Abstract
Background: Circular RNAs (circRNAs) as a new class of non-coding RNAs that are associated with cancer progression and can serve as potential markers for cancer diagnosis. However, the functions of circRNAs have not been completely clarified in non-small-cell lung cancer (NSCLC). Our study aimed to explore the expression profiles and apoptotic role of circRNAs in NSCLC. Methods: Forty-one NSCLC patients and twenty-six healthy subjects were recruited to our study. The levels of hsa_circ_0102533 in tumor tissues and whole blood were identified by circRNA microarray and RT-qPCR. The CCK-8 and apoptosis assays were performed in NSCLC cell lines after they were transfected with si-circRNA and si-NC. Results: Compared with the control group, hsa_circ_0102533 expression was significantly increased in tumor tissues and whole blood from NSCLC patients. ROC analysis showed that hsa_circ_0102533 had a higher diagnostic power for the detection of cancer in stage I-II NSCLC patients [AUC: 0.774 (95% CI: 0.624-0.923)] than in stage III-IV NSCLC patients [0.728 (95% CI: 0.588-0.869)]. Furthermore, the knockout of hsa_circ_0102533 by siRNA significantly inhibited proliferation and induced apoptosis in NSCLC cell lines. Conclusion: Hsa_circ_0102533 possesses an oncogenic property in the carcinogenesis of NSCLC and might be an early diagnostic biomarker for NSCLC detection.
Keywords: Non-small cell lung cancer, circRNA, apoptosis, biomarker, early diagnosis
Introduction
Lung cancer, the most common cancer, is becoming increasingly common worldwide [1]. Non-small-cell lung cancer (NSCLC) is the most common type of lung cancer, accounting for approximately 85% of all lung cancer cases [2]. The 5-year overall survival rate of NSCLC patients is less than 15%, a result of a low ratio of early diagnosis, with nearly 75% of NSCLC patients diagnosed at stages III and IV [3,4]. Despite increased understanding of the disease’s molecular mechanisms and clinical characteristics, the 5-year overall survival rate has seen no significant improvement in the clinical management of NSCLC [5,6]. Emerging evidence shows that reliable biomarkers for the early detection of NSCLC may be a promising measurement for improving the prognosis of patients with NSCLC [7]. Therefore, exploring novel and specific biomarkers for the early detection of NSCLC is urgently needed.
Circular RNAs (circRNAs) represent a new class of non-coding RNAs and are widely expressed in the eukaryotic transcriptome [8,9]. CircRNAs are characterized by a covalently closed continuous loop by back-splicing, without 5’ to 3’ polarity or a polyadenylated tail [10], which endow circRNAs with a stable structure to counteract RNA exonucleolytic digestion, highlighting that circRNAs are strong candidates for novel molecular biomarkers for disease diagnosis. In fact, accumulating evidence suggests that circRNAs are stably present in the whole blood, serum, and plasma of mammals and can serve as novel, non-invasive biomarkers in various diseases, including major depressive disorder [11], pre-eclampsia [12], diabetes mellitus [13] and malignancies [14]. In lung cancer, a few circRNAs have been reported in carcinogenesis and tumor progression [15-17]. For example, overexpressed hsa_circ_0013958 is associated with lymphatic metastasis and TNM stage [15]. Hsa_circ_0007385 functions as an oncogene in NSCLC tumorigenesis by regulating the proliferation, migration and invasion of NSCLC cells [16]. Hsa_circ_0014130 is significantly up-regulated in NSCLC tissues and shows good diagnostic potential using the area under the receiver operating characteristic curve (AUC) [17]. All of these circRNAs have been suggested as potential non-invasive biomarkers for the early detection and screening of lung cancer [15-17]. However, the circRNA expression profiles in whole blood have not been clarified in NSCLC patients.
In the present study, circRNA microarrays were performed to investigate the differential expression profiles of circRNAs in whole blood and NSCLC tissues from NSCLC patients. The up-regulation of hsa_circ_0102533 in whole blood and NSCLC tissues was confirmed by a circRNA microarray and a real-time reverse transcription quantitative polymerase chain reaction (RT-qPCR). In addition, we also found that hsa_circ_0102533 is associated with the proliferation and apoptosis of NSCLC cells in vitro. We finally validated that hsa_circ_0102533, as an oncogene involved in NSCLC cell proliferation and apoptosis, could serve as a blood-based biomarker for NSCLC patient screening.
Material and methods
Patients and specimens
Forty-one NSCLC patients and twenty-six healthy subjects were recruited from the Second Hospital of Anhui Medical University (Anhui, China) from January 2015 to June 2017. Forty-one pairs of tumor tissues and matched adjacent non-tumorous tissues were collected from the NSCLC patients who had undergone surgery. At the same time, whole blood from Forty-one NSCLC patients and twenty-six healthy subjects was collected. All tumor tissue and whole blood samples were immediately stored at -80°C in an ultra-low temperature refrigerator. 10 ml of blood samples from NSCLC patients were collected with ethylenediaminetetraacetic acid (EDTA)-containing tubes (Becton, Dickinson and Company). Human samples were obtained with written informed consent from all patients. The study was approved by the Ethics Committee of the Second Hospital of Anhui Medical University (Anhui, China).
Cell culture
Normal human bronchial epithelial cells (NHBE) and five NSCLC cell lines (A549, H1299, H1792, SK-MES-1 and SPC-A1) were purchased from the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China) and maintained in Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (Thermo Scientific HyClone, Beijing, China), 100 U/ml penicillin and 100 mg/ml streptomycin in a humidified incubator (Thermo, USA), 5% CO2, 95% air atmosphere.
Human circRNA microarray analysis
Total RNA was extracted from three pairs of tumor tissues and matched adjacent non-tumorous tissues from the NSCLC patients, as well as in whole blood from three NSCLC patients and three healthy subjects. CircRNA was enriched with Rnase R to digest linear RNA (Epicentre, Madison, WI, USA). RNA was labelled with Arraystar Human circRNA Array (8×15 K, Arraystar, Rockville, MD, USA) and was scanned using a Agilent Scanner G2505C (Jamul, CA, USA).
Real-time reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
RNA was extracted using TRIzol (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA) according to the manufacturer’s instructions. Moloney Murine Leukemia Virus reverse transcriptase (Promega Corporation, Madison, WI, USA) was used to synthesize cDNA. Divergent primers were designed to amplify the head-to-tail splicing of circRNA using ABI7300 System (Applied Biosystems, Foster City, CA, USA) with SYBR Select Master Mix (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized to normalize the expression of hsa_circ_0102533. The PCR primers were used in this study as follows: hsa_circ_0102533, forward 5’-CCGACCTGTGAAATTCTGGGA-3’ and reverse 5’-GCAGGCTGCAATACTGTGAAG-3’; GAPDH, forward 5’-GCACCGTCAAGCTGAGAAC-3’ and reverse 5’-TGGTGAAGACGCCAGTGGA-3’. The 2-ΔΔCt method was used to calculate the expression of hsa_circ_0102533 [18].
Small interfering RNA transfection
The small interfering RNA (si-RNA) was utilized for cell transfection and was synthesized by RiboBio (Guangzhou, China). The targeted sequence of si-RNA for hsa_circ_0102533 (si-0102533) was 5’-AATCCAGAGTGAGCGATTACA-3’. Cells were transfected with si-0102533 for 48 h at 37°C using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol.
Cell viability detection by CCK-8
A CCK-8 assay was performed as previously described [19]. After being transfected with si-0102533 or si-NC, the human NSCLC cells (1×104) were seeded in a 96-well plate for 24 hours, 48 hours, and 72 hours, and cell viability was measured using a CCK-8 Cell Proliferation/Viability Assay Kit (Dojindo Japan). Absorbance was recorded at 450 nm using Elx800 Reader (Bio-Tek Instruments Inc., Winooski, VT, USA).
Flow cytometry for apoptosis
NSCLC Cell apoptosis assay was determined as previously described [20]. After being transfected with si-0102533 or si-NC for 48 hours, a cell apoptosis assay was performed by flow cytometry analysis. An Annexin V-FITC apoptosis detection kit was purchased from Invitrogen (Carlsbad, Calif, USA). The samples were analyzed using a flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA).
Caspase-3 activity assay
The NSCLC cells (2×106) were lysed using a NP-40 buffer (Beyotime Institute of Biotechnology, Haimen, China) at ice-bath conditions for 15 min and centrifuged at 12000 g for 10 min at 4°C, and then the supernatant was collected. The levels of caspase-3 in the supernatant were measured using a caspase-3 kit (Beyotime Institute of Biotechnology, Haimen, China). The absorbance was measured at 405 nm using an ELISA reader (MD SpectraMax M5; Molecular Devices, LLC, Sunnyvale, CA, USA) according to the manufacturer’s protocol.
Statistical analysis
Data were presented as the mean ± SEM. The statistical analysis was performed using IBM SPSS Statistics Version 19.0 (SPSS Inc., Chicago, IL, USA) and GraphPad Prism Version 7.0 (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t-test was used to analyze two-group differences. Inter-group differences were analyzed by one-way analysis of variance, followed by a post hoc Tukey test for multiple comparisons. Receiver operating characteristic (ROC) curves and the area under the ROC curve (AUC) were used to assess the ability of using whole blood hsa_circ_0102533 as diagnostic tools for NSCLC. Spearman’s rank analysis was used to identify the correlation of hsa_circ_0102533 levels between tumor tissues and whole blood. A Bland-Altman plot (difference plot) was used to analyze the agreement of hsa_circ_0102533 levels between tumor tissues and whole blood. A P value less than 0.05 was considered to indicate a statistically significant difference.
Results
CircRNAs expression profiles in tumor tissues or whole blood from NSCLC patients or healthy controls
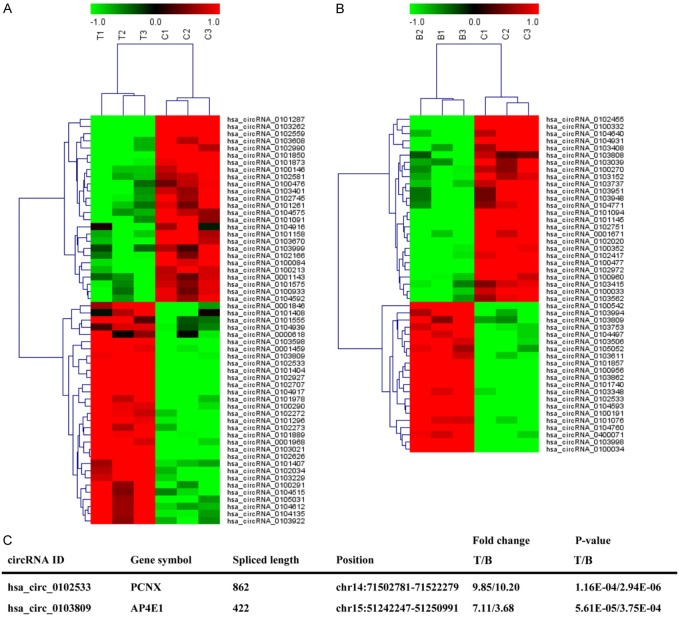
First, a human circRNA microarray analysis was used to analyze circRNA expression profiles in three pairs of NSCLC tumor tissues and corresponding nontumorous tissues as well as in the whole blood from three NSCLC patients and healthy controls. FDR ≤ 0.01 and |Log2fold change| ≥ 1 were performed as the criteria in filtering the abnormally expressed circRNAs, which were selected as candidate biomarkers for NSCLC patient detection. The results revealed that 57 circRNAs (31 up-regulated and 26 down-regulated) were differentially expressed in NSCLC tumor tissues compared with the corresponding nontumorous tissues (Figure 1A). In addition, 47 circRNAs were significantly differentially expressed in the whole blood from NSCLC patients and healthy controls, among which 21 circRNAs and 26 circRNAs were up-regulated and down-regulated, respectively (Figure 1B). Intriguingly, hsa_circ_0102533 and hsa_circ_0103809 (Figure 1C) were significantly increased in both tumor tissues and whole blood from NSCLC patients. Based on the fold change and P value, we focused on hsa_circ_0102533 in our study. We also found that a high expression of hsa_circ_0102533 was correlated with tumor type, TNM stages, lymph nodes metastasis and distant metastasis or recurrence but not correlated with gender, age, or tumor size (Table 1).
Figure 1.
CircRNAs expression profiles in tumor tissues and whole blood from NSCLC patients. The heat map represents significant differentially expressed circRNAs in three pairs of NSCLC tumor tissues and corresponding nontumorous tissues (A) or in whole blood from three NSCLC patients and three healthy controls (B). The characteristics of hsa_circ_0102533 and hsa_circ_0103809 are shown (C).
Table 1.
The associations between clinicopathological factors and hsa_circ_0102533 expression levels in the tumor tissues from NSCLC patients
| Variable | n | Low expression (n = 14) | High expression (n = 27) | P value |
|---|---|---|---|---|
| Age (Years) | 0.447 | |||
| ≤ 60 | 23 | 9 | 14 | |
| > 60 | 18 | 5 | 13 | |
| Gender | 0.443 | |||
| Male | 26 | 10 | 16 | |
| Female | 15 | 4 | 11 | |
| Tumor size (cm) | 0.771 | |||
| ≤ 3 | 12 | 5 | 7 | |
| > 3 | 29 | 9 | 20 | |
| Tumor type | 0.011 | |||
| Squamous cell carcinoma | 18 | 10 | 8 | |
| Adenocarcinoma | 23 | 4 | 19 | |
| TNM stages | 0.010 | |||
| I-II | 14 | 9 | 5 | |
| III-IV | 27 | 5 | 22 | |
| Distant metastasis or recurrence | 0.021 | |||
| Negative | 22 | 11 | 11 | |
| Positive | 19 | 3 | 16 | |
| Lymph nodes metastasis | 0.001 | |||
| Negative | 14 | 10 | 4 | |
| Positive | 27 | 4 | 23 |
Hsa_circ_0102533 as a candidate biomarker for NSCLC detection
To confirm the microarray data of hsa_circ_0102533 levels in tumor tissues and blood from NSCLC patients, hsa_circ_0102533 expression was identified by RT-qPCR analysis in forty-one pairs of NSCLC tumor tissues and their corresponding nontumorous tissues as well as in the whole blood from forty-one NSCLC patients and twenty-six healthy controls. The results demonstrated that hsa_circ_0102533 was markedly up-regulated in both tumor tissues (Figure 2A) and whole blood (Figure 2B) in NSCLC patients compared with the control group. Whole blood levels of hsa_circ_0102533 were significantly increased in stage I-II and stage III-IV NSCLC patients as compared to the healthy controls (Figure 2C). However, the expression of hsa_circ_0102533 had no obvious difference between stage I-II and stage III-IV NSCLC patients, suggesting that hsa_circ_0102533 expression in the blood was irrelevant to the TNM stage. To confirm whether hsa_circ_0102533 could serve as a blood-based tumor marker for NSCLC detection and validate its accuracy, a correlation analysis was performed between whole blood and tumor tissues from NSCLC patients. The expression levels of hsa_circ_0102533 were strongly correlated in whole blood and tumor tissues from NSCLC patients (r = 0.697, P < 0.001; Figure 2D). In addition, a Bland-Altman agreement analysis showed that the mean difference of hsa_circ_0102533 expression between whole blood and tumor tissues was 1.06 ± 2.26, indicating that hsa_circ_0102533 expression in whole blood was acceptable as blood-based tumor markers for NSCLC screening.
Figure 2.
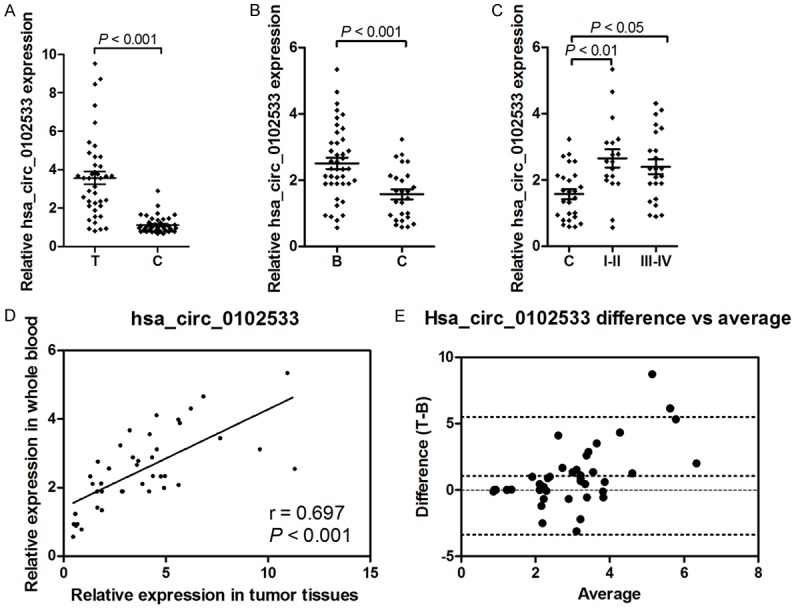
The expression of hsa_circ_0102533 was verified by RT-qPCR in tumor tissues and whole blood from NSCLC patients. The expression of hsa_circ_0102533 was measured by RT-qPCR in forty-one pairs of NSCLC tumor tissues and corresponding nontumorous tissues (A), as well as in the whole blood from forty-one NSCLC patients and twenty-six healthy controls (B). The expression of hsa_circ_0102533 was measured by RT-qPCR in twenty-six healthy controls, eighteen stage I-II and twenty-three stage III-IV NSCLC patients (C). Spearman’s rank analysis was used to identify the correlation of hsa_circ_0102533 expression between tumor tissues and whole blood from NSCLC patients (D). A Bland-Altman agreement analysis showed the mean difference of hsa_circ_0102533 expression between whole blood and tumor tissues (E).
Diagnostic significance of hsa_circ_0102533 in whole blood from NSCLC patients
To evaluate the performance of hsa_circ_0102533 in discriminating NSCLC patients from healthy subjects, the receiver operating characteristics (ROC) curves and the area under the ROC curves (AUC) were performed in forty-one NSCLC patients and twenty-six healthy subjects. The AUC for hsa_circ_0102533 was 0.744 [95% confidence interval (CI): 0.622-0.867; P = 0.001] in separating NSCLC patients from healthy subjects (Figure 3A). The AUC for hsa_circ_0102533 was 0.774 (95% CI: 0.624-0.923) in separating stage I-II NSCLC patients from healthy subjects (Figure 3B). The AUC for hsa_circ_0102533 was 0.728 (95% CI: 0.588-0.869) in separating stage III-IV NSCLC patients from healthy subjects (Figure 3C). These results indicated that hsa_circ_0102533 provided an effective diagnosis performance for the detection of NSCLC patients. Interestingly, hsa_circ_0102533 had a more effective diagnosis performance for detection of stage I-II NSCLC patients, suggesting that hsa_circ_0102533 might be an early detection marker for NSCLC patients.
Figure 3.
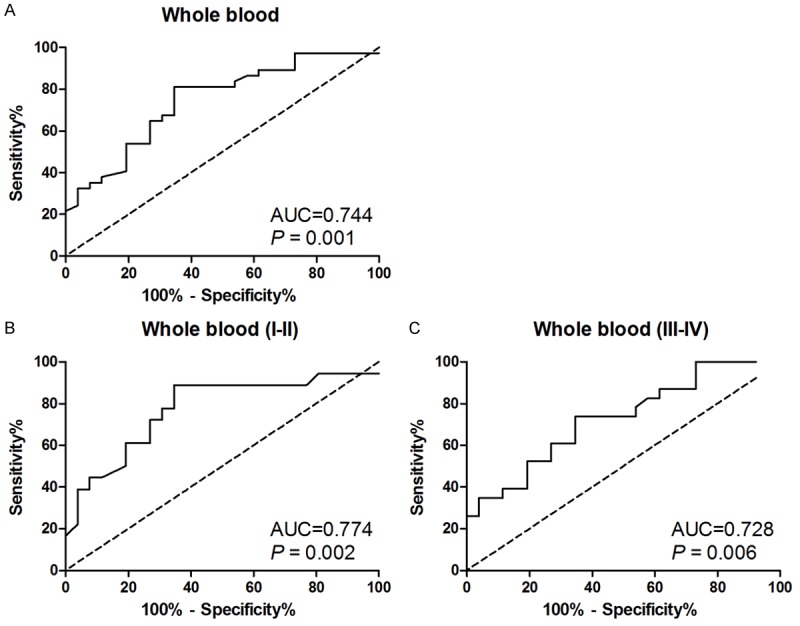
Evaluation of hsa_circ_0102533 for NSCLC diagnosis. Receiver operating characteristics (ROC) curves were drawn with the data of hsa_circ_0102533 expression in whole blood from forty-one NSCLC patients and twenty-six healthy controls (A). ROC curves were drawn with the data of hsa_circ_0102533 expression in whole blood from eighteen stage I-II NSCLC patients and twenty-six healthy controls (B). ROC curves were drawn with the data of hsa_circ_0102533 expression in whole blood from twenty-three stage III-IV NSCLC patients and twenty-six healthy controls (C).
Silencing of hsa_circ_0102533 inhibits proliferation and induces apoptosis in NSCLC cell lines
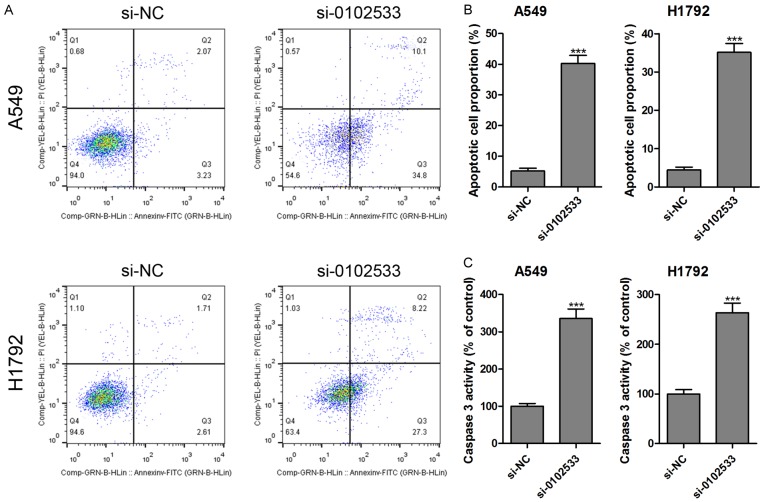
Based on above results, we found that hsa_circ_0102533 might be an oncogene involved in the progression and development of NSCLC. Therefore, we investigated the effect of hsa_circ_0102533 on proliferation and apoptosis in vitro. First, the expression of hsa_circ_0102533 was measured in NHBE cells and five NSCLC cell lines (A549, H1299, H1792, SK-MES-1 and SPC-A1), and the results showed that hsa_circ_0102533 was dramatically up-regulated in all of the NSCLC cell lines compared with the NHBE cells (Figure 4A). Hsa_circ_0102533 was higher in A549 and H1792 cell lines than in other cell lines (Figure 4A). Hence, the following experiments were performed using A549 and H1792 cells. After being transfected with hsa_circ_0102533 small interfering (si) RNA (si-0102533), the expression of hsa_circ_0102533 was significantly inhibited in A549 and H1792 cells (Figure 4B). More importantly, cell proliferation was significantly suppressed in si-0102533 transfected A549 (Figure 4C) and H1792 (Figure 4D) cells compared with si-NC transfection at 48 h and 72 h. Furthermore, the apoptotic cell proportion (Figure 5A and 5B) and caspase-3 activity (Figure 5C) were significantly increased in si-0102533 transfected A549 and H1792 cells compared with si-NC transfection.
Figure 4.
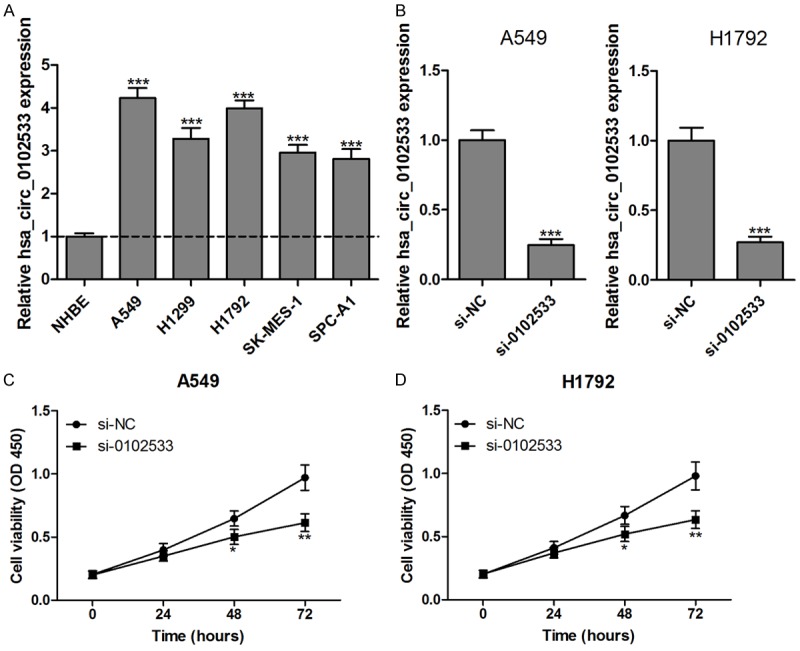
Hsa_circ_0102533 knockout inhibits proliferation in NSCLC cells. The levels of hsa_circ_0102533 in NHBE cells and five NSCLC cell lines (A549, H1299, H1792, SK-MES-1 and SPC-A1) were detected by RT-qPCR assay (A). Hsa_circ_0102533 expression was significantly reduced in A549 and H1792 cells after being transfected with si-0102533 (B). After being transfected with si-0102533 or si-NC, the cell viability was monitored by a CCK-8 assay in the A549 (C) and H1792 (D) cells. *P < 0.05; **P < 0.01; ***P < 0.001. n = 3 in each group.
Figure 5.
Hsa_circ_0102533 knockout induces apoptosis in NSCLC cells. After being transfected with si-0102533 or si-NC, cell apoptosis was analyzed using flow cytometry in A549 and H1792 cells (A and B), and the levels of caspase-3 in the supernatant of the A549 and H1792 cells were measured by caspase-3 kit (C). ***P < 0.001. n = 3 in each group.
Discussion
Mounting evidence suggests that non-coding RNAs, microRNAs, long non-coding RNAs, and circRNAs are detectable in body fluids, including whole blood, plasma, serum, and urine and can function as tools for cancer diagnosis [21-23]. CircRNAs as the potential stable molecular markers in disease diagnosis may be superior to microRNAs and long non-coding RNA, which we attribute to their stable structure, abundant expression, and tissue specificity [14].
In our study, NSCLC-related circRNA screening was performed based on different expression profiling between NSCLC tumor tissues and corresponding nontumorous tissues, as well as whole blood from NSCLC patients and healthy controls by human circRNA microarray analysis. The results demonstrated that the levels of hsa_circ_0102533 were significantly higher in both tumor tissues and whole blood from NSCLC patients than that of the controls. We further measured hsa_circ_0102533 expression in whole blood from NSCLC patients and healthy subjects by RT-qPCR, and the results showed that hsa_circ_0102533 was significantly increased in the whole blood from NSCLC patients compared with healthy subjects, providing strong evidence that hsa_circ_0102533 could be released into the blood and might be used as diagnostic marker for NSCLC patients.
On the other hand, hsa_circ_0102533 provided a higher diagnostic ability for the detection of stage I-II NSCLC patients [AUC: 0.774 (95% CI: 0.624-0.923)] than stage III-IV NSCLC patients [0.728 (95% CI: 0.588-0.869)], suggesting that whole blood hsa_circ_0102533 could serve as an early tumor marker for NSCLC detection.
Hsa_circ_0102533 is located in chr14:71502781-71522279, the spliced sequence length is 862 nucleotides, and its associated-gene symbol is pecanex (PCNX; circBase database, http://www.circbase.org/). Previous studies show that PCNX is upregulated in lung cancer tissues compared with the adjacent non-tumor tissues, and the 3’-untranslated Region (3’-UTR) of PCNX promotes cell proliferation and suppresses apoptosis in lung cancer cells [24]. In the present study, we found another possible molecular mechanism of PCNX-mediated proliferation and apoptosis in lung cancer cells and that PCNX-originated hsa_circ_0102533 is involved in these processes. We found that the inhibition of hsa_circ_0102533 by siRNA could inhibit proliferation and induce apoptosis in NSCLC cell lines, suggesting that hsa_circ_0102533 showed an oncogenic property in the initiation and development of NSCLC. Zhu et al. showed that the siRNA-mediated inhibition of hsa_circ_0013958 suppresses cell proliferation and invasion and enhances cell apoptosis in lung adenocarcinoma cells [15]. Jiang et al. also confirmed that hsa_circ_0007385 knockdown resulted in significant suppression of the proliferation, migration, and invasion of NSCLC cells in vitro [16]. These results [15,16] and our findings suggest that circRNAs play an important role in the carcinogenesis of NSCLC and may be new potential therapeutic targets to treat NSCLC.
However, there are some limitations to our study. First, the number of samples was limited, which may have biased the present findings. Second, the stability of circRNAs in extraneousness has not been performed in our study. Third, the molecular mechanism of hsa_circ_0102533 functioning as microRNA sponges has not been investigated. Furthermore, the reliability of the widespread use of circRNAs as a biomarker for cancer diagnosis still needs to be proven.
In conclusion, our findings suggest that hsa_circ_0102533 is involved in the carcinogenesis and progression of NSCLC and might be used as a potential blood-based biomarker for NSCLC detection at the early stage and serve as a new target for the treatment of NSCLC.
Acknowledgements
This study was supported by the National Natural Science Foundation of China (Grant Nos. 81100412 and 81670060), China Postdoctoral Science Foundation (Grant No. 2016M592038), the Key Program for the Excellent Youth Scholars of Anhui Province (Grant No. gxyqZD2016059) and the Provincial Natural Science Foundation of Anhui (Grant No. 1508085MH192).
Disclosure of conflict of interest
None.
References
- 1.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Zhao J, Li L, Wang Q, Han H, Zhan Q, Xu M. CircRNA expression profile in early-stage lung adenocarcinoma patients. Cell Physiol Biochem. 2017;44:2138–2146. doi: 10.1159/000485953. [DOI] [PubMed] [Google Scholar]
- 3.Wang Y, Zhao H, Gao X, Wei F, Zhang X, Su Y, Wang C, Li H, Ren X. Identification of a three-miRNA signature as a blood-borne diagnostic marker for early diagnosis of lung adenocarcinoma. Oncotarget. 2016;7:26070–26086. doi: 10.18632/oncotarget.8429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhou X, Wen W, Shan X, Zhu W, Xu J, Guo R, Cheng W, Wang F, Qi LW, Chen Y, Huang Z, Wang T, Zhu D, Liu P, Shu Y. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8:6513–6525. doi: 10.18632/oncotarget.14311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun Y, Mei H, Xu C, Tang H, Wei W. Circulating microRNA-339-5p and -21 in plasma as an early detection predictors of lung adenocarcinoma. Pathol Res Pract. 2018;214:119–125. doi: 10.1016/j.prp.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 6.Rosell R, Bivona TG, Karachaliou N. Genetics and biomarkers in personalisation of lung cancer treatment. Lancet. 2013;382:720–731. doi: 10.1016/S0140-6736(13)61715-8. [DOI] [PubMed] [Google Scholar]
- 7.Deffebach ME, Humphrey L. Lung cancer screening. Surg Clin North Am. 2015;95:967–978. doi: 10.1016/j.suc.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 8.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 9.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–338. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 10.Zhang HD, Jiang LH, Sun DW, Hou JC, Ji ZL. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25:1–7. doi: 10.1007/s12282-017-0793-9. [DOI] [PubMed] [Google Scholar]
- 11.Cui X, Niu W, Kong L, He M, Jiang K, Chen S, Zhong A, Li W, Lu J, Zhang L. hsa_circRNA_103636: potential novel diagnostic and therapeutic biomarker in Major depressive disorder. Biomark Med. 2016;10:943–952. doi: 10.2217/bmm-2016-0130. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YG, Yang HL, Long Y, Li WL. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of preeclampsia. BJOG. 2016;123:2113–2118. doi: 10.1111/1471-0528.13897. [DOI] [PubMed] [Google Scholar]
- 13.Zhao Z, Li X, Jian D, Hao P, Rao L, Li M. Hsa_circ_0054633 in peripheral blood can be used as a diagnostic biomarker of pre-diabetes and type 2 diabetes mellitus. Acta Diabetol. 2017;54:237–245. doi: 10.1007/s00592-016-0943-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meng S, Zhou H, Feng Z, Xu Z, Tang Y, Li P, Wu M. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16:94. doi: 10.1186/s12943-017-0663-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhu X, Wang X, Wei S, Chen Y, Chen Y, Fan X, Han S, Wu G. hsa_circ_0013958: a circular RNA and potential novel biomarker for lung adenocarcinoma. FEBS J. 2017;284:2170–2182. doi: 10.1111/febs.14132. [DOI] [PubMed] [Google Scholar]
- 16.Jiang MM, Mai ZT, Wan SZ, Chi YM, Zhang X, Sun BH, Di QG. Microarray profiles reveal that circular RNA hsa_circ_0007385 functions as an oncogene in non-small cell lung cancer tumorigenesis. J Cancer Res Clin Oncol. 2018;144:667–674. doi: 10.1007/s00432-017-2576-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Zeng X, Ding T, Guo L, Li Y, Ou S, Yuan H. Microarray profile of circular RNAs identifies hsa_circ_0014130 as a new circular RNA biomarker in non-small cell lung cancer. Sci Rep. 2018;8:2878. doi: 10.1038/s41598-018-21300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 19.Wang F, Ying HQ, He BS, Pan YQ, Deng QW, Sun HL, Chen J, Liu X, Wang SK. Upregulated lncRNA-UCA1 contributes to progression of hepatocellular carcinoma through inhibition of miR-216b and activation of FGFR1/ERK signaling pathway. Oncotarget. 2015;6:7899–7917. doi: 10.18632/oncotarget.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Wei Y, Yan Y, Wang H, Yang J, Zheng Z, Zha J, Bo P, Tang Y, Guo X, Chen W, Zhu X, Ge L. CircDOCK1 suppresses cell apoptosis via inhibition of miR196a5p by targeting BIRC3 in OSCC. Oncol Rep. 2018;39:951–966. doi: 10.3892/or.2017.6174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong YS, Wang XW, Zhou XL, Liu ZH, Yang TX, Shi WH, Xie HW, Lv J, Wu QQ, Cao XF. Identification of the long non-coding RNA POU3F3 in plasma as a novel biomarker for diagnosis of esophageal squamous cell carcinoma. Mol Cancer. 2015;14:3. doi: 10.1186/1476-4598-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mlcochova H, Hezova R, Stanik M, Slaby O. Urine microRNAs as potential noninvasive biomarkers in urologic cancers. Urol Oncol. 2014;32:41, e41–49. doi: 10.1016/j.urolonc.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 23.Schultz NA, Dehlendorff C, Jensen BV, Bjerregaard JK, Nielsen KR, Bojesen SE, Calatayud D, Nielsen SE, Yilmaz M, Hollander NH, Andersen KK, Johansen JS. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311:392–404. doi: 10.1001/jama.2013.284664. [DOI] [PubMed] [Google Scholar]
- 24.Li J, Tian H, Pan J, Jiang N, Yang J, Zhou C, Xu D, Meng X, Gong Z. Pecanex functions as a competitive endogenous RNA of S-phase kinase associated protein 2 in lung cancer. Cancer Lett. 2017;406:36–46. doi: 10.1016/j.canlet.2017.07.030. [DOI] [PubMed] [Google Scholar]