Abstract
Reactivation of Lin28 accelerates hair, cartilage, bone and mesenchyme regrowth after ear and digit injuries. However, the relationship of Lin28 to reparative dentin has been under investigation. The aim of the present study was to examine whether Lin28 participates in the reparative dentin process and lipopolysaccharide (LPS)-stimulated human dental pulp cells (HDPCs) and to identify the underlying signaling pathway mechanisms. The study established a wound-healing model of the dentin-dental pulp complex in vivo and LPS-induced dental pulp cell inflammation in vitro. In vivo, the results of hematoxylin and eosin staining demonstrated the obvious appearance of reparative dentin and odontoblast-like cells were arranged along the reparative dentin. Immunohistochemical examination demonstrated that Lin28 expression was increased by 72 h after cavity preparation but was decreased by 21 d after cavity preparation. In vitro, HDPCs were exposed to 100 ng/ml LPS for 24 h, and the expression of Lin28 was increased. Overexpression of Lin28 was associated with the downregulated expression of let-7b, let-7g and miR-98. These findings suggest that the wound-healing model was successfully established. Lin28 was involved in the reparative process of the dentin-dental pulp complex and HDPCs exposed to LPS, and Lin28/let-7 may be the underlying mechanism.
Keywords: Dentin-dental pulp complex, wound-healing model, Lin28, lipopolysaccharide, let-7 families
Introduction
Dental pulp is considered a source of mesenchymal stem cells that are suitable for tissue engineering applications [1,2]. It is known that dental pulp stem cells have the potential to differentiate into several cell types, including odontoblasts, neural progenitors, osteoblasts, chondrocytes, and adipocytes [3,4]. Damage to the dentin dental pulp by microbial irritants and chemical and mechanical stimuli leads to the deposition of reparative dentin [5,6]. Reparative dentin formation is essential for maintaining the integrity of the dentin structure during disease or trauma [7-9].
Lin28 is an RNA-binding protein that was first described in a Caenorhabditis elegans screen for heterochronic genes that regulate developmental timing, and the protein plays an important role in stem cell biology, development, metabolism and dysregulation in many human diseases [10,11]. Recently, Shyh-Chang and George demonstrated that Lin28 can enhance tissue repair in mice [12,13]. Conditional reactivation of Lin28 in adult mice markedly accelerates the wound-healing process in the injured tissue of transgenic mice. Lin28 reactivation accelerates the regrowth of mesenchyme, cartilage, and bone following finger and ear injuries and improves hair regrowth by promoting anagen in hair follicles [13]. Lin28 might be capable of promoting repair in other postnatal tissues as well; however, whether it participates in the dentin-dental pulp healing process remains unknown.
The mammalian let-7 family comprises twelve members, including let-7a-1, let-7a-2, and let-7a-3; let-7b; let-7c; let-7d; let-7e; let-7f-1 and let-7f-2; let-7g; let-7i; and miR-98 [14-18]. Lin28 stops the mature processing of let-7 at both the pri- and pre-miRNA steps. During embryogenesis, mature let-7 rises as Lin28 levels decline during ESC differentiation, fetal development and aging [11,18,19]. Ranging from embryonic development to cancer and metabolic processes, the Lin28/let-7 axis is now recognized as being central to maintaining a proper cell fate and to coordinating proliferation, growth, and energy utilization at the cellular level as well as metabolism, growth, developmental timing, and tissue homeostasis in organisms [20,21].
Lipopolysaccharide (LPS), a major component of the cell wall of gram-negative bacteria, serves a key role in pulpal, periodontal and periapical disease through the production of various proinflammatory-cytokines, including interleukin (IL)-1β, IL-6, IL-8 and tumor necrosis factor-α (TNF-α) [22,23]. Human dental pulp cells (HDPCs) also have the ability to produce proinflammatory-mediators such as IL-1β and IL-8 and express adhesion molecules such as vascular cell adhesion molecule-1 (VCAM-1) and intercellular adhesion molecule-1 (ICAM-1) in response to LPS [24]. The purpose of this in vitro study was to evaluate the effect of LPS on Lin28 expression in HDPCs.
The present study investigated the dentin-dental pulp complex for dentin regeneration in an animal model. Responses of odontoblasts and pulp cells to cavity preparation were investigated in the upper first molar teeth of rats. By constructing the wound-healing model of the dentin-dental pulp complex in vivo and LPS-induced dental pulp inflammation in vitro, we examined whether Lin28 can participate in the repair process and investigated the molecular mechanism of the repair process with the aim of gaining novel insights into the treatment of dentin-dental pulp damage.
Materials and methods
Construction of the wound-healing model
Fifty-six adult Sprague-Dawley (SD) rats (28 male and 28 female), weighing 180-220 g, were obtained from Xi’an Jiaotong University Animal Care (Xi’an, China) and were approved for use by a committee at Xi’an Jiaotong University (Xi’an, China). The rats were kept for approximately one month at room temperature with free access to food and were randomly divided into seven groups; one control and six experimental. Maxillary bones were collected in each of the groups at intervals of 0 h, 72 h, 7 d, 14 d and 21 d after cavity preparation.
Tissue preparation
The animals were anesthetized by intraperitoneal injection of Nembutal (40 mg/kg of body weight). A groove-shaped cavity was prepared on the mesial surface of the upper first molar by using an air turbine with a tungsten carbide bur (0.5 mm in diameter). The cavity depth was ~0.5 mm (~2/3 of the dentin) and was left without any further treatments such as air drying or filling. The control animals were left without any treatment. The whole procedure was carried out by an experienced clinician, so that the size of the cavity prepared by the bur was standard. The animals were perfused with physiological saline until the pouring liquid became clear and the limbs of the rat became white, and then were perfused with 4% paraformaldehyde. The maxillae were removed and immersed in 4% paraformaldehyde for an additional 48 h at 4°C, and then decalcified in 10% disodium ethylenediaminetetraacetic acid (EDTA) for one month (pH 7.4). The specimens were dehydrated and embedded in paraffin. Sagittal sections were ~5 μm thick.
Hematoxylin and eosin staining
Sagittal sections were approximately 5 μm thick. The sections were dewaxed by xylene I, II for 20 min and immersed in 100% alcohol I, II for 10 min, after which they were immersed in 95%, 80% and 70% alcohol for 5 min. Hematoxylin was used to stain the sections for 5 min, followed by eosin staining for 15 s at room temperature. Finally, the sections were dehydrated and sealed by neutral balsam.
Immunohistochemical staining
After deparaffination, the sections were washed three times in phosphate-buffered saline (PBS) and then treated with 3% H2O2 in PBS for 30 min at room temperature to inhibit endogenous peroxidase. Before incubation with primary antibodies, normal goat serum was used for blocking non-specific binding at room temperature for 20 min. Sections were incubated overnight at 4°C with Lin28 (1:50 dilution; ab46020; Abcam, Cambridge, UK) primary antibodies. After rewarming for 1 h at 37°C, sections were incubated with goat anti-rabbit IgG conjugated with peroxidase polymer (Boster Bioengineering, Wuhan, China) for 40 min at 37°C. Detection was performed by 3,3’-diaminobenzidine (DAB) and hematoxylin. These sections were observed under a light microscope.
LPS-induced dental pulp inflammation
Extracted third molars or premolars were collected from patients (18-25 yr old) at Stomatology Hospital, Xi’an Jiaotong University. Informed consent was obtained from the patients, and the study was performed with the approval of the ethics committee of Xi’an Jiaotong University. The tooth was split with a hammer, and the dental pulp tissue was separated from the tooth, minced into small pieces by surgical scissors, and digested with 3 mg/ml collagenase type I (Sigma, St Louis, MO, USA) for 30 min at 37°C. The tissue pieces were seeded into a 35-mm2 cell culture plate (Corning Incorporated, Corning, NY, USA) and cultured in α-minimum essential medium (α-MEM, HyClone, Logan, Utah, USA) supplemented with 20% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA). After the cells reached 70-80% confluence, the HDPCs migrated from the tissue were collected and subcultured. The cells obtain-ed between passages three and five were used in the subsequent experiments.
Enzyme-linked immunosorbent assay
Cells were incubated with 10 ng/ml, 100 ng/ml, 1 μg/ml and 10 μg/ml E. coli LPS (Sigma, St. Louis, MO, USA) for 24 h. The medium of each sample was collected and assayed. The levels of IL-1β and IL-8 were determined by means of an enzyme-linked immunosorbent assay kit (ELISA, cat. No. EHC008. Shenzhen, China) according to the manufacturer’s protocol and calibrated spectrophotometrically with a standard curve. The experiments were performed in triplicate.
Cell proliferation assay
The proliferation activity of HDPCs was measured using a commercial CCK-8 kit (Dojindo, Dongjing, Japan) according to the manufacturer’s protocol. Cells were plated in 96-well plates at a density of 4×103 cells/well in α-MEM supplemented with 10% FBS at 37°C in 5% CO2. When the cells reached 40% confluence, the HDPCs were treated with 10 ng/ml, 100 ng/ml, 1 μg/ml and 10 μg/ml E. coli LPS for 24 h. Then, 10 μl of CCK-8 solution was added into each well and the cells were further cultured for 4 h in a humidified atmosphere. The absorbance of each group (n=5) was measured at a test wavelength of 450 nm on 96-well plates using a microculture plate reader and was directly proportional to the number of living cells. The proliferation assays were performed three times. The background absorbance of the medium without cells was subtracted.
Real-time RT-PCR
Total RNA was isolated from the cells with TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The quantity and quality of the isolated RNA was tested using a NanoDrop 2000 spectrophotometer measuring the absorbance at 260/280 nm. Reverse transcription was performed in a 10 µl RT reaction containing 500 ng of total RNA, while the mature miRNA was reverse transcribed using miRNA-specific primers, according to the manufacturer’s protocol (Takara, Dalian, China). qRT-PCR was performed with SYBR-Green Real-Time PCR Master Mix Plus (Takara, Dalian, China) in a 20 μl reaction mixture using an FTC-3000TM System (Funglyn Biotech Incorporated, Toronto, Canada). The thermocycling conditions were 95°C for 30 s for initial denaturation, followed by 40 cycles of 95°C for 5 s, 60°C for 30 s and 72°C for 30 s. The primers used are listed in Table 1. Each measurement was performed in triplicate for each sample and a dissociation curve analysis was conducted for each PCR. U6 and β-actin were used as controls for the miRNA and mRNA levels, respectively. Relative quantitation was calculated using the 2-ΔΔCt [25] method.
Table 1.
Primer pairs used for quantitative real-time PCR
| Gene | Primer name | Sequence (5’-3’) |
|---|---|---|
| Lin28 | Forward primer | GGAGCCAAGCCACTACATTC |
| Reverse primer | GGGTGAGCATAGAGCAGCAG | |
| IL-8 | Forward primer | TTTCAGAGACAGCAGAGCACACAA |
| Reverse primer | CACACAGAGCTGCAGAAATCAGG | |
| IL-1β | Forward primer | CCAGGGACAGGATATGGAGCA |
| Reverse primer | TTCAACACGCAGGACAGGTACAG | |
| β-actin | Forward primer | AGCGAGCATCCCCCAAAGTT |
| Reverse primer | GGGCACGAAGGCTCATCATT | |
| Let-7a | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACTATA |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGTAGGTTG | |
| Let-7b | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACCAGGGAA |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGTAAGTTG | |
| Let-7c | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACGCTCCAA |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGTAAGTTG | |
| Let-7d | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACTATG |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTAGAGGTAGTAGGTTG | |
| Let-7e | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACTATA |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGGAGGTTG | |
| Let-7f | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACTAT |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGTAGATTG | |
| Let-7g | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACTGTA |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGTAGTTTG | |
| Let-7i | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACAGCA |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGTAGTTTG | |
| Mir-98 | RT-primer | GTCGTATCCAGTGCGTGTCGTGGAGTCGGCAATTGCACTGGATACGACAACAATA |
| Forward primer | ATCCAGTGCGTGTCGTG | |
| Reverse primer | TGCTTGAGGTAGTAAGTTG | |
| U6 | RT-primer | GCTTCGGCAGCACATATACTAAAAT |
| Forward primer | GCTTCGGCAGCACATATACTAAAAT | |
| Reverse primer | CGCTTCACGAATTTGCGTGTCAT |
Western blot analysis
Total protein was extracted from cells treated with 100 ng/ml LPS, in a growth medium for 48 h, using ice-cold RIPA buffer (Beyotime, Shanghai, China), containing 1 mM phenylmethanesulfonyl fluoride (PMSF). The protein concentrations were determined using a BCA™ protein assay kit (Thermo Scientific Pierce, Rockford, IL, USA). The total quantity of protein loaded onto each lane of the gel was 100-300 μg. The proteins were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) at 65 V for 45 min and 95 V for 1 h and transferred to polyvinylidene difluoride (PVDF) membranes (Pall Gelman Laboratory Corporation, Ann Arbor, MI, USA) at 250 mA for 1 h. After blocking with 5% bovine serum albumin (BSA) in Tris-buffered saline Tween-20 (TBST) 2 h at room temperature, the membranes were incubated with rabbit anti-human Lin28 (1:1,000) (ab46020; Abcam, Cambridge, UK) or mouse anti-human β-actin (1:10,000) (Trans HC201, Beijing, China) antibodies overnight at 4°C. β-Actin was used as an internal control. Next, the membranes were incubated with horseradish peroxidase conjugated anti-rabbit (1:5000) (Trans HS101) or anti-mouse (1:10000) (Trans HS201) secondary antibodies in TBST for 2 h at room temperature with gentle agitation. Proteins were detected using an enhanced chemiluminescence kit (Millipore, Billerica, MA, USA). Densitometry analyses were performed, and the values for target proteins were normalized to those of β-actin.
Statistical analysis
Statistical analysis was performed with SPSS 17.0 software (SPSS, Incorporated, Chicago, IL, USA). The data are expressed as the mean ± standard deviation. All the experiments were performed at least three times. One-way analysis of variance was used to compare between groups. The Student’s t-test was used to compare between groups. P < 0.05 was considered to indicate a statistically significant difference.
Results
Histological change in the injured dentin-pulp complex
The odontoblast layer, the cell-free zone, the cell-rich zone, and the subjacent pulp tissue could be seen clearly in the control teeth after cavity preparation (Figure 1A, 1B). The predentin became clearly thick and had neonatal blood vessels by 72 h after cavity preparation (Figure 1C). The inflammatory responses under the cavity were obvious. The cells were obviously proliferated with reparative dentin formation by 7 d after cavity preparation (Figure 2A, 2B). The cells proliferated markedly. A large amount of reparative dentin formed in the area by 14 d after cavity preparation (Figure 2C, 2D). Reparative dentin had formed corresponding to the cavity preparation. The newly differentiated odontoblasts were arrayed in an orderly fashion along the reparative dentin, which was mineralized intensely. The dentinal tubules were arrayed in a disorderly fashion by 21 d after cavity preparation (Figure 2E, 2F).
Figure 1.
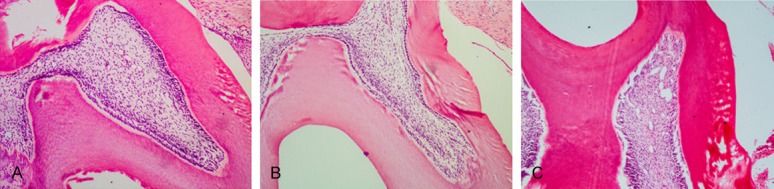
Histochemical staining of the injured dentin-dental pulp complex by 0 h and 72 h and the control teeth. (A, B) The odontoblast layer, the cell-free zone, the cell-rich zone and the subjacent pulp tissue could be seen clearly in the control teeth (A, ×100) after cavity preparation and 0 h (B, ×100). (C) The blood vessels expanded by 72 h, and the predentin became thick (C, ×100).
Figure 2.
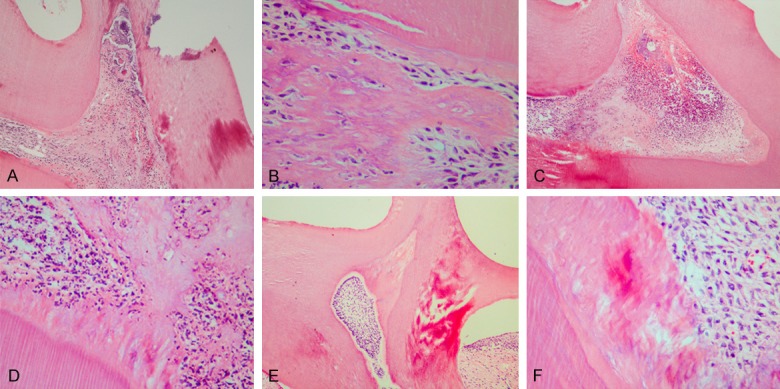
Histochemical staining of the injured dentin-dental pulp complex by 7 d, 14 d and 21 d. (A, B) The cells were obviously proliferated with the formation of reparative dentin by 7 d (A, ×100; B, ×400). (C, D) A large amount of reparative dentin had formed by 14 d (C, ×100; D, ×400). (E, F) The newly differentiated odontoblasts were arrayed in an orderly fashion along the reparative dentin by 21 d (E, ×100; F, ×400).
Expression of Lin28 in the injured dentin-dental pulp complex
The protein expression of Lin28 in the injured dentin-dental pulp complex was examined. Compared with that in the control teeth (Figure 3A), the expression of Lin28 by 72 h after cavity preparation showed a significant difference (P < 0.05), while the expression of Lin28 by 0 h showed no difference (P > 0.05) (Figure 3B). Immunohistochemical staining revealed a higher level of activity of Lin28 in the odontoblasts and the pulp cells adjacent to the cavity by 72 h, compared with those of the control teeth (P < 0.05) (Figure 3C, 3D). Compared with the expression in control teeth, the expression of Lin28 by 7 d, 14 d and 21 d after cavity preparation was significantly different (P < 0.05). By 7 d after cavity preparation, cells contributing to the dentin formation exhibited a positive reaction for Lin28 (P < 0.05) (Figure 4A). By 14 d after cavity preparation, the reparative dentin had weak expression, while the odontoblasts beneath the reparative dentin were stained (P < 0.05) (Figure 4B). However, a lower level of activity of Lin28 was observed in the odontoblasts and pulp cells by 21 d after cavity preparation (P < 0.05) (Figure 4C, 4D), compared with the expression of Lin28 expression by 7 d and 14 d.
Figure 3.
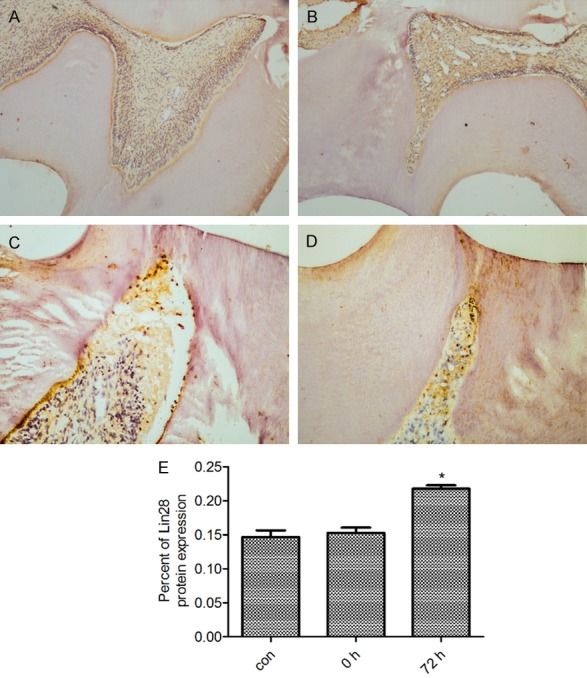
Immunolocalization of the expression of Lin28 protein by 0 h and 72 h and in the control teeth after cavity preparation. (A, B) Weak immunoreactivity expression of Lin28 protein was observed by 0 h (B, ×100) and in the control teeth (A, ×100). (C, D) A higher level of activity of Lin28 was observed in the odontoblasts and the pulp cells adjacent to the cavity by 72 h (C, ×100; D, ×200). (E) The percent of Lin28 protein expression by 0 h, 72 h and in the control teeth after cavity preparation. *P < 0.05 vs the normal dentin-dental pulp. The data are presented as the mean ± SEM.
Figure 4.
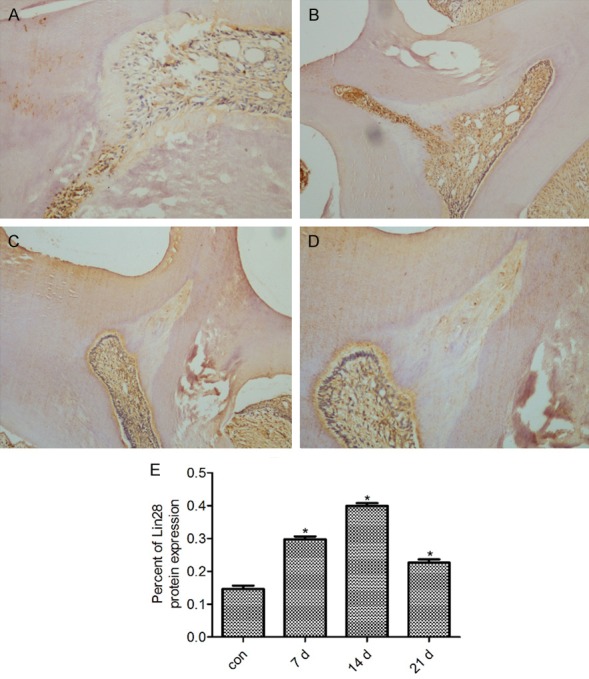
Immunolocalization of expression of Lin28 protein by 7 d, 14 d and 21 d after cavity preparation. (A) Some cells contributing to the dentin formation showed a positive reaction for Lin28 by 7 d (A, ×100). (B) The reparative dentin exhibited weak expression, while the odontoblasts beneath the reparative dentin were stained positively by 14 d (B, ×200). (C, D) A lower level of activity of Lin28 was observed in the odontoblasts and pulp cells by 21 d than was observed by 7 d and 14 d after cavity preparation (C, ×100; D, ×200). (E) The percent of Lin28 protein expression by 7 d, 14 d and 21 d after cavity preparation. *P < 0.05 vs the normal dentin-dental pulp. The data are presented as the mean ± SEM.
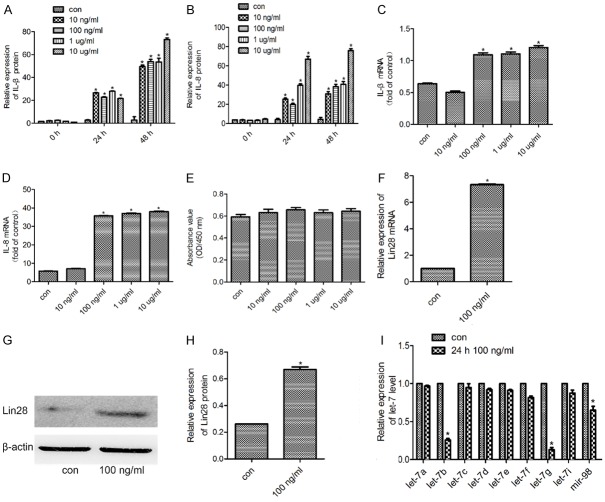
Effects of LPS on the expression of inflammatory cytokines
To evaluate the effect of LPS on inflammatory cytokines expression in HDPCs, we exposed HDPCs to various concentration of LPS for 24 h, collected the supernatant and performed ELISA. Compared with HDPCs without LPS, HDPCs exposed to 10 ng/ml, 100 ng/ml, 1 μg/ml and 10 μg/ml LPS for 24 h and 48 h showed induced upregulation of IL-1β and IL-8 expression (P < 0.05) (Figure 5A, 5B). Compared with HDPCs without LPS, HDPCs exposed to 100 ng/ml, 1 μg/ml and 10 μg/ml LPS for 24 h showed increased expression levels of both IL-1β and IL-8 mRNA (Figure 5C, 5D).
Figure 5.
The expression of Lin28 and let-7 families in normal HDPCs and HDPCs treated with LPS. A, B. The amounts of IL-1β and IL-8 in culture supernatants were determined by ELISA. C, D. The mRNA expression of IL-1β and IL-8 were determined by qPCR. E. The CCK-8 assay was used to assess the viability of normal cells and cells treated with LPS. F. Lin28 mRNA was examined by qPCR in normal cells and in cells treated with 100 ng/ml LPS for 24 h. G, H. Lin28 expression was determined in normal cells and in cells treated with 100 ng/ml LPS for 24 h by western blot. I. Expression of let-7 family miRNAs was determined by qPCR. β-Actin expression was used as a control. *P < 0.05 vs cells without LPS. The data are presented as the mean ± SEM.
Effects of LPS on the viability of HDPCs
A CCK-8 assay was used to study the impact of LPS on cell viability. The viability of cells was determined by measuring the optical density (OD) value at 450 nm. HDPCs were cultured for 24 h and the OD values were compared with those of the control cells. The results indicated that the viability of the cells exposed to 10 ng/ml, 100 ng/ml, 1 μg/ml and 10 μg/ml LPS for 24 h exhibited little difference from those of the control group (P > 0.05) (Figure 5E).
Effects of LPS on the expression of Lin28 in LPS-induced HDPCs
To examine the effect of LPS on the expression of Lin28 in HDPCs, we performed Western blot and qRT-PCR analyses. In accordance with the mRNA expression level (Figure 5F), the Western blot analysis demonstrated that the 100 ng/ml group expressed a higher protein level than the control group did (Figure 5G, 5H).
Expression of the let-7 miRNA family in LPS-induced HDPCs
The mRNA expression levels of members of the let-7 family, which are known to be regulated by Lin28, were examined. The results demonstrated that the expression of let-7b, let-7g and mir-98 were reduced in the LPS-induced group compared with the control, whereas let-7a, let-7c, let-7d, let-7e, let-7f and let-7i were increased (Figure 5I).
Discussion
Dentinogenesis occurs after intense injuries by a deposition of a protective layer of reparative dentine that is secreted by odontoblast-like cells at the injured dentin-dental pulp interface [26,27]. However, the differentiation of dental pulp cells and the formation of reparative dentin have complex mechanisms at the molecular level and requires further research. The results of the present study demonstrated that Lin28 was expressed in the injured dentin-dental pulp complex.
In the present study, the operative procedures of cavity preparation were similar to those described previously [28-30]. In vivo, the investigation performed on animals provided direct evidence that Lin28 participates in the repair process of the dentin-dental pulp complex. By 72 h, obvious inflammatory responses were observed. Reparative dentin formed 7 d after cavity preparation, which was consistent with the results of a previous study. By 14 d, Lin28 immunostaining was more visible around the zone of reparative dentin. By 21 d after cavity preparation, the newly differentiated odontoblast-like cells were located beneath the reparative dentin. Lin28 was seen in cells lining this irregular matrix. Lin28 protein expression levels were obviously increased by 7 d, 14 d and 21 d after cavity preparation, which was the precise time of reparative dentin formation. The level of activity of Lin28 was reduced by 21 d after cavity preparation, which revealed that the reparative dentin reached stability and that the proliferation process weakened.
The present in vitro study utilized LPS extracted from Escherichia coli O111:B4 LPS at a concentration of 100 ng/ml, as this concentration has been demonstrated to induce inflammatory reactions such as upregulation of IL-6, IL-8, IL-β, TNF-α, matrix metalloproteinase (MMP)-2, MMP-9, ICAM-1 and VCAM-1 in HDPCs [31,32]. It was observed that 100 ng/ml LPS stimulated HDPCs and upregulated the expression of inflammatory factors, such as IL-8 and IL-β, consistent with previous research. Moreover, it was demonstrated that Lin28 expression was increased while let-7b, let-7g and mir-98 were downregulated in the inflammatory cells, and let-7a, let-7c, let-7d, let-e, let-7f and let-7i had no significant difference, compared with the levels in control cells.
In conclusion, the present study demonstrated the participation of Lin28 in dentin regeneration in animal models. Additionally, the results indicated that let-7 may be the targets of Lin28 in the formation of reparative dentin and may constitute a very useful tool for pulp therapies. Future studies should focus on the specific regulatory mechanisms in order to provide novel directions for curing tooth diseases and promoting the development of effective therapies for other injuries.
Acknowledgements
This study was supported by the Science and Technology Planning Project of Shaanxi Province, China (grant No. 2015SF-153).
Disclosure of conflict of interest
None.
References
- 1.Gronthos S, Mankani M, Brahim J, Robey PG, Shi S. Postnatal human dental pulp stem cells (DPSCs) in vitro and in vivo. Proc Natl Acad Sci U S A. 2000;97:13625–13630. doi: 10.1073/pnas.240309797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steiner R, Fischer-Colbrie R, Bletsa A, Laimer J, Troger J. Secretoneurin and PE-11 immunoreactivity in the human dental pulp. Arch Oral Biol. 2018;86:13–17. doi: 10.1016/j.archoralbio.2017.11.005. [DOI] [PubMed] [Google Scholar]
- 3.Casagrande L, Cordeiro MM, Noer SA, Noer JE. Dental pulp stem cells in regenerative dentistry. Odontology. 2011;99:1–7. doi: 10.1007/s10266-010-0154-z. [DOI] [PubMed] [Google Scholar]
- 4.Nuti N, Corallo C, Chan BM, Ferrari M, Gerami-Naini B. Multipotent differentiation of human dental pulp stem cells: a literature review. Stem Cell Rev. 2016;12:511–523. doi: 10.1007/s12015-016-9661-9. [DOI] [PubMed] [Google Scholar]
- 5.de Santana DA, Fonseca GF, Ramalho LM, Rodriguez TT, Aguiar MC. Effect of low-level laser therapy (lambda780 nm) on the mechanically damaged dentin-pulp complex in a model of extrusive luxation in rat incisors. Lasers in Medical Science. 2017;32:1995–2004. doi: 10.1007/s10103-017-2295-6. [DOI] [PubMed] [Google Scholar]
- 6.Matsui M, Kobayashi T, Tsutsui TW. CD146 positive human dental pulp stem cells promote regeneration of dentin/pulp-like structures. Hum Cell. 2018;31:127–138. doi: 10.1007/s13577-017-0198-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W, Ju J. Odontoblast-targeted Bcl-2 overexpression promotes dentine damage repair. Arch Oral Biol. 2012;57:285–292. doi: 10.1016/j.archoralbio.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng Y, Wang XY, Wang YM, Liu XY, Zhang CM, Hou BX, Wang SL. Dentin regeneration using deciduous pulp stem/progenitor cells. J Dent Res. 2012;91:676–682. doi: 10.1177/0022034512449834. [DOI] [PubMed] [Google Scholar]
- 9.Spahr A, Lyngstadaas SP, Slaby I, Haller B, Boeckh C, Tsoulfidou F, Hammarstrom L. Expression of amelin and trauma-induced dentin formation. Clin Oral Investig. 2002;6:51–57. doi: 10.1007/s00784-001-0139-y. [DOI] [PubMed] [Google Scholar]
- 10.Rowe RG, Wang LD, Coma S, Han A, Mathieu R, Pearson DS, Ross S, Sousa P, Nguyen PT, Rodriguez A, Wagers AJ, Daley GQ. Developmental regulation of myeloerythroid progenitor function by the Lin28b-let-7-Hmga2 axis. J Exp Med. 2016;213:1497–1512. doi: 10.1084/jem.20151912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balzeau J, Menezes MR, Cao S, Hagan JP. The LIN28/let-7 pathway in cancer. Front Genet. 2017;8:31. doi: 10.3389/fgene.2017.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reddien PW. Lin28: time for tissue repair. Cell. 2013;155:738–739. doi: 10.1016/j.cell.2013.10.025. [DOI] [PubMed] [Google Scholar]
- 13.Shyh-Chang N, Zhu H, Yvanka de Soysa T, Shinoda G, Seligson MT, Tsanov KM, Nguyen L, Asara JM, Cantley LC, Daley GQ. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 2013;155:778–792. doi: 10.1016/j.cell.2013.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu Y, Gao L, Shi Z, You F, Zhang J, Li W. Characterization and expression of lin-28a involved in lin28/let-7signal pathway during early development of P. olivaceus. Fish Physiol Biochem. 2017;44:451–463. doi: 10.1007/s10695-017-0445-1. [DOI] [PubMed] [Google Scholar]
- 15.Manier S, Powers JT, Sacco A, Glavey SV, Huynh D, Reagan MR, Salem KZ, Moschetta M, Shi J, Mishima Y, Roche-Lestienne C, Leleu X, Roccaro AM, Daley GQ, Ghobrial IM. The LIN28B/let-7 axis is a novel therapeutic pathway in multiple myeloma. Leukemia. 2017;31:853–860. doi: 10.1038/leu.2016.296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang M, Lee KH, Lee HS, Jeong CW, Ku JH, Kim HH, Kwak C. Concurrent treatment with simvastatin and NF-kappaB inhibitor in human castration-resistant prostate cancer cells exerts synergistic anti-cancer effects via control of the NF-kappaB/LIN28/let-7 miRNA signaling pathway. PLoS One. 2017;12:e0184644. doi: 10.1371/journal.pone.0184644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jun-Hao ET, Gupta RR, Shyh-Chang N. Lin28 and let-7 in the metabolic physiology of aging. Trends Endocrinol Metab. 2016;27:132–141. doi: 10.1016/j.tem.2015.12.006. [DOI] [PubMed] [Google Scholar]
- 18.Zhu H, Shyh-Chang N, Segrè AV, Shinoda G, Shah SP, Einhorn WS, Takeuchi A, Engreitz JM, Hagan JP, Kharas MG, Urbach A, Thornton JE, Triboulet R, Gregory RI DIAGRAM Consortium; MAGIC Investigators. Altshuler D, Daley GQ. The Lin28/let-7 axis regulates glucose metabolism. Cell. 2011;147:81–94. doi: 10.1016/j.cell.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McDaniel K, Huang L, Sato K, Wu N, Annable T, Zhou T, Ramos-Lorenzo S, Wan Y, Huang Q, Francis H, Glaser S, Tsukamoto H, Alpini G, Meng F. The let-7/Lin28 axis regulates activation of hepatic stellate cells in alcoholic liver injury. J Biol Chem. 2017;292:11336–11347. doi: 10.1074/jbc.M116.773291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thornton JE, Gregory RI. How does Lin28 let-7 control development and disease? Trends Cell Biol. 2012;22:474–482. doi: 10.1016/j.tcb.2012.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, Iliopoulos D, Gregory RI. Lin28A and Lin28B inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–1079. doi: 10.1016/j.cell.2011.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim DS, Shin MR, Kim YS, Bae WJ, Roh DH, Hwang YS, Kim EC. Anti-inflammatory effects of glutamine on LPS-stimulated human dental pulp cells correlate with activation of MKP-1 and attenuation of the MAPK and NFkappaB pathways. Int Endod J. 2015;48:220–228. doi: 10.1111/iej.12303. [DOI] [PubMed] [Google Scholar]
- 23.Lim W, Bae H, Bazer FW, Song G. C-C motif chemokine ligand 23 abolishes ER stress- and LPS-induced reduction in proliferation of bovine endometrial epithelial cells. J Cell Physiol. 2018;233:3529–3539. doi: 10.1002/jcp.26210. [DOI] [PubMed] [Google Scholar]
- 24.Jung JY, Woo SM, Kim WJ, Lee BN, Nor JE, Min KS, Choi CH, Koh JT, Lee KJ, Hwang YC. Simvastatin inhibits the expression of inflammatory cytokines and cell adhesion molecules induced by LPS in human dental pulp cells. Int Endod J. 2017;50:377–386. doi: 10.1111/iej.12635. [DOI] [PubMed] [Google Scholar]
- 25.Rao X, Lai D, Huang X. A new method for quantitative real-time polymerase chain reaction data analysis. J Comput Biol. 2013;20:703–711. doi: 10.1089/cmb.2012.0279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orhan EO, Maden M, Senguuven B. Odontoblast-like cell numbers and reparative dentine thickness after direct pulp capping with platelet-rich plasma and enamel matrix derivative: a histomorphometric evaluation. Int Endod J. 2012;45:317–325. doi: 10.1111/j.1365-2591.2011.01977.x. [DOI] [PubMed] [Google Scholar]
- 27.Murakami Masuda Y, Wang X, Yokose S, Yamada Y, Kimura Y, Okano T, Matsumoto K. Effect of glypican-1 gene on the pulp cells during the reparative dentine process. Cell Biol Int. 2010;34:1069–1074. doi: 10.1042/CBI20090062. [DOI] [PubMed] [Google Scholar]
- 28.Wang X, He H, Wu X, Hu J, Tan Y. Promotion of dentin regeneration via CCN3 modulation on Notch and BMP signaling pathways. Biomaterials. 2014;35:2720–2729. doi: 10.1016/j.biomaterials.2013.12.029. [DOI] [PubMed] [Google Scholar]
- 29.Hwang YC, Hwang IN, Oh WM, Park JC, Lee DS, Son HH. Influence of TGF-beta1 on the expression of BSP, DSP, TGF-beta1 receptor I and Smad proteins during reparative dentinogenesis. J Mol Histol. 2008;39:153–160. doi: 10.1007/s10735-007-9148-8. [DOI] [PubMed] [Google Scholar]
- 30.Tziafas D. Mechanisms controlling secondary initiation of dentinogenesis: a review. Int Endod J. 1994;27:61–74. doi: 10.1111/j.1365-2591.1994.tb00233.x. [DOI] [PubMed] [Google Scholar]
- 31.Dodd CA, Filipov NM. Manganese potentiates LPS-induced heme-oxygenase 1 in microglia but not dopaminergic cells: role in controlling microglial hydrogen peroxide and inflammatory cytokine output. Neurotoxicology. 2011;32:683–692. doi: 10.1016/j.neuro.2011.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim JC, Lee YH, Yu MK, Lee NH, Park JD, Bhattarai G, Yi HK. Anti-inflammatory mechanism of PPARgamma on LPS-induced pulp cells: role of the ROS removal activity. Arch Oral Biol. 2012;57:392–400. doi: 10.1016/j.archoralbio.2011.09.009. [DOI] [PubMed] [Google Scholar]