Abstract
Uveal autoantigen with coiled-coil domains and ankyrin repeats (UACA/Nucling), has been reported to be upregulated in various cancers. However, its expression and function have not been studied in hepatocellular carcinoma (HCC). In the present study, expression of UACA was detected by reverse transcription-quantitative polymerase chain reaction (RT-qPCR) and the results revealed that UACA was upregulated in 23 cases of HCC compared with paired corresponding non-tumor liver tissues. In addition, the upregulation of UACA in HCC was further validated by analyzing the datasets from The Cancer Genome Atlas-Liver Hepatocellular Carcinoma (TCGA-LIHC) and GSE36376. Furthermore, knockdown of UACA suppressed the proliferative and invasive ability as well as inducing senescence of HCC cells. Besides, the expression level of UACA was positively associated with Hif1α (hypoxia-inducible factor 1α) in HCC datasets from TCGA-LIHC and GSE54236. Moreover, treatment with CoCl2 led to the increased expression and the localization alteration of UACA in HCC cells. In summary, UACA is upregulated in HCC and knockdown of UACA ameliorated malignant behaviors of HCC cells, and UACA was correlated with and under control of Hif1α.
Keywords: UACA, hepatocellular carcinoma, knockdown, proliferation, invasion, senescence, HIF1α
Introduction
Hepatocellular carcinoma (HCC) is the second most common cause of the cancer-related death worldwide. The increasing morbidity and mortality make it a major health challenge [1-5]. Despite the advances in therapeutic strategies including surgical resection, orthotropic liver transplantation, and radiofrequency thermal ablation, the clinical outcomes are still unsatisfactory [6]; one of the main reasons is the rapid progression of HCC [7]. However, the underlying mechanism still requires further elucidation. Therefore, urgent need exists to identify the novel molecular marker which facilitates the development of hepatocellular carcinoma, which may help to understand the molecular mechanism of HCC and find the potential therapeutic targets.
Uveal autoantigen with coiled-coil domains and ankyrin repeats (UACA/Nucling) was first identified as a novel autoantigen in patients with panuveitis and the prevalence of IgG anti-UACA auto-antibodies in Vogt-Koyanagi-Harada (VKH) patients was dramatically higher than that in healthy controls [8]. Growing evidence revealed the involvement of UACA in various pathologic conditions including Graves’ disease [9] and mammary gland involution [10]. However, the function of UACA in cancers differed according to its expression levels and its regulatory patterns. In non-small-cell lung carcinoma, both mRNA and protein level of UACA was down-regulated in non-small-cell lung cancer (NSCLC) cells and tumors. Particularly, stage IA NSCLC tumors showed a significantly lower level of UACA compared with higher stage tumors [11]. In addition, lack of UACA mediated Apaf-1 nuclear entry in NSCLC cells may suppress apoptosis and increase genomic instability [12-14]. Nevertheless, the UACA expression was markedly higher in lung adenocarcinoma and squamous cell carcinoma, and this upregulation was independent of tumor grade [11,15]. Moreover, UACA evaluation impaired endoplasmic reticulum stress and GRP78 translocation to the cell surface, thus blocking the sensitivity of cancer cells to apoptosis [15,16]. However, the role of UACA in HCC progression has not been investigated.
In this study, we detected UACA level in 23 pairs of HCC tissues and the corresponding non-tumor tissues and found that UACA was upregulated in HCC tissues, and then we used online datasets (TCGA-LIHC and GSE36376) to confirm this upregulation. Next, we observed that knockdown of UACA interfered the malignant behaviors of HCC. Further investigation using datasets from GCBI revealed that Hif1α modulates UACA in hypoxia condition. Taken together, these findings suggested that UACA plays a significant role in the progression of HCC.
Materials and methods
Patients and tissue samples
Twenty-three pairs of HCC tissues and matching adjacent non-tumor tissues were enrolled in this research. All of them were obtained from patients who initially underwent hepatectomies and diagnosed with HCC between January 2014 and December 2015 at Nanfang Hospital. None of the patients received anti-cancer therapies prior to surgery. All eligible patients provided informed consent before collection of HCC specimens and corresponding adjacent non-tumor specimens. The research was reviewed and approved by the ethics committee of Nanfang Hospital affiliated to Southern Medical University.
Cell culture
SK-Hep1 and MHCC97H human HCC cell lines were obtained from the Cell Bank of Type Culture Collection (Chinese Academy of Sciences, Shanghai, China). Cells were cultured in Dulbecco’s Modified Eagle’s Medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (Gibco). Cells were incubated in a humidified chamber at 37°C and 5% CO2.
Western blotting
This procedure has been previously described [17]. Cells were harvested and solubilized in SDS-sample buffer containing 5% β-mercaptoethanol to obtain total cell homogenate preparations. Thirty micrograms of total protein from each sample was resolved on a 12% tris gel and transferred to polyvinylidene difluoride membranes. The blots were probed with various antibodies, including anti-GAPDH (Rui Antibody Biotech, Beijing, China), anti-HIF1α (R&D Systems, MN, USA), anti-UACA (Proteintech, Wuhan, China), and anti-HSP70 (Rui Antibody Biotech).
Immunofluorescence staining
Cells were prepared for immunofluorescence staining as previously described [17]. Slides were fixed in 4% paraformaldehyde (15 min, room temperature), incubated in blocking solution (phosphate-buffered saline [PBS], 2% bovine serum albumin). Anti-UACA antibody (Proteintech) were added (22°C, 1 h) followed by three rinses with PBS then addition of the secondary antibodies conjugated to Cy3 (Beyotime Biotechnology, Shanghai, China). Staining was visualized using BX63 Automatic Fluorescence Microscope (Olympus). A 100x oil objective was used for the imaging.
RNA extraction, reverse transcription (RT) and real-time PCR
Total RNA was extracted from tissues or cultured cells using TRIzol reagent (Invitrogen, Carlsbad, CA). For qRT-PCR, RNA was reverse transcribed to cDNA by using a Reverse Transcription Kit (Takara, Dalian, China). Real-time PCR analyses were performed with Power SYBR Green (Takara). Results were normalized to the expression of β-actin. The PCR primers for UACA or β-actin were as follows: UACA-sense, 5’-CATCCTTATACATGGAGTTGATATTACAA-3’ and anti-sense, 5’-TGTCCGCCCGTCTACATC-3’; β-actin-sense, 5’-TCAAGATCATTGCTCCTCCTGA-3’ and anti-sense, 5’-CTCGTCATACTCCTGCTTGCTG-3’. qRT-PCR and data collection were performed on LightCycler® 480II. Expression data were normalized to the geometric mean of the housekeeping gene β-actin to control the variability in expression levels and calculated as 2-[(Ct of gene) - (Ct of β-actin)], where Ct represents the threshold cycle for each transcript.
Expression datasets
Gene expression profile TCGA-LIHC (https://tcga-data.nci.nih.gov/tcga/) and GSE36376 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36376) were downloaded from TCGA (The Cancer Genome Atlas-Liver), which contains about 351 HCC cases and 240 HCC cases respectively. These datasets comprise follow-up information for patients and gene expression data.
Transient transfection of siRNA
Cells were transfected with siRNA when they reached 70%-80% confluence using RNAiMAX (Invitrogen) according to the manufacturer’s protocol. Cells were cultured for 48 h before analysis. The oligos corresponding to UACA or HIF1α were termed siRNA-UACA or siRNA-HIF1α. The siRNA-UACA and siRNA-HIF1α was purchased from RiboBio (Guangzhou, China).
Cell counting kit (CCK)-8 assay
Cells were seeded at a density of 1000/well in 96-well plates (Nest Biotechnology, Wuxi, China). CCK-8 solution (Dojindo Laboratories, Osaka, Japan) (10 μl/well) was added after 24, 48, 72, 96, or 120 h. Cells were incubated in the dark for 2 h at 37°C. After shaking the plates for 5 s, the absorbance at 450 nm was measured. Cell viability was evaluated in triplicate samples from three independent experiments.
5-ethynyl-20-deoxyuridine (EdU) incorporation assay
The assay was performed according to the EdU assay kit (Ribobio, Wuhan, China) manufacturer’s instructions. 5×103 HCC cells were seeded per well in 96-well plates and cultured overnight. The newly synthesized DNA of the cells was assessed by The EdU incorporation rates; this rate was showed as the ratio of EdU positive cells (red cells) to total Hoechst33342 positive cells (blue cells). The assay was performed in triplicate.
SA-β-gal assay
Cells were washed twice with PBS, fixed for 3-5 min in 2% formaldehyde/0.2% glutaraldehyde at room temperature, and washed again with PBS. The cells were incubated at 37°C overnight without CO2 in fresh senescence-associated-beta-gal (SA-β-Gal) stain solution under construction of protocols. At least 300 cells were counted under a confocal microscope in five random fields. Three independent experiments were performed.
Statistical analysis
All data were analyzed using SPSS 16.0 software (Abbott Laboratories, Chicago, IL, USA). Two independent sample Student’s t test was used to assess the significance of differences between two groups. One-way ANOVA and Dunnett’s multiple comparisons test was used for multiple-group comparisons. Multi-way classification ANOVA was used to evaluate the results of the CCK-8 assay. Each experiment was repeated at least three times and error bars represent mean ± SEM. All statistical tests were two-sided. P < 0.05 was considered significant. Single, double and triple asterisks indicate statistical significance: *P < 0.05; **P < 0.01; ***P < 0.001.
Results
UACA is upregulated in HCC tissues
To examine the mRNA expression level in HCC tissues, we used 23 pairs of fresh HCC samples and corresponding adjacent non-tumor samples to measure the expression of UACA by RT-qPCR. The results showed that in comparison with non-tumor tissues, UACA expression was increased in the HCC tissues (Figure 1A). To validate the upregulation of UACA in HCC, TCGA-LIHC (The Cancer Genome Atlas-Liver Hepatocellular Carcinoma) and GSE36376 datasets were analyzed. In the unpaired TCGA-LIHC cohort, UACA was significantly higher in HCC tissues than in non-tumor liver tissues (P < 0.001; Figure 1B), and the similar results showed in the paired TCGA-LIHC cohort (P < 0.001; Figure 1C). Furthermore, the expression of UACA was significantly upregulated in tumors compared with the non-tumors from the GSE36376 datasets (P < 0.001; Figure 1D). Next, the protein expression of UACA in clinical specimens from The Human Protein Atlas was further investigated. The results demonstrated that the immunohistochemistry staining of UACA was more intense in HCC tissues than in the normal liver tissues (Figure 1E). In summary, UACA expression is upregulated in HCC tissues both in mRNA and protein level.
Figure 1.
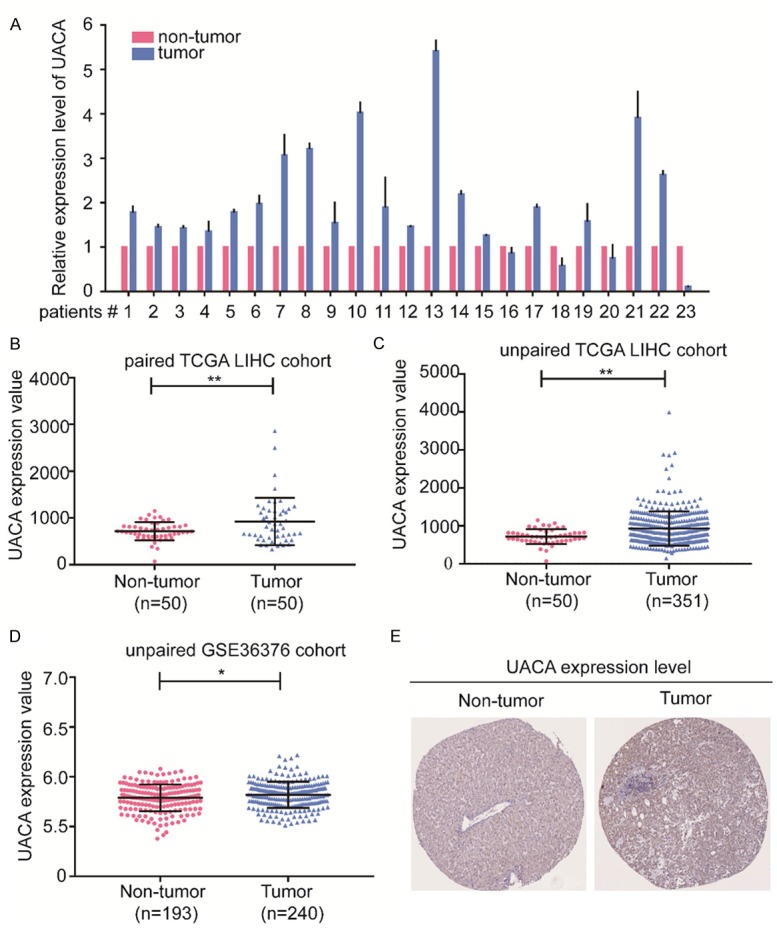
UACA is upregulated in HCC tissues. (A) UACA mRNA expression was analyzed by RT-qPCR in 23 pairs of HCC tissues and matched adjacent non-tumor liver tissues. β-actin expression served as an internal control. (B-D) The UACA expression level of HCC samples (n=50) and non-tumor tissues (n=50) in datasets including unpaired TCGA-LIHC (B), paired TCGA-LIHC (C) and GSE3376 (D). *P < 0.05, **P < 0.01. (E) Representative immunohistochemical staining image of UACA in normal liver and HCC tissues obtained from the Human Protein Atlas. Student’s t test was used for the comparison between two groups. RT-qPCR, reverse transcription-quantitative polymerase chain reaction; HCC, hepatocellular carcinoma; TCGA-LIHC, The Cancer Genome Atlas-Liver Hepatocellular Carcinoma.
Knockdown of UACA suppresses proliferation, inhibits invasion and promotes senescence in HCC cells
To explore the biological function of UACA in the development and progression of HCC, we investigated the effect of UACA knockdown on cell proliferation, invasion and senescence. Firstly, we silenced UACA by using siRNA and the effectiveness of silencing lasted for 6 days (Figure 2A). By performing CCK-8 assay, we found that the cell growth was significantly impaired in UACA-silencing cells (Figure 2B). However, the knockdown of UACA did not enhance the percentage of EdU+ cell (Figure 2C). As the EdU+ cells represent the cells going through S phase, we hypothesized that UACA might not affect the proliferation by interrupting S phase. These results suggested that knockdown of UACA inhibited the proliferation of HCC cells, which might not affect the S phase of cell cycle. Next, we further investigated the impact of UACA on the invasion of HCC cell lines. Data from the transwell invasion assay demonstrated that the downregulation of UACA attenuated the cell invasion ability (Figure 3A). Interestingly, we also found that the cells went through more cellular senescence in UACA-silencing cells as detected by SA-β-gal assay (Figure 3B). These findings indicated that UACA aproliferation and invasion and inhibits senescence of HCC cells.
Figure 2.
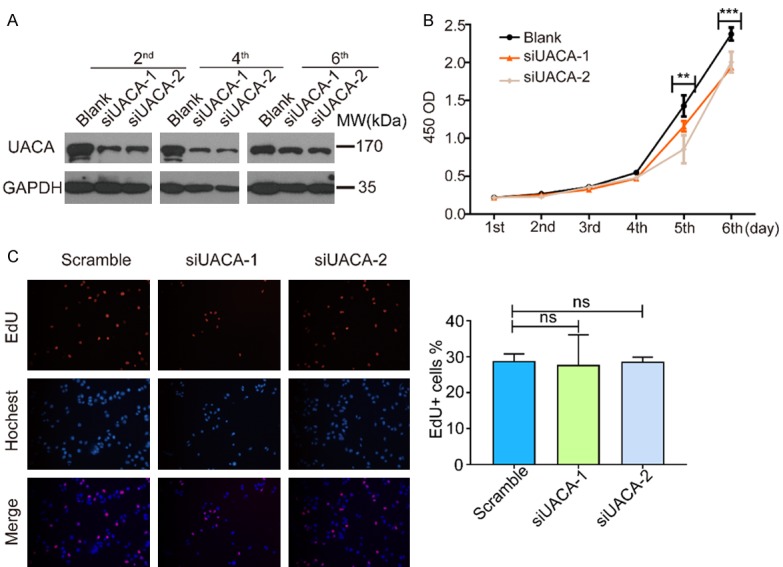
Knockdown of UACA leads to decrease in proliferation but not EdU-positive ratio in HCC cells. A. SK-Hep1 cells were transfected with UACA siRNA. After the indicated period of time, the cells were lysed and western blot was used to detect the level of UACA. GADPH was used as the internal control. B. CCK-8 assay was carried out on SK-Hep1 cells from the same batch with panel. Data was collected from three independent experiments. Two-way ANOVA was used to compare the OD values among three groups. **P < 0.01, ***P < 0.001. C. SK-Hep1 cells were transfected with siRNA-UACA and EdU assay was performed. Representative images are shown. The quantification was shown on the right. At least 10 fields were counted and the average EdU+% from three independent experiments were used for statistical analysis. One-way ANOVA and then Dunnett’s multiple comparisons test was used to do the comparison among the three groups and between two groups. ns, not significant.
Figure 3.
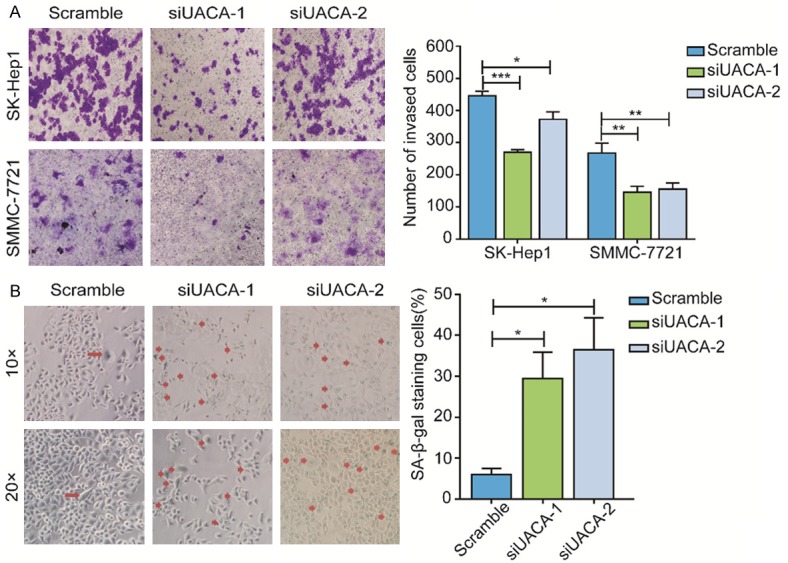
Knockdown of UACA inhibitsinvasion and promotes senescence of HCC cell lines. (A) SK-Hep1 cells were transfected with negative control or UACA siRNA (48 h) then transwell invasion assay was performed. The representative images were shown (magnification, ×40). (B) SK-Hep1 cells were treated as described in (A) and detection of senescent cells were measured by the presence of SA-β-Galactosidase activity. The invasive/senescent cells were stained in blue which is indicated by red arrows (magnification, ×40). The senescent cells per field were counted in at least 10 fields, and the average numbers from three independent experiments were used for the quantification which is shown in the right panel. One-way ANOVA and then Dunnett’s multiple comparisons test were used to do the comparison among the three groups and between two groups. **P < 0.01, ***P < 0.001.
The expression of UACA is positively correlated with HIF1α in HCC
To understand the mechanisms underlying the upregulation and functions of UACA in HCC, we identified transcription factors and interacting proteins of UACA by using the online tool from Gene-Cloud of Biotechnology Information (GCBI) website (https://www.gcbi.com.cn) (Figure 4A, 4B). Among all the candidate transcriptional factors, we found hypoxia-inducible factor subunit HIF2A and active heterodimer complexes HIF1AARNT, which are the key regulators in response to oxygen deprivation. Intriguingly, we also found HIF1α (hypoxia inducible factor 1 alpha subunit) was one of the interactors of UACA. Given that hypoxic microenvironment is common in solid tumors and HIF-mediated hypoxia response is involved in angiogenesis, glycolysis, cellular proliferation and metastasis, next we focused to investigate the relationship between HIF1α and UACA. Moreover, a positive correlation was found between UACA and HIF1α expression in the datasets from TCGA-LIHC (Figure 4C) and GSE54236 (Figure 4D) in HCC.
Figure 4.
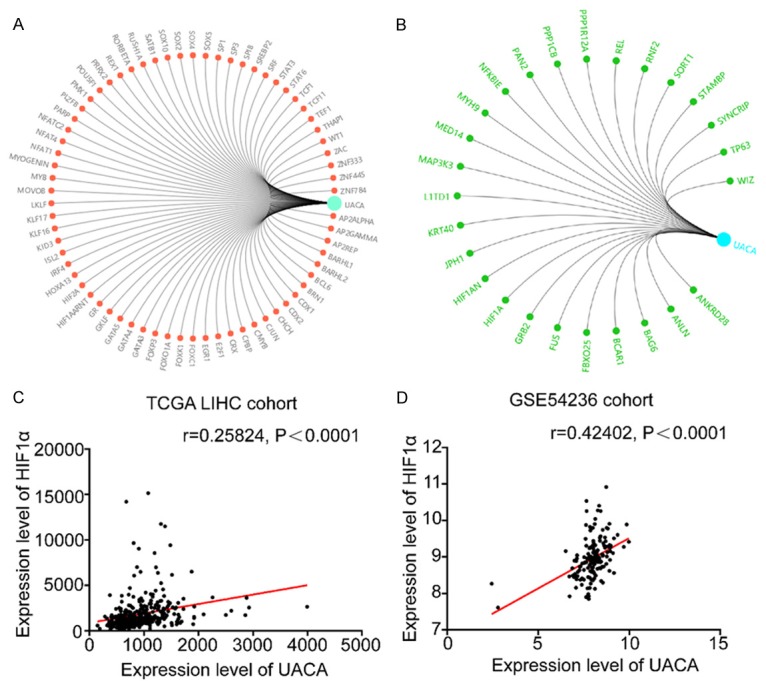
The expression of UACA is positively related to HIF1α in HCC. A. Transcriptional factors of UACA predicted in GCBI website. B. Proteins interacting with UACA predicted in GCBI website. C, D. UACA is positively associated with HIF1α in TCGA-LIHC and GSE54236. Linear regression was used to analyze the relation between UACA and HIF1α. GCBI, Gene-Cloud of Biotechnology Information; TCGA-LIHC, the Cancer Genome Atlas-Liver Hepatocellular Carcinoma.
Hypoxia induces the upregulation of UACA in HCC cell lines
To further validate the effect of hypoxia on UACA, we treated UACA-silencing or control HCC cells with CoCl2, a reagent which mimics a hypoxic environment. We observed that the expression of UACA was increased in hypoxic condition, which was rescued by HIF1α knockdown (Figure 5A). Next, we assessed the change of mRNA expression level of UACA by RT-qPCR. Similar to the alteration of protein level, hypoxia led to the upregulation of UACA at mRNA expression level and si-HIF1α lowered the UACA level (Figure 5B). Since the location of UACA is dynamic according to its function, next we monitored the locations of UACA under both normoxia and hypoxia by immunofluorescence staining (Figure 5C). Under normal oxygen condition, UACA localized predominantly in nucleus and around nucleus membrane, which is consistent with the prior studies [14]. Notably, UACA diffused in the cytoplasm when HCC cells were treated with CoCl2.
Figure 5.
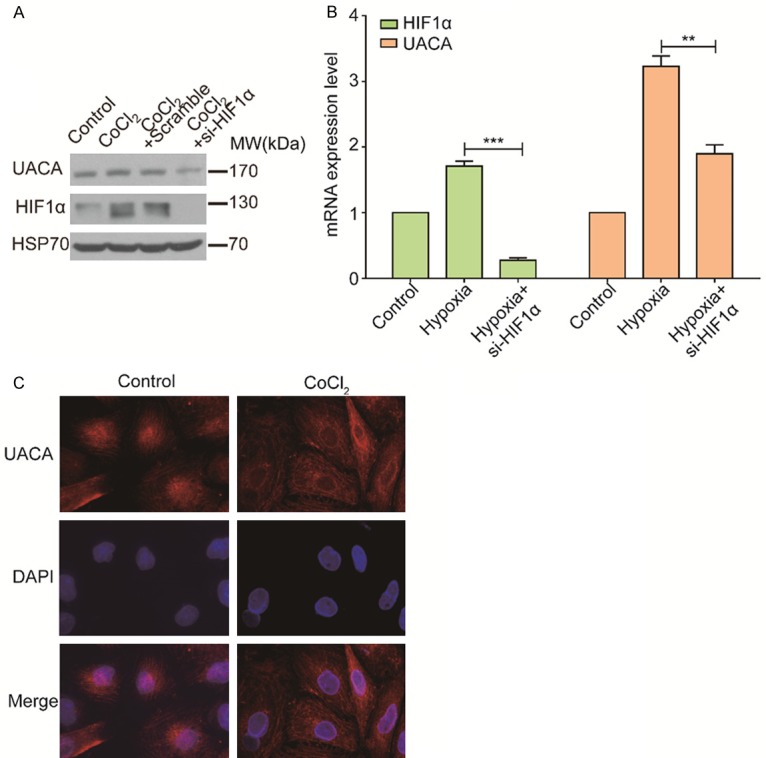
Hypoxia induces the upregulation of UACA in HCC cell lines. (A) MHCC97H cells was transfected with scramble or si-HIF1α for 48 h and incubated with or without CoCl2 for 24 h. Western blot was used to measure the expression level of UACA with different treatments. GAPDH was used as an internal control. (B) MHCC97H cells were treated as (A), and mRNA expression level was measured by RT-qPCR. One-way ANOVA and Tukey’s multiple comparisons test was used to do the comparison among three groups. **P < 0.01, ***P < 0.001. (C) MHCC97H cells were treated with or without CoCl2 for 24 h and followed by immunofluorescence assay to detect the translocation of UACA. UACA is in red, and the nucleus is stained with DAPI. RT-qPCR, reverse transcription-quantitative polymerase chain reaction.
Discussion
Our present study showed that UACA is upregulated in HCC and correlated with HCC progression (Figure 1). Knockdown of UACA resulted in reduced proliferation, inhibited invasion, and promoted senescence (Figures 2 and 3). Though knockdown of UACA significantly suppressed proliferation in HCC cells, there was no difference in the percentage of EdU+ cells between UACA-silencing cells and control cells (Figure 2C). Since EdU+ cells represent the cells in the S phase of cell cycle, we assumed that knockdown of UACA might inhibit proliferation without affecting DNA replication. However, UACA knockdown caused more senescent cells (Figure 3B). Since cellular senescence suppresses cancer by irreversibly arresting cell proliferation [18,19], we postulated that UACA silencing-mediated proliferation inhibition is probably the consequence of increased senescence. Consistent with our findings, UACA was reported to be evaluated in lung squamous cell carcinoma and lung adenocarcinoma, and UACA inhibits apoptosis by sequestering Par-4 and preventing it from translocating GRP78 to the cell surface [15]. Furthermore, suppression of UACA expression by siRNA sensitized cells to apoptosis. Our study uncovers another consequence of UACA inhibition, which provides a novel insight that the down-regulation of UACA brings benefit for HCC treatment.
Mechanistic study revealed that hypoxia may regulate UACA expression and localization. Hypoxia is a hallmark of cancer and it influences cancer cell function. Much clinical evidence indicates the prevalence of hypoxia in solid tumors, including cancers of the breast, uterine cervix, vulva, head & neck, prostate, rectum, pancreas, lung, brain and liver. We found a positive relation between HIF1α and UACA in HCC by analyzing datasets from TCGA-LIHC and GSE54236 (Figure 4C, 4D). Moreover, members of hypoxia inducible factor family can not only bind to the promoter of UACA but also interact with UACA protein as predicted by GCBI bioinformatics tool. Then the results of western blot and RT-qPCR proved that hypoxia stimulated the expression of UACA, and block of HIF1α lowered the UACA level (Figure 5A, 5B). These findings indicate that hypoxia may upregulate UACA expression at transcriptional level or post-translational level. Since hypoxia is associated with the development of an aggressive phenotype in cancers, so we assume that UACA may be one of the targets of HIF1α to promote progression in HCC.
Notably, from Figure 4B we can see that besides HIF1α, UACA might also interact with NFKBIE (NF-kappa-B inhibitor epsilon) which binds to components of NF-kappa-B and traps the complex in the cytoplasm and prevents it from activating genes in the nucleus. In support of our findings, UACA was shown to be a suppressor of NF-κB by preventing its translocation into the nucleus, which supported our finding [20,21]. Also, immunofluorescence staining showed the perinuclear and nuclear localization of UACA, which is consistent with previous reported that UACA interacts with Apaf-1 and induces its translocation into the nucleus upon proapoptotic stress [14], and CoCl2 treatment led to even UACA distributing in cytoplasm. As one of the endogenous regulators of apoptosome apparatus, UACA cytoplasmic distribution may cause deficiency in the apoptosome pathway, thus contributing to tumorigenesis and progression of HCC [12,22].
In summary, our study illustrates that a novel oncogene UACA, is upregulated in HCC and knockdown of UACA inhibited the malignant behaviors of HCC, which may be regulated by hypoxia and cooperates with HIF1α in the development of HCC. These findings indicate that UACA is a critical molecular for HCC progression and potentially is an effective therapeutic target for HCC targets.
Acknowledgements
This work was supported by the National Nature Science Foundation of China (Grant No. 81472711, 81672756, 81702390 and 91540111), Guangdong Province Universities and Colleges Pearl River Scholar Funded Scheme (2015), and the Natural Science Foundation of Guangdong Province (Grant No. 2017A030310105).
Disclosure of conflict of interest
None.
References
- 1.Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class, and effects on patient prognosis. Gastroenterology. 2017;152:745–761. doi: 10.1053/j.gastro.2016.11.048. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends--an update. Cancer Epidemiol Biomarkers Prev. 2016;25:16–27. doi: 10.1158/1055-9965.EPI-15-0578. [DOI] [PubMed] [Google Scholar]
- 3.Inokawa Y, Inaoka K, Sonohara F, Hayashi M, Kanda M, Nomoto S. Molecular alterations in the carcinogenesis and progression of hepatocellular carcinoma: tumor factors and background liver factors. Oncol Lett. 2016;12:3662–3668. doi: 10.3892/ol.2016.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.El-Deiry WS. Impact of genetic targets on cancer therapy. New York: Springer; 2013. [DOI] [PubMed] [Google Scholar]
- 5.Niu ZS, Niu XJ, Wang WH. Genetic alterations in hepatocellular carcinoma: an update. World J Gastroenterol. 2016;22:9069–9095. doi: 10.3748/wjg.v22.i41.9069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsoulfas G, Kawai T, Elias N, Ko SC, Agorastou P, Cosimi AB, Hertl M. Long-term experience with liver transplantation for hepatocellular carcinoma. J Gastroenterol. 2011;46:249–256. doi: 10.1007/s00535-010-0302-9. [DOI] [PubMed] [Google Scholar]
- 7.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62:394–399. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 8.Yamada K, Senju S, Nakatsura T, Murata Y, Ishihara M, Nakamura S, Ohno S, Negi A, Nishimura Y. Identification of a novel autoantigen UACA in patients with panuveitis. Biochem Biophys Res Commun. 2001;280:1169–1176. doi: 10.1006/bbrc.2001.4189. [DOI] [PubMed] [Google Scholar]
- 9.Ohkura T, Taniguchi S, Yamada K, Nishio N, Okamura T, Yoshida A, Kamijou K, Fukata S, Kuma K, Inoue Y, Hisatome I, Senju S, Nishimura Y, Shigemasa C. Detection of the novel autoantibody (anti-UACA antibody) in patients with Graves’ disease. Biochem Biophys Res Commun. 2004;321:432–440. doi: 10.1016/j.bbrc.2004.06.162. [DOI] [PubMed] [Google Scholar]
- 10.Dang HV, Sakai T, Pham TA, Tran DH, Yorita K, Shishido Y, Fukui K. Nucling, a novel apoptosis-associated protein, controls mammary gland involution by regulating NF-kappaB and STAT3. J Biol Chem. 2015;290:24626–24635. doi: 10.1074/jbc.M115.673848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moravcikova E, Krepela E, Prochazka J, Rousalova I, Cermak J, Benkova K. Down-regulated expression of apoptosis-associated genes APIP and UACA in non-small cell lung carcinoma. Int J Oncol. 2012;40:2111–2121. doi: 10.3892/ijo.2012.1397. [DOI] [PubMed] [Google Scholar]
- 12.Sakai T, Liu L, Teng X, Mukai-Sakai R, Shimada H, Kaji R, Mitani T, Matsumoto M, Toida K, Ishimura K, Shishido Y, Mak TW, Fukui K. Nucling recruits Apaf-1/pro-caspase-9 complex for the induction of stress-induced apoptosis. J Biol Chem. 2004;279:41131–41140. doi: 10.1074/jbc.M402902200. [DOI] [PubMed] [Google Scholar]
- 13.Rousalova I, Banerjee S, Sangwan V, Evenson K, McCauley JA, Kratzke R, Vickers SM, Saluja A, D’Cunha J. Minnelide: a novel therapeutic that promotes apoptosis in non-small cell lung carcinoma in vivo. PLoS One. 2013;8:e77411. doi: 10.1371/journal.pone.0077411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Sakai T, Sano N, Fukui K. Nucling mediates apoptosis by inhibiting expression of galectin-3 through interference with nuclear factor kappaB signalling. Biochem J. 2004;380:31–41. doi: 10.1042/BJ20031300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burikhanov R, Shrestha-Bhattarai T, Qiu S, Shukla N, Hebbar N, Lele SM, Horbinski C, Rangnekar VM. Novel mechanism of apoptosis resistance in cancer mediated by extracellular PAR-4. Cancer Res. 2013;73:1011–1019. doi: 10.1158/0008-5472.CAN-12-3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burikhanov R, Shrestha-Bhattarai T, Hebbar N, Qiu S, Zhao Y, Zambetti GP, Rangnekar VM. Paracrine apoptotic effect of p53 mediated by tumor suppressor Par-4. Cell Rep. 2014;6:271–277. doi: 10.1016/j.celrep.2013.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu L, Dong Z, Liang J, Cao C, Sun J, Ding Y, Wu D. As an independent prognostic factor, FAT10 promotes hepatitis B virus-related hepatocellular carcinoma progression via Akt/GSK3beta pathway. Oncogene. 2014;33:909–920. doi: 10.1038/onc.2013.236. [DOI] [PubMed] [Google Scholar]
- 18.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demaria M, O’Leary MN, Chang J, Shao L, Liu S, Alimirah F, Koenig K, Le C, Mitin N, Deal AM, Alston S, Academia EC, Kilmarx S, Valdovinos A, Wang B, de Bruin A, Kennedy BK, Melov S, Zhou D, Sharpless NE, Muss H, Campisi J. Cellular senescence promotes adverse effects of chemotherapy and cancer relapse. Cancer Discov. 2017;7:165–176. doi: 10.1158/2159-8290.CD-16-0241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin P, Shin SH, Chun YS, Shin HW, Shin YJ, Lee Y, Kim D, Nam DH, Park JW. Astrocyte-derived CCL20 reinforces HIF-1-mediated hypoxic responses in glioblastoma by stimulating the CCR6-NF-kappaB signaling pathway. Oncogene. 2018;37:3070–3087. doi: 10.1038/s41388-018-0182-7. [DOI] [PubMed] [Google Scholar]
- 21.Hirai K, Furusho H, Hirota K, Sasaki H. Activation of hypoxia-inducible factor 1 attenuates periapical inflammation and bone loss. Int J Oral Sci. 2018;10:12. doi: 10.1038/s41368-018-0015-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Teng X, Sakai T, Liu L, Sakai R, Kaji R, Fukui K. Attenuation of MPTP-induced neurotoxicity and locomotor dysfunction in Nucling-deficient mice via suppression of the apoptosome pathway. J Neurochem. 2006;97:1126–1135. doi: 10.1111/j.1471-4159.2006.03833.x. [DOI] [PubMed] [Google Scholar]


