Abstract
Viral and cellular RNA-binding proteins regulate numerous key steps in the replication of diverse virus genera. Viruses efficiently co-opt the host cell machinery for purposes such as transcription, splicing and subcellular localization of viral genomes. Though viral RNAs often need to resemble cellular RNAs to effectively utilize the cellular machinery, they still retain unique sequence and structural features for recognition by viral proteins for processes such as RNA polymerization, RNA export and selective packaging into virus particles. While beneficial for virus replication, distinct features of viral nucleic acids can also be recognized as foreign by several host defense proteins. Development of the crosslinking immunoprecipitation coupled with sequencing (CLIP) approach has allowed the study of viral and cellular RNA binding proteins that regulate critical aspects of viral replication in unprecedented detail. By combining immunoprecipitation of covalently crosslinked protein-RNA complexes with high-throughput sequencing, CLIP provides a global account of RNA sequences bound by RNA-binding proteins of interest in physiological settings and at nucleotide resolution. Here, we describe the step-by-step application of the CLIP methodology within the context of two cellular splicing regulatory proteins, hnRNP A1 and hnRNP H1 that regulate HIV-1 splicing. In principle, this versatile protocol can be applied to many other viral and cellular RNA-binding proteins.
Keywords: RNA-binding proteins, protein-RNA interactions, CLIP, sequencing
INTRODUCTION
Viruses containing RNA genomes as their genetic material (i.e. RNA viruses and retroviruses) cause a number of diseases ranging from the common cold to AIDS, and are major contributors to the global infectious disease burden. To replicate and propagate their RNA genomes, these viruses must efficiently utilize host cell machinery to facilitate a number of processes including viral RNA transcription, splicing, transport between subcellular compartments and translation. Viral RNAs also must retain unique sequence and structural features in order for recognition by viral RNA binding proteins (RBPs) that mediate RNA polymerization and export, selective genome packaging, RNA localization in virions, and RNA stability during early stages of infection. While beneficial for virus replication, the distinct features of viral RNAs can also be recognized as foreign by several host defense proteins that mount antiviral defenses through the production of cytokines upon binding to viral RNAs. In sum, a myriad of protein-RNA interactions play crucial roles in the life cycle of viruses, and as such identifying these interactions is crucial to understanding their pathogenesis.
Development of the CLIP approach has revolutionized the study of protein-RNA interactions [1–4]. Traditionally, RBP-RNA interactions have been studied by methods that required the a priori knowledge of the binding site, such as in vitro binding assays or purification of protein-RNA complexes from cell lysates with downstream analysis of bound RNA by Q-RT-PCR. Coupling of protein-RNA complex isolation with microarray analysis, and more recently, high throughput sequencing provided a more global picture of the bound RNAs, but suffered from high background and low specificity due to purification of non-specific RNAs or multiple RBPs in complex with their bound RNAs [5, 6]. While in vitro approaches such as SELEX [7, 8], RNA-compete [9], RNA-bind-n-Seq [10], and RNA-Map [11] can provide detailed nucleotide resolution information of the target site and the biochemical properties of these interactions, they cannot determine which sequence the RBP of interest will bind to at physiologically relevant concentrations. What makes CLIP a powerful approach is that it addresses all of the shortcomings of these previous approaches by yielding nucleotide resolution information of the RNA molecules bound by the RBP of interest in physiological settings, ranging from virus particles to animal tissues, at a global scale.
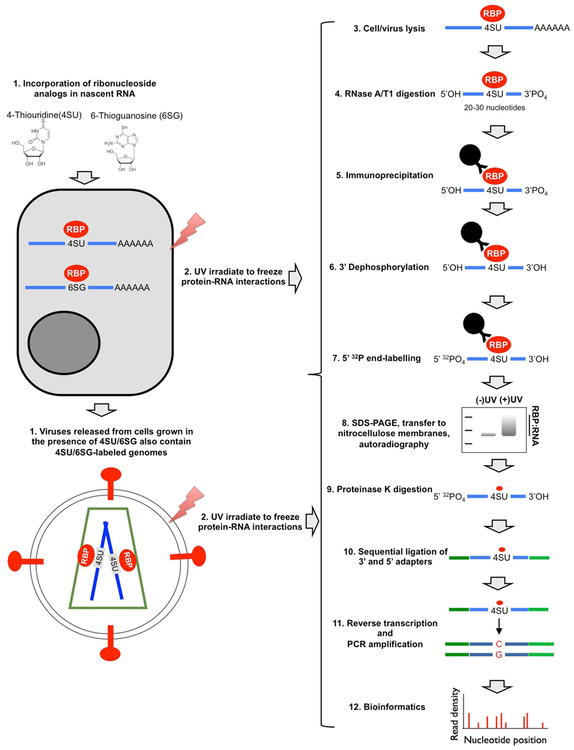
The key steps of the existing CLIP methodologies are (Fig. 1): (1) Protein-RNA complexes are covalently crosslinked in live cells/tissues/virions typically by UV crosslinking; (2) Cells/tissues/virions are lysed and treated with limited amounts of RNases leaving small fragments of RNA molecules (~20–50 nucleotides) protected by the protein of interest; (3) Protein-RNA complexes are immunoprecipitated, and non-specific RNAs and proteins are removed by stringent washes. Because the protein-RNA complexes are covalently crosslinked, these stringent conditions, in principle, do not affect purification of target protein-RNA adducts. (4) The purified protein-RNA complexes are radioactively labeled and separated by SDS-PAGE. (5) Bound RNA is isolated by Proteinase K treatment. (5) Eluted RNA is ligated to adapters, reverse transcribed, and the resulting cDNA is PCR amplified and subjected to sequencing. (6) Sequencing reads are processed and mapped to reference genomes. Depending on the method used, the resulting library contains nucleotide substitutions or deletions [12] at the site of crosslinking, which allows mapping of the site of protein-RNA interactions at nucleotide resolution.
Figure 1-.
Schematic diagram of steps involved in PAR-CLIP experiments.
Various versions of CLIP have been developed and the details of these alternative approaches have been reviewed elsewhere [6, 13]. The approach we commonly use depends on efficient UV crosslinking mediated by ribonucleoside analogs, including 4-thiouridine (4SU) and 6-thioguanosine (6SG), as in the original PAR-CLIP protocol [4]. In PAR-CLIP experiments, cells are grown in the presence of ribonucleoside analogs for up to 16 hours and UV-crosslinked at a longer wavelength (365 nm). With the majority of the viral and cellular RNA-binding proteins we studied in the past few years, this method significantly enhanced the amount of protein-RNA adducts obtained [14–17]. In addition, PAR-CLIP allows accurate nucleotide resolution mapping of target RNA sites due to mutations introduced by the reverse transcriptase enzyme (T-to-C for 4SU and G-to-A for 6SG) precisely at the site of crosslinking during cDNA synthesis. On the other hand, use of ribonucleoside analogs may inadvertently enrich RNA elements with distinct nucleotide composition or alter RNA structure [18], which may subsequently affect protein binding. Thus, we routinely validate our findings with the use of both ribonucleoside analogs and HITS CLIP, which utilizes conventional UV-crosslinking.
For library generation, we follow a combination of PAR-CLIP and HITS-CLIP protocols [1–3]. As reviewed elsewhere [5, 6, 13], this is the stage where most variant CLIP protocols differ. While the HITS-CLIP and many other protocols call for ligation of adapters while the protein-RNA complexes are on beads, the solution-phase PAR-CLIP library generation protocol was significantly more efficient in our experience with 3’ and 5’ adapter ligations routinely functioning at >90% efficiency. Another common alternative is the utilization of a two-part cleavable adapter introduced into cDNA during reverse transcription in the iCLIP approach [19]. This is followed by circularization and restriction enzyme digestion, which allows the recovery of a larger fraction of truncated cDNAs a result of reverse transcriptase stalling at crosslinking sites. Due to this enrichment, iCLIP can yield higher complexity libraries and has been proposed to perform better than previous approaches in identification of the precise site of crosslinking [19–21].
All CLIP approaches are technically challenging with numerous labor-intensive steps. In addition, the loss of the starting material at several inefficient steps is a major drawback. This problem is further exacerbated if the initial protein-RNA complexes are not abundant due to low levels of expression, low crosslinking or immunoprecipitation efficiencies. These problems can often lead to a final library with insufficient complexity and over-enrichment of environmental contaminating sequences. Below we provide a step-by-step guide to successfully execute CLIP experiments and notes where an alternative approach may be more suitable.
METHODS
2.1. UV-crosslinking of protein-RNA complexes, lysis and RNase treatment
Covalent crosslinking of protein-RNA complexes will need to be optimized based on the virus and RBP under study. As the PAR-CLIP approach depends on the efficient incorporation of the ribonucleoside analogs 4SU or 6SG into nascent RNA, duration of incubation will depend partly on how fast a given virus replicates. In the case of HIV-1, we typically transfect cells with proviral plasmids or infect cells, and initiate 4SU/6SG treatment 14 hours prior to harvest as detailed below. Lysis and RNase A treatment will likewise depend on the RBP of interest. The example below is designed to determine where hnRNP A1 and hnRNP H1 proteins bind to on the HIV-1 viral RNAs.
-
Plate one 15-cm plate of HEK293T cells stably expressing 3xHA-tagged hnRNP A1 and hnRNP H1 (long and short isoforms) proteins such that they are ~90% confluent on the day of transfection. Transfect each plate with 30 μg of HIV-1 proviral plasmids.
Note: If the yield of protein RNA complexes is high, one can use lower number of cells. For example, we have successfully implemented CLIP using cells plated in a single 10-cm dish. Lysis buffer volume should be adjusted below accordingly.
-
One day post-transfection and 14 hours prior to UV crosslinking, change media on plates with 100 μM 4-SU or 6-SG containing media.
Note: A potential disadvantage of the PAR-CLIP protocol is the cellular toxicity that may be induced by 4SU treatment depending on the cell type, the dose and incubation time [22]. Thus, optimal conditions that allow efficient protein-RNA crosslinking without major toxicity should be determined on a case-by-case basis.
-
Wash cells with 15 mL of ice-cold PBS and irradiate the dish containing cells uncovered at an energy setting of 500 mJ in a Boekel UV-crosslinking chamber equipped with UV368 nm bulbs. For analysis of virion incorporated RNA binding proteins, cell culture supernatants containing virions can be processed as detailed in steps 7–11.
Note: PAR-CLIP also works well with UV306 nm bulbs.
Add 5 mL of PBS to each plate and collect cells using a cell scraper. Pellet cells by centrifugation at 500 × g for 5 min and discard the PBS. Cells can be flash-frozen and stored at this stage.
-
Lyse the cell pellet in 2.5 mL of 1xRIPA-lysis buffer. Place samples on ice for 10 min.
Note: Alternative lysis buffers can be used. For example, we have utilized NP40-lysis buffer in previous work for HIV-1 Gag and MA proteins [15, 23], and 1xRIPA buffer for HIV-1 integrase [14] and cellular APOBEC3 and hnRNP proteins [16, 17].
Clear lysates by centrifugation at 14000 rpm, 4 °C for 10 min and collect the supernatants.
Cell culture supernatants containing virions can be processed as follows: Pellet cellular debris by centrifugation at 500 × g for 5 min and filter the supernatant through a 0.2 μm filter.
Add 13 mL of 20% sucrose solution in ultracentrifuge tubes and layer on top by 25 mL of cleared cell culture supernatant. Pellet the virions by ultracentrifugation at 27000 rpm, 4 °C, for 90 min.
Aspirate the supernatant and resuspend the virions in a total of volume of 500 μL 1XPBS.
In a 6-well cell-culture dish UV-crosslink the virions uncovered twice at the above setting. Mix in between the two irradiations.
Collect the virions and add 125 μL of 5x RIPA or NP40 lysis buffer.
-
Add RNase A and DNase I to lysates at a final concentration of 20 U/mL and 60 U/mL, respectively. Incubate samples at 37 °C for 5 min and transfer to ice.
Note: RNase A treatment should be evaluated carefully for each protein to avoid extensive or insufficient digestion of RNA. In principle, protein-RNA complexes should run above where one would expect to see naked protein. Instead of RNase A, RNase T1 can also be utilized.
2.2. Immunoprecipitation, alkaline phosphatase treatment, end-labeling:
Because the quality of CLIP experiments depend on the yield of protein-RNA complexes, it is extremely important to use an antibody that pulls down high levels of protein-RNA complexes. We have successfully immunoprecipitated endogenous proteins as well as proteins tagged with three copies of a HA-tag (3xHA) in CLIP experiments. Note that the below example is conducted with hnRNP proteins containing a 3xHA tag.
-
For each cell lysate, prepare 60 μL of Protein G-conjugated Dynabeads by washing twice with 1 mL citrate-phosphate buffer. Resuspend the beads in two bead volumes of citrate phosphate buffer.
Note: For virus samples, 40 μL of Protein G-conjugated Dynabeads will be sufficient.
Add 5–10 μg of antibody per 100 μL of beads. Incubate on a rotating wheel at room temperature for 45 min.
Wash beads twice with 1 mL and resuspend in one bead volume of citrate-phosphate buffer.
Add one bead volume of antibody-conjugated Dynabeads to cell lysates and incubate on a rotating wheel at 4 °C for 1 hr.
Collect beads on a magnetic stand in microcentrifuge tubes. Wash beads twice with 1 mL of IP wash buffer, LiCl buffer, NaCl buffer, KCl buffer and dephosphorylation buffer. Briefly spin the beads and remove the remaining buffer.
Resuspend beads in 1 bead volume of dephosphorylation buffer containing calf intestinal alkaline phosphatase at a final concentration of 0.5 U/μL. Incubate for 10 min at 37°C in a Thermomixer programmed to mix at 1400 rpm for 20 seconds every 2 min.
Wash beads twice with 1 mL of phosphatase wash buffer. Incubate on a rotating wheel for 5 min between washes.
Wash beads twice with 1 mL of PNK buffer. Briefly spin the beads and remove the remaining wash buffer.
Resuspend beads in one bead volume of 1x PNK buffer containing 0.5 μCi/μL γ−32P-ATP and 1 U/μL T4 PNK. Incubate at 37°C for 40 min in a Thermomixer programmed to mix at 1400 rpm for 20 seconds every 2 min.
Add non-radioactive ATP at a final concentration of 100 μM and incubate as above at 37°C for an additional 10 min.
Wash beads once with 1 mL of PNK Buffer, LiCl buffer, KCl buffer and NaCl buffer as described above. Briefly spin the beads and remove the remaining wash buffer.
Resuspend beads in 50 μL of 1x NuPAGE SDS-PAGE loading buffer and elute protein-RNA complexes by incubation at 72 °C for 10 min in a Thermomixer set to constantly mix at 1400 rpm. Collect the eluates.
2.3. Separation of protein-RNA adducts and purification of RNA
Run 45 μL of the eluate on a Novex Bis-Tris 4–12% polyacrylamide gel and transfer to a nitrocellulose membrane. Use the remaining eluate to test for the efficiency of immunoprecipitation by western blotting.
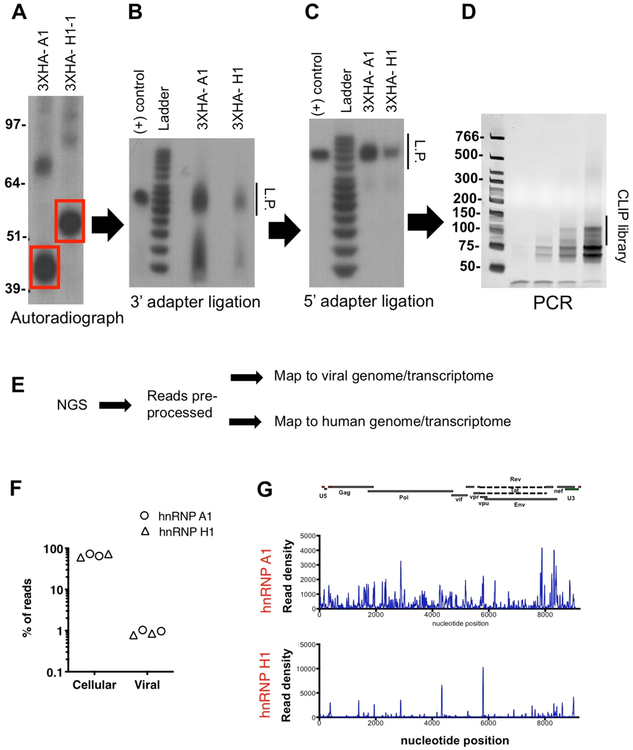
Place the membrane in a plastic wrap and expose to autoradiography film until the protein-RNA adducts can be visualized (Fig. 2A).
Using a clean scalpel or razor blade, cut a region of the membrane directly above the expected the molecular weight of the protein of interest. Cut the membrane further into smaller pieces and place in low-retention microcentrifuge tubes.
Add 400 μL of 1x Proteinase K buffer containing 2 mg/mL Proteinase K to each sample. Incubate for 30 min in a Themomixer set to 55 °C with constant agitation at 1100 rpm.
Supplement with an additional 400 μg Proteinase K and incubate an additional 15 min as above.
Lower the temperature to 37 °C and add 1 volume of phenol:chlorofom:isoamyl alcohol. Vortex and incubate at 37 °C in a Themomixer as above for 10 min.
Centrifuge samples at 14000 rpm, 3 min, RT.
Collect the supernatants and add 100 μL 3 M sodium-acetate, 1 μL GlycoBlue reagent and 1 mL ethanol:isopropanol (1:1). Mix well and place samples in −20 °C.
Figure 2-. CLIP with hnRNP A1 and hnRNP H1 proteins.
HEK293T cells were transfected with plasmids expressing 3xHA-tagged hnRNP A1 and hnRNP H1 (long and short isoforms) proteins. (A) Immunoprecipitated and end-labeled protein-RNA adducts were separated by SDS-PAGE, transferred to nitrocellulose membranes and visualized by autoradiography as detailed in the protocol. (B) Separation of 3’ adapter ligation products on a 15% TBE-Urea gel is shown. (C) Separation of 5’ adapter ligation products on a 15% TBE-Urea gel is shown. (D) Separation of PCR products (cycles 6, 9, 12 and 15) on a 6% TBE-Urea gel is shown.
2.4. Adapter ligations
While most CLIP approaches depend on radioactive labeling of RNA molecules bound by the RBP of interest or ligating the adapters to radioactively labeled adapters, adapters containing an infrared fluorescent dye can be used instead [24].
Pellet RNA by centrifugation at 14000 rpm, 4 °C for 30 min.
Wash with 500 μL 80% ethanol. Centrifuge as above for 5 min.
Air-dry RNA pellet and resuspend in 8 μL water.
In parallel, bring 1 pmole of the end-labeled positive control RNA up to 8 μL with water.
Add 100 pmoles of 3’ adapter, 2 μL DMSO and 5 μL PEG8000 (50%). Mix well and denature RNA by incubation at 72 °C for 2 min. Place samples immediately on ice.
To each sample add 2 μL 10x T4 RNA ligase 2 buffer without ATP, 1 μL SuperaseIN (20U/μL), 1 μl 2 mg/ml ultrapure BSA and 1 μL of T4 RNA ligase 2, truncated K227Q (200U/μL). Mix well.
Incubate samples overnight at 16 °C.
Add 20 μL of 2X Novex TBE-Urea loading buffer to each sample and incubate at 72°C for 2 min. Transfer samples on ice.
Load samples on a 15% TBE-Urea gel, while leaving at least one empty well between each sample to avoid cross-contamination. Run at 180V, 70 min.
Place gel in plastic wrap and expose to an autoradiography film (Fig. 2B).
Cut a gel piece corresponding to the ligated RNA products, including the ligated positive control RNA. Crush the gel into smaller pieces by passing it through a 0.5 mL microcentrifuge tube with holes on the bottom by centrifugation. Alternatively, a Teflon pestle can be used.
Add 3 volumes of 0.4M NaCl supplemented with 200 units/mL SuperaseIN.
Incubate samples overnight in a Thermomixer set to constant shaking at 1400 rpm, 4°C.
Pass gel slurry through a Costar spin column. Add 1 μL GlycoBlue reagent and 2.5V of ethanol:isopropanol (1:1). Place samples on ice for 20 min.
Precipitate RNA as above and resuspend in 5 μL of ultrapure water. Add 20 pmol of barcoded 5’ adapter, 5 μL PEG8000 (50%) and 2 μL DMSO. Incubate samples at 72 °C for 2 min and immediately transfer on ice.
To each sample add ligation mix containing 2 μL 10x T4 RNA ligase 1 buffer, 1 μL SuperaseIN, 1 μL of 2 mg/mL ultrapure BSA, 2 μL ATP (10 mM), 1 μL T4 RNA Ligase 1 (10U/μL).
Incubate samples overnight at 16 °C.
If barcoded adapters are used, samples can be pooled at this stage. After adding 20 μL of 2X Novex TBE-Urea loading buffer to each sample, incubate at 72°C for 5 min. Transfer samples on ice and combine them as desired.
Load samples on a 15% TBE-Urea gel as above and expose to an autoradiography film (Fig. 2C). Process samples as in 11–15. Resuspend pelleted RNA in 10 μL of ultrapure water.
2.5. Reverse transcription and PCR amplification of CLIP library
Use the SuperScript III First Strand Synthesis System as detailed by the manufacturer. To 8 μL of RNA, add 1 μL of reverse transcription primer (10 μM) and 1 μL of dNTP (10mM). Incubate at 65 °C for 5 min. Transfer samples on ice.
Add 4 μL MgCl2 (25 mM), 2 μL 10x Reverse Transcription Buffer, 2 μL DTT (0.1M), 1 μL RNaseOUT (40U/μL) and 1 μl SSIII RTase (200U/μL).
-
Reverse transcribe according to manufacturer’s instructions.
Note: CLIP has an inherent bias against identification of protein binding events on structured RNA elements due to stalling of RT at these sites. Use of a thermostable reverse transcriptase may allow for higher reaction temperatures may help to resolve potential RNA secondary structures [24].
Set up a 100 μL PCR reaction containing 10 μL of cDNA, 50 pmol of forward and reverse primers, 20 μL 5x Phusion High Fidelity Buffer, 2 μL dNTPs (10 mM), 2 μL Phusion polymerase (2U/μL). Run PCR for a total of 15 cycles programmed at 98 °C 15 seconds, 55 °C 30 seconds, 72 °C 15 seconds. Take 20 μL aliquots at the end of 6–9-12-15th cycles.
Run samples on a 6% TBE-Urea gel. Stain with EtBr in 1xTBE buffer and excise a region corresponding to the CLIP library (Fig. 2D).
Weigh the gel and add 1–2 volumes of diffusion buffer.
Incubate at 50°C for 30 min in a Thermomixer set to constant agitation at 1400 rpm.
Collect the supernatant and extract CLIP DNA library by Qiagen gel extraction kit.
As CLIP libraries are derived from short RNA sequences, they can be sequenced on an Illumina platform for 50-cycles.
2.6. Data analysis
In parallel with the development of variant CLIP approaches, several publicly available data analysis tools have been developed. Pipelines that can perform the majority of necessary analysis include the PARCLIPsuite [25], CLIPZ [26], CIMS [27] and CLIP-seq tools [28]. For a more detailed review of these tools and algorithms we refer the readers to detailed recent reviews [29–32]. We perform our data on a local high performance cluster utilizing some of these tools as well as in-house scripts, for which a basic knowledge of UNIX operating system and other programming languages is necessary. CLIP data analyses can be summarized in four major steps: (1) Preprocessing of sequencing reads, (2) Mapping of reads to reference genomes, (3) Subjecting mapped reads to cluster-finding algorithms to define binding sites, (4) Analysis of binding sites for enrichment of certain features including where within a gene body the binding site is located, and the presence of distinct motifs or nucleotide composition. Below we outline the basic analysis of the CLIP-seq data, including sample command line instructions:
-
Sequencing data is initially received as a FASTQ file, with the following format:
@SN1063:846:HVLCCBCX2:1:1108:1225:2068 1:N:0:0 ATCCTATCCCTTTAGCAGCAAGGTCCATATCTGACTTTTTGTTATCGTAT + #<<DDHHHIHIIIIHIIIIIIIIIIIIIIIIIIIIHIIIIIIIHIIIIHH @SN1063:846:HVLCCBCX2:1:1108:1100:2075 1:N:0:0 TAGCTGTCAAACAGGTGCCGTCGTATGCCGTCTTCTGCTTGAAAAAAAAA + #<DDDIHIIIIIIIIHHIHIIIIIIIIIIIIIIIIIIIIIIIIIIIIIIG @SN1063:846:HVLCCBCX2:1:1108:1233:2083 1:N:0:0 CGACTTTCCGATTTCCGAATGGGACACACCCTCACCAATCGTATGCCGTC + #<<DDGHHIIGIIEGHHEEEHHIHIHIEE<<<EGFEF@E@EHCGHE<CGC @SN1063:846:HVLCCBCX2:1:1108:1203:2183 1:N:0:0 CGACTTTCCGATTTCCGCCTGGGATGACCCAGCCTCACCAATCGTATGCC + #<<DDGHHIIGIIEGHHEEEHHIHIHIEE<<<EGFEF@E@EHCGHE<CGC
The data is organized in sets of four lines, where line 1 is a sequence identifier providing information about the supercomputing cluster and the sequencing run, line 2 is the sequence itself, line 3 is a separator (usually a ‘+’ character), and line 4 gives the quality score of each nucleotide represented in Phred +33 encoded ASCII characters. We have pointed out the 5’ adapter and 3’ adapter sequences in red and blue, respectively.
-
Raw reads obtained from the sequencing facility can be processed prior to mapping to human or viral genomes using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Alternative tools include PRINSeq1,2, BBMap, and FastQC for quality control reporting. Note that often times the 3’ adapter is only partially present in the sequence and therefore we typically search for the first eight nucleotides of the adapter string. At this stage one can also specify the minimum length cutoff, typically 15 nucleotides, for accurate mapping of the resulting reads in subsequent steps. Running the following command will search for the adapter string TCGTATGC, discarding reads that contain ambiguous nucleotides, lack the 3’ adapter sequence or are shorter than 23 nucleotides (15 nucleotides+8 nucleotides of 5’ adapter):
fastx_clipper -c -a TCGTATGC -l 23 -i INFILE.fastq -o OUTFILE
The resulting file will resemble the output below:@SN1063:846:HVLCCBCX2:1:1108:1225:2068 1:N:0:0 ATCCTATCCCTTTAGCAGCAAGGTCCATATCTGACTTTTTGTTA + #<<DDHHHIHIIIIHIIIIIIIIIIIIIIIIIIIIHIIIIIIIH @SN1063:846:HVLCCBCX2:1:1108:1233:2083 1:N:0:0 CGACTTTCCGATTTCCGAATGGGACACACCCTCACCAA + #<<DDGHHIIGIIEGHHEEEHHIHIHIEE<<<EGFEF@ @SN1063:846:HVLCCBCX2:1:1108:1203:2183 1:N:0:0 CGACTTTCCGATTTCCGCCTGGGATGACCCAGCCTCACCAA + #<<DDGHHIIGIIEGHHEEEHHIHIHIEE<<<EGFEF@E@E
-
Samples are separated based on their 5’ barcode sequences denoted by the first three nucleotides. This requires as input a file containing the various barcode sequences (i.e. the barcodes.txt file below) and generates a number of separate files containing the reads corresponding to a particular barcode. An example set of barcodes is provided below:
BC1 ATC BC2 CGA
Running the following command will generate two files from the above sequence:cat OUTFILE | /usr/local/bin/fastx_barcode_splitter.pl --bcfile barcodes.txt --bol --mismatches 0 --prefix OUTFILE_split
Barcode 1 file:@SN1063:846:HVLCCBCX2:1:1108:1225:2068 1:N:0:0 ATCCTATCCCTTTAGCAGCAAGGTCCATATCTGACTTTTTGTTA + #<<DDHHHIHIIIIHIIIIIIIIIIIIIIIIIIIIHIIIIIIIH
Barcode 2 file:@SN1063:846:HVLCCBCX2:1:1108:1233:2083 1:N:0:0 CGACTTTCCGATTTCCGAATGGGACACACCCTCACCAA + #<<DDGHHIIGIIEGHHEEEHHIHIHIEE<<<EGFEF@ @SN1063:846:HVLCCBCX2:1:1108:1203:2183 1:N:0:0 CGACTTTCCGATTTCCGCCTGGGATGACCCAGCCTCACCAA + #<<DDGHHIIGIIEGHHEEEHHIHIHIEE<<<EGFEF@E@E
If necessary, fastx_collapser can be run on this data to convert identical reads into a single sequence while maintaining read counts, which is useful for mitigating PCR overamplification issues. Collapsing will change the data from a FASTQ format to a FASTA format, where the first line is a descriptor of the sequence and the second is the sequence itself.
-
The final step is to remove the 5’ adapter using the fastx_trimmer tool. The below command discards the first 8 nucleotides of each sequence:
fastx_trimmer -f 9 -v -i OUTFILE collapsed -o OUTFILE_trimmed
Barcode 1 file:@SN1063:846:HVLCCBCX2:1:1108:1225:2068 1:N:0:0 CCTTTAGCAGCAAGGTCCATATCTGACTTTTTGTTA + IHIIIIHIIIIIIIIIIIIIIIIIIIIHIIIIIIIH
Barcode 2 file:@SN1063:846:HVLCCBCX2:1:1108:1233:2083 1:N:0:0 CGATTTCCGAATGGGACACACCCTCACCAA + HEEEHHIHIHIEE<<<EGFEF@E@EHCGHE @SN1063:846:HVLCCBCX2:1:1108:1203:2183 1:N:0:0 CGATTTCCGCCTGGGATGACCCAGCCTCACCAA + IIGIIEGHHEEEHHIHIHIEE<<<EGFEF@E@E
- Next, the output reads from the previous step must be aligned to the genome of interest. This can be done using aligners such as STAR, Bowtie, BWA, GSNAP, and SHRiMP. In order to use such aligners, one must have the genome on hand, usually in FASTA format. These can be downloaded from UCSC, Ensembl or NCBI. Prior to use, the genome usually must be indexed for use by the aligner so that it can be easily loaded into memory, which decreases the runtime. For our purposes, we will use the Bowtie aligner. Once the genome FASTA is downloaded, it is then built using the bowtie-build command. For example, to build an index with the basename ‘hg19_bowtie’ from the hg19.fa (human genome) FASTA file, the command would be:
bowtie-build hg19.fa hg19_bowtie
- The reads can then be mapped to the genome using the ‘bowtie’ command, specifying the index, input file, output file format, number of mismatches, and other parameters. The default output is a BOWTIE file, though it is more common to output alignments as SAM files, both of which provide the mapping location of a sequence, quality of mapping, reference sequence name, other such information. A software package known as SAMtools can then used to convert the SAM file to BAM file, a compressed binary representation of the SAM file. The BAM file can be sorted and indexed for quicker access to the alignments by other programs. Following alignment SAMtools [33] and BEDtools [34] can be used to further process the data. An example set of commands is given below, where index is the basename of the Bowtie index, read_file is the reads file from preprocessing in FASTQ format, and sam_file is the name of the SAM file to be output:
bowtie $index -q $read file -v 1 -m 10 --best --strata -S $sam_file # map allowing one mismatch, suppress alignments if they map to >10 positions samtools view -bS $sam_file > $bam_file #converts SAM to BAM samtools sort $bam_file -o $bam_file_sorted #sorts the bam file samtools index $bam_file_sorted $bam_file_index #indexes the BAM file
The SAM or BOWTIE outputs can be used for peak calling, which determines which areas in the genome are significantly enriched in aligned reads. Software that can be used for this includes PARalyzer [35], PIPE-CLIP [36], and Piranha [37]. We typically use PARalyzer, which yields a series of statistically significant clusters and the location, sequence, chromosome, and the number of counts for each. Following annotation, motif searches within clusters can be performed by the cERMIT algorithm [38]. Additionally, one can use software such as HOMER [39] to characterize the peak sequences and carry out de novo motif discovery analysis. The output of PARalyzer can be used directly with HOMER to characterize the motifs to which the protein of interest is binding.
4. FUTURE APPLICATIONS
Application of the CLIP methods to questions in virology will undoubtedly continue to increase, given the large number of RBPs that are known and continuing to emerge as key regulators of numerous processes in virus replication and host defenses against viruses. CLIP, when combined with other structural probing tools such as SHAPE and DMS-seq, can also provide information about the underlying structural elements.
HIGHLIGHTS.
RNA-binding proteins regulate numerous key steps of RNA and DNA virus replication.
Crosslinking immunoprecipitation coupled with sequencing (CLIP) is a powerful approach to identify the RNA-targets of RNA-binding proteins.
CLIP provides nucleotide resolution information of RBP binding sites on RNAs.
CLIP captures protein-RNA interactions in physiologically relevant settings.
ACKNOWLEDGEMENTS:
This work was supported by NIH grants P50 GM103297 (the Center for HIV RNA Studies) and GM122458 to S.B.K.
APPENDIX 1: Equipment list
Ultracentrifuge for virus pelleting (A note on the fact that one can do the same without ultra, at least for retroviruses)
Boekel crosslinker equipped with UV368 nm bulbs.
Magnetic stand (Life Technologies).
Thermomixer (Eppendorf, Hamburg, GE).
Centrifuges
Power supplies for electrophoresis
APPENDIX 2-. Supply list
4-thiouridine (Sigma-Aldrich Chemical Company, St. Louis, MO, USA): dissolve in water to a final concentration of 100 μM
Phosphate-buffered saline (PBS), without calcium and magnesium.
Ultracentrifuge tubes, thinwall, Ultra-clear ™, 38.5 mL (Beckman Coulter, Brea, CA, USA).
20% sucrose solution (w/v): Prepare in 1x PBS, filter and store at 4 °C.
Complete, Mini, EDTA-free protease inhibitor cocktail (Roche, Basel, CH).
NP-40 lysis buffer: 50 mM HEPES, pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% NP-40, supplemented with 0.5 mM DTT and complete EDTA-free protease inhibitor cocktail.
RIPA buffer: 50 mM Tris pH7.4, 1% NP-40, 0.25% Na-deoxycholate, 0.1% SDS, 150 mM NaCl, 1mM EDTA, supplemented with 0.5 mM DTT and complete EDTA-free protease inhibitor cocktail.
RNase A (Thermo-Fisher Scientific, Pittsburgh, PA, USA).
DNase I recombinant, RNase-free (Roche).
Citrate-phosphate buffer: 4.7 g/L citric acid, 9.2 g/L Na2HPO4, pH 5.0.
Dynabeads® Protein G (Life Technologies, Carlsbad, CA, USA).
Calf intestinal alkaline phosphatase (New England Biolabs, Ipswich, MA, USA).
T4 Polynucleotide kinase (New England Biolabs).
ATP, [γ−32P], 3000Ci/mmol, 10mCi/ml (Perkin Elmer, Waltham, MA, USA).
ATP (New England Biolabs)
Fisherbrand ™ siliconized low-retention microcentrifuge tubes (Thermo-Fisher).
IP wash buffer: 50 mM HEPES-KOH, pH 7.5, 300 mM KCl, 0.05% NP-40, supplemented with 0.5 mM DTT.
LiCl buffer: 250 mM LiCl, 10 mM Tris pH 8.0, 1 mM EDTA, 0.5% NP-40, 0.5% Na-deoxycholate, supplemented with 0.5 mM DTT.
NaCl buffer: 50 mM Tris pH 7.4, 1 M NaCl, 1 mM EDTA, 0.1% SDS, 0.5% Na-deoxycholate, 1% NP-40, supplemented with 0.5 mM DTT.
KCl buffer: 50 mM HEPES-KOH, pH 7.5, 500 mM KCl, 0.05% NP-40, supplemented with 0.5 mM DTT.
Dephosphorylation buffer: 50 mM Tris-HCl, pH 7.9, 100 mM NaCl, 10 mM MgCl2, supplemented with 1 mM DTT.
Phosphatase wash buffer: 50 mM Tris-HCl, pH 7.5, 20 mM EGTA, 0.5% NP-40, supplemented with 1 mM DTT.
PNK buffer: 50 mM Tris-HCl, pH 7.5, 50 mM NaCl, 10 mM MgCl2, supplemented with 1 mM DTT.
NuPAGE® protein sample buffer (4x).
NuPAGE® Novex® 4–12% Bis-Tris protein gels (Life Technologies).
NuPAGE® MOPS or MES SDS Running buffer (20X, Life Technologies).
Nitrocellulose membranes (GE Healthcare, Little Chalfont, UK).
Tris-Glycine transfer buffer (10x): 250 mM Tris, 1.92 M glycine. Prepare 1x buffer containing 20% ethanol.
Autoradiography cassettes and film.
Proteinase K, recombinant, PCR Grade (Roche).
Proteinase K buffer (2x): 200 mM Tris-HCl, pH 7.5, 100 mM NaCl, 20 mM EDTA, 100 mM NaCl, 2% SDS.
Glycoblue co-precipitant (Life Technologies).
3M Sodium acetate, pH 5.2.
Ethanol:isopropanol (1:1) mixture.
Acid phenol:chlorofom: isoamyl alcohol (125:24:1, Sigma-Aldrich).
80% ethanol.
Nuclease free water.
SuperaseIN (Life Technologies).
UltraPure ™ BSA (Life Technologies).
DMSO (Sigma-Aldrich).
50% PEG8000 (New England Biolabs).
T4 RNA Ligase 2, truncated K227Q (New England Biolabs).
T4 RNA Ligase 1 (New England Biolabs).
6% and 15% Novex® TBE-Urea gels (Life Technologies).
Novex® TBE-Urea sample buffer (2x, Life Technologies).
TBE running buffer (10X): 890 mM Tris, 890 mM boric acid, 20 mM EDTA, pH 8.3.
Corning® Costar® SpinX® centrifuge tube filters cellulose acetate membrane, pore size 0.22 μm, sterile (Corning, Corning, NY, USA).
Low Molecular Weight Marker 10–100 nt (Affymetrix, Santa Clara, CA).
SuperScript ® III First-Strand Synthesis System (Life Technologies)
Phusion® High-Fidelity DNA Polymerase (New England Biolabs)
Quick-Load® Low Molecular Weight DNA Ladder (New England Biolabs)
Diffusion buffer: 0.5 M ammonium acetate, 10 mM magnesium acetate, 1 mM EDTA, pH 8.0, 0.1% SDS.
QIAquick Gel Extraction Kit (Qiagen, Hilden, GE)
APPENDIX 3-. Adapter sequences:
3’ adapter: 5’adenylated/TCG TAT GCC GTC TTC TGC TTG-3’dideoxyC
5’ barcoded adapters:
rGUU CAG AGU UCU ACA GUC CGA CGA UC AGU NNN UC
rGUU CAG AGU UCU ACA GUC CGA CGA UC GAU NNN UC
rGUU CAG AGU UCU ACA GUC CGA CGA UC GUG NNN UC
rGUU CAG AGU UCU ACA GUC CGA CGA UC ACG NNN UC
rGUU CAG AGU UCU ACA GUC CGA CGA UC UAG NNN UC
rGUU CAG AGU UCU ACA GUC CGA CGA UC AUC NNN UC
Positive control primer: rAUAGCUACGAUUGCA
RT/Reverse PCR primer: CAAGCAGAAGACGGCATACGA
Forward PCR primer:
AATGATACGGCGACCACCGACAGGTTCAGAGTTCTACAGTCCGA
With the development of new sequencing systems, these adapters will need to be modified to incorporate necessary elements for cluster formation and sequencing.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Licatalosi DD, et al. , HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature, 2008. 456(7221): p. 464–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ule J, et al. , CLIP identifies Nova-regulated RNA networks in the brain. Science, 2003. 302(5648): p. 1212–5. [DOI] [PubMed] [Google Scholar]
- 3.Ule J, et al. , CLIP: a method for identifying protein-RNA interaction sites in living cells. Methods, 2005. 37(4): p. 376–86. [DOI] [PubMed] [Google Scholar]
- 4.Hafner M, et al. , Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell, 2010. 141(1): p. 129–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ule J, Hwang HW, and Darnell RB, The Future of Cross-Linking and Immunoprecipitation (CLIP). Cold Spring Harb Perspect Biol, 2018. 10(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee FCY and Ule J, Advances in CLIP Technologies for Studies of Protein-RNA Interactions. Mol Cell, 2018. 69(3): p. 354–369. [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C and Gold L, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science, 1990. 249(4968): p. 505–10. [DOI] [PubMed] [Google Scholar]
- 8.Ellington AD and Szostak JW, In vitro selection of RNA molecules that bind specific ligands. Nature, 1990. 346(6287): p. 818–22. [DOI] [PubMed] [Google Scholar]
- 9.Ray D, et al. , Rapid and systematic analysis of the RNA recognition specificities of RNA-binding proteins. Nat Biotechnol, 2009. 27(7): p. 667–70. [DOI] [PubMed] [Google Scholar]
- 10.Lambert N, et al. , RNA Bind-n-Seq: quantitative assessment of the sequence and structural binding specificity of RNA binding proteins. Mol Cell, 2014. 54(5): p. 887–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buenrostro JD, et al. , Quantitative analysis of RNA-protein interactions on a massively parallel array reveals biophysical and evolutionary landscapes. Nat Biotechnol, 2014. 32(6): p. 562–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang C and Darnell RB, Mapping in vivo protein-RNA interactions at single-nucleotide resolution from HITS-CLIP data. Nat Biotechnol, 2011. 29(7): p. 607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bieniasz PD and Kutluay SB, CLIP-related methodologies and their application to retrovirology. Retrovirology, 2018. 15(1): p. 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kessl JJ, et al. , HIV-1 Integrase Binds the Viral RNA Genome and Is Essential during Virion Morphogenesis. Cell, 2016. 166(5): p. 1257–1268 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kutluay SB, et al. , Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell, 2014. 159(5): p. 1096–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.York A, et al. , The RNA Binding Specificity of Human APOBEC3 Proteins Resembles That of HIV-1 Nucleocapsid. PLoS Pathog, 2016. 12(8): p. e1005833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kutluay SB, et al. , Genome wide analysis of hnRNP binding to HIV-1 RNA reveals a key role for hnRNP H1 in alternative viral mRNA splicing. J Virol, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Testa SM, et al. , Thermodynamics of RNA-RNA duplexes with 2- or 4-thiouridines: implications for antisense design and targeting a group I intron. Biochemistry, 1999. 38(50): p. 16655–62. [DOI] [PubMed] [Google Scholar]
- 19.Konig J, et al. , iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol, 2010. 17(7): p. 909–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto Y, et al. , Analysis of CLIP and iCLIP methods for nucleotide-resolution studies of protein-RNA interactions. Genome Biol, 2012. 13(8): p. R67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haberman N, et al. , Insights into the design and interpretation of iCLIP experiments. Genome Biol, 2017. 18(1): p. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burger K, et al. , 4-thiouridine inhibits rRNA synthesis and causes a nucleolar stress response. RNA Biol, 2013. 10(10): p. 1623–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutluay SB and Bieniasz PD, Analysis of HIV-1 Gag-RNA Interactions in Cells and Virions by CLIP-seq. Methods Mol Biol, 2016. 1354: p. 119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zarnegar BJ, et al. , irCLIP platform for efficient characterization of protein-RNA interactions. Nat Methods, 2016. 13(6): p. 489–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garzia A, et al. , Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods, 2017. 118–119: p. 24–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khorshid M, Rodak C, and Zavolan M, CLIPZ: a database and analysis environment for experimentally determined binding sites of RNA-binding proteins. Nucleic Acids Res, 2011. 39(Database issue): p. D245–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore MJ, et al. , Mapping Argonaute and conventional RNA-binding protein interactions with RNA at single-nucleotide resolution using HITS-CLIP and CIMS analysis. Nat Protoc, 2014. 9(2): p. 263–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maragkakis M, et al. , CLIPSeqTools--a novel bioinformatics CLIP-seq analysis suite. RNA, 2016. 22(1): p. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bottini S, et al. , Recent computational developments on CLIP-seq data analysis and microRNA targeting implications. Brief Bioinform, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Uhl M, et al. , Computational analysis of CLIP-seq data. Methods, 2017. 118–119: p. 60–72. [DOI] [PubMed] [Google Scholar]
- 31.Liu Q, et al. , Assessing Computational Steps for CLIP-Seq Data Analysis. Biomed Res Int, 2015. 2015: p. 196082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reyes-Herrera PH and Ficarra E, Computational Methods for CLIP-seq Data Processing. Bioinform Biol Insights, 2014. 8: p. 199–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H, et al. , The Sequence Alignment/Map format and SAMtools. Bioinformatics, 2009. 25(16): p. 2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quinlan AR and Hall IM, BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics, 2010. 26(6): p. 841–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Corcoran DL, et al. , PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome Biol, 2011. 12(8): p. R79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen B, et al. , PIPE-CLIP: a comprehensive online tool for CLIP-seq data analysis. Genome Biol, 2014. 15(1): p. R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Uren PJ, et al. , Site identification in high-throughput RNA-protein interaction data. Bioinformatics, 2012. 28(23): p. 3013–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Georgiev S, et al. , Evidence-ranked motif identification. Genome Biol, 2010. 11(2): p. R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Heinz S, et al. , Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell, 2010. 38(4): p. 576–89. [DOI] [PMC free article] [PubMed] [Google Scholar]