Abstract
Melanoma, a malignant tumor of melanocytes, is considered to be the most aggressive of skin cancers and its incidence keeps increasing worldwide. miR-134 and CTHRC1 have been demonstrated to be involved in the occurrence and development of various tumors. However, their roles are still elusive in the progression of melanoma. qRT-PCR and western blot (WB) were used to examine the expressions of miR-134 and CTHRC1 in clinical specimens of melanoma patients and melanoma cell lines. Dual-luciferase reporter assay was applied to verify the target interaction between miR-134 and CTHRC1. The mRNA and protein expressions of CTHRC1 were measured by qRT-PCR and WB after treatment by miR-134 inhibitor and mimic. Subsequently, CCK8, colony formation assay, and flow cytometry were utilized to assess the influences of miR-134 and CTHRC1 on cell growth of melanoma. Cell migration and invasion experiments were performed to evaluate the effects of miR-134 and CTHRC1 on metastasis of melanoma. It was shown that CTHRC1 was up-regulated and miR-134 was down-regulated in melanoma patients and cell lines. CTHRC1 was demonstrated to be a direct target of miR-134. Ultimately, we also found that up-regulated miR-134 expression and down-regulated CTHRC1 expression could suppress cell proliferation and cell colony formation, promote apoptosis, delay the cell cycle, and hinder cell migration and invasion. Our findings suggest that miR-134 could inhibit the growth and metastasis of melanoma by negatively regulating CTHRC1.
Keywords: miR-134, CTHRC1, melanoma, cell metastasis, cell migration
Introduction
Melanoma is the most lethal form of skin cancer and is the second leading cancer in adolescents and young adults aged 15-29 years [1]. Historically, melanoma was a rare cancer, but in the last 50 years, the annual incidence has increased as rapidly as 4~6% in many regions of fair-skinned populations, such as New Zealand, Northern Europe, North America and Australia, which is faster than any other cancer [2]. Currently, surgery remains the main and definitive treatment for early-stage melanoma to significantly enhance the proportion and duration of objective responses and to provide extended prolongation of patient survival [3]. However, melanoma, as a highly aggressive malignancy, easily tends to metastasize beyond its primary site, thereby making it difficult to treat the advanced stages of melanoma [4,5]. Although there are 2 FDA-approved chemotherapy drugs (i.e., fotemustine and dacarbazine) which were used in the past for the treatment of metastatic melanoma, they are still not satisfactory curative approaches to improve the survival rate and ameliorate life quality of patients [6]. Therefore, seeking new high sensitivity and specificity biomarkers that run through the steps of initiation, promotion, and progression of melanoma is an urgent need for establishment of early screening or identification strategies and in-depth investigating the molecular basis, pathogenesis, and biological features of melanoma occurrence and development are also of utmost importance for better selection of new effective agents for targeted treatment. Accumulating evidence has demonstrated that epigenetic events, including methylation of the promoter regions, histone modification, chromatin remodeling, the positioning of nucleosomes, and non-coding RNAs (ncRNAs) aberrant changes, were closely associated with the initiation and progression of melanoma [7,8]. Hence, in this study, we primarily focused on the genesis mechanism of melanoma.
microRNAs (miRNAs) are characterized as a group of short (generally 18-22 nucleotides in length), endogenous, single stranded, and non-protein-coding RNA molecules that are involved in the post-transcriptional regulation of gene expression and exert significant roles in various human physiological and pathological conditions [9]. By base-pairing with imperfect complementary sites in the 3’-untranslated region (3’-UTR) of the message RNA (mRNA), miRNAs modulate target genes by translational inhibition and mRNA destabilization [10]. Given the tremendous impact of miRNA-guided gene regulation on almost all aspects of cellular processes, such as cell development, differentiation, proliferation, apoptosis, cell fate determination, and signal transduction, it is not surprising that abnormal miRNA expression is intimately related to the molecular mechanisms of many human tumors, including bladder cancer, lung cancer, hepatocellular carcinoma, gastric cancer, and melanoma [11]. Initial data on miRNA expression in malignant melanoma samples were included in an article published by Lu et al who showed the miRNA expression profiles to reflect the developmental lineage and differentiation state of melanoma [12]. Subsequently, Zhang et al found that miR-610 functioned as a tumor suppressor in inhibiting tumor growth of melanoma by targeting lipoprotein receptor-related protein 6 (LRP6), which might represent a novel potential therapeutic target and prognostic marker for melanoma [13]. Cirilo et al verified that miR-195 acted as an anti-proliferative miRNA in human melanoma cells by negatively regulating Prohibitin 1 [14]. Thus, these publications supported that miRNAs play a crucial role in melanoma cancer development.
Recent research has manifested that miR-134 is essential for human carcinoma and participates in tumor cell proliferation, apoptosis, metastasis, drug resistance, as well as cancer diagnosis, treatment, and prognosis [15]. For example, Su et al demonstrated that miR-134 could target Kirsten rat sarcoma viral oncogene homolog (KRAS) to suppress breast cancer cell proliferation, migration, and invasion [16]; Qin et al found that miR-134 could restrain non-small cell lung cancer (NSCLC) growth by targeting the epidermal growth factor receptor (EGFR) [17]. However, the detailed function of miR-134 in melanoma is not elucidated. In our previous study, we predicted the potential targets of miR-134 by TargetScan and miRDB online software. We discovered that collagen triple helix repeat containing-1 (CTHRC1) might be a target gene of miR-134. Nevertheless, CTHRC1, mainly expressed in adventitial fibroblasts and neointimal smooth muscle cells of balloon-injured vessels, has been confirmed to enhance the migratory and invasive ability of various tumor cells [18]. For instance, Wang et al revealed that CTHRC1 in a hepatocellular carcinoma cell line activated cell migration and invasiveness in vitro, and promoted tumor metastasis in a lung metastasis mouse model [19]. Therefore, the functional roles of miR-134 and CTHRC1 were investigated in melanoma growth and metastasis in the present study.
Materials and methods
Patients and tissue samples
The study was performed following the guidelines set forth in the Declaration of Helsinki. All patients with histologically confirmed melanoma gave their informed consent, and experiments were approved by the Clinical Management Committee of Binzhou Medical University Hospital. Tissue samples were obtained from melanoma patients (7 males, 11 females; age range, 47-67 years) that underwent surgical resection at the Department of Oncology of Binzhou Medical University Hospital between January 2016 and December 2017. In addition, 18 matched adjacent non-tumorous normal tissues 5 cm away from the cancer lesion were also collected from these patients. None of the patients received any pre-operative chemotherapy or radiotherapy. All tissue samples were immediately frozen in liquid nitrogen and stored at -80°C for further examination by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) assay and Western blot (WB) analysis for detection of CTHRC1 and miR-134.
Cancer cell lines and culture
The melanoma cell lines (BT549, MB-231, MB-486, MCF7 and SK-BR-3) and 293T cell line used in the present study were supplied by American Type Culture Collection (ATCC). All cells were maintained in a medium containing Dulbecco’s modified Eagle’s medium (DMEM, USA), 10% fetal bovine serum (FBS; Gibco, USA), and 80 U/mL penicillin and 80 µg/mL streptomycin under an incubator at 37°C, with 90% humidity and 5% CO2.
The medium was replaced every 2-3 days, and the melanoma cell lines were digested with 0.25% trypsin/EDTA solution (Invitrogen, USA) and passaged. The third passage was selected for the examination of CTHRC1 and miR-134 expression using qRT-PCR and WB. However, the 293T cells were mainly applied to the dual-luciferase reporter assay.
RNA extraction and qRT-PCR
Total RNA containing miRNA was isolated from clinical specimens and cultured cells using TRIzol Reagent (Tiangen, China) following the manufacturer’s protocols. The concentration and purity of RNA was determined spectrophotometrically. The target gene (i.e., CTHRC1) and miRNA (i.e., miR-134) were reverse transcribed to complementary DNA (cDNA) with M-MLV reverse transcriptase Kit (Promega, USA) and the miRcute miRNA First-Strand cDNA Synthesis Kit (TIANGEN, China) as recommended by the manufacturer, respectively, using 200 ng of RNA from each sample. Then, real-time PCR was carried out using SYBR Green I Fluorescent Assay kit (TAKARA, Japan) and quantified in an ABI PRISM® 7500 Sequence Detection System (Applied Biosystems, USA). The temperature protocol for real-time PCR was as follows: pre-denaturation at 95°C for 2 min, then 40 cycles: 95°C, 15 s; 60°C for 32 s, with data collection after each cycle, followed by a melting curve. The sequences of primers used for this study were including: CTHRC1, forward: 5’-ATAATGGAATGTGCTTACAAGG-3’ and reverse: 5’-TTCCCAAGATCTATGCCATAAT-3’; GAPDH (as an internal control for GTHRC1), forward: 5’-GAGAAGGCTGGGGCTCATTT-3’ and reverse: 5’-AGTGATGGCATGGACTGTGG-3’ (reverse); U6 (as a loading control for miR-134), forward, 5’-GCGCGTCGTGAAGCGTTC-3’; universal reverse primer, 5’-GTGCAGGGTCCGAGGT-3’. Normalized to the corresponding endogenous control, the relative quantification values of CTHRC1 and miR-134 were computed by the 2-ΔΔCt method.
Preparation of cell extracts and analysis by WB
Total protein was extracted from clinical specimens and cell lines samples using a radioimmunoprecipitation (RIPA) buffer (Beyotime, China) and collected following centrifugation at 13000 rmp at 4°C for 20 min. The protein concentration was measured using a BCA kit (Thermo Fisher Scientific, USA) in accordance with the manufacturer’s instructions. A total of 2 µg protein/per lane was separated by 10% sodium dodecyl sulfate-polyacrylamide gels for electrophoresis (SDS-PAGE) followed by transfer onto a polyvinylidene difluoride (PVDF) membrane (Thermo Fisher Scientific, USA). The membranes were blocked with 5% nonfat dry milk suspended in Tris-buffered saline containing 0.1% Tween-20 (TBST) for 1 h at room temperature and then incubated with primary antibodies against CTHRC1 (1:1000 dilution; Abcam, USA) and GAPDH (as an endogenous control, 1:500 dilution; Abcam, USA) overnight at 4°C. extensive washing with TBST, the membranes were further incubated with horseradish peroxidase (HRP)-conjugated anti-immunoglobulin G (1:5000 dilution; Zhongshan Gold Bridge Biological Technology Co., China) for 2 h at room temperature. Finally, the bands were visualized using an enhanced chemiluminescence kit (Beyotime, China) and the images were captured by an ChemiDoc™ XRS imaging system (Bio-Rad Laboratories, USA). The relative protein expression levels were analyzed by Quantity One software version 4.62 (Bio-Rad Laboratories, USA).
Dual-luciferase reporter assay
The human CTHRC1 3’-UTR oligonucleotides containing the wild-type (WT) or mutant (Mut) miR-134 binding sites were designed according to the target interaction prediction between CTHRC1 and miR-134 and then synthesized by Ribobio Co. (Guangzhou, China). The synthesized fragments were inserted into the downstream of the firefly luciferase reporter genes in the psiCHECK-2 plasmid (Promega, USA) between the Xho I and Not I sites to generate the recombinant vectors WT-CTHRC1 and Mutant-CTHRC1 and the insertions were verified by DNA sequencing. For dual-luciferase reporter assay, 293T cells were seeded in 96-well culture plates and cultured for 24 h and transfected with the WT or Mut CTHRC1 3’-UTR construct along with miR-134 mimic, miR-134 inhibitor, negative control (NC) plasmid, or NC inhibitor using Lipofectamine 2000 (Thermo Fisher Scientific, USA) following the manufacturer’s recommendation. Firefly and renilla luciferase activities were determined at 48 h post-transfection using the Dual-Luciferase Reporter Assay kit (Promega).
Cell counting Kit-8 (CCK8) test
The CCK8 test was used according to the manufacturer’s manual to evaluate the proliferation of MB-231 cells which were treated with NC plasmid, miR-134 mimic, CTHRC1 over-expression plasmid and CTHRC1-siRNA purchased from Sangon Biotech, Shanghai, China. Briefly, the transfected MB-231 cells were seeded in 96-well plates at a density of 5.0×104 cells/well. At the indicated time points (0 day, 1 day, 2 day and 3 day), the cells were incubated with 10 μl CCK-8 reagent at 37°C for another 2 h. Eventually, the optical density (OD) was measured at an absorbance wavelength of 450 nm on an enzyme immunoassay analyzer (Bio-Rad, USA). The experiments were performed in triplicate.
Colony formation assay
MB-231 cells were plated into 35 mm plates at a density of 3000 cells. When cells were reached 80% confluence, NC plasmid, miR-134 mimic, CTHRC1 over-expression plasmid, and CTHRC1-siRNA were transfected into cells with Lipofectamine 2000 following the manufacturer’s recommendation. The culture medium was replaced with fresh DMEM every 3 days and two weeks later, colonies were fixed with 4% paraformaldehyde (Solarbio, China) and stained with 1% crystal violet (Sigma, USA) for 30 min. The number of clearly visible colonies (diameter > 50 µm) in three random microscopic fields were counted, and the images were captured under a microscope and analyzed using Image J software.
Flow cytometry
Approximately 5×105 MB-231 cells per well were plated in 6-well plates and cells grown to 80% confluence were transfected with NC plasmid, miR-134 mimic, CTHRC1 over-expression plasmid and CTHRC1-siRNA. After 48 h incubation, the cells with different treatments were harvested for cell apoptosis and cell cycle measurement by flow cytometry.
Annexin V FITV/propidium iodide (PI) double staining was conducted to determine cell apoptosis. The collected cells were washed with PBS at 4°C twice and added 250 μl Annexin V binding buffer followed by thorough mixing. Then, the cells were stained with 5 μl of AnnexinV-FITC and 5 μl of PI per sample for 15 minutes at room temperature in the dark. Finally, samples were analyzed within 1 h on a FACScan flow cytometer. In the apoptosis scatter diagram, the left lower quadrant (Annexin V-negative/PI-negative) was recorded as normal cells; the right lower quadrant (Annexin V-positive/PI-negative) was considered as early apoptotic cells; the right upper quadrant (Annexin V-positive/PI-positive) was scored as advanced cell apoptosis and necrosis; and the left upper quadrant (Annexin V-negative/PI-positive) was regarded as dead cells.
Cell cycle was analyzed by staining with PI. The collected cells were similarly washed with cold PBS and fixed with 1 ml ice-cold 75% ethanol at -20°C overnight. After centrifugation at 3000 rpm for 5 min, the supernatant was removed and cells were resuspended in 0.5 ml PI for 30 min incubation. The samples were examined with a fluorescence activated cell sorting (FACS) analyzer FACSVerse (BD Biosciences, USA) and the DNA content distribution curve was obtained by measuring the DNA content of each cell in the population.
Cell invasion and migration measurement
Cell invasion and migration were studied using transwell inserts (8-μm pore filter, 24-well cell clusters; Millipore, USA). For cell invasion experiment, approximately 2×105 MB-231 cells were seeded in the upper chamber with 2 ml serum free DMEM culture and transfected with NC plasmid, miR-134 mimic, CTHRC1 over-expression plasmid or CTHRC1-siRNA, whereas fresh medium containing 20% FBS was plated in the lower chambers as a chemoattractant. After 24 h of transfection at 37°C with 5% CO2, the upper chamber was removed, the medium was discarded and the non-invasive cells attached to on the apical side of each transwell membranes were carefully scraped off with a sterile cotton swab. The invasive cells on the other side of the membrane were fixed with 4% paraformaldehyde at room temperature for 30 min, stained with 0.5% crystal violet (dissolved in 10% acetic acid) for 15 min, rinsed with PBS three times and dried at 80°C for 30 min. Finally, the number of invasive cells went through Matrigel were obtained from five randomly selected fields per membrane under a light microscope (×200 magnification; Nikon, Japan). However, for cell migration experiment, assays were conducted as above except that the MB-231 cells in each group was adjusted to 1×105 cells/well and cultivated on top of uncoated (Matrigel-free) filters, and the other procedures were similar to invasion experiment.
Statistical analysis
Statistical testing was conducted with the assistance of Statistical Product and Service Solutions (SPSS) 17.0 software (IBM SPSS, USA). All of the results are depicted as mean ± standard deviation (SD) from at least three biological repeated experiments. Differences between 2 groups were analyzed via the Student’s t-test and differences among > 2 groups were analyzed using one-way analysis of variance followed by post hoc Tukey analysis. A two-sided P value of less than 0.05 was considered a significant difference.
Results
The different expression of CTHCR1 and miR-134 in carcinoma tissues and para-carcinoma tissues of melanoma patients
To investigate the role of CTHCR1 and miR-134 during the development of melanoma, we first compared the mRNA and protein levels of CTHCR1 between carcinoma tissues and para-carcinoma tissues of melanoma patients. As shown in Figure 1A, 1B, CTHCR1 mRNA and protein expressions in carcinoma tissues were expressed significantly higher than that in para-carcinoma tissues. However, the expression of miR-134 in carcinoma tissues was lower than that in para-carcinoma tissues (Figure 1C). These results indicated that dysregulated expressions of CTHCR1 and miR-134 might play an important role in progression of melanoma.
Figure 1.
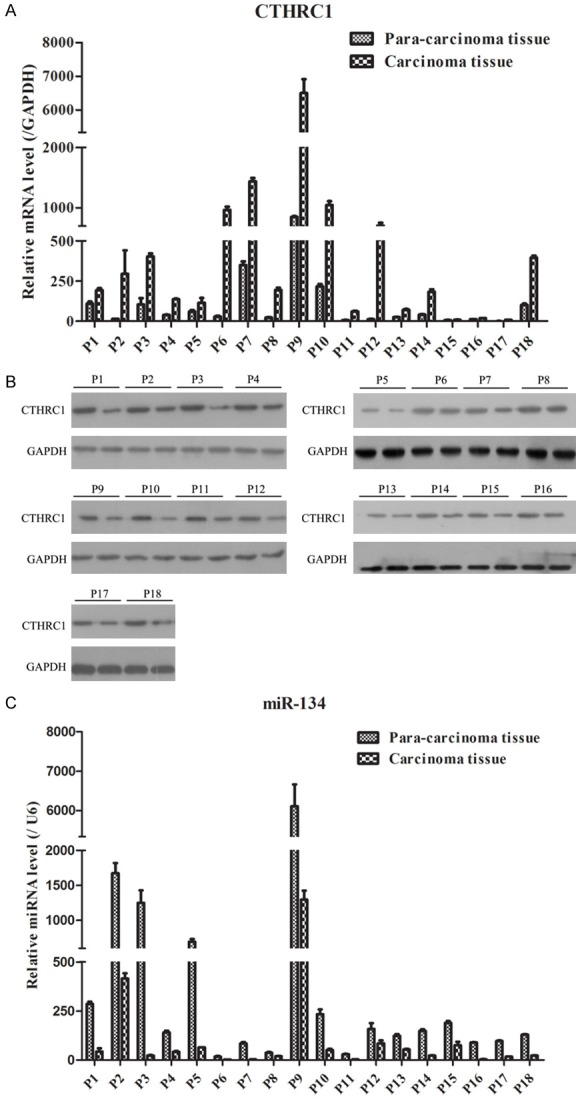
CTHRC1 and miR-134 expression in tumor tissues of melanoma patients. A. qRT-PCR examination showed that CTHRC1 mRNA expression was remarkably increased in carcinoma tissues. B. WB showed that CTHRC1 protein expression was markedly elevated in carcinoma tissues. C. qRT-PCR analysis revealed that miR-134 was significantly decreased in carcinoma tissues.
CTHCR1 and miR-134 expressions in human melanoma cell lines
In order to further confirm the above results, we subsequently examined the expressions of CTHCR1 and miR-134 under an in vitro level. Five human melanoma cell lines were harvested to measure. As illustrated in Figure 2A, 2B, it was found that the mRNA and protein levels of CTHRC1 were lowest in MB-231 cells and highest in BT549 cells. Nevertheless, the miR-134 expression presented the highest in MB-231 cells and lowest in BT549 cells. Therefore, these data suggested that miR-134 might negatively regulate CTHCR1.
Figure 2.
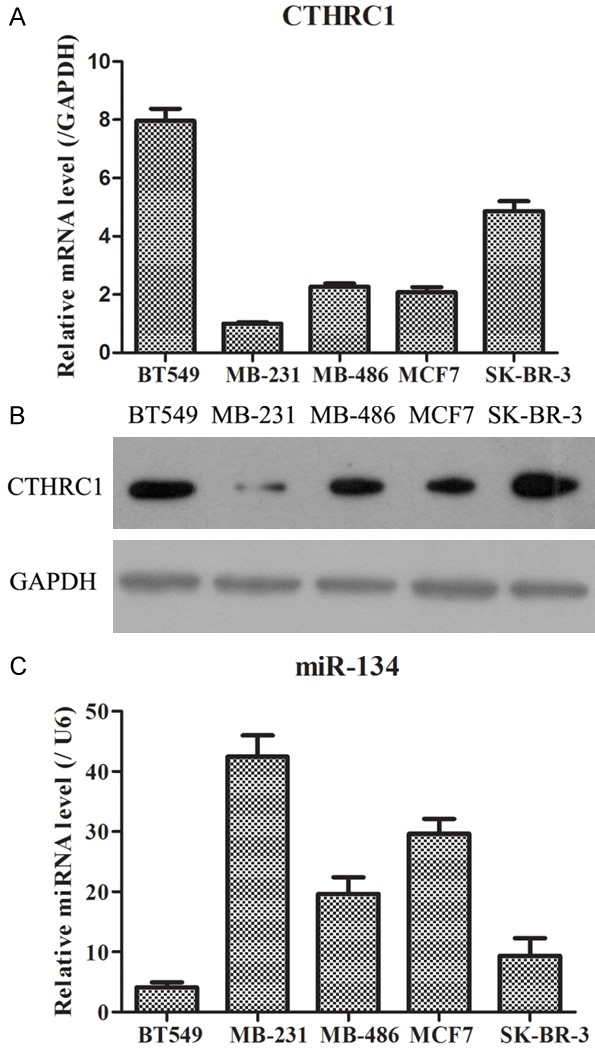
CTHRC1 and miR-134 expression in melanoma cell lines. A. Relative mRNA expression of CTHRC1 in five melanoma cell lines, as determined using qRT-PCR. B. Relative protein expression of CTHRC1 in five melanoma cell lines, as identified by WB. C. Relative expression of miR-134 in five melanoma cell lines, as measured by qRT-PCR.
CTHCR1 might be the direct target of miR-134
In order to determine the fundamental molecular mechanisms of CTHCR1 and miR-134 in melanoma cells, we performed a literature search for representative target genes of miR-134 through bioinformatic algorithms. As a result, CTHCR1 was shown to a be potential target site of miR-134 (Figure 3A). The complementary sequences between CTHRC1 and miR-134 were displayed and according to these sequences, we also constructed the WT-CTHRC1 3’-UTR and Mut-CTHRC1 3’-UTR vector. Furthermore, the data of dual-luciferase reporter system exhibited that the luciferase activity was notably decreased in WT-CTHRC1 vector transfected with miR-155-5p mimics compared to the other 4 groups, while the luciferase activity was not markedly changed in Mutant-CTHRC1 vector transfected with the 5 plasmids (Figure 2A). Thus, this result implied that CTHCR1 may be the direct target of miR-134. To confirm this prediction, we checked the influence of miR-134 on the expression of CTHRC1 by detecting the mRNA and protein levels after transfecting miR-134 inhibitor and mimics. It was observed that knockdown and overexpression of miR-134 could apparently promote and inhibit the expression of CTHRC1 in mRNA and protein levels. Thereby, these results demonstrated that miR-134a could directly regulate CTHCR1 by targeting its 3’-UTR.
Figure 3.
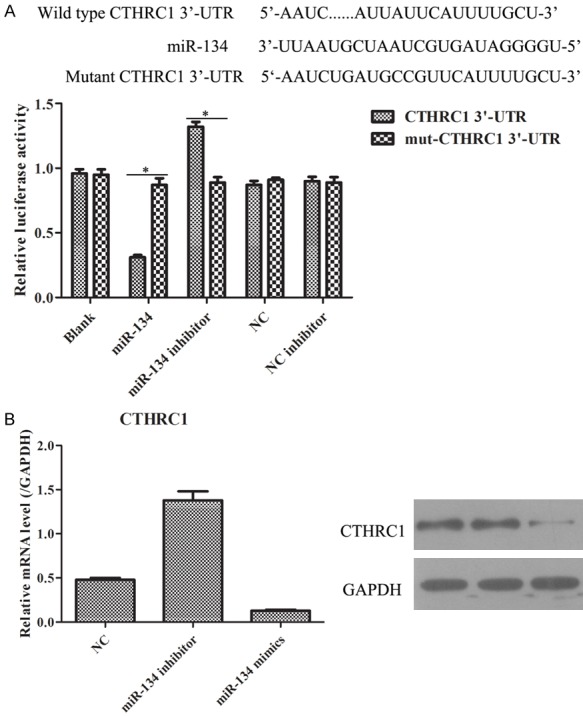
CTHRC1 acts as a direct target downstream of miR-134. A. Schematic diagram of the presumed binding site of miR-134 and on the 3’-UTR of CTHRC1 is shown on the upper panel, and the result of dual-luciferase assay is shown on the lower panel. *P < 0.05. B. The mRNA and protein expression levels of CTHRC1 were detected by qRT-PCR and WB.
miR-134 suppressed the growth of MB-231 cells by negatively regulating CTHRC1
In order to further investigate the biological function of miR-134 and its target CTHRC1, we assessed the ability of cell growth in MB-231 cells treated with NC plasmid, miR-134 mimic, CTHRC1 overexpression plasmid, or CTHRC1-siRNA. The MTT assay revealed that upregulation of CTHRC1 remarkably enhanced the proliferation of MB-231 cells as compared with NC group, whereas there were no significant differences in OD450 values between miR-134 mimic and CTHRC1-siRNA group (Figure 4A). Additionally, colony formation assay also uncovered that CTHRC1 over-expression elevated the formation of colonies (Figure 4B). Meanwhile, increased miR-134 and decreased CTHRC1 expressions reduced the formation of colonies (Figure 4B). Hence, these results implied that miR-134 could inhibit MB-231 cells growth by targeting CTHRC1.
Figure 4.
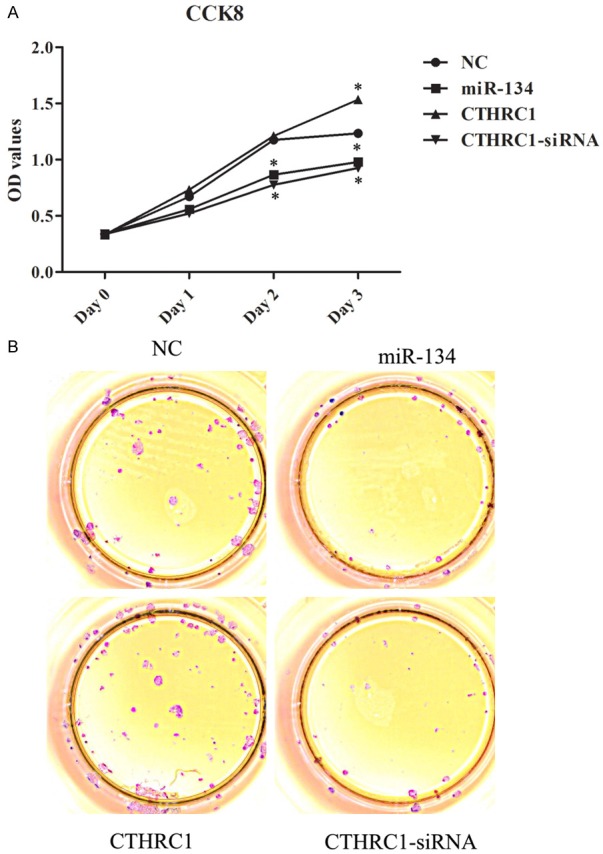
The influences of miR-134 and CTHRC1 in cell growth of MB-231 cells. A. Transfection with miR-134 mimics or CTHRC1-siRNA notably reduced the cell proliferation of MB-231 cells. *P < 0.05. B. Transfection with miR-134 mimics or CTHRC1-siRNA conspicuously reduced colony formation of MB-231 cells.
miR-134 promoted MB-231 cell apoptosis and induced cell cycle arrest in G1/G0 phase, probably by CTHCR1
To confirm the effects of miR-134 on cell apoptosis and cell cycle in MB-231 cells, flow cytometry was carried out. Our data showed that the late apoptosis (Annexin V+/PI+) was remarkably unregulated in cells transfected with miR-134 mimics or CTHCR1-siRNA plasmid in comparison with NC and CTHCR1 groups (Figure 5A). Furthermore, cell cycle analysis presented that the percentage of cells in G1 phase was increased from 62.38% to 78.05% and 77.12% in cells transfected with miR-134 mimics and CTHCR1-siRNA plasmid, respectively, when compared with NC group, whereas the percentage of cells in G1 phase was reduced from 62.38% to 60.34% in cells transfected with CTHCR1-overexpression plasmid (Figure 5B). Together with the above confirmation in this study that CTHCR1 may be a direct target of miR-134, these findings indicated the involvement of miR-130a in mediating cell apoptosis and G1 cell cycle arrest, probably by targeting CTHCR1.
Figure 5.
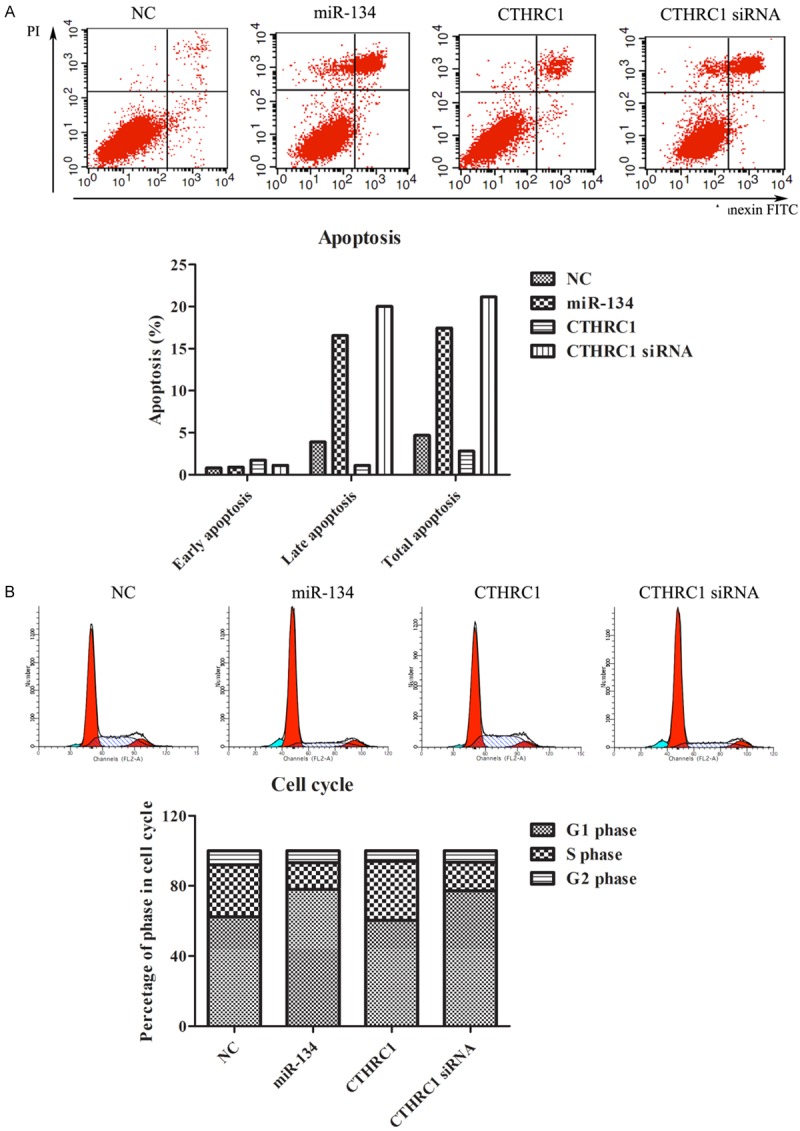
The effects of miR-134 and CTHRC1 on apoptosis and cell cycle of MB-231 cells. A. Overexpression of miR-134 or knockdown of CTHRC1 promoted apoptosis of MB-231 cells. B. Overexpression of miR-134 or knockdown of CTHRC1 enhanced cell cycle arrest in G1/G0 phase.
miR-134 limited the migratory and invasive ability of MB-231 cells by targeting CTHCR1
In order to further elucidate whether miR-134 may regulate MB-231 cells migration and invasion through CTHCR1, transwell assays were carried out in this study. As presented in Figure 6A, 6B, the number of migratory and invasive cells in miR-134 group and CTHCR1-siRNA group was dramatically less than those in NC group, but the number of migratory and invasive cells in CTHRC1 overexpression group was sharply greater than those in NC group. Collectively, these results implied that the regulatory mechanism of miR-134 in MB-231 cell metastasis might be by targeting CTHRC1.
Figure 6.
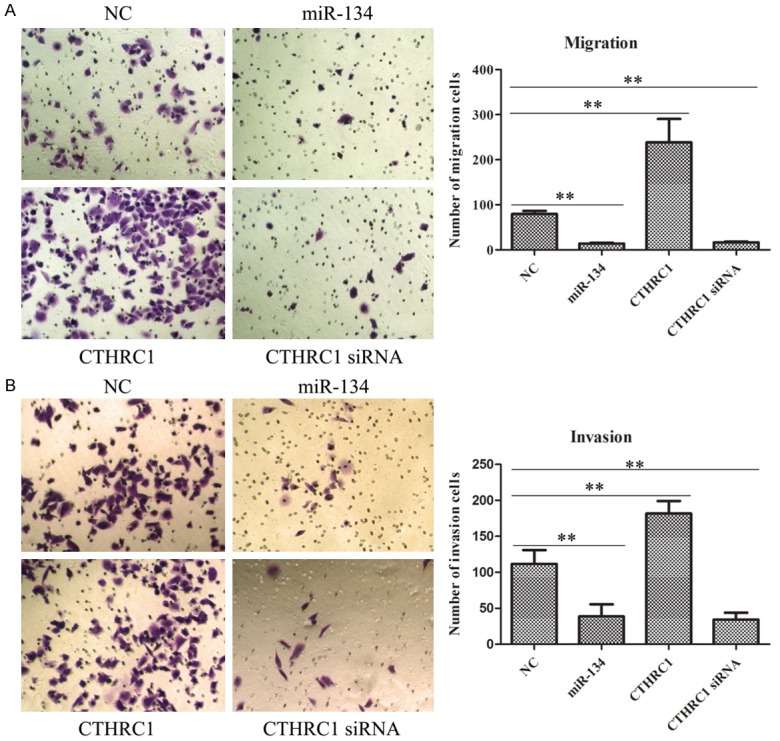
The roles of miR-134 and CTHRC1 in cell migration and invasion of MB-231 cells. A. Up-regulation of miR-134 or down-regulation of CTHRC1 dramatically suppressed the migratory ability of MB-231 cells. *P < 0.05. B. Up-regulation of miR-134 or down-regulation of CTHRC1 sharply inhibited the invasive ability of MB-231 cells. *P < 0.05.
Discussion
Melanoma, one of the most lethal human skin malignancies at present, represents a worldwide public health dilemma [20]. Moreover, it has had a relatively steady increase, a high metastatic potential, and poor prognosis in the past decades [21]. Thus, it is important to find novel biomarkers to develop effective therapeutic strategies against this malignancy. Previous studies showed CTHRC1 was overexpressed in many cancers, including melanoma, to contribute to tumor formation [18]. Nevertheless, to date, the detailed molecular mechanism remains unclear. Additionally, recent work has revealed the existence of miRNAs, which have critical functions across various biological processes, such as cell growth, proliferation, apoptosis, metastasis, cell cycle progression, and differentiation [9]. Furthermore, evidence has been provided in various studies highlighting the strong correlation between miR-134 and tumor progression [15]. For example, miR-134 promoted p21 expression and inhibited cyclin D1, cyclin D2, and CDK4 protein expression in SPC-A1 and A549 cells, which suggested that miR-134 could suppress NSCLC cell proliferation [22]. However, the role of CTHRC1 and miR-134 in melanoma are still unknown. Therefore, in this study, we firstly examined the expressions of CTHRC1 and miR-134 in vivo and in vitro levels. The results exhibited that CTHRC1 and miR-134 expressions were significantly increased and decreased, respectively. Moreover, the highest CTHRC1 expression and lowest miR-134 expression occurred in MB-231 cells which were chosen as the following experiment cells. Subsequently, based on the inverse expression of CTHRC1 and miR-134, dual-luciferase assay was used to detect the interaction between CTHRC1 and miR-134. The data revealed that there may be a target relationship between CTHRC1 and miR-134. Furthermore, the knockdown and overexpression of miR-134 could up-regulate and down-regulate CTHRC1 expression, respectively, in mRNA and protein levels. Hence, these data further verified that CTHRC1 was the direct target of miR-134.
Accumulating research has reported that miR-134 functioned as an intrinsic suppressor in various cancers [15]. For instance, upregulation of miR-134 inhibited the EGFR-correlated signal pathway in NSCLC and eventually leaded to cell proliferation arrest [17]. miR-134 was found to notably promote cell apoptosis in glioblastoma tumor [23]. Overexpressed miR-134 could also stop cell metastasis in endometrial tumor [24]. The related results of these studies were identical with our results which displayed that overexpression of miR-134 by transfecting with a miR-134 mimics in MB-231 cells could remarkably reduce the cell proliferation and the colonies formation, enhance the apoptosis rate, delay the cell cycle in G1/G0 phase, and hinder the cell migration and invasion. In addition, CTHRC1 has been shown to be involved in vascular remodeling, bone formation and morphogenesis; moreover, it also acts as a prognostic factor in many human aggressive tumors, including NSCLC, hepatocellular carcinoma, and ovarian cancer [18]. In this study, we applied the CTHRC1 mimics and CTHRC1-siRNA to transfect into MB-231 cells and further explored the effects of CTHRC1 on the cell growth and metastasis. Cell growth is strictly regulated by cell proliferation, and apoptosis, as well as the cell cycle [25]. Dysregulation of cell proliferation, apoptosis, and cell cycle originating from miRNA changes is often implicated in occurrence and development of cancer. Furthermore, metastasis, which is a key step in cancer progression, is usually the major cause of morbidity and mortality in cancer patients [26,27]. Thus, the examination of cell growth and metastasis could further evaluate the role of CTHRC1. However, it was found that knock-down of CTHRC1 expression had similar effects to the miR-134 mimics group in MB-231 cells on cell proliferation, colony formation, apoptosis, cell cycle, cell migration, and invasion. Nevertheless, these effects were mostly reversed in MB-231 cells with CTHRC1 over-expression treatment. This suggests the influence of CTHRC1 in cell growth and metastasis may be regulated by miR-134.
To sum up, in this paper, we showed for the first time that CTHRC1 and miR-134 were abundantly expressed in melanoma patients and cell lines. Moreover, the abnormal expression of the two markers was inverse. Additionally, it was also demonstrated that miR-134 could negatively interact with CTHRC1 to regulate cell biologic behavior, including cell proliferation, coloniy formation, apoptosis, cell cycle, and cell migration and invasion. Collectively, these results provide a new direction in melanoma diagnosis and prognosis, as well as therapeutic strategy.
Acknowledgements
This study was supported by Shandong Pro-vincial Key Research and Development Program (No 2016GSF201110).
Disclosure of conflict of interest
None.
References
- 1.Barr RD, Ries LA, Lewis DR, Harlan LC, Keegan TH, Pollock BH, Bleyer WA US National Cancer Institute Science of Adolescent and Young Adult Oncology Epidemiology Working Group. Incidence and incidence trends of the most frequent cancers in adolescent and young adult Americans, including “nonmalignant/noninvasive” tumors. Cancer. 2016;122:1000–8. doi: 10.1002/cncr.29867. [DOI] [PubMed] [Google Scholar]
- 2.Austin MT, Xing Y, Hayes-Jordan AA, Lally KP, Cormier JN. Melanoma incidence rises for children and adolescents: an epidemiologic review of pediatric melanoma in the United States. J Pediatr Surg. 2013;48:2207–13. doi: 10.1016/j.jpedsurg.2013.06.002. [DOI] [PubMed] [Google Scholar]
- 3.Marconcini R, Spagnolo F, Stucci LS, Ribero S, Marra E, Rosa F, Picasso V, Guardo LD, Cimminiello C, Cavalieri S, Orgiano L, Tanda E, Spano L, Falcone A, Queirolo P Italian Melanoma Intergroup(IMI) Current status and perspectives in immunotherapy for metastatic melanoma. Oncotarget. 2018;9:12452–12470. doi: 10.18632/oncotarget.23746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pappo AS. Melanoma in children and adolescents. Eur J Cancer. 2003;39:2651–61. doi: 10.1016/j.ejca.2003.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Saiyed FK, Hamilton EC, Austin MT. Pediatric melanoma: incidence, treatment, and prognosis, pediatric health, medicine and therapeutics. Pediatric Health Med Ther. 2017;8:39–45. doi: 10.2147/PHMT.S115534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daponte A, Signoriello S, Maiorino L, Massidda B, Simeone E, Grimaldi AM, Caraco C, Palmieri G, Cossu A, Botti G, Petrillo A, Lastoria S, Cavalcanti E, Aprea P, Mozzillo N, Gallo C, Comella G, Ascierto PA Southern Italy Cooperative Oncology. Phase III randomized study of fotemustine and dacarbazine versus dacarbazine with or without interferon-alpha in advanced malignant melanoma. J Transl Med. 2013;11:38. doi: 10.1186/1479-5876-11-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar D, Leung EY, Baguley BC, Finlay GJ, Askarian-Amiri ME. Epigenetic regulation in human melanoma: past and future. Epigenetics. 2015;10:103–21. doi: 10.1080/15592294.2014.1003746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fattore L, Costantini S, Malpicci D, Ruggiero CF, Ascierto PA, Croce CM, Mancini R, Ciliberto G. MicroRNAs in melanoma development and resistance to target therapy. Oncotarget. 2017;8:22262–22278. doi: 10.18632/oncotarget.14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wei F, Yang S, Wang S. MicroRNAs: a critical regulator under mechanical force. Histol Histopathol. 2018;33:335–342. doi: 10.14670/HH-11-924. [DOI] [PubMed] [Google Scholar]
- 10.Hao J, Duan FF, Wang Y. MicroRNAs and RNA binding protein regulators of microRNAs in the control of pluripotency and reprogramming. Curr Opin Genet Dev. 2017;46:95–103. doi: 10.1016/j.gde.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Ding J, Yang J, Guo X, Zheng Y. MicroRNA roles in the nuclear factor kappa B signaling pathway in cancer. Front Immunol. 2018;9:546. doi: 10.3389/fimmu.2018.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA, Downing JR, Jacks T, Horvitz HR, Golub TR. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–8. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 13.Zhang G, Ai D, Yang X, Ji S, Wang Z, Feng S. MicroRNA-610 inhibits tumor growth of melanoma by targeting LRP6. Oncotarget. 2017;8:97361–97370. doi: 10.18632/oncotarget.22125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cirilo PD, de Sousa Andrade LN, Correa BR, Qiao M, Furuya TK, Chammas R, Penalva LO. MicroRNA-195 acts as an anti-proliferative miRNA in human melanoma cells by targeting Prohibitin 1. BMC Cancer. 2017;17:750. doi: 10.1186/s12885-017-3721-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pan JY, Zhang F, Sun CC, Li SJ, Li G, Gong FY, Bo T, He J, Hua RX, Hu WD, Yuan ZP, Wang X, He QQ, Li DJ. miR-134: a human cancer suppressor? Mol Ther Nucleic Acids. 2017;6:140–149. doi: 10.1016/j.omtn.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su X, Zhang L, Li H, Cheng P, Zhu Y, Liu Z, Zhao Y, Xu H, Li D, Gao H, Zhang T. MicroRNA-134 targets KRAS to suppress breast cancer cell proliferation, migration and invasion. Oncol Lett. 2017;13:1932–1938. doi: 10.3892/ol.2017.5644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin Q, Wei F, Zhang J, Wang X, Li B. miR-134 inhibits non-small cell lung cancer growth by targeting the epidermal growth factor receptor. J Cell Mol Med. 2016;20:1974–83. doi: 10.1111/jcmm.12889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang N, Cui Y, Liu J, Zhu X, Wu H, Yang Z, Ke Z. Multidimensional roles of collagen triple helix repeat containing 1 (CTHRC1) in malignant cancers. J Cancer. 2016;7:2213–2220. doi: 10.7150/jca.16539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Lee M, Yu G, Lee H, Han X, Kim D. CTHRC1 activates pro-tumorigenic signaling pathways in hepatocellular carcinoma. Oncotarget. 2017;8:105238–105250. doi: 10.18632/oncotarget.22164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng CY, Yen H, Hsiao HY, Su SC. Phytochemicals in skin cancer prevention and treatment: an updated review. Int J Mol Sci. 2018:19. doi: 10.3390/ijms19040941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Belter B, Haase-Kohn C, Pietzsch J. Biomarkers in malignant melanoma: recent trends and critical perspective. In: Ward WH, Farma JM, editors. Cutaneous Melanoma: Etiology and Therapy. Brisbane (AU): Codon Publications; 2017. [PubMed] [Google Scholar]
- 22.Sun CC, Li SJ, Li DJ. Hsa-miR-134 suppresses non-small cell lung cancer (NSCLC) development through down-regulation of CCND1. Oncotarget. 2016;7:35960–35978. doi: 10.18632/oncotarget.8482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Y, Kim J, Mueller AC, Dey B, Yang Y, Lee DH, Hachmann J, Finderle S, Park DM, Christensen J, Schiff D, Purow B, Dutta A, Abounader R. Multiple receptor tyrosine kinases converge on microRNA-134 to control KRAS, STAT5B, and glioblastoma. Cell Death Differ. 2014;21:720–34. doi: 10.1038/cdd.2013.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao Y, Liu T, Huang Y. MicroRNA-134 suppresses endometrial cancer stem cells by targeting POGLUT1 and Notch pathway proteins. FEBS Lett. 2015;589:207–14. doi: 10.1016/j.febslet.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 25.Lynch MP, Nawaz S, Gerschenson LE. Evidence for soluble factors regulating cell death and cell proliferation in primary cultures of rabbit endometrial cells grown on collagen. Proc Natl Acad Sci U S A. 1986;83:4784–8. doi: 10.1073/pnas.83.13.4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown RB, Razzaque MS. Phosphate toxicity and tumorigenesis. Biochim Biophys Acta. 2018;1869:303–309. doi: 10.1016/j.bbcan.2018.04.007. [DOI] [PubMed] [Google Scholar]
- 27.Venkatesh V, Nataraj R, Thangaraj GS, Karthikeyan M, Gnanasekaran A, Kaginelli SB, Kuppanna G, Kallappa CG, Basalingappa KM. Targeting notch signalling pathway of cancer stem cells. Stem Cell Investig. 2018;5:5. doi: 10.21037/sci.2018.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]


