions and populations, the current mortality of patients with pancreatic cancer is nearly identical to its incidence [2]. The estimated 5-year survival rate for pancreatic cancer remains at less than 5% [3], ranging from 20% for the localized stages to less than 1% for the advanced stages [1,4]. According to population and family-based studies, both environmental and inherited factors contribute to the development of pancreatic cancer, such as smoking [5], long-standing diabetes [6], increased body mass index (BMI) [7], alcohol consumption [8], pancreatitis [9] and inherited genetic changes [10]. Individuals with a history of chronic pancreatitis have an increased risk of pancreatic cancer. Surgical resection is the only option of curative treatment. Nevertheless, due to lack of symptoms until advanced stages, only 15%-20% of patients are diagnosed early enough to be considered for a potentially curative treatment [4]. However, due to its location in the retroperitoneum, the pancreas is difficult to assess [4]. The relapse rate after surgery is far from being acceptable, even with adjuvant therapies such as chemotherapy or chemo-radiotherapy [11,12]. Therefore, the median overall survival of patients with pancreatic cancer remains short.
Bile acids (BAs) are physiological detergent molecules that synthesized from cholesterol in the liver [13]. In the gallbladder, BAs form mixed micelles with phospholipids and cholesterol to increase cholesterol solubility and decrease bile acid toxicity. In the small intestine, BAs facilitate the intestinal digestion and absorption of dietary cholesterol, fat and other lipophilic nutrients. In addition, BAs can activate intracellular ligand-activated nuclear receptors and cell surface G protein coupled receptors to regulate various cellular processes involved in metabolism, immune response, and cell growth [14,15]. However, excess accumulation of BA in cholestasis can lead to tissue inflammation and injury, and increase the risk of tumorigenesis [14,16]. Patients with cholestasis develop fibrosis, cirrhosis, and eventually liver failure and even hepatocellular carcinomas [14]. Although pancreatic cancer originates from ductal cells, whereas pancreatitis predominantly affects pancreatic acinar cells, BA reflux-related chronic inflammation can induce dedifferentiation of acinar cells into progenitor duct-like cells, which are the sources of pancreatic cancer [17]. Since BA also acts as a regulatory molecule in metabolism, which is associated with risk factors (obesity, diabetes and hypertriglyceridemia) of pancreatic cancer, BA is very likely involved in pancreatic cancer carcinogenesis.
The G-protein coupled bile acid receptor-1 (Gpbar-1, or TGR5), also known as membrane-type receptor for bile acids (M-BAR), is the first identified G-protein coupled receptor specific for bile acids [18]. In human, the TGR5 gene is located on chromosome 2q35, and highly conserved in mammals [18]. Since been discovered in 2002, it has been found to be ubiquitously expressed in humans and animals, and to activate various intracellular signaling pathways upon interaction with bile acid. TGR5 is expressed in the liver sinusoidal endothelial cells [19], gallbladder epithelial cells, and Kupffer cells [20], and highly expressed in the ileum and colon and in non-traditional bile acid target organs including white and brown adipose, spleen, kidney, pancreas, lung, macrophages and the central nervous system [21]. It has been reported that TGR5 is expressed at the apical pole of pancreatic acinar cells and mediates bile acid-induced pancreatitis [22,23]. Recently, TGR5 has been reported to connect bile acids (BAs) with cancers [24,25]. The interaction between BA and TGR5 induces cancer cell proliferation via upregulation of NOX5-S expression [24].
In this study, we assessed TGR5 expression in pancreatic cancer specimens and paired adjacent normal tissues. We then examined the relationship between TGR5 expression and clinicopathological features as well as patient survival.
Materials and methods
Ethical statement
The study was approved by the Ethical Committee of Shanghai General Hospital, and all patients gave written informed consent at the time of the diagnosis for the use of tumor samples for research.
Patients
The patient cohort involved 95 patients treated at Shanghai for pancreatic cancer between 2004 and 2008. Patients were included or excluded according to the following criteria: (1) Definitive pathological diagnosis of pancreatic cancer; (2) No preoperative anticancer treatment, such as chemotherapy or radiotherapy; (3) No perioperative death; (4) Curative resection with negative surgical margin as confirmed by a pathologist; (5) Availability of appropriate paraffin-embedded tissues; (6) Complete clinicopathological and follow-up data.
Tissue samples
All slides were cut into 4 μm thick from a pre-existing pancreatic cancer tissues and adjacent normal tissues maintained by Shanghai ZuoCheng Bio Co., Ltd (Shanghai, China), and stained by hematoxylin and eosin (HE) and TGR5 primary antibody (Novus Biologcials, Minneapolis, MN, USA). Patient clinical information including age, gender, pathological grade, tumor size, nerve invasion, lymph node metastasis, tumor stage, and TGR5 status was collected.
Immunohistochemical staining
Immunohistochemistry for TGR5 expression was performed as previously described [26]. Briefly, formalin-fixed, paraffin-embedded cancer tissue sections were deparaffinized with xylene and rehydrated with ethanol. These tissue slides were then incubated in 3% hydrogen peroxidase solution (H2O2; Dako, Glostrup, Denmark) for 15 min. For antigen retrieval, these slides were heated in a water bath at 98°C with Target Retrieval Solution pH 9 (EDTA, pH 9, Gene Tech, Shanghai, China). Sections were saturated with protein block serum-free (Dako, Glostrup, Denmark) for 20 min. Without washing, slides were incubated with the primary antibody, a rabbit polyclonal anti-TGR5 antibody (1:1000; Novus Biologcials, Minneapolis, MN, USA), overnight at 4°C, followed by the secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA, USA) for 30 min, the streptavidin-horse radish peroxidase (Dako, Glostrup, Denmark) was added for an additional 30 min. Peroxidase activity was revealed by addition of daminobenzidine substrate (DAB; Dako, Glostrup, Denmark), and then the slides were counterstained with hemotoxylin (Dako, Glostrup, Denmark), dehydrated and cover slipped as per normal laboratory protocol.
Immunohistochemistry scoring
All sections were independently reviewed by two pathologists, who were blinded to all clinical and pathologic information. The staining was scored according to the intensity and percentage of the stained cells. The intensity of TGR5 staining was graded as 1 (no stain), 2 (weak stain), 3 (moderate stain), and 4 (strong stain). The percentages of stained cells were classified into four categories: 0 (no stain), 1 (<25%), 2 (25%-50%), and 3 (>50%). The final scores were calculated as the staining intensity time the percentage of positive cells. For statistical analyses, TGR5 positive expression was defined as a score >3.
Cell culture
Human pancreatic cancer cell lines MiaCaPa-2, SW1990, PANC-1, AsPC-1 and pancreatic duct epithelial cell HPDE6 were purchased from the Chinese Academy of Science (Shanghai, China) and cultured in Dulbecco’s Modified Eagles’s Medium (DMEM) (Thermo Scientific Inc., Beijing, China) containing 10% fetal bovine serum (FBS) (Sijiqing, Hangzhou, China), 100 U/mL penicillin, and 100 μg/mL streptomycin at 37°C, 5% CO2.
Western blot analysis
Cells were washed twice with ice-cold PBS and lysed using standard RIPA buffer containing proteinase inhibitors (Beyotime, Jiangsu, China). Following quantification, protein samples were heated to 95°C for 5 min, then separated in a SDS-polyacrylamide gel and transferred to PVDF membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% non-fat milk in TBS buffer for 1h and then incubated with TGR5 primary antibody (Novus Biologicials, Minneapolis, MN, USA) overnight at 4°C. After washing with TBST buffer for 4*10 min, the membranes were incubated with HRP-conjugated secondary antibody (Bio-Rad, Hercules, CA, USA) for 2 h at room temperature. The immune reaction was visualized using ECL Plus substrates (Roche, Basel, Switzerland).
Statistical analysis
Statistical analyses were performed using SPSS statistics software version 19 (SPSS) and GraphPad Prism v5.0 (Graphpad Software). The Wilcoxon test was used to compare TGR5 expression in paired tumor tissue samples and adjacent tissues. The Mann-Whitney U test and Kruskal-Wallis test were used to perform statistical analysis of tissue TGR5 levels between unpaired groups and multiple comparison groups respectively. Survival curves were constructed with the Kaplan-Meier method and compared using log-rank test. Cox proportional hazards regression analysis was used for univariate and multivariate analyses of prognostic values. Two-sided P value < 0.05 was considered statistically significant.
Results
TGR5 expression in pancreatic cancer and adjacent tissues
TGR5 was found in the membrane and cytoplasm of tumor tissues and adjacent tissues by immunohistochemical staining (Figure 1A-C). For pancreatic cancer samples, 62 cases (81.6%; upper panel) displayed positive, whereas 28 cases (36.8%; lower panel) of adjacent tissue samples showed positive (Table 1). The rate of TGR5 high expression was significantly increased in pancreatic cancer compared to adjacent tissues (P<0.001).
Figure 1.

TGR5 expression in pancreatic cancer compared with adjacent tissues. A-C. Upper panel: TGR5 expression in pancreatic cancer; lower panel: TGR5 expression in adjacent tissues. Scale bar: 400 μm, 100 μm. Expression of TGR5 was higher in pancreatic cancer. D. TGR5 expression in human pancreatic duct epithelial cell HPDE6 and pancreatic cancer cells. Expression of TGR5 was higher in pancreatic cancer cells.
Table 1.
Increased TGR5 expression in pancreatic cancer
| Cases | TGR5 expression | P value | ||
|---|---|---|---|---|
|
| ||||
| Negative | Positive | |||
| Pancreatic cancer | 76 | 14 (18.4%) | 62 (81.6%) | <0.001 |
| Adjacent tissue | 76 | 48 (63.2%) | 28 (36.8%) | |
We also investigated the expression of TGR5 in human pancreatic duct epithelial cell HPDE6 and pancreatic cancer cells. As shown in Figure 1D, we found that TGR5 was elevated in cancer cell lines compared to HPDE6 cells.
Correlation between tissue TGR5 expression level and patient baseline clinical characteristics
In total, 95 patients (60 male, 35 female) with pancreatic cancer were enrolled in this study. The median age at diagnosis was 61 years (range, 34-85 years). The relation of TGR5 expression with patient clinical pathological parameters is shown in Table 2. TGR5 expression was significantly increased in patients with a lymph node metastasis (P=0.021) and advanced stage (P=0.011) (Figure 2A-C; Table 2). However, it was not correlated with age (P=0.960), gender (P=0.842), pathological grade (P=0.945), tumor size (P=0.669), and nerve invasion (P=0.669) (Table 2).
Table 2.
Correlation between TGR5 expression and clinicopathological features of patients with pancreatic cancer
| Variables | Numbers | TGR5 expression | P value | |
|---|---|---|---|---|
|
| ||||
| Negative | Positive | |||
| Age (years) | 0.960 | |||
| ≤61 | 48 | 9 | 39 | |
| >61 | 47 | 9 | 38 | |
| Gender | 0.842 | |||
| Male | 60 | 11 | 49 | |
| Female | 35 | 7 | 28 | |
| Tumor size (cm) | 0.669 | |||
| ≤4 | 57 | 10 | 47 | |
| >4 | 38 | 8 | 30 | |
| Pathological grade | 0.945 | |||
| Well differentiated | 11 | 2 | 9 | |
| Poorly differentiated | 84 | 16 | 68 | |
| Lymph node metastasis | 0.021 | |||
| Yes | 36 | 3 | 33 | |
| No | 49 | 14 | 35 | |
| Nerve invasion | 0.669 | |||
| Yes | 38 | 8 | 30 | |
| No | 57 | 10 | 47 | |
| TNM stage | 0.011 | |||
| I | 43 | 13 | 30 | |
| II-IV | 52 | 5 | 47 | |
Figure 2.
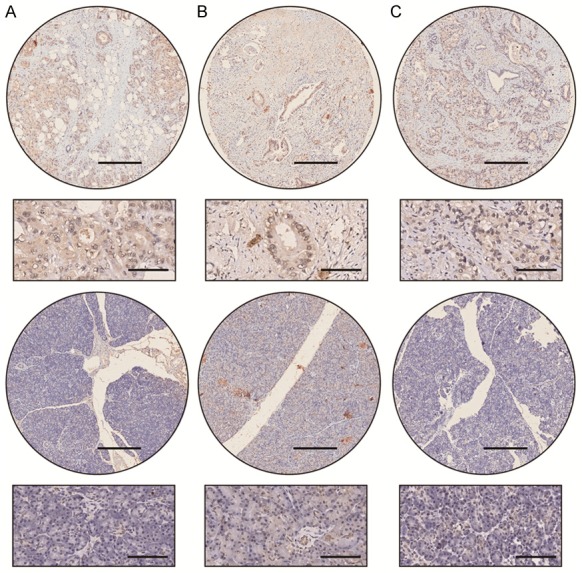
Different expression of TGR5 in patients with or without lymph node metastatic status. A-C. Upper panel: TGR5 expression in patients with lymph node metastatic status; lower panel: TGR5 expression in patients without lymph node metastatic status. Scale bar: 400 μm, 100 μm. Expression of TGR5 was higher in tissue with lymph node metastasis.
Relation between TGR5 expression and prognosis
The median follow-up time was 11 months (range, 0.6-87 months). TGR5-high pancreatic cancer patients had significantly shorter values than those of TGR5-low patients (22.5 months vs. 58.2 months). The 5-year survival rate of TGR5-high pancreatic cancer patients is 11.4% compared with 66.7% of TGR5-low pancreatic cancer patients. The overall survival for TGR5-high pancreatic cancer patients was significantly shorter than those with TGR5-low patients (P<0.001) (Figure 3).
Figure 3.

Kaplan-Meier survival curves of pancreatic cancer patients based on the TGR5 expression level. Patients with high TGR5 expression level had a significantly poorer survival than those with low TGR5 expression level (P<0.001, log-rank test).
Univariate and multivariate analyses
TGR5 expression and patient characteristics including age, gender, pathological grade, tumor size, nerve invasion, lymph node metastasis, and tumor stage were included to perform univariate and multivariate analyses. Univariate analysis showed that TGR5 expression (P<0.001), lymph node metastasis (P=0.002), and tumor stage (P=0.008) were significantly related to overall survival. Cox proportional hazards regression analysis confirmed that TGR5 expression was an independent predictor of the overall survival of patients with pancreatic cancer (P=0.019) (Table 3).
Table 3.
Univariate and multivariate survival analysis of clinicopathological variables in pancreatic cancer patients
| Variables | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% Cl | P value | HR | 95% Cl | P value | |
| TGR5 expression (positive vs. negative) | 0.24 | 0.10-0.55 | <0.001 | 0.34 | 0.14-0.84 | 0.019 |
| Age (years) (>61 vs. ≤61) | 0.80 | 0.50-1.27 | 0.346 | |||
| Gender (male vs. female) | 1.09 | 0.67-1.78 | 0.718 | |||
| Tumor size (cm) (>4 vs. ≤4) | 1.16 | 0.72-1.88 | 0.548 | |||
| Pathological grade (poorly vs. well) | 0.58 | 0.27-1.27 | 0.174 | |||
| Nerve invasion (yes vs. no) | 0.83 | 0.52-1.33 | 0.445 | |||
| Lymph node metastasis (yes vs. no) | 0.44 | 0.26-0.73 | 0.002 | |||
| TNM stage (II-IV vs. I) | 0.52 | 0.32-0.85 | 0.008 | |||
Discussion
In the present study, we investigated the prevalence and significance of TGR5 expression in pancreatic cancer. Among these pancreatic cancer patients, TGR5 expression was particularly higher in cancer tissues compared with adjacent normal tissues. High expression of TGR5 had a forward correlation to lymph node metastasis and advanced tumor grade. Patients with high TGR5 expression, lymph node metastasis, and advanced tumor stage had decreased overall survival. Cox proportional hazards model analysis indicated that TGR5 expression was a strong independent prognostic factor for pancreatic cancer patients.
TGR5 is a novel G protein-coupled cell-surface bile acid receptor and plays an important role in tumorigenesis [24]. It has been reported that TGR5 can suppress gastric cancer cell proliferation and migration through antagonizing STAT3 signaling pathway [27]. Meanwhile, BAs can conjugate TGR5 to activate c-Jun-N terminal kinase (JNK) and enhance apoptosis in hepatocytes [28]. Moreover, TGR5 expression was evaluated as a prognostic biomarker in various malignancies such as liver cancer, esophageal carcinoma, and colon cancer [26]. Patients with high TGR5 expression tend to have earlier tumor stage and better disease-specific survival rate [29]. However, TGR5 also performs as a tumor promoter. In esophageal adenocarcinoma, patients with high expression of TGR5 exhibited significantly worse overall survival compared with the patients with low expression [26]. In non-small-cell lung carcinoma (NSCLC), TGR5 was aberrantly expressed positively correlated with an advanced clinical stage in NSCLC patients, and TGR5 knockdown prevented JAK2 and STAT3 phosphorylation and repressed the expression of STAT3 target genes, thus inhibiting cell proliferation, migration and invasion in NSCLC [30]. Also, some reports suggested that TGR5 activation can promote cholangiocarcinoma progression by regulating proliferation, migration, and mitochondrial energy metabolism [31]. In our study, we found that TGR5 maybe a pro-tumoral factor in pancreatic cancer. Patients with high TGR5 expression were more likely to have advanced tumor stage, poor prognosis, and lymph node metastasis.
In conclusion, our results revealed that TGR5 is a useful prognostic marker in pancreatic cancer and further studies are needed to understand the molecular mechanisms underlying its role in pancreatic cancer development to support its potential clinical applications.
Acknowledgements
This study was supported in part by the funding of the National Natural Science Foundation of China (No.81502648) (to Jin Cheng), (No.81572951) (to Qian Huang) and (No.8140130232) (to Jing-Jing Ma).
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.Mitsuhashi K, Nosho K, Sukawa Y, Matsunaga Y, Ito M, Kurihara H, Kanno S, Igarashi H, Naito T, Adachi Y, Tachibana M, Tanuma T, Maguchi H, Shinohara T, Hasegawa T, Imamura M, Kimura Y, Hirata K, Maruyama R, Suzuki H, Imai K, Yamamoto H, Shinomura Y. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget. 2015;6:7209–7220. doi: 10.18632/oncotarget.3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wolfgang CL, Herman JM, Laheru DA, Klein AP, Erdek MA, Fishman EK, Hruban RH. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348. doi: 10.3322/caac.21190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lamarca A, Feliu J. Pancreatic biomarkers: could they be the answer? World J Gastroenterol. 2014;20:7819–7829. doi: 10.3748/wjg.v20.i24.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosetti C, Lucenteforte E, Silverman DT, Petersen G, Bracci PM, Ji BT, Negri E, Li D, Risch HA, Olson SH, Gallinger S, Miller AB, Bueno-de-Mesquita HB, Talamini R, Polesel J, Ghadirian P, Baghurst PA, Zatonski W, Fontham E, Bamlet WR, Holly EA, Bertuccio P, Gao YT, Hassan M, Yu H, Kurtz RC, Cotterchio M, Su J, Maisonneuve P, Duell EJ, Boffetta P, La Vecchia C. Cigarette smoking and pancreatic cancer: an analysis from the International Pancreatic Cancer Case-Control Consortium (Panc4) Ann Oncol. 2012;23:1880–1888. doi: 10.1093/annonc/mdr541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chari ST, Leibson CL, Rabe KG, Ransom J, de Andrade M, Petersen GM. Probability of pancreatic cancer following diabetes: a population-based study. Gastroenterology. 2005;129:504–511. doi: 10.1053/j.gastro.2005.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Arslan AA, Helzlsouer KJ, Kooperberg C, Shu XO, Steplowski E, Bueno-de-Mesquita HB, Fuchs CS, Gross MD, Jacobs EJ, Lacroix AZ, Petersen GM, Stolzenberg-Solomon RZ, Zheng W, Albanes D, Amundadottir L, Bamlet WR, Barricarte A, Bingham SA, Boeing H, Boutron-Ruault MC, Buring JE, Chanock SJ, Clipp S, Gaziano JM, Giovannucci EL, Hankinson SE, Hartge P, Hoover RN, Hunter DJ, Hutchinson A, Jacobs KB, Kraft P, Lynch SM, Manjer J, Manson JE, McTiernan A, McWilliams RR, Mendelsohn JB, Michaud DS, Palli D, Rohan TE, Slimani N, Thomas G, Tjonneland A, Tobias GS, Trichopoulos D, Virtamo J, Wolpin BM, Yu K, Zeleniuch-Jacquotte A, Patel AV Pancreatic Cancer Cohort Consortium (PanScan) Anthropometric measures, body mass index, and pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan) Arch Intern Med. 2010;170:791–802. doi: 10.1001/archinternmed.2010.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lucenteforte E, La Vecchia C, Silverman D, Petersen GM, Bracci PM, Ji BT, Bosetti C, Li D, Gallinger S, Miller AB, Bueno-de-Mesquita HB, Talamini R, Polesel J, Ghadirian P, Baghurst PA, Zatonski W, Fontham E, Bamlet WR, Holly EA, Gao YT, Negri E, Hassan M, Cotterchio M, Su J, Maisonneuve P, Boffetta P, Duell EJ. Alcohol consumption and pancreatic cancer: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4) Ann Oncol. 2012;23:374–382. doi: 10.1093/annonc/mdr120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duell EJ, Lucenteforte E, Olson SH, Bracci PM, Li D, Risch HA, Silverman DT, Ji BT, Gallinger S, Holly EA, Fontham EH, Maisonneuve P, Bueno-de-Mesquita HB, Ghadirian P, Kurtz RC, Ludwig E, Yu H, Lowenfels AB, Seminara D, Petersen GM, La Vecchia C, Boffetta P. Pancreatitis and pancreatic cancer risk: a pooled analysis in the International Pancreatic Cancer Case-Control Consortium (PanC4) Ann Oncol. 2012;23:2964–2970. doi: 10.1093/annonc/mds140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jacobs EJ, Chanock SJ, Fuchs CS, Lacroix A, McWilliams RR, Steplowski E, Stolzenberg-Solomon RZ, Arslan AA, Bueno-de-Mesquita HB, Gross M, Helzlsouer K, Petersen G, Zheng W, Agalliu I, Allen NE, Amundadottir L, Boutron-Ruault MC, Buring JE, Canzian F, Clipp S, Dorronsoro M, Gaziano JM, Giovannucci EL, Hankinson SE, Hartge P, Hoover RN, Hunter DJ, Jacobs KB, Jenab M, Kraft P, Kooperberg C, Lynch SM, Sund M, Mendelsohn JB, Mouw T, Newton CC, Overvad K, Palli D, Peeters PH, Rajkovic A, Shu XO, Thomas G, Tobias GS, Trichopoulos D, Virtamo J, Wactawski-Wende J, Wolpin BM, Yu K, Zeleniuch-Jacquotte A. Family history of cancer and risk of pancreatic cancer: a pooled analysis from the Pancreatic Cancer Cohort Consortium (PanScan) Int J Cancer. 2010;127:1421–1428. doi: 10.1002/ijc.25148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Oettle H, Post S, Neuhaus P, Gellert K, Langrehr J, Ridwelski K, Schramm H, Fahlke J, Zuelke C, Burkart C, Gutberlet K, Kettner E, Schmalenberg H, Weigang-Koehler K, Bechstein WO, Niedergethmann M, Schmidt-Wolf I, Roll L, Doerken B, Riess H. Adjuvant chemotherapy with gemcitabine vs observation in patients undergoing curative-intent resection of pancreatic cancer: a randomized controlled trial. JAMA. 2007;297:267–277. doi: 10.1001/jama.297.3.267. [DOI] [PubMed] [Google Scholar]
- 12.Neoptolemos JP, Stocken DD, Friess H, Bassi C, Dunn JA, Hickey H, Beger H, Fernandez-Cruz L, Dervenis C, Lacaine F, Falconi M, Pederzoli P, Pap A, Spooner D, Kerr DJ, Buchler MW European Study Group for Pancreatic Cancer. A randomized trial of chemoradiotherapy and chemotherapy after resection of pancreatic cancer. N Engl J Med. 2004;350:1200–1210. doi: 10.1056/NEJMoa032295. [DOI] [PubMed] [Google Scholar]
- 13.Russell DW, Setchell KD. Bile acid biosynthesis. Biochemistry. 1992;31:4737–4749. doi: 10.1021/bi00135a001. [DOI] [PubMed] [Google Scholar]
- 14.Li T, Apte U. Bile acid metabolism and signaling in cholestasis, inflammation, and cancer. Adv Pharmacol. 2015;74:263–302. doi: 10.1016/bs.apha.2015.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T, Chiang JY. Bile acid signaling in metabolic disease and drug therapy. Pharmacol Rev. 2014;66:948–983. doi: 10.1124/pr.113.008201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fan M, Wang X, Xu G, Yan Q, Huang W. Bile acid signaling and liver regeneration. Biochim Biophys Acta. 2015;1849:196–200. doi: 10.1016/j.bbagrm.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinho AV, Rooman I, Reichert M, De Medts N, Bouwens L, Rustgi AK, Real FX. Adult pancreatic acinar cells dedifferentiate to an embryonic progenitor phenotype with concomitant activation of a senescence programme that is present in chronic pancreatitis. Gut. 2011;60:958–966. doi: 10.1136/gut.2010.225920. [DOI] [PubMed] [Google Scholar]
- 18.Duboc H, Tache Y, Hofmann AF. The bile acid TGR5 membrane receptor: from basic research to clinical application. Dig Liver Dis. 2014;46:302–312. doi: 10.1016/j.dld.2013.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keitel V, Reinehr R, Gatsios P, Rupprecht C, Gorg B, Selbach O, Haussinger D, Kubitz R. The G-protein coupled bile salt receptor TGR5 is expressed in liver sinusoidal endothelial cells. Hepatology. 2007;45:695–704. doi: 10.1002/hep.21458. [DOI] [PubMed] [Google Scholar]
- 20.Keitel V, Donner M, Winandy S, Kubitz R, Haussinger D. Expression and function of the bile acid receptor TGR5 in Kupffer cells. Biochem Biophys Res Commun. 2008;372:78–84. doi: 10.1016/j.bbrc.2008.04.171. [DOI] [PubMed] [Google Scholar]
- 21.Kawamata Y, Fujii R, Hosoya M, Harada M, Yoshida H, Miwa M, Fukusumi S, Habata Y, Itoh T, Shintani Y, Hinuma S, Fujisawa Y, Fujino M. A G protein-coupled receptor responsive to bile acids. J Biol Chem. 2003;278:9435–9440. doi: 10.1074/jbc.M209706200. [DOI] [PubMed] [Google Scholar]
- 22.Muili KA, Jin S, Orabi AI, Eisses JF, Javed TA, Le T, Bottino R, Jayaraman T, Husain SZ. Pancreatic acinar cell nuclear factor kappaB activation because of bile acid exposure is dependent on calcineurin. J Biol Chem. 2013;288:21065–21073. doi: 10.1074/jbc.M113.471425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng HY, Chen YC. Role of bile acids in carcinogenesis of pancreatic cancer: An old topic with new perspective. World J Gastroenterol. 2016;22:7463–7477. doi: 10.3748/wjg.v22.i33.7463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong J, Behar J, Wands J, Resnick M, Wang LJ, DeLellis RA, Lambeth D, Souza RF, Spechler SJ, Cao W. Role of a novel bile acid receptor TGR5 in the development of oesophageal adenocarcinoma. Gut. 2010;59:170–180. doi: 10.1136/gut.2009.188375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao W, Tian W, Hong J, Li D, Tavares R, Noble L, Moss SF, Resnick MB. Expression of bile acid receptor TGR5 in gastric adenocarcinoma. Am J Physiol Gastrointest Liver Physiol. 2013;304:G322–327. doi: 10.1152/ajpgi.00263.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pang C, LaLonde A, Godfrey TE, Que J, Sun J, Wu TT, Zhou Z. Bile salt receptor TGR5 is highly expressed in esophageal adenocarcinoma and precancerous lesions with significantly worse overall survival and gender differences. Clin Exp Gastroenterol. 2017;10:29–37. doi: 10.2147/CEG.S117842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo C, Su J, Li Z, Xiao R, Wen J, Li Y, Zhang M, Zhang X, Yu D, Huang W, Chen WD, Wang YD. The G-protein-coupled bile acid receptor Gpbar1 (TGR5) suppresses gastric cancer cell proliferation and migration through antagonizing STAT3 signaling pathway. Oncotarget. 2015;6:34402–34413. doi: 10.18632/oncotarget.5353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang JI, Yoon JH, Myung SJ, Gwak GY, Kim W, Chung GE, Lee SH, Lee SM, Kim CY, Lee HS. Bile acid-induced TGR5-dependent c-Jun-N terminal kinase activation leads to enhanced caspase 8 activation in hepatocytes. Biochem Biophys Res Commun. 2007;361:156–161. doi: 10.1016/j.bbrc.2007.07.001. [DOI] [PubMed] [Google Scholar]
- 29.Chen MC, Chen YL, Wang TW, Hsu HP, Lai MD. Membrane bile acid receptor TGR5 predicts good prognosis in ampullary adenocarcinoma patients with hyperbilirubinemia. Oncol Rep. 2016;36:1997–2008. doi: 10.3892/or.2016.5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu X, Chen B, You W, Xue S, Qin H, Jiang H. The membrane bile acid receptor TGR5 drives cell growth and migration via activation of the JAK2/STAT3 signaling pathway in non-small cell lung cancer. Cancer Lett. 2018;412:194–207. doi: 10.1016/j.canlet.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 31.Erice O, Labiano I, Arbelaiz A, Santos-Laso A, Munoz-Garrido P, Jimenez-Aguero R, Olaizola P, Caro-Maldonado A, Martin-Martin N, Carracedo A, Lozano E, Marin JJ, O’Rourke CJ, Andersen JB, Llop J, Gomez-Vallejo V, Padro D, Martin A, Marzioni M, Adorini L, Trauner M, Bujanda L, Perugorria MJ, Banales JM. Differential effects of FXR or TGR5 activation in cholangiocarcinoma progression. Biochim Biophys Acta. 2018;1864:1335–1344. doi: 10.1016/j.bbadis.2017.08.016. [DOI] [PubMed] [Google Scholar]


