Abstract
CAMP responsive element binding protein 5 (CREB5) has been reported to be overexpressed in several types of human cancers and has crucial roles in regulating cell growth, proliferation, differentiation, and the cell cycle. However, the expression and function of CREB5 in hepatocellular carcinoma (HCC) remains unclear. The purpose of this study was to investigate the role of CREB5 in HCC, and its prognostic significance. We measured the expression of CREB5 in 91 specimens of paraffin-embedded HCC tissue by immunohistochemistry and performed a clinicopathological analysis. Gain of function and loss of function assays were used to evaluate the effect of CREB5 on cell proliferation in vitro. We found that up-regulation of CREB5 was associated with a poor prognosis, and CREB5 status was an independent prognostic factor. The overexpression of CREB5 increased the proliferation of SMMC-7721 cells, but the knockdown of CREB5 had the opposite effect. The results indicate that CREB5 may be useful when determining a treatment strategy for patients with HCC.
Keywords: CREB5, HCC, proliferation, prognosis
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common cancer in the world and represents the third leading cause of cancer mortality worldwide with a 30% to 40% 5-year postoperative survival rate [1-3]. The high mortality of HCC is mainly related to HCC metastasis, which is a major cause of tumor recurrence after tumor resection [4-7]. However, the molecular mechanism of HCC metastasis remains largely unknown. Therefore, it is critical to identify the mechanisms underlying HCC migration and invasion in order to develop novel therapeutic strategies.
CAMP responsive element binding protein 5 (CREB5) belongs to the CRE (cAMP response element)-binding protein family. The protein specifically binds to CRE as a homodimer or heterodimer with c-Jun or CRE-BP1 to function as a CRE-dependent transactivator [8,9]. Previous studies suggested that CREB5 might play a key role in the tumorigenesis of colorectal cancer, lung cancer, and ovarian cancer [10-12]. However, the expression and function of CREB5 in HCC remains unknown. Thus, the purpose of this study was to examine the expression of CREB5 in HCC and determine its prognostic significance.
Materials and methods
HCC patient samples
Ninety-one patients with a median follow-up time of 18 months with HCC who underwent hepatectomy at the First Affiliated Hospital of Sun Yat-sen University were included in the study. All patients were diagnosed with primary liver cancer, and none had received preoperative radiotherapy or chemotherapy. The study was carried out under the approval of the Committees for Ethical Review of Research involving Human Subjects of the First Affiliated Hospital of Sun Yat-sen University (Guangzhou, China). All patients, or their guardians or caregivers, provided written informed consent for the use of clinical specimens in medical research. Of the 91 patients (80 men and 11 women; mean age, 51 years; range, 21 to 73 years), 70 were serum hepatitis B virus (HBV)-positive, and 53 had an elevated alpha-fetoprotein level (AFP; ≥ 200 ng/mL) (Table 1).
Table 1.
Relationship of CREB5 expression with pathological parameters of HCC
| Variables | Low CREB5 Group | High CREB5 Group | P |
|---|---|---|---|
| Age (yrs) | |||
| ≤ 50 | 35 | 12 | 0.625 |
| > 50 | 35 | 9 | |
| Sex | |||
| Male | 60 | 20 | 446 |
| Female | 10 | 1 | |
| HBsAg | |||
| Positive | 58 | 19 | 0.508 |
| Negative | 12 | 2 | |
| Tumor size (cm) | |||
| ≤ 10 cm | 57 | 12 | 0.039 |
| > 10 cm | 13 | 9 | |
| Tumor number | |||
| Solitary | 57 | 14 | 0.227 |
| Multiple | 13 | 7 | |
| Vascular invasion | |||
| Yes | 9 | 4 | 0.488 |
| No | 61 | 17 | |
| Encapsulation | |||
| Intact | 46 | 15 | 0.793 |
| Non-intact | 24 | 6 | |
| Edmonson | |||
| I-II | 51 | 17 | 0.573 |
| III-IV | 19 | 4 | |
| AFP (μg/l) | |||
| ≤ 400 | 40 | 13 | 0.803 |
| > 400 | 30 | 8 | |
| TNM stage | |||
| I-II | 59 | 10 | 0.001 |
| III | 11 | 11 |
Immunocytochemistry
Paraffin-embedded sections of HCC tissue or adjacent non-cancerous liver tissue were cut and dehydrated in xylene, followed by microwave antigen retrieval. The sections were incubated with an anti-ELMO3 antibody and then analyzed using the Evision/HRP Kit (Dako, CA, USA). The degree of immunostaining was reviewed and scored by two observers according to the proportion of positively stained tumor cells and the intensity of staining. Tumor cell proportion was scored as follows: 0 (no positive tumor cells), 1 (< 35% positive tumor cells), 2 (35-70% positive tumor cells), and 3 (> 70% positive tumor cells). Staining intensity was graded according to the following criteria: 0 (no staining), 1 (weak staining = light yellow), 2 (moderate staining = yellow brown), and 3 (strong staining = brown). The staining index was calculated as the product of the staining intensity score and the proportion of positive tumor cells. Using this method of assessment, we evaluated CREB5 expression by determining the staining index, with scores of 0, 1, 2, 3, 4, 6, or 9. An optimal cutoff value was identified: a staining index score of > 5 was used to define tumors with high CREB5 expression, and a staining index score of < 5 was used to define low CREB5 expression.
HCC cell line
The HCC cell line SMMC-7721 was obtained from the Cell Bank of the Chinese Academy of Medical Sciences (Shanghai, China). The cells were maintained in high-glucose Dulbecco’s modified Eagle’s media (DMEM) (Gibco, Australia) supplemented with 10% fetal bovine serum (Gibco, Australia).
Plasmid constructs and siRNA transfection
Full-length CREB5 cDNA was amplified and cloned into the pReciever M68 expression vector (FulenGen, Guangzhou, China). The expression plasmids were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
Oligonucleotide siRNA duplex was synthesized by Shanghai Gene Pharma (Shanghai, China). The siRNA sequence of CREB5 was: 5’-GGUGGAAAUGGACUGGCUTT-3’. The transfection of siRNA in the HCC cells was carried out with Lipofectamine 2000 (Invitrogen), according to the manufacturer’s instructions.
Cell cycle analysis
To evaluate the cell cycle, 1×106 cells were harvested and washed in PBS, then fixed in 75% alcohol for 30 min at 4°C. After washing in cold PBS three times, the cells were resuspended in 1 ml of PBS solution with 40 ug of propidium iodide (Sigma) and 100 ug of RNase A (Sigma) for 30 min at 37°C. Samples were then analyzed for their DNA content by FACSCalibur (Becton Dickinson, Mountain View, CA). The proliferative index (PIx), a measure of the number of cells in a tumor that are dividing (proliferating), was calculated with the following formula: PIx = (S+G2M)/(G0/G1+S+G2M)]×100%.
Colony formation assays
For colony formation assays, 500 cells were plated onto 6-well plates and incubated at 37°C. When the cells had grown to form visible colonies, the colonies were washed once with PBS and fixed with 4% paraformaldehyde for 20 min. The cells were stained with crystal violet, and images were collected by a video camera.
Western blot analysis
Tissues or harvested cultured cells were homogenized in a lysis buffer (50 mmol/L Hepes pH 7.5, 150 mmol/L NaCl, 10% glycerol, 1% Triton X-100, 1.5 mmol/L MgCl2, 1 mmol/L EGTA, 10 mmol/L NaF, 10 mmol/L Na4P2O7, 1 mmol/L Na3VO5, 1 mmol/L phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin and 20 μg/ml aprotinin). Protein was quantified by the Bradford assay (Bio-Rad, Hercules, CA). Equal amounts of protein were separated on SDS-polyacrylamide gels and transferred onto nitrocellulose membranes (Amersham Biosciences, Piscataway, NJ). After blocking in 5% skim milk for 1 h at room temperature, membranes were incubated with a primary antibody at 4°C overnight, followed by a horseradish peroxidase-conjugated secondary antibody, and analysis by a chemiluminescence assay (Amersham Biosciences, Piscataway, NJ). Quantification of the Western blot data were performed by measuring the intensity of the hybridization signals using and image analysis program (Fluor-ChemTM 8900, Alpha Inotech).
Statistical analysis
The differences between the two groups were examined using Student’s t test. A value of P < 0.05 was considered statistically significant. Data were expressed as the mean ± standard deviation from at least three experiments. Comparisons between groups were performed with two-tailed paired Student’s t test. The chi-square test was used to analyze the relationship between the CREB5 expression and the clinicopathological features. Bivariate correlations between variables were determined by Spearman’s correlation coefficients. Survival curves were plotted using the Kaplan-Meier method and compared using the log-rank test. Survival data were evaluated using univariate and multivariate Cox regression analyses. All statistical analyses were performed using SPSS 11.0 software.
Results
High CREB5 expression is associated with a poor prognosis of patients with HCC
We detected the expression of the CREB5 protein by immunohistochemistry in 91 pairs of HCC tissue and matched adjacent non-cancerous tissue samples after liver resection (Figure 1). Cancer tissue from 63 of the 91 (69.2%) patients exhibited higher CREB5 protein expression as compared to the expression of CREB5 in the adjacent non-cancerous tissues. The expression of CREB5 was significantly elevated (a staining index score > 5) in tumor tissue samples compared with the matched non-tumor tissues (P < 0001). In the 91 cases examined, high CREB5 expression was detected in 21 (23.08%) HCC specimens, but only 10 (11.00%) of the non-tumor specimens (Table 1).
Figure 1.
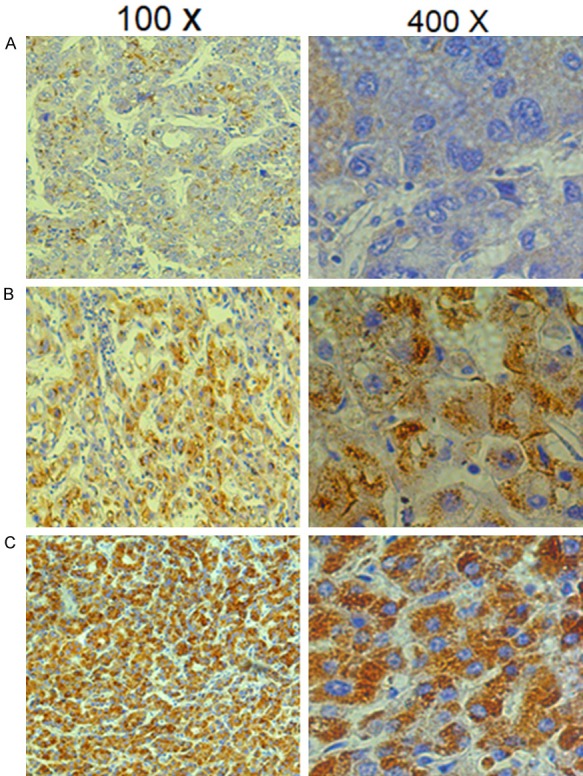
Representative immunohistochemical staining of CREB5 in tumor tissues. The micrographs showed weak (A), medium (B), and strong (C) expression of CREB5 in liver cancer tissues. (Magnification: left panel 100×, right panel 400×).
A clinical association analysis using the Pearson chi-square test revealed that CREB5 expression in the HCC tumors was significantly associated with a poor prognosis. All 91 patients were included in the overall survival (OS) analysis. A Kaplan-Meier analysis revealed the association of CREB5 expression with short OS times (P = 0.004) (Figure 2A). The 1-, 3-, and 5-year survival rates decreased from 89%, 54%, and 47%, respectively, in the low CREB5 expression group to 52%, 29%, and 24%, respectively, in the high CREB5 expression group. The Kaplan-Meier analysis also revealed an association of CREB5 expression and disease-free survival (DFS) (P = 0.017) (Figure 2B). The 1-, 3-, and 5-year DFS rates decreased from 56%, 33%, and 30%, respectively, in the low CREB5 expression group to 19%, 19%, and 19%, respectively, in the high CREB5 expression group.
Figure 2.
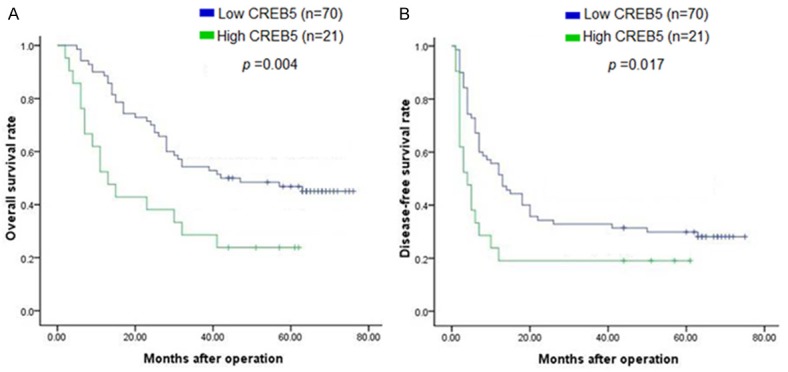
Kaplan-Meier survival curves of hepatocellular carcinoma (HCC) patients based on CREB5 expression. HCC patients with high CREB5 expression had a significantly shorter (A) overall survival (P = 0.004) and (B) disease-free survival (P = 0.017) than those with low CREB5 expression.
Univariate Cox regression analyses revealed that tumor size, tumor number, vascular invasion, encapsulation, Edmonson grade, TNM stage, and CREB5 expression were all predictors of worse OS and DFS of HCC patients (Table 2). In addition, multivariate Cox regression analysis revealed that a higher level of CREB5 expression and a higher Edmonson grade were independent prognostic factors for DFS, whereas only CREB5 expression was an independent prognostic factors for OS (Table 3).
Table 2.
Univariate analysis of different prognostic factors in 91 HCC patients by cox regression analysis
| Variables | n | DFS | OS | ||||||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| 1-yr | 3-yr | 5-yr | P | 1-yr | 3-yr | 5-yr | P | ||
| Age (yrs) | |||||||||
| ≤ 50 | 47 | 44% | 30% | 26% | 0.622 | 79% | 49% | 41% | 0.524 |
| > 50 | 44 | 49% | 25% | 24% | 76% | 45% | 36% | ||
| HBsAg | |||||||||
| Positive | 77 | 45% | 29% | 24% | 0.484 | 78% | 47% | 38% | 0.726 |
| Negative | 14 | 61% | 30% | 30% | 78% | 48% | 39% | ||
| Tumor size (cm) | |||||||||
| ≤ 10 cm | 69 | 25% | 9% | 7% | 0.000 | 85% | 55% | 47% | 0.000 |
| > 10 cm | 22 | 55% | 35% | 32% | 59% | 25% | 15% | ||
| Tumor number | |||||||||
| Solitary | 71 | 56% | 34% | 30% | 0.001 | 83% | 55% | 47% | 0.000 |
| Multiple | 20 | 27% | 12% | 6% | 67% | 29% | 18% | ||
| Vascular invasion | |||||||||
| Yes | 13 | 10% | 3% | 3% | 0.000 | 86% | 55% | 46% | 0.000 |
| No | 78 | 56% | 35% | 30% | 40% | 13% | 7% | ||
| Encapsulation | |||||||||
| Intact | 61 | 61% | 36% | 32% | 0.000 | 86% | 55% | 45% | 0.001 |
| Non-intact | 30 | 24% | 17% | 13% | 63% | 34% | 27% | ||
| Edmonson | |||||||||
| I-II | 68 | 53% | 31% | 28% | 0.026 | 79% | 50% | 43% | 0.036 |
| III-IV | 23 | 30% | 16% | 14% | 73% | 38% | 24% | ||
| AFP (μg/l) | |||||||||
| ≤ 400 | 53 | 56% | 34% | 32% | 0.012 | 84% | 50% | 44% | 0.108 |
| > 400 | 38 | 37% | 23% | 17% | 71% | 44% | 32% | ||
| TNM stage | |||||||||
| I-II | 69 | 59% | 36% | 33% | 0.000 | 87% | 57% | 49% | 0.000 |
| CREB5 expression | |||||||||
| High | 21 | 56% | 33% | 30% | 0.017 | 89% | 54% | 47% | 0.004 |
| Low | 70 | 19% | 19% | 19% | 52% | 29% | 24% | ||
Table 3.
Multivariate analysis of different prognostic factors in 91 HCC patients using a Cox regression analysis
| Variables | DFS | OS | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| HR | 95% CI | P | HR | 95% CI | P | |
| Tumor size | 1.014 | 0.456-2.253 | 0.973 | 1.050 | 0.467-2.363 | 0.906 |
| Tumor number | 0.882 | 0.375-2.075 | 0.774 | 0.933 | 0.358-2.435 | 0.888 |
| Vascular invasion | 1.666 | 0.744-3.726 | 0.214 | 1.672 | 0.602-4.638 | 0.324 |
| Encapsulation | 0.767 | 0.424-1.387 | 0.380 | 0.916 | 0.491-1.709 | 0.783 |
| Edmonson | 1.772 | 1.008-3.116 | 0.047 | 1.358 | 0.738-2.499 | 0.325 |
| AFP | 1.262 | 0.729-2.185 | 0.405 | |||
| TNM stage | 1.786 | 0.592-5.384 | 0.303 | 1.795 | 0.503-6.399 | 0.367 |
| CREB5 expression | 1.865 | 1.017-3.422 | 0.044 | 1.962 | 1.008-3.817 | 0.047 |
HR, hazard ratio; CI, confidence interval; DFS, Disease-free survival; OS, overall survival; HCC, hepatocellular carcinoma.
A Spearman correlation analysis showed significant associations between CREB5 expression and tumor size and TNM stage.
CREB5 promotes cellular proliferation
After identifying the significant association between CREB5 expression and tumor size, we overexpressed CREB5 in SMMC-7721 cells to better understand the role of CREB5 in HCC. Stable CREB5 and control transfectants were generated and examined for CREB5 protein levels (Figure 3C). We analyzed cell cycle kinetics in SMMC7721 cells transfected with either CREB5 or control. We further analyzed cell proliferation with a clone formation assay. The CREB5-overexpressing cells showed a statistically significant increase in proliferation when compared to the control cells (Figure 3A, 3B). In order to confirm these findings, we next transfected specific-stranded RNA oligonucleotides against CREB5 to knockdown the CREB5 levels in the SMMC-7721 cells. When CREB5-specific oligonucleotides were used, a rapid down-regulation of CREB5 was detected (Figure 4C). The decrease in the CREB5 level induced G0/G1 cell cycle arrest in the SMMC7721 cells and a concomitant decrease in proliferation (Figure 4A, 4B). Using the clone formation assay, we found that the over-expression of CREB5 increased the CREB5er of the colonies as compared with the control (Figure 3D). The knockdown of CREB5 decreased the CREB5er of the colonies as compared with the control (Figure 4D).
Figure 3.
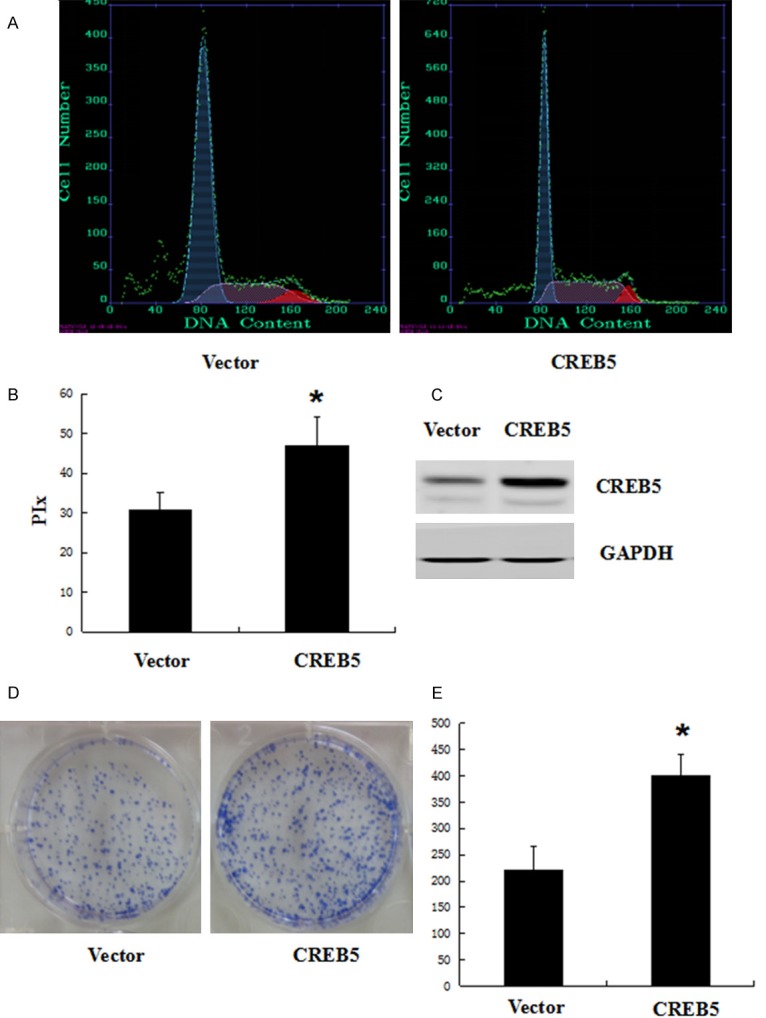
Overexpression of CREB5 promoted SMMC-7721 cell cycle progression and colony formation. SMMC-7721 cells were transfected with an empty vector or Ad-CMV-HA-CREB5. At 48 h after transfection , the cell cycle was analyzed by FACSCalibur (A). (B) The proliferative index (PIx) of CREB5-overexpressed cells was greater than that of the control cells (P < 0.05). The amount of CREB5 was determined by antibodies specific for CREB5 (C). For colony formation assays, we transfected the CREB5 overexpression construct into SMMC-7721 cells. Overexpressed CREB5 promoted colony formation in SMMC-7721 cells (D). Quantitative results of the colony-formation assay are also shown; *P < 0.05 (E).
Figure 4.
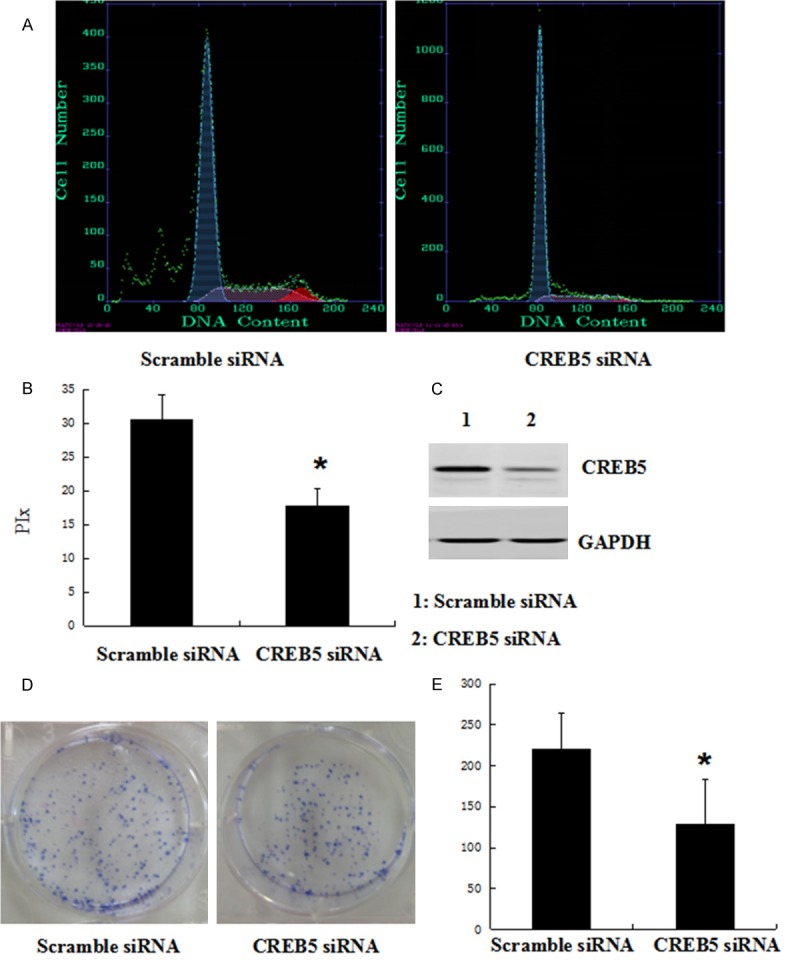
Knockdown of CREB5 inhibited SMMC-7721 cell cycle progression and colony formation. The SMMC-7721 cells were transfected with scramble siRNA or CREB5 siRNA. At 48 h after transfection, the amount of CREB5 and p21 was determined by antibodies specific for CREB5 (C). (A) The cell cycle was analyzed by FACSCalibur. (B) The proliferative index (PIx) was decreased compared to the control group (P < 0.05). (D) The knockdown of CREB5 in SMMC-7721 cells inhibited colony formation. Quantitative results of the colony-formation assay are shown; *P < 0.05 (E).
Discussion
CREB5 belongs to the CRE-binding protein family. The protein specifically binds to CRE as a homodimer or heterodimer with c-Jun or CRE-BP1 to function as a CRE-dependent transactivator. CREB regulates a number of genes with diverse functions, including proliferation, survival, memory, and learning [8,13-20]. Deregulation or aberrant expression of some of these genes has been associated with cancer. Seo et al. reported that CREB expression levels in non-small cell lung cancer cells were highly dependent on elevated CREB activity for their survival and cell growth, and the inhibition of CREB effectively suppressed the growth of non-small cell lung cancer cells [12]. He et al. reported that CREB5 was significantly overexpressed in epithelial ovarian cancer and was significantly correlated with aggressive features and an unfavorable prognosis [10].
In this study, the expression of CREB5 was detected in 91 specimens of paraffin-embedded HCC tissues. The immunohistochemistry results showed that the positive staining of CREB5 was observed in the cytoplasms of the hepatocytes. However, we did not perform corresponding HE staining of IHC, which should be addressed in a later study. Then, we studied the clinicopathological significance of CREB5 in HCC. We found tumor size, tumor number, vascular invasion, encapsulation, Edmonson grade, TNM stage, and CREB5 expression to be predictors of poorer OS and DFS in HCC patients. Besides CREB5 expression, all of the other factors are recognized indicators of HCC progression. Furthermore, a Spearman correlation analysis showed that CREB5 expression was significantly correlated with tumor size and TNM stage. These data indicate that CREB5 expression is an independent prognostic indicator for survival in patients with HCC.
In addition, we further explored the possible role of CREB5 in the occurrence and development of HCC. We found that forced expression of CREB5 in SMMC-7721 cells promoted the progression of the cell cycle in vitro. The depletion of CREB5 in the SMMC-7721 cells had the opposite effect. The results of the clone formation assay indicated that CREB5 expression is positively correlated with cell proliferation in SMMC-7721 cells.
In conclusion, this is the first study to assess the role of CREB5 in HCC. The results suggest that CREB5 expression may be a useful prognostic indicator in patients with HCC. However, further study of the mechanisms by which CREB5 affects cell proliferation of HCC is required.
Acknowledgements
This work was supported by grant 81702416 (to Dr. Jian Wu) from the National Natural Science Foundation of China.
Disclosure of conflict of interest
None.
References
- 1.Mendizabal M, Reddy KR. Current management of hepatocellular carcinoma. Med Clin North Am. 2009;93:885–900. viii. doi: 10.1016/j.mcna.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Said A, Wells J. Management of hepatocellular carcinoma. Minerva Med. 2009;100:51–68. [PubMed] [Google Scholar]
- 3.Taieb J, Barbare JC, Boussaha T, Cunha AS, Baere TD, Rosmorduc O, Zucman-Rossi J, Franco D. [Management of hepatocellular carcinoma. Where are we now? What’s next?] Bull Cancer. 2009;96:19–34. doi: 10.1684/bdc.2009.0790. [DOI] [PubMed] [Google Scholar]
- 4.Chu KK, Cheung TT. Update in management of hepatocellular carcinoma in Eastern population. World J Hepatol. 2015;7:1562–71. doi: 10.4254/wjh.v7.i11.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bodzin AS, Busuttil RW. Hepatocellular carcinoma: advances in diagnosis, management, and long term outcome. World J Hepatol. 2015;7:1157–67. doi: 10.4254/wjh.v7.i9.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pang TC, Lam VW. Surgical management of hepatocellular carcinoma. World J Hepatol. 2015;7:245–52. doi: 10.4254/wjh.v7.i2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allaire M, Nault JC. Molecular targets for HCC and future treatments. J Hepatol. 2017;66:234–5. doi: 10.1016/j.jhep.2016.07.034. [DOI] [PubMed] [Google Scholar]
- 8.Long X, Li Y, Qiu S, Liu J, He L, Peng Y. MiR-582-5p/miR-590-5p targeted CREB1/CREB5-NFkappaB signaling and caused opioid-induced immunosuppression in human monocytes. Transl Psychiatry. 2016;6:e757. doi: 10.1038/tp.2016.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Huang J, Jiang M. CREB5 computational regulation network construction and analysis between frontal cortex of HIV encephalitis (HIVE) and HIVE-control patients. Cell Biochem Biophys. 2011;60:199–207. doi: 10.1007/s12013-010-9140-x. [DOI] [PubMed] [Google Scholar]
- 10.He S, Deng Y, Liao Y, Li X, Liu J, Yao S. CREB5 promotes tumor cell invasion and correlates with poor prognosis in epithelial ovarian cancer. Oncol Lett. 2017;14:8156–61. doi: 10.3892/ol.2017.7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi L, Ding Y. Involvement of the CREB5 regulatory network in colorectal cancer metastasis. Yi Chuan. 2014;36:679–84. doi: 10.3724/SP.J.1005.2014.0679. [DOI] [PubMed] [Google Scholar]
- 12.Seo HS, Liu DD, Bekele BN, Kim MK, Pisters K, Lippman SM, Wistuba II, Koo JS. Cyclic AMP response element-binding protein overexpression: a feature associated with negative prognosis in never smokers with non-small cell lung cancer. Cancer Res. 2008;68:6065–73. doi: 10.1158/0008-5472.CAN-07-5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miao Y, Cui L, Chen Z, Zhang L. Gene expression profiling of DMU-212-induced apoptosis and anti-angiogenesis in vascular endothelial cells. Pharm Biol. 2016;54:660–6. doi: 10.3109/13880209.2015.1071414. [DOI] [PubMed] [Google Scholar]
- 14.Koga Y, Hisada T, Ishizuka T, Utsugi M, Ono A, Yatomi M, Kamide Y, Aoki-Saito H, Tsurumaki H, Dobashi K, Yamada M. CREB regulates TNFalpha-induced GM-CSF secretion via p38 MAPK in human lung fibroblasts. Allergol Int. 2016;65:406–13. doi: 10.1016/j.alit.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 15.Liang Y, Liu Y, Hou B, Zhang W, Liu M, Sun YE, Ma Z, Gu X. CREB-regulated transcription coactivator 1 enhances CREB-dependent gene expression in spinal cord to maintain the bone cancer pain in mice. Mol Pain. 2016:12. doi: 10.1177/1744806916641679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang YW, Chen X, Ma R, Gao P. Understanding the CREB1-miRNA feedback loop in human malignancies. Tumour Biol. 2016;37:8487–502. doi: 10.1007/s13277-016-5050-x. [DOI] [PubMed] [Google Scholar]
- 17.Wang G, Cheng Z, Liu F, Zhang H, Li J, Li F. CREB is a key negative regulator of carbonic anhydrase IX (CA9) in gastric cancer. Cell Signal. 2015;27:1369–79. doi: 10.1016/j.cellsig.2015.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Balogh A, Nemeth M, Koloszar I, Markó L, Przybyl L, Jinno K, Szigeti C, Heffer M, Gebhardt M, Szeberényi J, Müller DN, Sétáló G Jr, Pap M. Overexpression of CREB protein protects from tunicamycin-induced apoptosis in various rat cell types. Apoptosis. 2014;19:1080–98. doi: 10.1007/s10495-014-0986-z. [DOI] [PubMed] [Google Scholar]
- 19.Su WH, Yao Shugart Y, Chang KP, Tsang NM, Tse KP, Chang YS. How genome-wide SNP-SNP interactions relate to nasopharyngeal carcinoma susceptibility. PLoS One. 2013;8:e83034. doi: 10.1371/journal.pone.0083034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gubbay O, Rae MT, McNeilly AS, Donadeu FX, Zeleznik AJ, Hillier SG. cAMP response element-binding (CREB) signalling and ovarian surface epithelial cell survival. J Endocrinol. 2006;191:275–85. doi: 10.1677/joe.1.06928. [DOI] [PubMed] [Google Scholar]


