Abstract
Precision medicine, applying knowledge of breast cancer’s molecular subtypes, has improved the disease’s prognosis. However, recurrence and chemoresistance are critical issues for breast cancer patients. PTPN4, a new potential therapeutic target, has not been studied sufficiently in breast cancer, and the potential role of PTPN4 in the prognosis of breast cancer patients is still unknown. In our study, data from 140 invasive breast cancer patients were retrospectively collected to identify the association between PTPN4 expression and clinical outcomes of these patients. The expressions of PTPN4 were detected by immunohistochemical analysis. Breast hyperplasia tissues showed higher expression of PTPN4. We found that PTPN4 expression was lower in breast cancer patients with relapse than in patients without relapse. Patients with an increased PTPN4 level had a significantly longer relapse-free survival and overall survival time. Decreased PTPN4 expression was an independent factor associated with relapse-free survival and overall survival, as shown by multivariate Cox regression analysis. The study found that PTPN4 is an attractive prognostic biomarker for predicting the clinical outcome and effective disease management of breast cancer patients.
Keywords: Breast cancer, PTPN4, relapse, immunohistochemistry
Introduction
Breast cancer is the leading cause of cancer-related deaths in Chinese women [1]. Targeted treatment for breast cancer dramatically improves the survival outcome based on specific molecular expressions. The estrogen receptor (ER) positive type (luminal type) represents about 70% of all breast cancers, which is sensitive to endocrine therapy and promises a good prognosis. The patients with human epidermal growth factor receptor 2 (HER2) overexpression are susceptible to trastuzumab, but triple negative breast cancer patients who lack targeted biomarkers have the poorest outcomes [2]. In general, breast cancer patients have achieved an improved prognosis with systemic therapy, but the clinical benefits are limited owing to therapeutic resistance and recurrence [3-6]. We need to explore other appropriate targets to improve overall survival in breast cancer patients.
Accumulating evidence shows that the hyperactivation of the protein tyrosine phosphorylation contributes to migration, aggressiveness and metastasis in breast cancer [7-10]. Breast cancer cells with different subtypes exhibit distinct tyrosine phosphorylation signatures. Elevated tyrosine phosphorylation of the epidermal growth factor receptor (EGFR), focal adhesion kinase (FAK) and the Src kinase family (SFKs) are found in TNBC. Zins K uncovered that tyrosine phosphorylations of extracellular-signal regulated kinase (ERK) and FAK regulated by IL-34 participate in the migration and poor prognosis of luminal and HER2-overexpression breast cancer [11]. Targeting tyrosine phosphorylation of the β4 integrin/FAK complex triggered by EGFR/Scr might be a valuable therapeutic target for TNBC [12].
Recently studies have found that some potential targeted drugs can overcome chemoresistance in breast cancer by regulating the network of tyrosine phosphorylation, such as lapatinib, apatinib, and dasatinib [13-16]. These results found that targeting tyrosine phosphorylation might be a promising therapeutic strategy, which improves prognosis in breast cancer.
Protein tyrosine phosphatases (PTPs) have both oncogenic and anti-oncogenic roles in breast cancer [17-20] by balancing the status of tyrosine phosphorylation. PTPN4 (human protein tyrosine phosphatase, non-receptor type 4), is one of the PTPs, the role of which is still unknown in cancer. A few studies have demonstrated that PTPN4 might be a tumor suppressor gene [21,22]. However, the association of PTPN4 expression with clinical parameters in breast cancer is yet to be studied. In our study, we aim to determine the correlation of PTPN4 expression with prognosis in breast cancer patients. PTPN4 might be a useful biomarker for predicting clinical outcomes in breast cancer.
Materials and methods
Patients and tissue samples
The 140 cases of breast cancer tissues were collected from March 2005 to December 2013 at the Affiliated Houjie Hospital of Guangdong Medical University (Dongguan, Guangdong, PR China). All the tissue samples were selected based on the availability of archived formalin-fixed paraffin-embedded tissue (FFPE) blocks for immunohistochemical analysis. In addition, we picked 30 breast hyperplasia tissues from nearby carcinoma tissues. From the 140 invasive breast cancer samples, we detected the different expression levels of PTPN4 by IHC analysis, as shown in Table 1. All the 140 cases of breast cancer were initial tumors and were given surgical treatment and chemotherapy, of which 38 cases had a relapse and 29 cases died from the cancer through September 2016. This study was carried out in accordance with the recommendations of human biomedical research guidelines by the Ethics Research Committee of Houjie Hospital. The protocol was approved by the Ethics Research Committee of Houjie Hospital.
Table 1.
Demographic and characteristics of PTPN4 in breast cancer patients (n=140)
| Variable | Total n=140 | Low PTPN4 | High PTPN4 | P |
|---|---|---|---|---|
| Age | ||||
| <60 | 93 | 31 (33.3%) | 62 (66.7%) | 1.0000 |
| ≥60 | 47 | 16 (34.0%) | 31 (66.0%) | |
| Position | ||||
| Left | 71 | 23 (32.4%) | 48 (67.6%) | 0.8583 |
| Right | 69 | 24 (34.8%) | 45 (65.2%) | |
| Histological type | ||||
| Ductal | 130 | 42 (32.3%) | 88 (67.7%) | 0.3033 |
| Non-ductal | 10 | 5 (50.0%) | 5 (50.0%) | |
| Stage | ||||
| I | 33 | 8 (24.2%) | 25 (75.8%) | 0.3781 |
| II | 61 | 21 (34.4%) | 40 (65.5%) | |
| III | 46 | 18 (39.1%) | 28 (60.9%) | |
| Grade | ||||
| G1 | 8 | 4 (50.0%) | 4 (50.0%) | 0.5542 |
| G2 | 101 | 32 (31.7%) | 69 (68.3%) | |
| G3 | 31 | 11 (35.5%) | 20 (64.5%) | |
| Tumor size | ||||
| T1 | 57 | 19 (33.3%) | 38 (66.7%) | 0.5921 |
| T2 | 81 | 81 (34.6%) | 53 (65.4%) | |
| T3 | 2 | 0 (0.0%) | 2 (100%) | |
| LN metastasis | ||||
| Yes | 65 | 26 (40.0%) | 39 (60.0%) | 0.1535 |
| No | 75 | 21 (28.0%) | 54 (72.0%) | |
| ER | ||||
| Positive | 79 | 23 (29.1%) | 56 (70.9%) | 0.2127 |
| Negative | 61 | 24 (39.3%) | 37 (60.7%) | |
| PR | ||||
| Positive | 60 | 19 (31.7%) | 41 (68.3%) | 0.7204 |
| Negative | 80 | 28 (35.0%) | 52 (65.0%) | |
| Her2 | ||||
| Positive | 29 | 8 (27.6%) | 21 (72.4%) | 0.5129 |
| Negative | 111 | 39 (35.1%) | 72 (64.9%) | |
| Relapse | ||||
| Yes | 38 | 19 (50.0%) | 19 (50.0%) | 0.0158 |
| No | 102 | 28 (27.5%) | 74 (72.5%) | |
| Menopause | ||||
| Yes | 69 | 21 (30.4%) | 48 (69.6%) | 0.4774 |
| No | 71 | 26 (36.6%) | 45 (63.4%) |
LN: lymph node.
Hematoxylin and eosin (HE) staining and immunohistochemical analysis
We performed hematoxylin and eosin (HE) staining according to the clinical pathological guidelines. The histological characteristics were reviewed by two senior pathologists according to Breast tumors, Pathology and Genetics (WHO 2012). A total of 140 formalin-fixed, paraffin-embedded human specimens and 30 breast hyperplasia specimens were analyzed. First, the tissues were deparaffinized and sectioned. Next, the sections were subjected to a sodium citrate buffer for heat-induced antigen retrieval, and then immersed in a 3% hydrogen peroxide solution to inhibit endogenous peroxidase activity. Third, the sections were then incubated with the human PTPN4 monoclonal antibody (LifeSpan Biosciences, Seattle, WA, USA, LS-C406706) at 1:40 dilution overnight, and incubated with a secondary antibody. Finally, the sections were subjected to the Liquid DAB Substrate Chromogen System according to the manufacturer’s instructions. Specific staining of the PTPN4 protein was localized to the cytoplasms of the cancer cells. The protein’s expression level was evaluated in at least 500 tumor cells in at least 5 high-power microscopy fields. All experiments were performed in accordance with the approved guidelines and regulations of Affiliated Houjie Hospital of Guangdong Medical University.
mRNA level of PTPN4 in different tissues and analysis of clinical mRNA microarrays for the detection of correlations between PTPN4 and patient survival
The mRNA levels of PTPN4 were plotted in different adenocarcinomas by performing a GEPIA online database search (http://gepia.cancer-pku.cn/). We investigated the relationship of PTPN4 with clinical subtypes and relapse in breast cancer patients using the R2 online database (http://r2.amc.nl). We chose specific databases according to our research (Tumor Breast-Bergh-159-MAS5.0-u133a for subtypes and Tumor Breast-Wang-286-MAS5.0-u133a for relapse). The correlations between PTPN4 and overall survival and relapse-free survival were plotted with the online database of Tumor Breast-Bergh-159-MAS5.0-u133a.
Statistical analysis
Breast cancer relapse was defined as a regional or distant relapse in any other position all over the body. Overall survival (OS) was defined as the period from initial diagnosis to death attributable to any cause. Relapse-free survival (RFS) was defined as the time from initial diagnosis to relapse. A chi-Square test was used to compare the PTPN4 expression with various clinicopathologic variables. The data between groups was analyzed with an unpaired t-test. We identified risk factors involving recurrence by analysis of logistic regression. The Kaplan-Meier method was performed to plot the survival curves, and the differences between the survival curves were analyzed by a log-rank test. Cox multivariate regression analysis was used to determine the independent factors contributing to relapse in breast cancer patients. All the statistical analyses were performed using IBM SPSS 20 software (SPSS, Chicago, IL, USA), and P<0.05 was considered statistically significant.
Results
Patient characteristics and demographics
Samples from 140 patients with invasive breast cancer (median age, 54 years; range, 29-87 years) were used in the present study. To create a control group, we randomly selected 30 cases of breast hyperplasia tissues nearby the carcinoma from the 140 samples to determine the expressions of PTPN4. The clinical characteristics, such as age, tumor position, tumor size, ER and Her2 status, are described in Table 1.
Association between PTPN4 expression and clinicopathological parameters
The HE staining and immunohistochemical analysis of breast hyperplasia and breast cancers with or without relapse are shown in Figure 1. The data show that the expression of PTPN4 was higher in the breast hyperplasia tissues than that in the carcinoma tissues. And a decreased PTPN4 expression was seen in recurrent breast cancer tissues (Figure 2A). We also found that the expression of PTPN4 was higher in breast hyperplasia tissues than that in breast carcinoma, although the level was uncertain in different adenocarcinomas (Figure 2C). Correlations between the PTPN4 high or low expression cases and the clinicopathological parameters are summarized in Table 1. PTPN4 expression was correlated with relapse (P=0.0158) in breast cancer patients.
Figure 1.
HE and IHC staining of breast hyperplasia, breast cancer and relapsed breast cancer tissues (×200). A. The HE staining of the three group tissues, breast hyperplasia (left), invasive breast cancer (middle), and relapsed breast cancer (right). B. Immunohistochemical analysis of PTPN4 in three group tissues, breast hyperplasia (left), invasive breast cancer (middle) and relapsed breast cancer (right).
Figure 2.
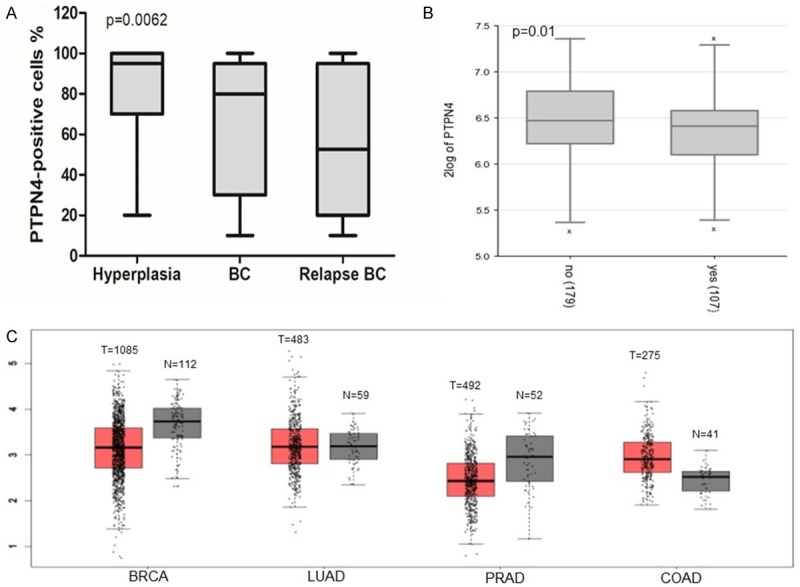
Expression level of PTPN4 in breast tissues. A. The percentage of PTPN4 positive cells in breast hyperplasia and breast cancer tissues without relapse or with relapse. B. The PTPN4 mRNA levels in breast cancer patients without relapse or with relapse were plotted using R2: Genomics Analysis and Visualization Platform. C. The mRNA levels of PTPN4 in different adenocarcinomas including breast, lung, prostate and colon were plotted using GEPIA online data (http://gepia.cancer-pku.cn/).
Expression levels of PTPN4 in clinical subtypes of breast cancer patients
We detected the expression levels of PTPN4 in luminal group, Her2 overexpression and triple negative breast cancer patients. The data suggested that there was no difference among molecular subtypes as shown in Figures 3 and 4A. The data online was consistent with our study. The mRNA levels in breast cancer patients with different subtypes were plotted using a microarray analysis and the visualization platform R2 (http://r2.amc.nl) (Figure 4B).
Figure 3.
Immunohistochemical analysis of PTPN4 in different subtypes of breast cancer tissues (×200). A. The IHC staining of PTPN4 in breast cancer tissues without relapse, luminal breast cancer (left), Her2 overexpression breast cancer tissues (middle) and TNBC tissues (right). B. IHC analysis of PTPN4 in breast cancer tissues with relapse, luminal breast cancer (left), Her2 overexpression breast cancer tissues (middle) and TNBC tissues (right).
Figure 4.

PTPN4 expression in different subtypes of breast cancer tissues. A. The percentage of PTPN4 positive cells in the different clinical subtypes of breast cancer tissues. B. The PTPN4 mRNA levels in breast cancer patients with different subtypes were plotted using microarray analysis and the visualization platform R2 (right, http://r2.amc.nl).
PTPN4 predicts relapse in breast cancer patients
Immunohistochemical analysis showed that the expression of PTPN4 in breast cancer was higher than that in recurrent breast cancer tissues (Figure 1). We found that the percentage of PTPN4 positive cells in recurrent breast cancer tissues was lower compared with carcinoma tissues without relapse (Figures 2 and 3). The analysis from the online database showed the similar results in breast cancer patients with or without relapse (Figure 2B). We identified the relapse-related risk factors by logistic regression analysis, and the analysis suggested decreased expression of PTPN4 was correlated with relapse in breast cancer patients (Table 2).
Table 2.
Logistic regression analysis of factors predicting recurrence in breast cancer patients
| Parameters | B | Wald | Sig. | Exp (B) | 95% CI |
|---|---|---|---|---|---|
| Age (≥60 y) | -0.584 | 0.715 | 0.398 | 0.558 | 0.144-2.159 |
| Position (left) | 0.021 | 0.002 | 0.960 | 1.022 | 0.440-2.375 |
| Histological type (Ductal) | 1.473 | 1.403 | 0.236 | 4.364 | 0.381-49.944 |
| Grade | -0.342 | 0.099 | 0.753 | 0.711 | 0.085-5.957 |
| Stage | 0.575 | 0.401 | 0.526 | 1.777 | 0.300-10.527 |
| Tumor size | -0.453 | 0.601 | 0.438 | 0.636 | 0.202-1.998 |
| LN metastasis (+) | 0.872 | 2.284 | 0.131 | 2.391 | 0.772-7.408 |
| ER (+) | -1.275 | 5.093 | 0.024 | 0.280 | 0.092-0.846 |
| PR (+) | 1.069 | 3.481 | 0.062 | 2.913 | 0.947-8.959 |
| Her2 (+) | 0.559 | 1.137 | 0.286 | 1.749 | 0.626-4.888 |
| Menopause (+) | 0.746 | 1.348 | 0.246 | 2.110 | 0.598-7.438 |
| PTPN4 (low) | 1.013 | 5.084 | 0.024 | 2.753 | 1.142-6.641 |
LN: lymph node.
Survival analysis
Among the 140 cases of breast cancer, 29 cases have died till September 2016. The mean follow-up time of the patients was 108 months (26-132 (OS) and the range was 10-132 (DFS)). Kaplan-Meier analysis was used to compare the outcomes with the grouped patients. The results in our study showed that patients with lower expression of PTPN4 had a significant association with a worse OS (P=0.0334) and RFS (P=0.0172) in breast cancer (Figure 5A). We also investigated the correlation between PTPN4 and the survival of breast cancer patients using Microarray data and Kaplan-Meier plots from OncoLnc database online (http://www.oncolnc.org/), which showed similar results (Figure 5B).
Figure 5.

PTPN4 predicts survival in breast cancer. A. Kaplan-Meier analysis of the correlation between PTPN4 and OS (left) and RFS (right) in breast cancer patients. B. Microarray data and Kaplan-Meier plots from OncoLnc database online (http://www.oncolnc.org/) were used to assess the associations between PTPN4 gene expression and patient survival of breast cancer (OS, left; RFS, right).
Prognostic factors
By Cox regression analysis, we assessed multivariates including age, position, histological type, tumor grade, tumor stage, tumor size, lymph node metastasis, ER status, PR status, Her2 status, menopause status, and PTPN4 expression, to evaluate whether PTPN4 expression in breast cancer patients was an independent predictor of OS and RFS. PTPN4 expression (OS: P=0.047; RFS: P=0.028) was a significant prognostic factor for breast cancer patients (Table 3). We demonstrated that low PTPN4 expression was a significant independent factor for poor outcome of OS and RFS in breast cancer patients.
Table 3.
Multivariate survival analysis for OS and DFS in breast cancer
| Parameters | OS | RFS | ||
|---|---|---|---|---|
|
|
|
|||
| P | HR (95% CI) | P | HR (95% CI) | |
| Age (≥60 y) | 0.209 | 2.128 (0.655-6.913) | 0.318 | 1.643 (0.620-4.356) |
| Position (left) | 0.640 | 1.199 (0.560-2.564) | 0.334 | 1.361 (0.728-2.546) |
| Histological type (Ductal) | 0.284 | 3.596 (0.346-37.390) | 0.584 | 1.580 (0.308-8.104) |
| Grade | 0.989 | 0.988 (0.194-5.048) | 0.529 | 1.678 (0.335-8.389) |
| Stage | 0.960 | 1.047 (0.176-6.221) | 0.571 | 1.487 (0.377-5.867) |
| Tumor size | 0.304 | 1.726 (0.609-4.889) | 0.901 | 1.050 (0.485-2.273) |
| LN metastasis | 0.105 | 2.274 (0.842-6.137) | 0.073 | 2.170 (0.932-5.045) |
| ER | 0.011 | 0.299 (0.118-0.755) | 0.011 | 0.378 (0.178-0.802) |
| PR | 0.467 | 0.705 (0.275-1.808) | 0.507 | 1.279 (0.619-2.642) |
| Her2 | 0.985 | 0.991 (0.412-2.383) | 0.889 | 1.050 (0.522-2.118) |
| Menopause | 0.477 | 1.543 (0.467-5.098) | 0.591 | 1.303 (0.497-3.412) |
| PTPN4 (low) | 0.047 | 2.289 (1.010-5.190) | 0.028 | 2.113 (1.086-4.111) |
DFS: disease-free survival; HR: hazard ratio; CI: confidence interval; LN: lymph node.
Discussion
Lacking reliable biomarkers in breast cancer patients for the prediction of recurrence is still a difficult problem, which affects overall survival or relapse-free survival. Previous studies revealed an association between PTPN4 expression and cell death, but the role of PTPN4 in breast cancer prognosis is unclear. In this study we aimed to determine the correlation between PTPN4 expression and relapse in breast cancer patients.
In our study, we demonstrated that PTPN4 expression was decreased in breast cancer tissues. Further, we revealed that decreased PTPN4 expression predicted poor RFS and OS. Multivariate survival analysis showed that low expression of PTPN4 was an independent prognostic factor for poor survival in breast cancer patients. In our study, we also determined that the level of PTPN4 was significantly lower in breast cancer tissues with relapse than that in tissues without relapse. The results strongly suggested that PTPN4 might be a tumor suppressor in breast cancer and uncovered a potential clinical value of PTPN4 in breast cancer recurrence, although we found there was no differences in PTPN4 expression among the molecular subtypes of breast cancer.
Previous studies showed that anti-oncogenic PTPs played vital roles in cell growth-related processes through their dephosphorylate oncogenic receptor protein in breast cancer [17,21]. Zhu explored whether miR-183 could inhibit PTPN4 to regulate the death of cancer stem-like cells in non-small cell lung cancer, which were responsible for tumor growth and invasion leading to recurrence and metastasis [22]. The anti-cancer role of PTPN4 in gastric cancer depended on the cancer cell type and the mutant status [23]. However, the mechanism of PTPN4 that correlates with tumorigenesis and prognosis remains to be elucidated.
With current studies on tumor environment, a knowledge of tumor infiltrating lymphocytes (TILs) is essential for understanding mechanisms of breast cancer progression [24]. Studies showed that T-cell marker positive cells (CD3+ and CD8+) contributed to the prediction of recurrence and therapeutic response in triple negative and HER2-overexpression breast cancers [25,26]. Moreover, PTPN4 functions in T cell development triggering immune responses against the invasion of pathogens [27]. PTPN4 is an enzyme that leaves dephosphorylate tyrosine residues on some proteins involved in TCR (T cell receptor) signaling pathways [28].
Based on previous studies, we supposed that PTPN4, as a tumor suppressor, participated in tumorigenesis, chemotherapeutic response and progression through regulating immune responses and metabolization. The effect is dependent on its chemical structure. PTPN4 contains highly homologous N-terminal FERM-, central PDZ-, and C-terminal catalytic-domains. The PDZ domain plays a key role in the regulation of enzyme activity and the dynamic regulation of signaling pathways [29], and it will be a potential target for cancer treatment.
In conclusion, we demonstrated that recurrent breast cancers are characterized by decreased PTPN4 expression. PTPN4 might be a novel predictive biomarker to identify relapse in breast cancer patients. Increasing PTPN4 expression might be a promising strategy to improve the chemotherapeutic response and outcome of breast cancer patients, which has a great value for additional targeted therapy.
Acknowledgements
This work was funded by the Guangdong Natural Science Foundation of China (2017A030310326) and the Dongguan Social Development project (2015108101018). All the medical data and the tissues were obtained from the Department of Clinical Pathology, The Affiliated Houjie Hospital of Guangdong Medical University.
Disclosure of conflict of interest
None.
References
- 1.Zheng Y, Wu CX, Wu F. Status and trends of breast cancer mortality in Chinese females. Zhonghua Yu Fang Yi Xue Za Zhi. 2011;45:150–154. [PubMed] [Google Scholar]
- 2.Chen L, Linden HM, Anderson BO, Li CI. Trends in 5-year survival rates among breast cancer patients by hormone receptor status and stage. Breast Cancer Res Treat. 2014;47:609–616. doi: 10.1007/s10549-014-3112-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dawson SJ, Provenzano E, Caldas C. Triple negative breast cancers: clinical and prognostic implications. Eur J Cancer. 2009;45:27–40. doi: 10.1016/S0959-8049(09)70013-9. [DOI] [PubMed] [Google Scholar]
- 4.Nahta R, Esteva FJ. Herceptin: mechanisms of action and resistance. Cancer Lett. 2006;232:123–138. doi: 10.1016/j.canlet.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 5.Selli C, Dixon JM, Sims AH. Accurate prediction of response to endocrine therapy in breast cancer patients: current and future biomarkers. Breast Cancer Res. 2016;18:118. doi: 10.1186/s13058-016-0779-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kourea HP, Zolota V, Scopa CD. Targeted pathways in breast cancer: molecular and protein markers guiding therapeutic decisions. Curr Mol Pharmacol. 2014;7:4–21. doi: 10.2174/187446720701150105170830. [DOI] [PubMed] [Google Scholar]
- 7.Blume Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–365. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 8.Sun T, Aceto N, Meerbrey KL, Kessler JD, Zhou C, Migliaccio I, Nguyen DX, Pavlova NN, Botero M, Huang J, Bernardi RJ, Schmitt E, Hu G, Li MZ, Dephoure N, Gygi SP, Rao M, Creighton CJ, Hilsenbeck SG, Shaw CA, Muzny D, Gibbs RA, Wheeler DA, Osborne CK, Schiff R, Bentires-Alj M, Elledge SJ, Westbrook TF. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144:703–18. doi: 10.1016/j.cell.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HM, Xu YF, Ning SL, Yang DX, Li Y, Du YJ, Yang F, Zhang Y, Liang N, Yao W, Zhang LL, Gu LC, Gao CJ, Pang Q, Chen YX, Xiao KH, Ma R, Yu X, Sun JP. The catalytic region and PEST domain of PTPN18 distinctly regulate the HER2 phosphorylation and ubiquitination barcodes. Cell Res. 2014;24:1067–1090. doi: 10.1038/cr.2014.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Youssef G, Gillett C, Agbaje O, Crompton T, Montano X. Phosphorylation of NTRK1 at Y674/Y675 induced by TP53-dependent repression of PTPN6 expression: a potential novel prognostic marker for breast cancer. Mod Pathol. 2014;27:361–374. doi: 10.1038/modpathol.2013.129. [DOI] [PubMed] [Google Scholar]
- 11.Zins K, Heller G, Mayerhofer M, Schreiber M, Abraham D. Differential prognostic impact of interleukin-34 mRNA expression and infiltrating immune cell composition in intrinsic breast cancer subtypes. Oncotarget. 2018;9:23126–48. doi: 10.18632/oncotarget.25226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tai YL, Chu PY, Lai IR, Wang MY, Tseng HY, Guan JL, Liou JY, Shen TL. An EGFR/Srcdependent β4 integrin/FAK complex contributes to malignancy of breast cancer. Sci Rep. 2015;5:16408. doi: 10.1038/srep16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sridhar J, Sfondouris ME, Bratton MR, Nguyen TL, Townley I, Klein Stevens CL, Jones FE. Identification of quinones as HER2 inhibitors for the treatment of trastuzumab resistant breast cancer. Bioorg Med Chem Lett. 2014;24:126–131. doi: 10.1016/j.bmcl.2013.11.064. [DOI] [PubMed] [Google Scholar]
- 14.Chen YJ, Yeh MH, Yu MC, Wei YL, Chen WS, Chen JY, Shih CY, Tu CY, Chen CH, Hsia TC, Chien PH, Liu SH, Yu YL, Huang WC. Lapatinib-induced NF-kappaB activation sensitizes triple-negative breast cancer cells to proteasome inhibitors. Breast Cancer Res. 2013;15:R108. doi: 10.1186/bcr3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan M, Zhang J, Wang Z, Wang B, Zhang Q, Zheng C, Li T, Ni C, Wu Z, Shao Z, Hu X. Phosphorylated VEGFR2 and hypertension: potential biomarkers to indicate VEGF-dependency of advanced breast cancer in anti-angiogenic therapy. Breast Cancer Res Treat. 2014;143:141–151. doi: 10.1007/s10549-013-2793-6. [DOI] [PubMed] [Google Scholar]
- 16.Assi J, Srivastava G, Matta A, Chang MC, Walfish PG, Ralhan R. Transglutaminase 2 overexpression in tumor stroma identifies invasive ductal carcinomas of breast at high risk of recurrence. PLoS One. 2013;8:e74437. doi: 10.1371/journal.pone.0074437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, Davidson D, Martins Souza C, Zhong MC, Wu N, Park M, Muller WJ, Veillette A. Loss of PTPN12 stimulates progression of ErbB2-dependent breast cancer by enhancing cell survival, migration, and epithelial-to-mesenchymal transition. Mol Cell Biol. 2015;35:4069–4082. doi: 10.1128/MCB.00741-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hwang BM, Chae HS, Jeong YJ, Lee YR, Noh EM, Youn HZ, Jung SH, Yu HN, Chung EY, Kim JS. Protein tyrosine phosphatase controls breast cancer invasion through the expression of matrix metalloproteinase-9. BMB Rep. 2013;46:533–8. doi: 10.5483/BMBRep.2013.46.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alho I, Costa L, Bicho M, Coelho C. Low molecular weight protein tyrosine phosphatase isoforms regulate breast cancer cells migration through a RhoA dependent mechanism. PLoS One. 2013;8:e76307. doi: 10.1371/journal.pone.0076307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nunes-Xavier CE, Martín-Pérez J, Elson A, Pulido R. Protein tyrosine phosphatases as novel targets in breast cancer therapy. Biochim Biophys Acta. 2013;1836:211–226. doi: 10.1016/j.bbcan.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 21.Chun J, Li RJ, Cheng MS, Kim YS. Alantolactone selectively suppresses STAT3 activation and exhibits potent anticancer activity in MDAMB-231 cells. Cancer Lett. 2015;357:393–403. doi: 10.1016/j.canlet.2014.11.049. [DOI] [PubMed] [Google Scholar]
- 22.Zhu C, Deng X, Wu J, Zhang J, Yang H, Fu S, Zhang Y, Han Y, Zou Y, Chen Z, Lin S. MicroRNA-183 promotes migration and invasion of CD133(+)/CD326(+) lung adenocarcinoma initiating cells via PTPN4 inhibition. Tumour Biol. 2016;37:11289–11297. doi: 10.1007/s13277-016-4955-8. [DOI] [PubMed] [Google Scholar]
- 23.Wu CW, Chen JH, Kao HL, Li AF, Lai CH, Chi CW, Lin WC. PTPN3 and PTPN4 tyrosine phosphatase expression in human gastric adenocarcinoma. Anticancer Res. 2006;26:1643–1649. [PubMed] [Google Scholar]
- 24.Wang Y, Dong T, Xuan Q, Zhao H, Qin L, Zhang Q. Lymphocyte-activation gene-3 expression and prognostic value in neoadjuvanttreated triple-negative breast cancer. J Breast Cancer. 2018;21:124–133. doi: 10.4048/jbc.2018.21.2.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Althobiti M, Aleskandarany MA, Joseph C, Toss M, Mongan N, Diez-Rodriguez M, Nolan CC, Ashankyty I, Ellis IO, Green AR, Rakha EA. Heterogeneity of tumour infiltrating lymphocytes (TILs) in breast cancer and its prognostic significance. Histopathology. 2018 doi: 10.1111/his.13695. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 26.McIntire PJ, Irshaid L, Liu Y, Chen Z, Menken F, Nowak E, Shin SJ, Ginter PS. Hot spot and whole-tumor enumeration of CD8+ tumor-infiltrating lymphocytes utilizing digital image analysis is prognostic in triple-negative breast cancer. Clin Breast Cancer. 2018 doi: 10.1016/j.clbc.2018.04.019. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 27.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 28.Huai W, Song H, Wang L, Li B, Zhao J, Han L, Gao C, Jiang G, Zhang L, Zhao W. Phosphatase PTPN4 preferentially inhibits TRIF-dependent TLR4 pathway by dephosphorylating TRAM. J Immunol. 2015;194:4458–4465. doi: 10.4049/jimmunol.1402183. [DOI] [PubMed] [Google Scholar]
- 29.Park KW, Lee EJ, Lee S, Lee JE, Choi E, Kim BJ, Hwang R, Park KA, Baik J. Molecular cloning and characterization of a protein tyrosine phosphatase enriched in testis, a putative murine homologue of human PTPMEG. Gene. 2000;257:45–55. doi: 10.1016/s0378-1119(00)00351-6. [DOI] [PubMed] [Google Scholar]




