Abstract
The aberrant expression of microRNAs (miRNAs) has been found in various types of cancer and is associated with tumorigenesis and metastasis. However, the expression and function of miR-223 in nasopharyngeal carcinoma (NPC) remain unclear. The present study demonstrated that miR-223 was downregulated in NPC cell lines. The ectopic expression of miR-223 dramatically suppressed cell proliferation, invasion and epithelial-mesenchymal transition (EMT). Moreover, a luciferase reporter assay identified the structure-specific recognition protein (SSRP1) as a novel direct target of miR-223. SSRP1 expression was upregulated in NPC cell lines and the overexpression of miR-233 markedly reduced the expression of SSRP1. Furthermore, SSRP1 was involved in miR-223-regulated NPC cell proliferation, invasion, and EMT. Taken together, these results indicate that miR-223 functions as a tumor suppressor miRNA in NPC and that its suppressive effects are primarily mediated by repressing SSRP1 expression and inhibiting EMT.
Keywords: miR-223, nasopharyngeal carcinoma (NPC), EMT, SSRP1
Introduction
Nasopharyngeal carcinoma (NPC) is a non-lymphomatous squamous cell carcinoma derived from the epithelial cells of the nasopharynx, which is a relatively common malignant tumor in Southeast Asia, particularly in Southern China [1]. NPC is characterized by a highly malignant local invasion and early distant metastasis, and 30-40% of NPC patients will develop distant metastases with poor prognoses [2-4]. Despite improvements in the diagnosis and treatment of NPC and its five-year survival rate, distant metastasis is still one of the most common failure patterns [5-7]. Therefore, an improved understanding of the mechanisms that underlie the initiation and progression of NPC is of crucial significance to the development of novel therapeutic strategies.
MicroRNA (miRNA), an abundant group of endogenous non-coding single strand RNAs of 22 nucleotides, regulates the expression of genes at the post-transcriptional level by translational repression or degradation of target mRNA. In this manner, they participate in the regulation of a range of biological processes including cell proliferation, apoptosis, invasion, migration, differentiation, and so on [8-11]. Growing evidence proves that deregulated miRNAs contribute to cancer progression as a result of changes in the expression of their target genes in various cancers, such as breast cancer, lung cancer, pancreatic cancer, and nasopharyngeal carcinoma [12-17].
Recently, an increasing number of studies has demonstrated that the expression of miR-223 is deregulated in various cancers [18-20]. For example, miR-223 expression is upregulated in gastric cancer and miRNA-223 promotes gastric cancer invasion and metastasis by targeting the tumor suppressor EPB41L3 [21]. Zhang et al. reported that miR-223 also functions as an oncogene in human colorectal cancer cells [19]. Meanwhile, numerous studies have reported that miR-233 is frequently decreased in many malignancies, including osteosarcoma, cervical cancer, hepatocellular carcinoma, and pancreatic cancer, and functions as a tumor suppressor by inhibiting tumor cell growth, invasion, metastasis, and tumorigenesis, regulating cell apoptosis, the cell cycle, and the epithelial to mesenchymal transition [22-25]. However, little is known about the expression and function of miR-223 in nasopharyngeal carcinoma (NPC).
Structure-specific recognition protein (SSRP1), initially isolated from the expression screening of a human B-cell cDNA library for proteins that bind to cisplatin (cis-diamminedichloroplatinum (II))-modified DNA, contains a single DNA-binding high mobility group (HMG) domain. SSRP1 and a suppressor of Ty 16 (SPT16) can form a heterodimeric complex, and can facilitate chromatin transcription (FACT 1), which was reported to be overexpressed in many types of tumor cells and to function as a molecular target of a novel class of candidate anti-cancer agents named curaxins [26]. Recently, Koman et al. showed that SSRP1 and SPT16 were upregulated in mammary tumors and targeting the FACT complex suppresses mammary tumorigenesis in Her2/neu transgenic mice [27]. These indicated that SSRP1 may function as an oncogene in tumors.
In our study, we investigated the biological functions and molecular mechanisms of miR-223 in NPC. We found that miR-223 was downregulated in NPC cells. SSRP1 was upregulated in NPC cells and was identified as a direct target of miR-223. Furthermore, miR-223 suppressed NPC cell proliferation, invasion and epithelial-mesenchymal transition (EMT) by targeting SSRP1. Mechanistically, our results demonstrated that miR-223 functions as a tumor suppressor of NPC by targeting SSRP1.
Materials and methods
Cell culture and transfection
Cells from the human immortalized nasopharyngeal epithelial cell line (NP69 cells), were cultured in a keratinocyte/serum-free medium (Invitrogen, Grand Island, NY, USA) supplemented with bovine pituitary extract (BD Biosciences, San Diego, CA, USA). Human NPC cell lines (CNE-1, HNE-1, SUNE-1, 5-8F and 6-10B) were maintained in RPMI-1640 (Invitrogen) and supplemented with 10% FBS (Gibco, Grand Island, NY, USA). The CNE-1 cell line was derived from patients in China, but this cell line has been subcultured more than 100 times. HNE-1 is a relatively early-passage un-cloned NPC cell line, but, by passage 45, EBV DNA could no longer be detected in HNE-1 cells by Southern blot analysis [28]. The NPC cell line 6-10B was derived from the cell line SUNE-1 and has a low metastatic ability. The NPC cell line 5-8F cells were derived from the SUNE-1 cell, with high metastasis and high tumorigenicity.
5-8F cells were transfected with SSRP1 siRNAs including SSRP1-siRNA-471, SSRP1-siRNA-810, and SSRP1-siRNA-1457. SSRP1-siRNA-810 showed the greatest inhibitory effect on SSRP1 expression, so it was used for further study at SSRP1 knockdown. The transfections of 5-8F cells were divided in four groups: Group A, transfection with negative control siRNA (NC; 50 nM, GenePharma, Suzhou, China); Group B, transfection with miR-223 mimics (50 nM, GenePharma); Group C, co-transfection with the miR-223 mimics (50 nM) and SSRP-siRNA-810 (100 ng; GenePharma); Group D, transfection with SSRP1-siRNA-810 (100 ng; GenePharma). The transfections were performed using the Lipofectamine 2000 reagent (Invitrogen).
qRT-PCR
The PCR amplification for the quantification of miR-223 and U6 was performed using the TaqMan miRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) and the TaqMan Human MiRNA Assay Kit (Applied Biosystems, Foster City, CA, USA). The relative expression of miR-223 was shown as fold difference relative to U6. The PCR amplification for the quantification of the SSRP1 and GAPDH mRNAs was performed using an ABI PRISM 7300 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) and a SYBR®Premix Ex TaqTM ii (Perfect Real Time) Kit (Takara Bio, Shiga, Japan). The primers were as follows: miR-223 (MIMAT0000280): 5’-UGUCAGUUUGUCAAAUACCCCA-3’; miR-223 mimics, forward primer: 5’-UGUCAGUUUGUCAAAUACCCCA-3’ and reverse primer: 5’-UGGGGUAUUUGACAAACUGACA-3’; miR-223 negative control, forward primer: 5’-UUCUCCGAACGUGUCACGUTT-3’ and reverse primer: 5’-ACGUGACACGUUCGGAGAATT-3’. Homo-SSRP1, forward primer: 5’-TGACTACAAGATCCCCTACACC-3’ and reverse primer: 5’-GAGTTTGGCCTTGCTTGATTG-3’. β-actin, forward primer: 5’-CATTAAGGAGAAGCTGTGCT-3’ and reverse primer: 5’-GTTGAAGGTAGTTTCGTGGA-3’.
Western blot
Whole cell extracts were prepared with a cell lysis reagent (Sigma-Aldrich, St. Louis, MO, USA) according to the manual, and then, the protein was quantified by a BCA assay (Pierce, Rockford, IL, USA). Then, the protein samples were separated by SDS-PAGE (10%) and detected by Western blot using polyclonal (rabbit) anti-SSRP1, anti-E-cadherin, anti-N-cadherin and anti-Vimentin antibodies (Santa Cruz Bio-technology, Santa Cruz, CA, USA). Goat anti-rabbit IgG (Pierce, Rockford, IL, USA) secondary antibody conjugated to horseradish peroxidase and ECL detection systems (SuperSignal West Femto, Pierce) were used for detection.
Luciferase reporter assay
The 3’-UTR sequence of SSRP1 was amplified from normal human genomic DNA and subcloned into the pmirGLO luciferase reporter vector (Promega). 5-8F cells (3.5 × 104) were seeded in triplicate in 24-well plates and cotransfected with wild-type (WT) or mutant (Mut) 3’-UTR vectors and miR-223 mimics using Lipofectamine 2000. After 48 h, the cells were assayed for luciferase activity using the Dual-Luciferase Reporter Assay System (Promega) by following the manufacturer’s instructions. The firefly luciferase activities were normalized to Renilla luciferase activity. The firefly luciferase activity of the cells that were transfected with miRNA mimics or inhibitors is represented as the percentage of activity relative to that of cells that were transfected with negative controls. All experiments were performed in triplicate.
Cell proliferation assay and invasion assay
The 3-(4,5-dimethylthiazal-2-yl)-2,5-diphenyl-tetrazolium bromide (MTT) assay was used to estimate cell viability [29]. Briefly, cells were plated at a density of 1 × 104 cells per well in 96-well plates. After exposure to specific treatment, the cells were incubated with MTT at a final concentration of 0.5 mg/ml for 4 h at 37°C. After the removal of the medium, 150 mM DMSO solutions were added to dissolve the formazan crystals. The absorbance was read at 570 nm using a multi-well scanning spectrophotometer reader. Cells in the control group were considered 100% viable.
The capability of cell invasion was examined by transwell invasion assay. Cells were cultivated to 80% confluence on the 12-well plates. Then, we observed the procedures of cellular growth at 72 h. All the experiments were repeated in triplicate. The transwell migration chambers were used to evaluate cell invasion. Then cells invading cells across the membrane were counted under a light microscope.
Tumor xenograft in nude mice
The animal experiment was approved by the Ethical Committee for Animal Research of Tongji Medical College (protocol number: 2011-020). Nude mice (4-5 weeks old, male) were purchased from the Central Animal Facility of Tongji Medical College. 200 ml of 5-8F cells (1 × 106), which were transfected with miR-223 mimics and SSRP-siRNA-810 alone or in combination, were injected into the left side on the back of each mouse. The tumor volume was monitored on days 6, 12, 18 and 24. The tumor volume was calculated as v = a×b2/2.
Immunohistochemistry assay
Formalin-fixed and paraffin-embedded tissues of the xenografted NPC tumor were deparaffinized and rehydrated. Tissues were treated for 20 min at 100°C in an autoclave for antigen retrieval and blocked with a blocking reagent (Protein Block Serum-Free, Dako Cytomation, Glostrup, Denmark) to avoid non-specific reactions. Then, tissues in sections were incubated with the anti-Ki67 antibody (Dilution 1:400, ab16667, Abcam, Cambridge, UK) and the anti-caspase-3 antibody (Dilution 1:500, ab13847, Abcam) overnight at 4°C, followed by horseradish peroxidase (HRP)-labeled anti-rabbit IgG (Histofine, simple stain MAX-PO; Nichirei, Tokyo, Japan) for 30 min at room temperature. The sections were treated with a 3, 3’-diaminobenzidine tetrahydrochloride solution. All the sections were counterstained with HE.
Statistical analysis
Each experiment was repeated at least three times. Data were shown as the mean ± SD and analyzed using SPSS 18.0. Statistical comparisons between the groups were analyzed using Student’s t-test, and a two-tailed test. P < 0.05 was considered to indicate statistical significance.
Results
The expression of miR-223 and SSRP1 in NPC cell lines
We first employed qRT-PCR to detect miR-223 levels in the human immortalized nasopharyngeal epithelial cell line (NP69) and the human NPC cell lines (CNE-1, HNE-1, SUNE-1, 5-8F and 6-10B). The miR-223 expression was downregulated in all NPC cell lines as compared with the expression in NP69 (P < 0.01; Figure 1A). Moreover, miR-223 expression in the highly metastatic NPC cell lines (5-8F) was obviously lowest in the NPC cell lines and the miR-223 expression in the lowly metastatic NPC cell lines (6-10B) was obviously highest in NPC cell lines (Figure 1A). These data indicate that downregulated miR-223 expression confers an increased metastatic potential to the NPC cells.
Figure 1.
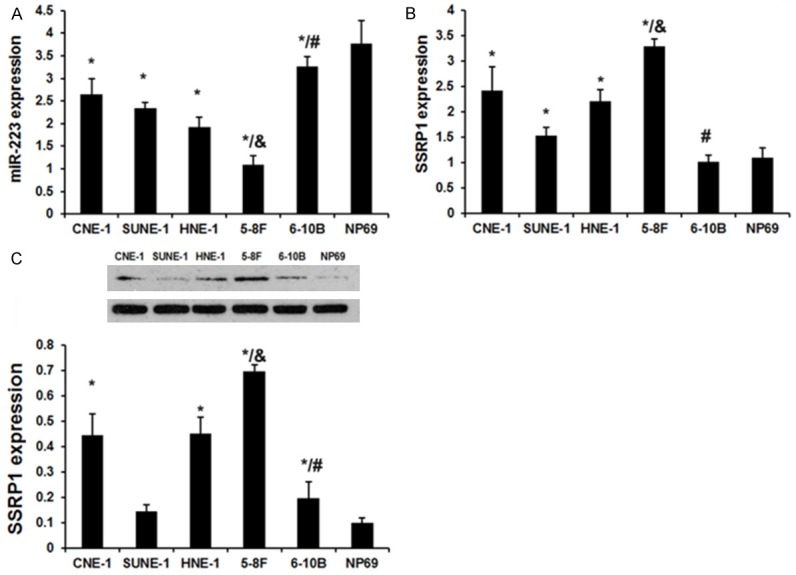
The expression of miR-223 and SSRP1 in NPC cell lines. A. qRT-PCR analysis revealed the miR-223 expression in the human immortalized nasopharyngeal epithelial cell line (NP69 cells) and the human NPC cell lines (CNE-1, HNE-1, SUNE-1, 5-8F and 6-10B). B. qRT-PCR analysis revealed the SSRP1 expression in the human immortalized nasopharyngeal epithelial cell line (NP69 cells) and the human NPC cell lines (CNE-1, HNE-1, SUNE-1, 5-8F and 6-10B). C. Western blot analysis revealed the SSRP1 expression in the human immortalized nasopharyngeal epithelial cell line (NP69 cells) and the human NPC cell lines (CNE-1, HNE-1, SUNE-1, 5-8F and 6-10B). Each bar represents the mean of three independent experiments. *P < 0.05 versus NP69 cell line. #P < 0.05 versus CNE-1, HNE-1, SUNE-1 and 5-8F cell line. &P < 0.05 versus CNE-1, HNE-1, SUNE-1 and 6-10B cell line.
We next assayed the SSRP1 expression levels in the human immortalized nasopharyngeal epithelial cell line (NP69) and the human NPC cell lines (CNE-1, HNE-1, SUNE-1, 5-8F and 6-10B). The SSRP1 expression was upregulated in all NPC cell lines as compared with the expression in NP69 (P < 0.01; Figure 1B). Moreover, SSRP1 expression in the highly metastatic NPC cell lines (5-8F) was obviously highest in the NPC cell lines and SSRP1 expression in the lowly metastatic NPC cell lines (6-10B) was obviously lowest in the NPC cell lines (Figure 1B). These data indicate that upregulated SSRP1 expression confers an increased metastatic potential to the NPC cells.
miR-223 directly targeted SSRP1
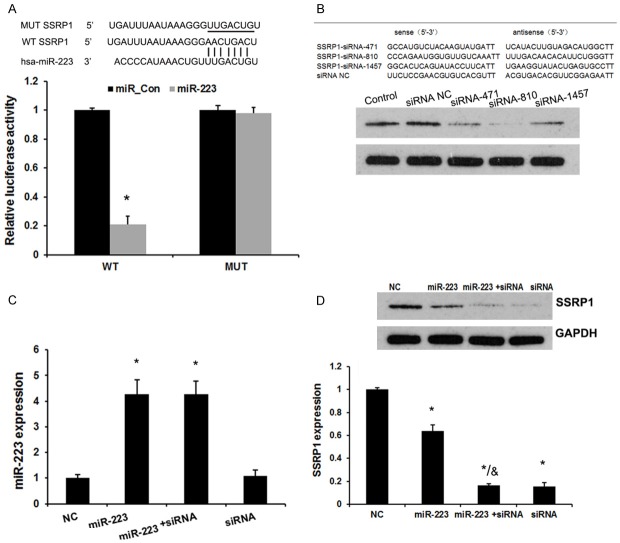
To elucidate whether SSRP1 is a potential downstream target gene of miR-223 in NPC cells, we constructed luciferase reporter vectors containing the wild-type (Wt) or mutant (Mt) miR-223 target sequences of the SSRP1 3’-UTR (Figure 2A). Over-expression of miR-223 significantly inhibited the luciferase activity of the Wt SSRP1 3’-UTR reporter gene but not the Mt reporter gene (Figure 2A).
Figure 2.
miR-223 directly targeted SSRP1. A. A representative diagram of the predicted wild-type (WT) or mutant (Mut) binding site of miR-223 in the 3’-untranslated region (UTR) of SSRP1 mRNA. The luciferase reporter plasmid containing the WT or Mut SSRP1 3’-UTR was co-transfected into 5-8F cells with miR-223 mimics. The luciferase activity of the cells was assayed at 48 h after transfection, and the values were normalized to the normal control values. *P < 0.05 (compared with the control). B. Western blot analysis examined the effects of three siRNAs and the negative control on SSRP1 expression. C. A qRT-PCR analysis revealed the effects of SSRP1 siRNA-810 and miR-223 mimics on the expression level of miR-223. D. A Western blot analysis revealed the effects of SSRP1 siRNA-810 and miR-223 mimics on the expression level of SSRP1. Error bars represent ± S.E. and *P < 0.05 versus control and NC. &P < 0.05 versus miR-223 group.
Next, we transfected SSRP1 siRNAs (SSRP1-siRNA-471, SSRP1-siRNA-810 and SSRP1-siRNA-1457) in 5-8F cells (Figure 2B). Western blot analysis revealed that the SSRP1 expression was at 51.1% with siRNA-471, 14.2% with siRNA-810, and 66.4% with siRNA-1457, compared with the SSRP1 negative control siRNA or the control (un-transfected) group (P < 0.05). Thus, the cells transfected with SSRP1 siRNA-810 were used for further experiments for SSRP1 knockdown. In addition, overexpression of miR-233 markedly reduced the expression of SSRP1 (P < 0.05, Figure 2D), but silenced SSRP1 did not affect miR-223 expression. These results demonstrated that SSRP1 is a direct target of miR-223 in NPC cells (P < 0.05, Figure 2C).
SSRP1 involves in miR-223-regulated NPC cell proliferation and invasion
To determine the role of SSRP1 and miR-223 in the NPC cell growth and metastasis, 5-8F cells were transiently co-transfected with SSRP1 siRNA (siRNA) and an miR-223 mimic (miR-223). Consistent with the effects induced by the overexpression of miR-223, the knockdown of SSRP1 significantly suppressed the cell viability (P < 0.05) and invasion (Figure 3A), but overexpression of miR-223 did not have further suppressive effects on cell growth and metastasis in SSRP1-siRNA -transfected 5-8F cells.
Figure 3.
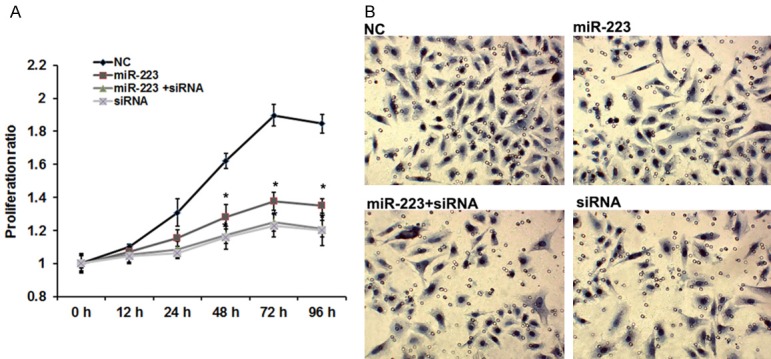
SSRP1 involved in miR-223-regulated NPC cell proliferation and invasion. The transfections of 5-8F cells were divided into four groups: Group A, transfection with negative control siRNA; Group B, transfection with miR-223 mimics; Group C, co-transfection with the miR-223 mimics and SSRP-siRNA-810; Group D, transfection with SSRP1-siRNA-810. Cell viability and invasion capacity were assessed following the cell transfections. A. The effects of miR-223 and SSRP1 on cell proliferation. B. The effects of miR-223 and SSRP1 on cell invasion. Error bars represent ± S.E. and *P < 0.05 versus NC.
MiR-223 inhibited EMT of NPC cells by suppressing SSRP1 expression
EMT has been identified as having a key role in the invasion of various cancer cells by the transformation of polarized and adherent epithelial cells into motile and invasive mesenchymal cells. Here, to explore proteins regulated by miR-223 in the EMT process, we investigated the expression of three EMT related proteins, E-cadherin, N-cadherin, and Vimentin, by Western blot. 5-8F cells were transfected with NC, miR-223 mimics, SSRP1 siRNA and co-transfected with miR-223 mimics and SSRP1 siRNA. The results indicated the expression of E-cadherin was increased in the miR-223 mimics group compared with NC and the un-transfected groups (Figure 4). Moreover, E-cadherin expression in the siRNA group was higher than it was in the miR-223 group (miR-223 mimics) and similar to that in the miR-223 + siRNA group. N-cadherin and Vimentin was downregulated significantly in the miR-223 group. Moreover, N-cadherin and Vimentin expression in the siRNA group were lower than they were in the miR-223 group (miR-223 mimics) and similar to its expression in the miR-223 + siRNA group. This indicated that miR-223 represses the expression of N-cadherin and Vimentin, while promoting the induction of E-cadherin by targeting SSRP1.
Figure 4.
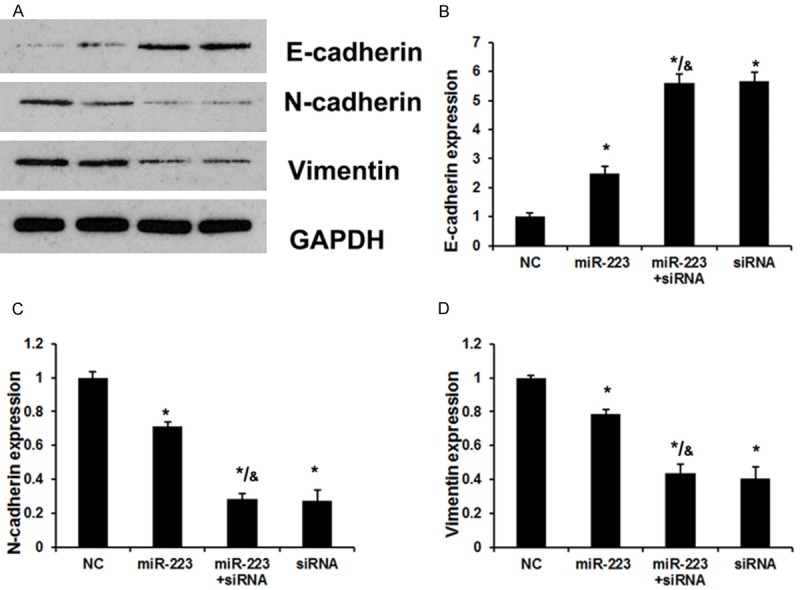
miR-223 induces EMT of NPC cells by suppressing SSRP1 expression. The transfections of 5-8F cells were divided in four groups: Group A, transfection with negative control siRNA; Group B, transfection with miR-223 mimics; Group C, co-transfection with the miR-223 mimics and SSRP-siRNA-810; Group D, transfection with SSRP1-siRNA-810. A Western blot assay was performed after the cell transfections. A. A Western blot analysis revealed the effects of miR-223 and SSRP1 on EMT-relative protein expression. B. The effects of miR-223 and SSRP1 on E-cadherin expression. C. The effects of miR-223 and SSRP1 on N-cadherin expression. D. The effects of miR-223 and SSRP1 on Vimentin expression. Error bars represent ± S.E. and *P < 0.05 versus NC. &P < 0.05 versus miR-223 group.
miR-223 inhibited the growth of xenografted NPC in nude mice by targeting SSRP1
This study evaluated the proliferation of xenografted NPC in nude mice on days 6, 12 18, and 24 by measuring tumor volume. As shown in Figure 5A, transfection with the miR-223 mimics suppressed the growth of xenografted NPC compared with NC (P < 0.05 on day 24). Likewise, SSRP1 knockdown notably inhibited the growth of xenografted NPC (P < 0.01 on day 24). Transfection with miR-223 mimics and SSRP1-siRNA-810 in combination showed the strongest inhibition of the xenografted NPC growth (P < 0.001 on day 24). The expressions of Ki67 and caspase-3 in the tumor tissues were commonly used to evaluate cell proliferation and apoptosis levels respectively, so this detected Ki67 and caspase-3 expressions in xenografted NPC using an immunohistochemistry assay. As indicated by Figure 5B, Ki67 expression was downregulated in groups in which cells were transfected with miR-223 mimics and SSRP1-siRNA-810, alone or in combination. However, caspase-3 expression was up-regulated in groups in which cells were transfected with miR-223 mimics and SSRP1-siRNA-810, alone or in combination (Figure 5C).
Figure 5.
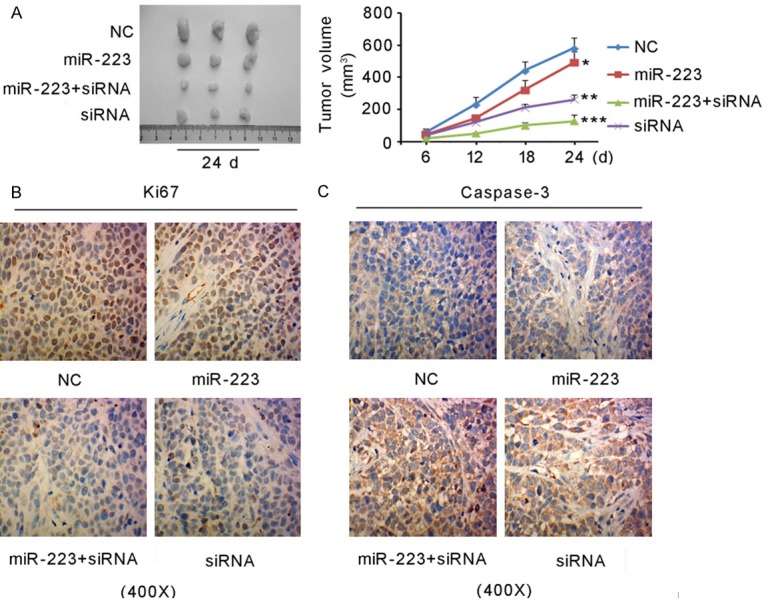
miR-223 inhibited the growth of xenografted NPC in nude mice by targeting SSRP1. The transfections of 5-8F cells were divided in four groups: Group A, transfection with negative control siRNA; Group B, transfection with miR-223 mimics; Group C, co-transfection with the miR-223 mimics and SSRP-siRNA-810; Group D, transfection with SSRP1-siRNA-810. The cells were then injected into nude mice to induce the generation of a xenografted tumor. A. tumor volume on days 6, 12, 18, and 24 after the injection. *P < 0.05, **P < 0.01, ***P < 0.001 versus and NC on day 24. B and C. An immunohistochemistry assay showed Ki67 and caspase-3 expression in the xenografted tumor.
Discussion
To date, multiple studies have observed the dysregulated expression of miRNAs in NPC, which are involved in NPC development and progression [4,30,31]. In this study, qRT-PCR demonstrated that the expression of miR-223 was significantly reduced in NPC cell lines. In addition, miR-223 expression was lowest in the highly metastatic NPC cell line (5-8F). These findings indicate that downregulation of miR-223 has an important role in the development and progression of NPC. miR-223 has been reported to be deregulated in various cancers [18-20]. miR-223 is up-regulated in some types of cancers, such as prostate cancer, colon cancer, and cutaneous squamous cell carcinoma [18-20]. And up-regulated miR-223 promotes cancer proliferation and metastasis [18-20]. However, miR-223 is also downregulated in breast cancer. miR-223 functions as a tumor suppressor in breast cancer by inhibiting stromal interaction molecule 1 [32]. The different roles of miR-223 in cancers are likely related to the different functions of the targets of miR-223.
The function of miR-223 in NPC has rarely been reported, so we explored the biological function of miR-223 in NPC. The ectopic expression of miR-223 significantly suppressed the proliferative and invasive capabilities of NPC cells in vitro. These results are consistent with observations in breast cancer [33], Lewis lung carcinoma [34] and cutaneous T-cell lymphoma [35]. The dysregulation of miR-223 has been proven to regulate tumor cell proliferation, apoptosis, migration and invasion, suggesting pivotal biological and pathological functions of miR-223. The present study demonstrates that miR-223 suppresses NPC cell proliferation and invasion, indicating an important role of miR-223 in the development and progression of NPC.
qRT-PCR and Western blot analysis demonstrated that SSRP1 is upregulated in NPC cell lines. In addition, we identified that SSRP1 is a novel direct target of miR-223 in NPC. Restoring the expression of miR-223 significantly reduces the expression of SSRP1. This means that SSRP1 expression levels in NPC cell lines are notably influenced by miR-223. A previous study found that miR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells [36]. The present study additionally discovered that the knockdown of SSRP1 phenocopies the suppressive effects of miR-223 on cell proliferative and invasion in NPC cells. These results indicate that SSRP1 is a functional target of miR-223, and that SSRP1 involves in miR-223-regulated NPC cell proliferation and invasion. In a previous study, it was found that silencing SSRP1 exerts an inhibitory effect on the proliferation and malignancy of human glioma cells via modulating the MAPK signaling pathway [37]. In addition, SSRP1 contributes to the malignancy of hepatocellular carcinoma and is negatively regulated by miR-497 [38]. There is more evidence indicating that SSRP1 meditates DNA repair mechanisms by SSRP1/PARP/XRCC1 and SSRP1/Ets-1/Pim-3 cascades, resulting in cancer resistance to ionizing radiation and DNA-targeting anti-cancer drugs [39,40].
EMT, a dynamic and reversible cellular process, is characterized by loss of cell polarity and intracellular junctions and the acquisition of mesenchymal features, which contributes to tumor development and metastasis [41]. Here, we determined the expression of an epithelial marker, E-cadherin, and a mesenchymal marker, vimentin, and N-cadherin in HCC cells by altering the expression of miR-223 and SSRP1. Interestingly, we demonstrated that miR-223 mimics and SSRP1 siRNA inhibited EMT and were associated with reduced expression of E-cadherin and the elevated expression of N-cadherin and vimentin in NPC cells. Taken together, miR-223 suppress the EMT of NPC cells by targeting SSRP1.
In the present study, we found that miR-223 was downregulated in NPC cell lines. The ectopic expression of miR-223 suppressed NPC cell growth, invasion and EMT in vitro. Furthermore, SSRP1 was identified as a direct target of miR-223. The upregulation of miR-223 may suppress the expression of SSRP1 and thus suppress the proliferation of NPC cells in vitro. Taken together, these results suggest that the downregulation of miR-223 may have an important role in the development and progression of NPC.
Acknowledgements
The present study was supported by grants from the National Science Fund of Wuhan (No. 201161038340-09) and the National Science Fund of Hubei Province (No. 201161038340-09).
Disclosure of conflict of interest
None.
References
- 1.Li YQ, He QM, Ren XY, Tang XR, Xu YF, Wen X, Yang XJ, Ma J, Liu N. MiR-145 inhibits metastasis by targeting fascin actin-bundling protein 1 in nasopharyngeal carcinoma. PLoS One. 2015;10:e0122228. doi: 10.1371/journal.pone.0122228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhen Y, Liu Z, Yang H, Yu X, Wu Q, Hua S, Long X, Jiang Q, Song Y, Cheng C, Wang H, Zhao M, Fu Q, Lyu X, Chen Y, Fan Y, Liu Y, Li X, Fang W. Tumor suppressor PDCD4 modulates miR-184-mediated direct suppression of C-MYC and BCL2 blocking cell growth and survival in nasopharyngeal carcinoma. Cell Death Dis. 2013;4:e872. doi: 10.1038/cddis.2013.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ye Y, Zhou Y, Zhang L, Chen Y, Lyu X, Cai L, Lu Y, Deng Y, Wang J, Yao K, Fang W, Cai H, Li X. EBV-miR-BART1 is involved in regulating metabolism-associated genes in nasopharyngeal carcinoma. Biochem Biophys Res Commun. 2013;436:19–24. doi: 10.1016/j.bbrc.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T, Sun Q, Liu T, Chen J, Du S, Ren C, Liao G, Yuan Y. MiR-451 increases radiosensitivity of nasopharyngeal carcinoma cells by targeting ras-related protein 14 (RAB14) Tumour Biol. 2014;35:12593–9. doi: 10.1007/s13277-014-2581-x. [DOI] [PubMed] [Google Scholar]
- 5.Huang GL, Chen ML, Li YZ, Lu Y, Pu XX, He YX, Tang SY, Che H, Zou Y, Ding C, He Z. Association of miR-146a gene polymorphism with risk of nasopharyngeal carcinoma in the centralsouthern Chinese population. J Hum Genet. 2014;59:141–4. doi: 10.1038/jhg.2013.135. [DOI] [PubMed] [Google Scholar]
- 6.Chua DT, Ma J, Sham JS, Mai HQ, Choy DT, Hong MH, Lu TX, Min HQ. Long-term survival after cisplatin-based induction chemotherapy and radiotherapy for nasopharyngeal carcinoma: a pooled data analysis of two phase III trials. J. Clin. Oncol. 2005;23:1118–24. doi: 10.1200/JCO.2005.12.081. [DOI] [PubMed] [Google Scholar]
- 7.Lai SZ, Li WF, Chen L, Luo W, Chen YY, Liu LZ, Sun Y, Lin AH, Liu MZ, Ma J. How does intensity-modulated radiotherapy versus conventional two-dimensional radiotherapy influence the treatment results in nasopharyngeal carcinoma patients? Int J Radiat Oncol Biol Phys. 2011;80:661–8. doi: 10.1016/j.ijrobp.2010.03.024. [DOI] [PubMed] [Google Scholar]
- 8.Li Z, Lei H, Luo M, Wang Y, Dong L, Ma Y, Liu C, Song W, Wang F, Zhang J, Shen J, Yu J. DNA methylation downregulated mir-10b acts as a tumor suppressor in gastric cancer. Gastric Cancer. 2015;18:43–54. doi: 10.1007/s10120-014-0340-8. [DOI] [PubMed] [Google Scholar]
- 9.Xiao X, Tang C, Xiao S, Fu C, Yu P. Enhancement of proliferation and invasion by MicroRNA-590-5p via targeting PBRM1 in clear cell renal carcinoma cells. Oncol Res. 2013;20:537–44. doi: 10.3727/096504013X13775486749335. [DOI] [PubMed] [Google Scholar]
- 10.Yin WZ, Li F, Zhang L, Ren XP, Zhang N, Wen JF. Down-regulation of microRNA-205 promotes gastric cancer cell proliferation. Eur Rev Med Pharmacol Sci. 2014;18:1027–32. [PubMed] [Google Scholar]
- 11.Yang X, Ni W, Lei K. miR-200b suppresses cell growth, migration and invasion by targeting Notch1 in nasopharyngeal carcinoma. Cell Physiol Biochem. 2013;32:1288–98. doi: 10.1159/000354527. [DOI] [PubMed] [Google Scholar]
- 12.Liu Z, Mai C, Yang H, Zhen Y, Yu X, Hua S, Wu Q, Jiang Q, Zhang Y, Song X, Fang W. Candidate tumour suppressor CCDC19 regulates miR-184 direct targeting of C-Myc thereby suppressing cell growth in non-small cell lung cancers. J Cell Mol Med. 2014;18:1667–79. doi: 10.1111/jcmm.12317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Q, Wang Y, Lu X, Zhao Z, Zhu L, Chen S, Wu Q, Chen C, Wang Z. MiR-125b regulates epithelial-mesenchymal transition via targeting Sema4C in paclitaxel-resistant breast cancer cells. Oncotarget. 2015;6:3268–79. doi: 10.18632/oncotarget.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gong B, Hu H, Chen J, Cao S, Yu J, Xue J, Chen F, Cai Y, He H, Zhang L. Caprin-1 is a novel microRNA-223 target for regulating the proliferation and invasion of human breast cancer cells. Biomed Pharmacother. 2013;67:629–36. doi: 10.1016/j.biopha.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Raimondo M, Guha S, Chen J, Diao L, Dong X, Wallace MB, Killary AM, Frazier ML, Woodward TA, Wang J, Sen S. Circulating micrornas in pancreatic juice as candidate biomarkers of pancreatic cancer. J Cancer. 2014;5:696–705. doi: 10.7150/jca.10094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duan HF, Li XQ, Hu HY, Li YC, Cai Z, Mei XS, Yu P, Nie LP, Zhang W, Yu ZD, Nie GH. Functional elucidation of miR-494 in the tumorigenesis of nasopharyngeal carcinoma. Tumour Biol. 2015;36:6679–89. doi: 10.1007/s13277-015-3356-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lu J, He ML, Wang L, Chen Y, Liu X, Dong Q, Chen YC, Peng Y, Yao KT, Kung HF, Li XP. MiR-26a inhibits cell growth and tumorigenesis of nasopharyngeal carcinoma through repression of EZH2. Cancer Res. 2011;71:225–33. doi: 10.1158/0008-5472.CAN-10-1850. [DOI] [PubMed] [Google Scholar]
- 18.Wei Y, Yang J, Yi L, Wang Y, Dong Z, Liu Z, Ouyang S, Wu H, Zhong Z, Yin Z, Zhou K, Gao Y, Yan B, Wang Z. MiR-223-3p targeting SEPT6 promotes the biological behavior of prostate cancer. Sci Rep. 2014;4:7546. doi: 10.1038/srep07546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang J, Luo X, Li H, Yue X, Deng L, Cui Y, Lu Y. MicroRNA-223 functions as an oncogene in human colorectal cancer cells. Oncol Rep. 2014;32:115–20. doi: 10.3892/or.2014.3173. [DOI] [PubMed] [Google Scholar]
- 20.Xu N, Zhang L, Meisgen F, Harada M, Heilborn J, Homey B, Grandér D, Ståhle M, Sonkoly E, Pivarcsi A. MicroRNA-125b down-regulates matrix metallopeptidase 13 and inhibits cutaneous squamous cell carcinoma cell proliferation, migration, and invasion. J Biol Chem. 2012;287:29899–908. doi: 10.1074/jbc.M112.391243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, Sun L, Ji G, Shi Y, Han Z, Han S, Nie Y, Chen X, Zhao Q, Ding J, Wu K, Daiming F. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011;9:824–33. doi: 10.1158/1541-7786.MCR-10-0529. [DOI] [PubMed] [Google Scholar]
- 22.Xu J, Yao Q, Hou Y, Xu M, Liu S, Yang L, Zhang L, Xu H. MiR-223/Ect2/p21 signaling regulates osteosarcoma cell cycle progression and proliferation. Biomed Pharmacother. 2013;67:381–6. doi: 10.1016/j.biopha.2013.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Wong QW, Lung RW, Law PT, Lai PB, Chan KY, To KF, Wong N. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–69. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 24.Wu L, Li H, Jia CY, Cheng W, Yu M, Peng M, Zhu Y, Zhao Q, Dong YW, Shao K, Wu A, Wu XZ. MicroRNA-223 regulates FOXO1 expression and cell proliferation. FEBS lett. 2012;586:1038–43. doi: 10.1016/j.febslet.2012.02.050. [DOI] [PubMed] [Google Scholar]
- 25.Ma J, Fang B, Zeng F, Ma C, Pang H, Cheng L, Shi Y, Wang H, Yin B, Xia J, Wang Z. Down-regulation of miR-223 reverses epithelial-mesenchymal transition in gemcitabine-resistant pancreatic cancer cells. Oncotarget. 2015;6:1740–9. doi: 10.18632/oncotarget.2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yarnell AT, Oh S, Reinberg D, Lippard SJ. Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J Biol Chem. 2001;276:25736–41. doi: 10.1074/jbc.M101208200. [DOI] [PubMed] [Google Scholar]
- 27.Koman IE, Commane M, Paszkiewicz G, Hoonjan B, Pal S, Safina A, Toshkov I, Purmal AA, Wang D, Liu S, Morrison C, Gudkov AV, Gurova KV. Targeting FACT complex suppresses mammary tumorigenesis in Her2/neu transgenicmice. Cancer Prev Res (Phila) 2012;5:1025–35. doi: 10.1158/1940-6207.CAPR-11-0529. [DOI] [PubMed] [Google Scholar]
- 28.Glaser R, Zhang HY, Yao KT, Zhu HC, Wang FX, Li GY, Wen DS, Li YP. Two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein-Barr virus that were derived from nasopharyngeal carcinomas. Proc Natl Acad Sci U S A. 1989;86:9524–8. doi: 10.1073/pnas.86.23.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu B, Che W, Xue J, Zheng C, Tang K, Zhang J, Wen J, Xu Y. SIRT4 prevents hypoxia-induced apoptosis in H9c2 cardiomyoblast cells. Cell Physiol Biochem. 2013;32:655–62. doi: 10.1159/000354469. [DOI] [PubMed] [Google Scholar]
- 30.Zhu X, Wang Y, Sun Y, Zheng J, Zhu D. MiR-155 up-regulation by LMP1 DNA contributes to increased nasopharyngeal carcinoma cell proliferation and migration. Eur Arch Otorhinolaryngol. 2014;271:1939–45. doi: 10.1007/s00405-013-2818-0. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Z, Ma J, Luan G, Kang L, Su Y, He Y, Luan F. MiR-506 suppresses tumor proliferation and invasion by targeting FOXQ1 in nasopharyngeal carcinoma. PLoS One. 2015;10:e0122851. doi: 10.1371/journal.pone.0122851. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 32.Yang Y, Jiang Z, Ma N, Wang B, Liu J, Zhang L, Gu L. MicroRNA-223 targeting STIM1 inhibits the biological behavior of breast cancer. Cell Physiol Biochem. 2018;45:856–866. doi: 10.1159/000487180. [DOI] [PubMed] [Google Scholar]
- 33.Pinatel EM, Orso F, Penna E, Cimino D, Elia AR, Circosta P, Dentelli P, Brizzi MF, Provero P, Taverna D. miR-223 is a coordinator of breast cancer progression as revealed by bioinformatics predictions. PLoS One. 2014;9:e84859. doi: 10.1371/journal.pone.0084859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nian W, Ao X, Wu Y, Huang Y, Shao J, Wang Y, Chen Z, Chen F, Wang D. miR-223 functions as a potent tumor suppressor of the Lewis lung carcinoma cell line by targeting insulin-like growth factor-1 receptor and cyclin-dependent kinase 2. Oncol Lett. 2013;6:359–66. doi: 10.3892/ol.2013.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGirt LY, Adams CM, Baerenwald DA, Zwerner JP, Zic JA, Eischen CM. miR-223 regulates cell growth and targets proto-oncogenes in mycosis fungoides/cutaneous T-cell lymphoma. J Invest Dermatol. 2014;134:1101–7. doi: 10.1038/jid.2013.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang W, Lan X, Li D, Li T, Lu S. miR-223 targeting MAFB suppresses proliferation and migration of nasopharyngeal carcinoma cells. BMC Cancer. 2015;15:461. doi: 10.1186/s12885-015-1464-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liao J, Tao X, Ding Q, Liu J, Yang X, Yuan FE, Yang JA, Liu B, Xiang GA, Chen Q. SSRP1 silencing inhibits the proliferation and malignancy of human glioma cells via the MAPK signaling pathway. Oncol Rep. 2017;38:2667–2676. doi: 10.3892/or.2017.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding Q, He K, Luo T, Deng Y, Wang H, Liu H, Zhang J, Chen K, Xiao J, Duan X, Huang R, Xia Z, Zhou W, He J, Yu H, Jiao X, Xiang G. SSRP1 contributes to the malignancy of hepatocellular carcinoma and is negatively regulated by miR-497. Mol Ther. 2016;24:903–14. doi: 10.1038/mt.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, Li C, Wei L, Teng Y, Nakajima S, Chen X, Xu J, Leger B, Ma H, Spagnol ST, Wan Y, Dahl KN, Liu Y, Levine AS, Lan L. SSRP1 cooperates with PARP and XRCC1 to facilitate single-strand DNA break repair by chromatin priming. Cancer Res. 2017;77:2674–2685. doi: 10.1158/0008-5472.CAN-16-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ai J, Li W, Zeng R, Xie Z, Liu H, Hou M, Tan G. Blockage of SSRP1/Ets-1/Pim-3 signalling enhances chemosensitivity of nasopharyngeal carcinoma to docetaxel in vitro. Biomed Pharmacother. 2016;83:1022–1031. doi: 10.1016/j.biopha.2016.08.022. [DOI] [PubMed] [Google Scholar]
- 41.Kan H, Guo W, Huang Y, Liu D. MicroRNA-520g induces epithelial-mesenchymal transition and promotes metastasis of hepatocellular carcinoma by targeting SMAD7. FEBS Lett. 2015;589:102–9. doi: 10.1016/j.febslet.2014.11.031. [DOI] [PubMed] [Google Scholar]