Abstract
This study mainly studied the correlation of RASEF expression and the clinical index of colorectal cancer by tissue microarray (TMAs, HCol-Adel180sur-06) containing tissue samples of 90 colorectal cancers. The results showed that RASEF was significantly highly expressed both in nuclei (3.07±1.95 vs 1.83±1.74, P=0.000) and cytoplasm (7.74±2.08 vs 5.83±1.97, P=0.000) compared to their para-carcinoma tissues, which was in line with the data of the Oncomine database. The correlation between RASEF expression and microsatellite instability, analyzed by Spearman’s correlation analysis showed that RASEF expression in colorectal cancer cytoplasm was correlated significantly with the mismatch repair genes MLH1 (P=0.037; r=0.227) and MSH6 (P=0.038; r=0.224). Additionally, high RASEF expression was associated with a significantly better prognosis (45.3% vs 8%, P=0.041), which was consistent with the data of the Human Protein Atlas. Subsequently, Cox analysis of multi-factor survival showed that RASEF expression was an independent predictive factor for colorectal cancer (P=0.001). Thus, we speculated that RASEF may be a suppressor gene, and may inhibit the development of colorectal cancer through participating in DNA repair processes.
Keywords: RASEF, high expression, colorectal cancer, prognosis
Introduction
Colorectal cancer is one of the most common gastrointestinal tumors with a high death rate and an apparent increase in incidence. Colorectal cancer incidence is mainly concentrated in North America and other developed countries [1]. Although the incidence and death rates of colorectal cancer declined about 3% per year inrecent years, death rate remains high. Therefore, the prevention, treatment, and prognosis of colorectal cancer remains a significant problem in the global public health field [2]. The characterization of the molecular mechanisms and independent predictive factors of colorectal cancer should improve diagnosis and treatment.
The RAS (Ras-related protein) superfamily of GTPases consists of five different subfamilies including RAS, RHO, RAB, and ARF families, and the closely related Gα family [3]. The Rabs formed the largest branch of the RAS superfamily, which act as indispensable regulators of vesicular transport pathways [4-6]. Traditionally, Rab1, 2, 3, and other more than 70 members have been numbered in order of discovery over the last decade [8]. Since the Rabs perform the necessary function of regulation in cellular vesicular transport, studies have demonstrated that Rabs played a key role in tumorigenesis and some cancer progression [6]. For example, in a cancer type-dependent manner, Rab25 could functions as an oncogene or a tumor suppressor as it regulates apical transport as well as recycling of vesicles to the plasma membrane [3]. Studies have revealed that several members’ abnormal expression might perform an important role in the occurrence, development, or metastasis of colorectal cancer [3,9,10]. Few studies have revealed the relationship of RASEF expression and human cancers, especially colorectal cancer.
The RASEF (RAS and EF-hand domain-containing protein), a member of Rab GTPase protein family, contains a Rab GTPase domain in the C-terminal region, a coiled coil motif in an internal region as well as 2 EF hand domains which were of importance for binding to calcium ions in the N-terminus [11,12]. However, little is known about RASEF and human cancers especially colorectal cancer, so unraveling the mechanism of tumor progression and further discovery of novel prognostic markers for prediction and treatment evaluation is urgently required.
Materials and methods
Clinical data
The colorectal cancer tissue microarray (HCol-Adel180sur-06) contained 90 colorectal cancer patients’ tissue samples and 90 paired para-carcinoma tissues were made by Shanghai Outdo Biotech Co., Ltd to research the expression of RASEF. The colorectal cancer tissue microarray contained well-documented clinicopathological information: among these patients, 47 were men, other 42 were women as well as 1 was lost to follow-up, while the age of these patients’ were range from 24 to 90 with a median age of 70, and the tumor size varied from 1.5 cm to 15 cm, with the median size of 5.9 cm. 8 of these colorectal cancer patients’ clinical grade were stage I, 48 were stage II, 30 were stage III, and 2 were stage IV. 4 patients’ clinical stage was not obtained.
All the colorectal cancer patients were clinicopathologically diagnosed as colorectal cancer and received no extra treatment before surgery. The operation time was from July 2006 to May 2007 and the eventual follow-up time was in August 2014, containing 87-97 months. The results of statistical analysis showed that during the follow-up time, 56 of the 90 patients died of colorectal cancer, though the other 34 patients were still alive, while the median follow-up time was about 92 months.
Immunohistochemistry
Two-step immunohistochemistry: first, tissue sections were treated with EDTA buffer to retrieval antigen; and then incubated with primary antibody which anti-RE (1:800, Proteintech) at 4°C overnight; the very next sections were incubated with secondary antibody (HRP-labeled anti-mouse antibody, DAKO). It was visualized using diaminobenzidine (DAB) system and hematoxylin re-dying after washed with PBS. The RE expression was observed and analyzed by pathologist under microscope, scored and grouped with positive staining rate and intensity, and the positive staining rate was defined according to the proportion of positively stained cancer cells: “Negative” is 0, “1%-25%” for 1, “26%-50%” for 2, “51-75%” for 3, “76-100%” for 4. The score of staining intensity was defined as: “Negative” is 0, “1+” for 1, “2+” for 2, “3+” for 3. Thus, patients were divided into two groups according to the scores of “positive staining rate score” multiplied by “staining intensity score”. It was divided into a high expression group when the score is equal or more than 6, while less than 6 wasconsidered low expression group.
Statistical analysis
We statistically analyzed the differential expressions of RASEF in colorectal cancer tissues and their adjacent tissues useing NPar Test. The RASEF mRNA expression in colorectal specimens was obtained from the public available cancer microarray database ONCOMINE (https://www.oncomine.org/). The correlations between RASEF expression and colorectal cancer patients’ clinical indexes were analyzed by Pearson’s correlation. The relationships between the overall survival time of colorectal cancer patients and RASEF expression were analyzed by the Kaplan-Meier method and the log-rank test. Finally, statistically significant variables in univariate analysis were included in Cox multivariate regression survival analysis. P<0.05 was considered significant.
Results
RASEF expression was significantly elevated in colorectal cancer
To characterize the role of RASEF in colorectal cancer, we analyzed RASEF mRNA expression in colorectal specimens from the public available cancer microarray database ONCOMINE (https://www.oncomine.org/). We found that RASEF expression was significantly increased in colorectal carcinoma (Figure 1A) compared to colon.
Figure 1.
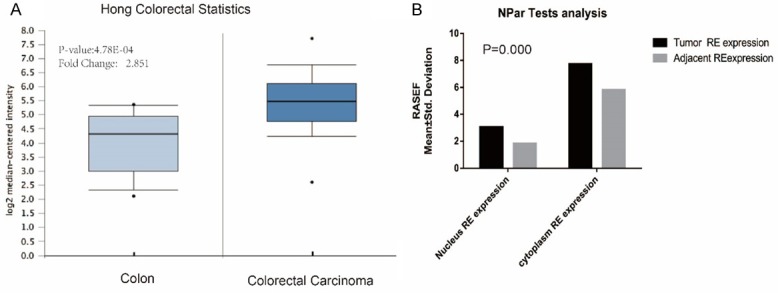
RASEF expression is significantly elevated in colorectal cancer. A. RASEF mRNA expression analysis using Oncomine. RASEF was significantly overexpressed in colorectal carcinoma compared with normal colon samples from the Hong Colorectal Statistics (P=4.78E-4). B. Similar results were observed in the colorectal microarray data (P=0.000). P<0.05 was considered significant.
Beyond that, we also evaluated the levels of RASEF protein in colorectal cancer tissue microarray and did statistical analysis of results of immunohistochemistry showing that the RASEF could expressed in both cytoplasm and nucleus, while the expression of RASEF between colorectal tissues and para-carcinoma tissues were significantly elevated both in cytoplasm of colorectal cancer (7.74±2.08 VS 5.83±1.97, P=0.000) and in the nucleus (3.07±1.95 VS 1.83±1.74, P=0.000). The results are shown in Figure 1.
The correlation between RASEF expression and colorectal cancer clinical parameters
The correlation between RASEF expression and colorectal cancer clinical parameters were analyzed by the Spearman’s correlation analysis, and the results showed that there was no significant correlation between the expression of RASEF and the clinical indicators of colon cancer. The results are shown in Table 1.
Table 1.
Correlation between RASEF expression and clinical indicators of colorectal cancer patients: Spearman’s correlation analysis
| Gender | Age | Tumor size | Pathological grading | T | N | M | Clinical grading | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Spearman’s rho | Cytoplasm RE expression | Tumor | Correlation Coefficient | -.035 | -.017 | .064 | .010 | -.081 | .005 | .125 | .054 |
| Sig. (2-tailed) | .752 | .878 | .562 | .927 | .480 | .960 | .251 | .628 | |||
| N | 85 | 81 | 85 | 86 | 79 | 86 | 86 | 84 | |||
| Adjacent | Correlation Coefficient | -.032 | -.017 | -.103 | -.107 | -.142 | .066 | -.156 | -.010 | ||
| Sig. (2-tailed) | .779 | .885 | .356 | .336 | .220 | .552 | .160 | .930 | |||
| N | 82 | 78 | 82 | 83 | 76 | 83 | 83 | 81 | |||
| Nucleus RE expression | Tumor | Correlation Coefficient | .024 | .088 | .154 | .162 | .156 | .007 | .010 | .055 | |
| Sig. (2-tailed) | .824 | .433 | .156 | .133 | .167 | .950 | .927 | .616 | |||
| N | 86 | 82 | 86 | 87 | 80 | 87 | 87 | 85 | |||
| Adjacent | Correlation Coefficient | -.067 | -.101 | -.001 | .242* | .100 | .027 | .146 | .068 | ||
| Sig. 2-tailed) | .549 | .378 | .992 | .027 | .389 | .808 | .188 | .547 | |||
| N | 82 | 78 | 82 | 83 | 76 | 83 | 83 | 81 | |||
**Correlation is significant at the 0.01 level (2-tailed).
Correlation is significant at the 0.05 level (2-tailed).
The correlation between RASEF expression and Microsatellite Instability
Spearman’s correlation analysis showed that RASEF expression in colorectal cancer cytoplasm was correlated significantly with the mismatch repair gene MLH1 (P=0.037; r=0.227) and MSH6 (P=0.038; r=0.224). The results are shown in Table 2.
Table 2.
The ccorrelation between RASEF expression and microsatellite instability: spearman’s correlation analysis
| MLH1 | MSH6 | MSH2 | PMS2 | ||||
|---|---|---|---|---|---|---|---|
| Spearman’s rho | Cytoplasm RE expression | Tumor | Correlation Coefficient | .227* | .224* | .201 | .209 |
| Sig. (2-tailed) | .037 | .038 | .068 | .056 | |||
| N | 85 | 86 | 83 | 84 | |||
| Adjacent | Correlation Coefficient | -.190 | -.113 | -.185 | -.074 | ||
| Sig. (2-tailed) | .090 | .310 | .102 | .516 | |||
| N | 81 | 83 | 79 | 80 | |||
| Nucleus RE expression | Tumor | Correlation Coefficient | -.116 | .069 | .013 | .047 | |
| Sig. (2-tailed) | .286 | .526 | .904 | .672 | |||
| N | 86 | 87 | 84 | 85 | |||
| Adjacent | Correlation Coefficient | -.274* | -.035 | -.144 | .033 | ||
| Sig. (2-tailed) | .013 | .750 | .204 | .769 | |||
| N | 81 | 83 | 79 | 80 | |||
**Correlation is significant at the 0.01 level (2-tailed).
Correlation is significant at the 0.05 level (2-tailed).
The relationships between the overall survival time of colorectal cancer patients and RASEF expression
Kaplan-Meier method and the log-rank test analysis results showed that patients with RASEF high expression had a significantly better prognosis (45.3% vs 8%, P=0.041), which was consistent with the data of the Human Protein Atlas (https://www.proteinatlas.org/). The analysis results were shown in Figure 2.
Figure 2.
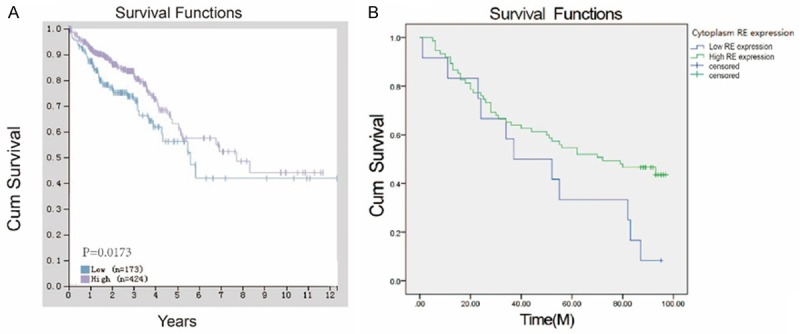
High RASEF was associated with a significantly better prognosis in colorectal cancer. A. High RASEF expression associated with better prognosis in colorectal carcinoma compared with low RASEF expression (P=1.73e-2) from the Human Protein Atlas. B. Similar results were observed in the correlation of RASEF expression in cancer cytoplasm and the prognosis of colorectal cancer from colorectal microarray data (P=0.041). P<0.05 was considered significant.
Subsequently, multivariate analyses of clinical factors associated with survival and RASEF expression in colorectal cancer demonstrated that RASEF expression was an independent predictive factor (P=0.001). Detailed results are shown (Table 3).
Table 3.
Cox multivariate regression analysis of the independent predictors of RASEF in colorectal cancer patients
| B | SE | Wald | Df | Sig. | Exp(B) | 95.0% Exp(B) confidence interval | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Lower limit | Upper limit | |||||||
| Cytoplasm RE expression in cancer | -1.191 | .361 | 10.897 | 1 | .001 | .304 | .150 | .616 |
| Nucleus RE expression in cancer | 1.743 | .548 | 10.113 | 1 | .001 | 5.713 | 1.952 | 16.723 |
| N | .624 | .369 | 2.865 | 1 | .091 | 1.866 | .906 | 3.844 |
| M | 1.636 | .860 | 3.621 | 1 | .057 | 5.137 | .952 | 27.709 |
| Clinical stage | .241 | .406 | .352 | 1 | .553 | 1.273 | .574 | 2.821 |
A. The degree of freedom was reduced because of the constant or linear variation. B. Constant or linear covariance. Clinical stages = T + N + M. **Correlation is significant at the 0.01 level (2-tailed). *Correlation is significant at the 0.05 level (2-tailed).
Discussion
Colorectal cancer is one of the most common gastrointestinal tumors with a high death rate and an apparent increase in incidence. The prevention, treatment, and prognosis of colorectal cancer remain unresolved issues in the global public health field. Therefore, finding independent predictive factors of colorectal cancer should have a significant sense on diagnosing and treating colorectal cancer.
The RAB protein is a small GTP binding protein that forms the largest branch of the RAS superfamily which plays a vital role in multiple types of tumorigenesis, as well as regulates the proliferation, migration, and invasion of tumor cells. It has been reported that Rab proteins’ expression have a closely connection with tumorigenesis and development progression like breast cancer [13,14], renal cancer [15], gastric cancer [16], liver cancer [17,18], ovarian cancer [19], glioblastoma multiforme [20], head and neck cancer [21], and esophageal squamous cell carcinoma [22]. An increasing number of studies report that some Rabs proteins’ expression have close correlation with colorectal cancer [10,23,24]. However, as a member of Rabs protein family, there was little research on the correlation between RASEF and human cancers. Studies reported that high RASEF expression could promote cell growth via enhanced ERK signaling and was associated with poor prognosis of NSCLC, so that RASEF could serve as a novel diagnostic biomarker and a therapeutic target for lung cancer.
Our experiment mainly studied the correlation between the RASEF expression and the clinical index of colorectal cancer. The results showed that RASEF was significantly highly expressed both in colorectal cancer nuclei (3.07±1.95 vs 1.83±1.74, P=0.000) and cytoplasm compared to their para-carcinoma tissues (7.74±2.08 vs 5.83±1.97, P=0.000), which was in line with data of Oncomine database and also previous studies reporting that RAB has important roles in ccRCC [1,3,11]. Spearman’s analysis on the correlation between RASEF expression and Microsatellite Instability showed that RASEF expression in colorectal cancer cytoplasm was significantly correlated with MLH1 and MSH6. Mismatch repair gene abnormality and microsatellite instability could serve as preliminary evidence of the tumorigenesis as most colorectal cancers have proficient MMR [25], which demonstrated that the high expression of RASEF might correlate with inhibiting tumorigenesis or the development of colorectal cancer through interacting with MLH1 and MSH6.
Moreover, patients with high RASEF expression had a significantly better prognosis (45.3% vs 8%, P=0.041), which was consistent with the data of the Human Protein Atlas (https://www.proteinatlas.org/). The Cox multi-factor survival analysis showed that RASEF expression was an independent predict factor for colorectal cancer (P=0.001). Thus, we speculate that RASEF may be serve as a suppressor gene, and may inhibit the process of colon carcinogenesis through participating in DNA repair processes. Further work focusing on the RASEF expression affection on colorectal cancer cellular function and metastasis ability should be done to support this.
In conclusion, we unveiled the prognostic significance of RASEF in colorectal cancer, and suggested RASEF expression to be a potential prognostic and therapeutic marker for this disease.
Acknowledgements
The study was supported by National Nature Science Foundation of China (to M.L.) [Grant number: 81372612] National Nature Science Foundation of China (to G.L.) [Grant number: 81302059] and Nature Science Foundation of Heilongjiang Province of China (to G.L.) [Grant number: LC2013C35].
Disclosure of conflict of interest
None.
References
- 1.Chang YC, Su CY, Chen MH, Chen WS, Chen CL, Hsiao M. Secretory RAB GTPase 3C modulates IL6-STAT3 pathway to promote colon cancer metastasis and is associated with poor prognosis. Mol Cancer. 2017;16:135. doi: 10.1186/s12943-017-0687-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 3.Goldenring JR, Nam KT. Rab25 as a tumour suppressor in colon carcinogenesis. Br J Cancer. 2011;104:33–36. doi: 10.1038/sj.bjc.6605983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. J Mol Biol. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- 5.Colicelli J. Human RAS superfamily proteins and related GTPases. Sci Stke. 2004;2004:RE13. doi: 10.1126/stke.2502004re13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferro-Novick S, Novick P. The role of GTP-binding proteins in transport along the exocytic pathway. Annu Rev Cell Biol. 1993;9:575–99. doi: 10.1146/annurev.cb.09.110193.003043. [DOI] [PubMed] [Google Scholar]
- 7.Wang S, Hu C, Wu F, He S. Rab25 GTPase: functional roles in cancer. Oncotarget. 2017;8:64591–64599. doi: 10.18632/oncotarget.19571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feng F, Jiang Y, Lu H, Lu X, Wang S, Wang L, Wei M, Lu W, Du Z, Ye Z, Yang G, Yuan F, Ma Y, Lei X, Lu Z. Rab27A mediated by NF-kappaB promotes the stemness of colon cancer cells via up-regulation of cytokine secretion. Oncotarget. 2016;7:63342–63351. doi: 10.18632/oncotarget.11454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu MH, Luo Y, Qin SL, Zhong M. Increased expression of Rab5A predicts metastasis and poor prognosis in colorectal cancer patients. Int J Clin Exp Pathol. 2015;8:6974–6980. [PMC free article] [PubMed] [Google Scholar]
- 10.Shintani M, Tada M, Kobayashi T, Kajiho H, Kontani K, Katada T. Characterization of Rab45/RASEF containing EF-hand domain and a coiled-coil motif as a self-associating GTPase. Biochem Biophys Res Commun. 2007;357:661–667. doi: 10.1016/j.bbrc.2007.03.206. [DOI] [PubMed] [Google Scholar]
- 11.Oshita H, Nishino R, Takano A, Fujitomo T, Aragaki M, Kato T, Akiyama H, Tsuchiya E, Kohno N, Nakamura Y, Daigo Y. RASEF is a novel diagnostic biomarker and a therapeutic target for lung cancer. Mol Cancer Res. 2013;11:937–951. doi: 10.1158/1541-7786.MCR-12-0685-T. [DOI] [PubMed] [Google Scholar]
- 12.Mitra S, Federico L, Zhao W, Dennison J, Sarkar TR, Zhang F, Takiar V, Cheng KW, Mani S, Lee JS, Mills GB. Rab25 acts as an oncogene in luminal B breast cancer and is causally associated with Snail driven EMT. Oncotarget. 2016;7:40252–40265. doi: 10.18632/oncotarget.9730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grismayer B, Solch S, Seubert B, Kirchner T, Schafer S, Baretton G, Schmitt M, Luther T, Kruger A, Kotzsch M, Magdolen V. Rab31 expression levels modulate tumor-relevant characteristics of breast cancer cells. Mol Cancer. 2012;11:62. doi: 10.1186/1476-4598-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, Jia Q, Zhang Q, Wan Y. Rab25 upregulation correlates with the proliferation, migration, and invasion of renal cell carcinoma. Biochem Biophys Res Commun. 2015;458:745–750. doi: 10.1016/j.bbrc.2015.01.144. [DOI] [PubMed] [Google Scholar]
- 15.Li Y, Chen S, Shan Z, Bi L, Yu S, Li Y, Xu S. miR-182-5p improves the viability, mitosis, migration, and invasion ability of human gastric cancer cells by down-regulating RAB27A. Biosci Rep. 2017;7 doi: 10.1042/BSR20170136. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Yang MQ, Du Q, Goswami J, Varley PR, Chen B, Wang RH, Morelli AE, Stolz DB, Billiar TR, Li J, Geller DA. Interferon regulatory factor 1-Rab27a regulated extracellular vesicles promote liver ischemia/reperfusion injury. Hepatology. 2018;67:1056–1070. doi: 10.1002/hep.29605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldenring JR, Shen KR, Vaughan HD, Modlin IM. Identification of a small GTP-binding protein, Rab25, expressed in the gastrointestinal mucosa, kidney, and lung. J Biol Chem. 1993;268:18419–18422. [PubMed] [Google Scholar]
- 18.Fan Y, Wang L, Han X, Liu X, Ma H. Rab25 is responsible for phosphoinositide 3-kinase/AKTmediated cisplatin resistance in human epithelial ovarian cancer cells. Mol Med Rep. 2015;11:2173–2178. doi: 10.3892/mmr.2014.2963. [DOI] [PubMed] [Google Scholar]
- 19.Ding B, Cui B, Gao M, Li Z, Xu C, Fan S, He W. Knockdown of ras-related protein 25 (Rab25) inhibits the in vitro cytotoxicity and in vivo antitumor activity of human glioblastoma multiforme cells. Oncol Res. 2017;25:331–340. doi: 10.3727/096504016X14736286083065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Amornphimoltham P, Rechache K, Thompson J, Masedunskas A, Leelahavanichkul K, Patel V, Molinolo A, Gutkind JS, Weigert R. Rab25 regulates invasion and metastasis in head and neck cancer. Clin Cancer Res. 2013;19:1375–1388. doi: 10.1158/1078-0432.CCR-12-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tong M, Chan KW, Bao JY, Wong KY, Chen JN, Kwan PS, Tang KH, Fu L, Qin YR, Lok S, Guan XY, Ma S. Rab25 is a tumor suppressor gene with antiangiogenic and anti-invasive activities in esophageal squamous cell carcinoma. Cancer Res. 2012;72:6024–6035. doi: 10.1158/0008-5472.CAN-12-1269. [DOI] [PubMed] [Google Scholar]
- 22.Dong W, Cui J, Yang J, Li W, Wang S, Wang X, Li X, Lu Y, Xiao W. Decreased expression of Rab27A and Rab27B correlates with metastasis and poor prognosis in colorectal cancer. Discov Med. 2015;20:357–367. [PubMed] [Google Scholar]
- 23.Bao J, Ni Y, Qin H, Xu L, Ge Z, Zhan F, Zhu H, Zhao J, Zhou X, Tang X, Tang L. Rab27b is a potential predictor for metastasis and prognosis in colorectal cancer. Gastroenterol Res Pract. 2014;2014:913106. doi: 10.1155/2014/913106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 25.Poynter JN, Siegmund KD, Weisenberger DJ, Long TI, Thibodeau SN, Lindor N, Young J, Jenkins MA, Hopper JL, Baron JA, Buchanan D, Casey G, Levine AJ, Marchand LL, Gallinger S, Bapat B, Potter JD, Newcomb PA, Haile RW, Laird PW. Molecular characterization of MSI-H colorectal cancer by MLHI promoter methylation, immunohistochemistry, and mismatch repair germline mutation screening. Cancer Epidemiol Biomarkers Prev. 2008;17:3208–3215. doi: 10.1158/1055-9965.EPI-08-0512. [DOI] [PMC free article] [PubMed] [Google Scholar]


