Abstract
microRNA-210 (miR-210) plays an important role in human disease, but its function in endometrial cancer (EC) is still unclear. Similarly, the nuclear factor I/X (NFIX) plays an important role in various biological functions of cells, but its function in EC is not yet known. In this study, we detected the expression of miR-210 from 66 EC patient tissues and 29 normal endometrium (NU) tissues by quantitative real-time PCR (RT-qPCR), as well as the expression of NIFX protein by western blot. We found that the expression of miR-210 in EC tissues was up-regulated and NIFX protein was down-regulated which was negatively correlated with NU tissues. The luciferase gene reporter system confirmed that miR-210 targeted inhibition of NIFX expression in HEC-1A cells. Up-regulation of miR-210 expression by transfection of miR-210-inhibitor could promote the proliferation, migration, and invasion of HEC-1A/HEC-1B cells. Taken together, we demonstrated that miR-210 could promote proliferation, migration and invasion by the negative regulation of NFIX expression in vitro, and that miR-210 promoted the progression of endometrial carcinoma by negative regulation NFIX expression.
Keywords: miR-210, NIFX, endometrial cancer, progression
Introduction
Endometrial cancer (EC) is one of the most common malignancies in the female reproductive tract. According to the American Cancer Society, there were 60,050 new cases of endometrial cancer in the United States and 10,470 deaths in 2015 [1]. In China, the incidence of endometrial cancer has also increased year by year, and is the second most common female reproductive system malignancies after cervical cancer [2]. microRNA (miRNA) is a group of short non-coding RNAs that do not encode proteins and are widely present in eukaryotes. They are approximately 19-25 nt in length, and their synthesis is an extremely complex biological process which mainly occur in the cytoplasm and nucleus [3]. miRNAs are incompletely complementarily paired with the 3’UTR region of the target gene mRNA through their “seed” sequence; this leads to the degradation of the target gene mRNA or inhibition of the expression of regulatory genes in the translation process, thereby regulating cell survival, differentiation, and response to the external environment [4]. In the process of tumor development and progression, miRNAs mainly play a role in two ways: one is that miRNAs directly regulate proliferation and apoptosis of cancer cells; the other way is to regulate through down-regulation of oncogene and tumor suppressor gene expression [5,6]. Finally, miRNA therapy is also being increasingly focused on cancer and various types of human diseases [7,8].
miR-210 was found to have abnormal expression in breast cancer [9], hepatocellular carcinoma [10], pancreatic cancer [11] and renal cancer [12], and is related to the progress and prognosis of various cancers [13], but its function in endometrial cancer (EC) remains unclear. Similarly, NIFX had also been found to be abnormally expressed in a variety of tumors [13], but there are no data on EC. In this study, we detected the expression of miR-210 and NIFX protein in 66 cases EC tissues and 29 cases normal endometrium (NU) tissues, and found that miR-210 promoted the progression of endometrial carcinoma through negative regulation the expression of NFIX.
Materials and methods
Patients tissue samples
Sixty-six cases of endometrial carcinoma tissues were collected at Tangshan Workers’ Hospital from patients with an average age of 47-73 years. According to the new revised standards of the International Obstetrics and Gynecology Union in 2009, there were 49 cases in FIGO I stage, 7 in II stage and 10 in III stage in this cohort. The other basic clinicodemographic information of these EC patients is shown in Table 1. EC patients who met the following conditions were excluded from this study: receiving treatment such as chemotherapy, radiotherapy, sex hormone therapy or immunotherapy before surgery; other comorbidities from cancer, infectious diseases or severe cardiovascular and cerebrovascular diseases; liver and kidney dysfunction; and incomplete clinical data.
Table 1.
Relationship between miR-210 or NFIX expression and clinical characteristics of EC patients
| Characteristic | n | miR-210 expression | NFIX protein expression | ||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| High | Low | P | High | Low | P | ||
| Age (years) | |||||||
| <50 | 21 | 15 | 6 | 0.06 | 10 | 11 | 0.942 |
| ≥50 | 45 | 21 | 24 | 21 | 24 | ||
| Family history of tumor | |||||||
| Negative | 55 | 28 | 27 | 0.185 | 25 | 30 | 0.581 |
| Positive | 11 | 8 | 3 | 6 | 5 | ||
| Menopausal status | |||||||
| Premenopausal | 37 | 23 | 14 | 0.160 | 20 | 17 | 0.193 |
| Postmenopausal | 29 | 13 | 16 | 11 | 18 | ||
| Complications | |||||||
| No | 45 | 28 | 17 | 0.067 | 20 | 25 | 0.574 |
| Yes | 21 | 8 | 13 | 11 | 10 | ||
| FIGO stage | |||||||
| I | 49 | 19 | 30 | <0.001 | 28 | 21 | 0.014 |
| II | 7 | 7 | 0 | 2 | 5 | ||
| III | 10 | 10 | 0 | 1 | 9 | ||
| Histologicl Grade | |||||||
| G1 | 28 | 4 | 24 | <0.001 | 22 | 6 | 0.000 |
| G2 | 28 | 22 | 6 | 8 | 20 | ||
| G3 | 10 | 10 | 0 | 1 | 9 | ||
| Lymph node metastasis | |||||||
| No | 56 | 26 | 30 | 0.002 | 30 | 26 | 0.011 |
| Yes | 10 | 10 | 0 | 1 | 9 | ||
Twenty-nine cases of normal endometrial tissues were also collected at Tangshan Workers’ Hospital, these NU tissues were all obtained from patients undergoing total hysterectomy for hysteromyoma. These patients were 45-69 years old and they also did not receive any sex hormone therapy before surgery. In addition, all patients participating in this study provided informed consent and were approved by the Tangshan Workers’ Hospital Ethics Association.
Quantitative real-time PCR
Quantitative Real-time PCR (RT-qPCR) was used to detect the expression of GAPDH mRNA, U6 and miR-210 in tissues and cells. Trizol was used to extract total RNA from the tissue or cell. The extracted RNA was reverse-transcribed into cDNA by using PrimeScript™RT Master Mix reverse transcription kit (RR036B, Takara, Kusatsu, Japan). PCR parameters were set at: 37°C/60 minutes, 85°C/5 seconds. Then a 20 μl Real-time fluorescence quantitative PCR (RT-qPCR) system was prepared according to the SYBR Green qPCR Master Mix kit instructions (638320, TakaRa, Kusatsu, Japan) and amplified using ABI 7500 fluorescence quantitative PCR instrument (Applied Biosystems, Maryland, USA). PCR parameters set: 95°C/30 s, [90°C/5 s, 65°C/30 s] -40 cycles.
PCR primers were as follows, U6-F: 5’-AUAAAUCCCUUUACACCUCTT-3’, U6-F: 5’-AAUAAAUCCCUUUACACCUCTT-3’; GAPDH-F: 5’-CTGGGCTACACTGAGCACC-3’, GAPDH-R: 5’-AAGTGGTCGTTGAGGGCAATG-3’; miR-210-F: 5’-ACACTCCAGCTGGGAGCCCCTGCCCACCGC-3’, miR-210-R: 5’-TGGTGTCGTGGAGTCG-3.
Western blot
Tissue or cell lysates were separated by SDS-page and then transferred to a PVDF membrane. The following primary antibodies were used: anti-cyclin D1 (ab1013415, 1:2000, ABCAM, Cambridge, UK), or anti-GAPDH (ab9484, 1:3000, ABCAM, Cambridge, UK). A second antibody was selected as follows: goat anti-rabbit (ab150077, 1:1000, ABCAM, Cambridge, UK), or goat anti-rat (ab150117, 1:1000, ABCAM, Cambridge, UK). Membranes were incubated with the primary antibody overnight at 4°C and the secondary antibody was incubated for 1 hour at room temperature.
Cells and cell transfection
HEC-1A (HTB-112, ATCC, VA, USA) and HEC-1B (HTB-113, ATCC, VA, USA) were cultured with DMEM medium (12491-15, ThermoFisher, CA, USA) which had 10% fetal bovine serum (10100-147, ThermoFisher, CA, USA) and 1% penicillin streptomycin (15640055, ThermoFisher, CA, USA).
miR-210-NC, miR-210-mimic and miR-210-inhibitor were designed and synthesized by Shenggong Bioengineering Co., Ltd. (Shanghai, China). These molecules were directly transfected into cells by Lipofectamine™ 2000 transfection reagent (11668019, Invitrogen, CA, USA). For wild type or mutation mRNA 3’-UTR of NIFX, they were first connected to pisCHECK2 (Promega, WI, USA) and then were directly transferred into cells by Lipofectamine™ 2000 transfection reagent.
Cell cloning assay
Finally, 4×103 cells/2 mL cells were inoculated into 6-well plates, and the culture medium was changed once every three day cells. After 3 weeks, the culture was stopped, the supernatant was removed, 4% formaldehyde was added for 15 minute, and then the cells were dyed with 0.25% crystal violet d for 25 minutes. At last, the culture plate was placed in a sterile, clean bench for drying and was imaged for cell counting.
Transwell
Corning BioCoat™ Matrigel transwell Invasion chamber (354480, Corning, USA) was used to evaluate the invasion ability of HEC-1A and HEC-1B. Briefly, 3×104 cells were added into the upper chamber with 200 µl DMEM medium without FBS. At the same time, 700 µl DMEM medium with 10% FBS was added into lower chamber. After 24 hours of incubation in a cell culture incubator, we removed the chamber, aspirated the medium, gently rinsed the chamber three times with 37°C pre-warmed PBS, gently wiped the upper non-migrating cells with a cotton swab, and added 1 ml 14% formaldehyde to the 24-well plate. We then put in incubator for 25-30 minutes. Once we removed the chamber, it was washed three times with PBS and air dried. We added 800 µl of 0.1% crystal violet to each well of the 24-well plate for staining. After 30 minutes, plates were placed in double distilled water and rinsed lightly. Afterwards, the plates were inverted and allowed to air dry. Five randomly selected fields were used for counting under a microscope at 200×.
Scratch test
Here, a concentration of 5×105 cells/2 mL was inoculated in 6-well plates. The next day, we scratched a horizontal line on the surface of the plate with a pipette tip and a ruler. The cells were softly washed three times with PBS, the cells were removed and serum-free medium was added. They were placed in one 37°C and 5% CO2 incubator and incubated for 24 hours; then images were taken.
Statistical analysis
In this study, SPSS 20.0 was used for statistical analysis of data. Measurement data was thought to be represented as mean-standard ± deviation, and count data was recorded as a percent. Differences between the two treatment groups were compared by chi-square test or student’s t test. Pearson method was used to analyze the correlation between the two indicators. P<0.05 was considered a significant difference.
Results
miR-210 was up-regulated in EC tissues
RT-qPCR was used to detect the expression of miR-210 in 66 cases endometrial cancer (EC) tissues and 29 cases normal endometriuml (NU) tissues. As shown in Figure 1A, the expression of miR-210 in EC tissues was higher than that in NU tissues (P<0.001). We further subdivided the 66 EC tissues, by comparing the expression of miR-210 in different groups. We found that the expression of miR-210 increased with the increase of FIGO stage (Figure 1B) and histological grade (Figure 1C). We observed miR-210 in EC patients with lymph node metastasis was higher than that in EC patients without lymph node metastasis (Figure 1D).
Figure 1.
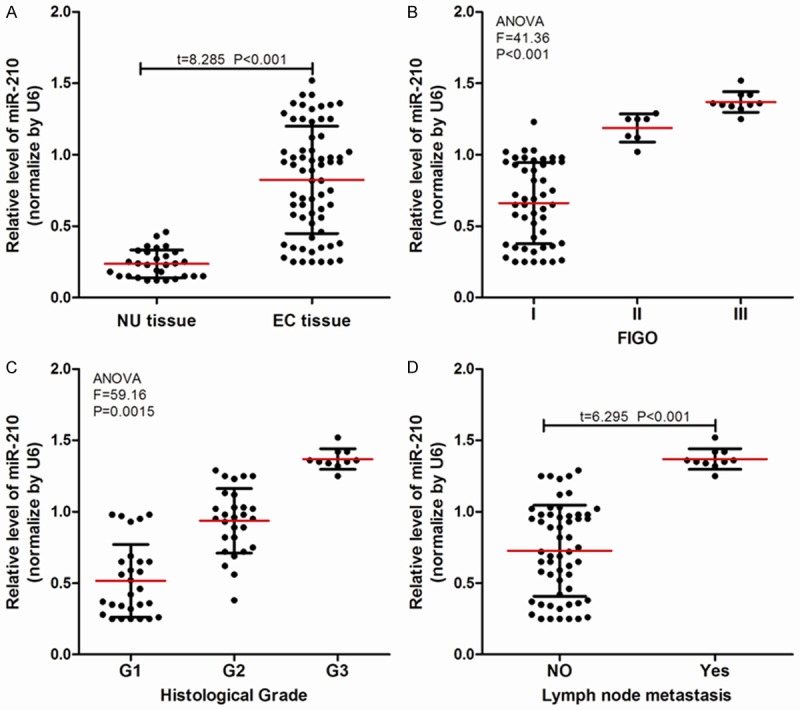
miR-210 expression in normal tissue and EC tissue. (A) miR-210 was highly expressed in endometrial carcinoma (EC) tissues (n=66) vs. normal endometrium (NU) tissues (n=29); (B-D) The expression of miR-210 in (B) different FIGO stages, (C) histologic grades, and (D) lymph node status.
In addition, we analyzed the relationship between miR-210 expression and the clinical characteristics of EC patients, and found that (Table 1) the expression of miR-210 in 66 cases EC patients had no significant correlation with age, family history of tumor, menopausal status or complications, but had a significant correlation with FIGO stage, histological grade and lymph node metastasis.
NIFX protein was down-regulated in EC tissues
Western blot was used to measure the expression of NFIX protein in 66 cases EC tissues and 29 cases of NU tissues, as shown in Figure 2A. The expression of NFIX protein in EC tissues was lower than that in NU tissues (P<0.001). Further, we also found that the expression of NFIX protein decreased with the increase in FIGO stage (Figure 2B) and histological grade (Figure 2C). NFIX protein in EC patients with lymph node metastasis was higher than that in patients without lymph node metastasis (Figure 2D).
Figure 2.
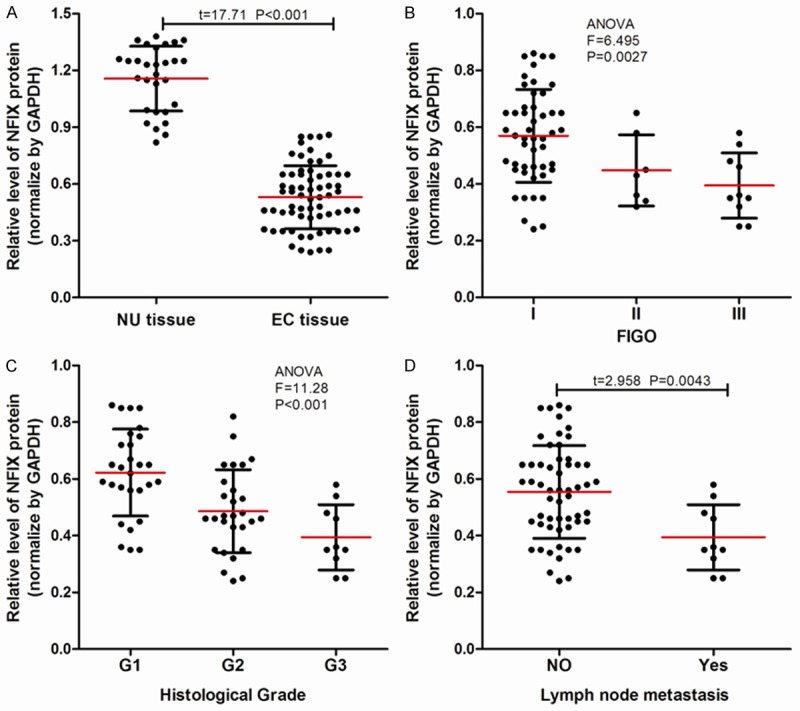
NIFX protein expression in normal tissue and EC tissue. (A) NIFX protein was minimally expressed in endometrial carcinoma (EC) tissues (n=66) vs. normal endometrium (NU) tissues (n=29); (B-D) The expression of NIFX protein in (B) different FIGO stages, (C) histologic grades, and (D) lymph node status.
Just as with miR-210, we also found that the expression of NFIX protein in 66 cases EC patients had no significant correlation with age, family history of tumor, menopausal status, or complications, but had a significant correlation with FIGO stage, histological grade, and lymph node metastasis (Table 1).
miR-210 inhibited the expression of NFIX
We analyzed the relationship between miR-210 expression and NIFX protein expression in 66 EC patients, and found that there was a significant positive correlation between miR-210 expression and NIFX protein expression in 66 EC patients (Figure 3A).
Figure 3.
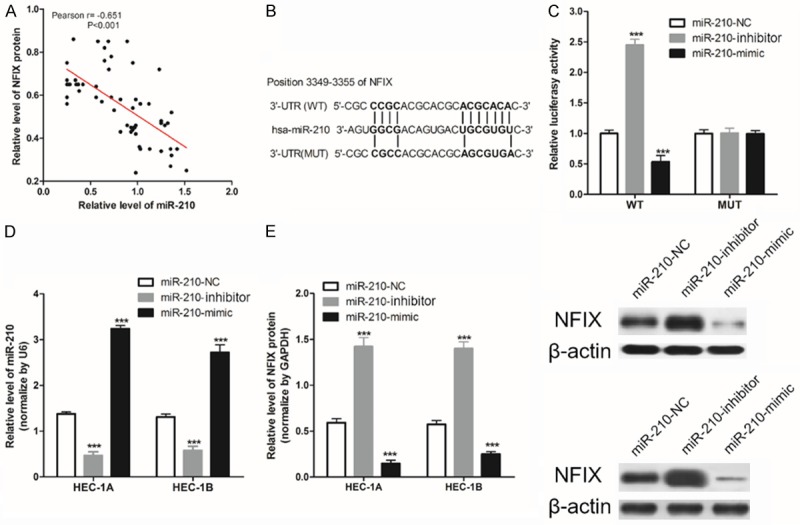
miR-210 targeted inhibition of NFIX expression in HEC-1A cells. A. The expression of NIFX protein was negatively related to the expression of miR-210 in 66 EC tissues; B. Construction of a WT-NFIX 3’UTR luciferase reporter vector, and a MUT-NFIX 3’UTR luciferase reporter vector with mutations on miR-210 binding sites of the NIFX 3’UTR; C. WT-NIFX/MUT-NIFX and miR-210-NC/miR-210-mimic/miR-210-inhibitor were transfected into HEC-1A cells, and luciferase activity was detected; D. RT-qPCR was used to measure the expression of miR-210 in HEC-1A/HEC-1B cells; E. Western blot was then used to detect the expression of NIFX protein in HEC-1A/HEC-1B cells. ***P<0.001 vs. miR-210-NC.
Moreover, we found that there are mutual binding sequences between them by analyzing the sequences of NFIX and miR-210 (Figure 3B). In order to confirm that miR-210 can regulate NIFX expression by binding to the NIFX 3’-UTR, we used the luciferase gene reporter system. Results showed that the transfection of miR-210-mimic significantly decreased WT type 3’-UTR luciferase activity (P<0.001) in HEA-1A, but did not work in MUT.
Further, as shown in Figure 3D, 3E, miR-210 could regulate the expression of NIFX protein in HEC-1A/HEC-1B cell. These results suggest that miR-210 targeted inhibition the expression of NFIX in EC cells.
miR-210 regulated the proliferation of EC cells in vitro
Tumor size played an important role in many tumor staging guidelines, and the proliferation capacity of tumor cells affects the progress of cancer. In this study, we used the clone formation assay to evaluate the proliferation of EC cells in vitro, and found that up-regulation of the expression of miR-210 by transfection of miR-210-mimic could increase the number of cell clones (Figure 4), and down-regulation of the expression of miR-210 by transfection of the miR-210-mimic could decrease the number of cell clones.
Figure 4.
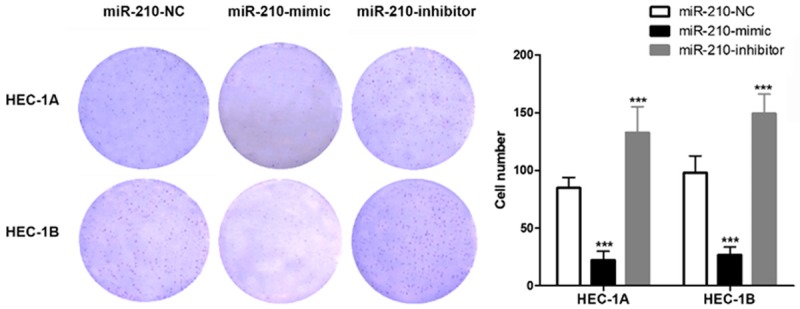
miR-210 regulated the proliferation of HEC-1A/HEC-1B cells in vitro.
miR-210 regulated the migration/invasion of EC cells in vitro
One of the most important biological characteristics of cancer cells is their mobility and invasiveness, and the invasion and metastasis of cancer cells are also the most important factors leading to treatment failure and even death of cancer patients [14]. In this study, we found that miR-210-mimic could increase the number of invading HEC-1A/HEC-1B cells, and miR-210-inhibitor decreased it (Figure 5A). The same trend also occurs in the number of migrating HEC-1A/HEC-1B cells (Figure 5B).
Figure 5.
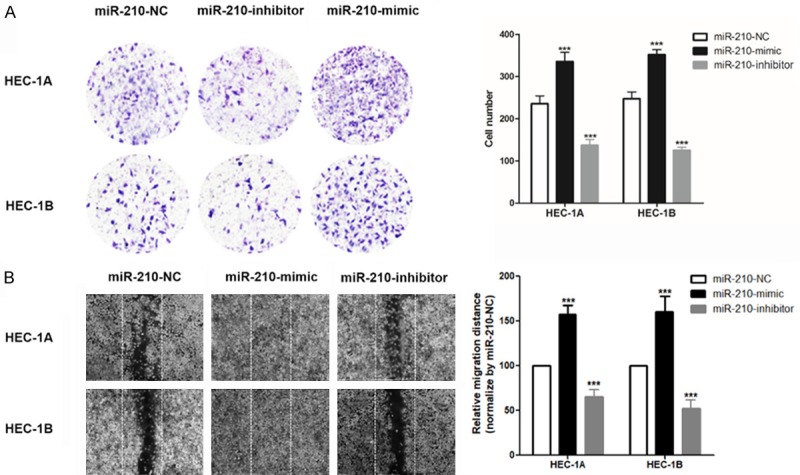
miR-210 regulated the migration/invasion of HEC-1A/HEC-1B cells in vitro. A. Transwell assay was used to evaluate the invasion ability of HEC-1A and HEC-1B cells; B. A scratch test was used to evaluate migration ability in HEC-1A and HEC-1B cells; ***P<0.001 vs. miR-210-NC.
Discussion
In this study, we found that the expression of miR-210 in 66 sample EC tissues was higher than that in 29 samples of NU tissues. Additionally, this expression was significantly correlated with FIGO stage, histologic grade, and lymph node metastasis. miR-210 is located on the human chromosome 11q15.5, and the target gene of miR-210 has reached 50 [15]. miR-210 regulates the expression of these genes and is widely involved in various physiological and pathological metabolic processes in vivo, such as the development of a variety of adult stem cells, embryonic development, cell growth and apoptosis, cell differentiation, angiogenesis, the regulation of the immune system, and the occurrence and development of tumors [16]. Previous research has shown that compared with normal tissues, the expression of miR-210 is increased in many tumors, such as melanoma [17], lung cancer [18], pancreatic cancer [11], breast cancer [9], and head and neck tumor [19], kidney tumors, etc. Such expression was also likely to be closely related to the occurrence and development of tumors. This suggested that the higher expression of miR-210 in EC tissue might be related to the occurrence and development of EC.
In addition, this study found that the expression of NFIX protein in 66 cases EC tissues were lower than in 29 cases NU tissues, and had a significant correlation with FIGO stage, histological grade and lymph node metastasis. Nuclear factor I (NFI) is a family of closely related transcription factors. They constitutively bind as dimers to specific sequences of DNA with high affinity, and so could be used as a transcription switch from embryonic to fetal muscle [20]. They also can regulate proliferation and migration during SVZ neural niche development [21]. NFIX is an important member of the NFI family and participated in the regulation of the proliferation and differentiation of radial glial cells in the embryonic forebrain, as well as the migration of neurons within the postnatal cerebellum [22]. Recent studies in the field of cancer found that NFIX was down-regulated in various tumor tissues [23] such as lung cancer [24] and esophageal cancer [25]. Moreover, down-regulating the expression of NFIX in esophageal cancer cells did not only increase the proliferation capacity of esophageal cancer cells, but also increased its invasion and migration ability, indicating that NFIX may be involved in regulating biological functions of tumor cells.
Further research has found that the expression of miR-210 and NIFX protein in EC tissues had a negative correlation. Additionally, the luciferase gene reporter system confirmed that miR-210 could target inhibition of NFIX expression in EC cells. miR-210 played an important regulatory role under hypoxic conditions, and in many cell types, hypoxia could induce cell cycle arrest through HIF-1α. One of its possible mechanisms was a change in the expression of miR-210, so as to regulate the progression of cell cycle by inhibiting the transcription factor E2F family member E2F3, myc antagonistic factor max network transcriptional repressor (MNT) to regulate the progression of the cell cycle [26]. Zhang et al. [27] pointed out that the myc antagonist MNT was a destroyer of myc/Max/Med network structure and could inhibit the expression of myc target gene, and miR-210 could also promote the progression of cell cycle by down-regulating MTT. In addition, Chen et al. [26] pointed out that miR-210 could positively or negatively regulate the myc pathway through the regulation of many key points in the cell cycle. This was facilitated through gene regulation of the c-myc signaling pathway, and thus participates in the proliferation and metabolism of tumor cells. In this study, we found up-regulation of the expression of miR-210 via transfection of a miR-210-mimic that could increase the number of EC cell clones, and down-regulated the expression of miR-210 by transfection of miR-210-mimic decreasing the number of cell clones. Previous studies have shown that down-regulating the expression of NFIX in tumor cells such as lung cancer or esophageal cancer can enhance the proliferation of cancer cells [24,25]. Combining our research suggested that miR-210 could enhance EC cell proliferation by negatively regulating the expression of NFIX protein.
The invasion and migration of cancer cells from the primary site to other tissues and organs of the patient are the main causes of failure and death in cancer patients [14]. In this study, we found miR-210 could positively regulate the invasion and migration ability of EC cells. Similarly, Zhang et al. [28] showed that miR-210 was down-regulated in breast cancer tissues and cells. Inhibition of miR-210 expression in breast cancer cells could significantly reduce its invasion and migration ability. In addition, many previous studies have also demonstrated that inhibition of miR-210 expression in vitro can reduce the invasion and migration abilities of hepatoma [29], gastric cancer [30], breast cancer [31], and colorectal cancer [32] cells.
Conclusion
miR-210 was up-regulated and NIFX down-regulated in EC tissues. The expression of miR-210 and NIFX protein in EC tissues had a negative correlation. miR-210 promoted the proliferation, migration, and invasion of HEC-1A/HEC-1B cells and the progression of endometrial carcinoma by negative regulation of the expression of NFIX.
Acknowledgements
This research was supported by Science and technology research project of Hebei higher education institutions Youth Fund (QN2018309) and Scientific research foundation of Tianjin Education Commission (NO.2017KJ225).
Disclosure of conflict of interest
None.
References
- 1.Miller KD, Siegel RL, Lin CC, Mariotto AB, Kramer JL, Rowland JH, Stein KD, Alteri R, Jemal A. Cancer treatment and survivorship statistics, 2016. CA Cancer J Clin. 2016;66:271–289. doi: 10.3322/caac.21349. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, Powers S, Cordon-Cardo C, Lowe SW, Hannon GJ. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bushati N, Cohen SM. microRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- 5.Joyce BT, Zheng Y, Zhang Z, Liu L, Kocherginsky M, Murphy R, Achenbach C, Musa J, Wehbe F, Just A. MicroRNA processing gene methylation and cancer risk. Cancer Epidemiol Biomarkers Prev. 2018;27:550–557. doi: 10.1158/1055-9965.EPI-17-0849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kwak PB, Iwasaki S, Tomari Y. The microRNA pathway and cancer. Cancer Sci. 2010;101:2309–2315. doi: 10.1111/j.1349-7006.2010.01683.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hosseinahli N, Aghapour M, Duijf PH, Baradaran B. Treating cancer with microRNA replacement therapy: a literature review. J Cell Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 8.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16:203. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 9.Jung EJ, Santarpia L, Kim J, Esteva FJ, Moretti E, Buzdar AU, Di Leo A, Le XF, Bast RC Jr, Park ST. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118:2603–2614. doi: 10.1002/cncr.26565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y, Zhang J, Xia T, Li G, Tian T, Wang M, Wang R, Zhao L, Yang Y, Lan K. MicroRNA-210 promotes cancer angiogenesis by targeting fibroblast growth factor receptor-like 1 in hepatocellular carcinoma. Oncol Rep. 2016;36:2553–2562. doi: 10.3892/or.2016.5129. [DOI] [PubMed] [Google Scholar]
- 11.Amponsah PS, Fan P, Bauer N, Zhao Z, Gladkich J, Fellenberg J, Herr I. microRNA-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017;388:107–117. doi: 10.1016/j.canlet.2016.11.035. [DOI] [PubMed] [Google Scholar]
- 12.McCormick R, Blick C, Ragoussis J, Schoedel J, Mole D, Young A, Selby P, Banks R, Harris A. miR-210 is a target of hypoxia-inducible factors 1 and 2 in renal cancer, regulates ISCU and correlates with good prognosis. Br J Cancer. 2013;108:1133–1142. doi: 10.1038/bjc.2013.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu Y, Wang Y, Xu Q, Zhou X, Qin Z, Chen C, Zhang Q, Tian Y, Zhang C, Li X. Prognostic evaluation of microRNA-210 in various carcinomas: Evidence from 19 studies. Medicine (Baltimore) 2017;96:e8113. doi: 10.1097/MD.0000000000008113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Valastyan S, Weinberg RA. Tumor metastasis: molecular insights and evolving paradigms. Cell. 2011;147:275–292. doi: 10.1016/j.cell.2011.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devlin C, Greco S, Martelli F, Ivan M. miR-210: More than a silent player in hypoxia. IUBMB Life. 2011;63:94–100. doi: 10.1002/iub.427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang X, Le QT, Giaccia AJ. MiR-210-micromanager of the hypoxia pathway. Trends Mol Med. 2010;16:230–237. doi: 10.1016/j.molmed.2010.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ono S, Oyama T, Lam S, Chong K, Foshag LJ, Hoon DS. A direct plasma assay of circulating microRNA-210 of hypoxia can identify early systemic metastasis recurrence in melanoma patients. Oncotarget. 2015;6:7053–7064. doi: 10.18632/oncotarget.3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grosso S, Doyen J, Parks SK, Bertero T, Paye A, Cardinaud B, Gounon P, Lacas-Gervais S, Noël A, Pouysségur J, Barbry P, Mazure NM, Mari B. MiR-210 promotes a hypoxic phenotype and increases radioresistance in human lung cancer cell lines. Cell Death Dis. 2013;4:e544. doi: 10.1038/cddis.2013.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gee HE, Camps C, Buffa FM, Patiar S, Winter SC, Betts G, Homer J, Corbridge R, Cox G, West CM. hsa-miR-210 is a marker of tumor hypoxia and a prognostic factor in head and neck cancer. Cancer. 2010;116:2148–2158. doi: 10.1002/cncr.25009. [DOI] [PubMed] [Google Scholar]
- 20.Messina G, Biressi S, Monteverde S, Magli A, Cassano M, Perani L, Roncaglia E, Tagliafico E, Starnes L, Campbell CE. Nfix regulates fetalspecific transcription in developing skeletal muscle. Cell. 2010;140:554–566. doi: 10.1016/j.cell.2010.01.027. [DOI] [PubMed] [Google Scholar]
- 21.Heng YH, Zhou B, Harris L, Harvey T, Smith A, Horne E, Martynoga B, Andersen J, Achimastou A, Cato K. NFIX regulates proliferation and migration within the murine SVZ neurogenic niche. Cereb Cortex. 2014;25:3758–3778. doi: 10.1093/cercor/bhu253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piper M, Harris L, Barry G, Heng YH, Plachez C, Gronostajski RM, Richards LJ. Nuclear factor one X regulates the development of multiple cellular populations in the postnatal cerebellum. J Comp Neurol. 2011;519:3532–3548. doi: 10.1002/cne.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin G, Kang TW, Yang S, Baek SJ, Jeong YS, Kim SY. GENT: gene expression database of normal and tumor tissues. Cancer Inform. 2011;10:149–157. doi: 10.4137/CIN.S7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rahman NI, Abdul Murad NA, Mollah MM, Jamal R, Harun R. NFIX as a master regulator for lung cancer progression. Front Pharmacol. 2017;8:540. doi: 10.3389/fphar.2017.00540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mao Y, Liu J, Zhang D, Li B. MiR-1290 promotes cancer progression by targeting nuclear factor I/X (NFIX) in esophageal squamous cell carcinoma (ESCC) Biomed Pharmacother. 2015;76:82–93. doi: 10.1016/j.biopha.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 26.Chen WY, Liu WJ, Zhao YP, Zhou L, Zhang TP, Chen G, Shu H. Induction, modulation and potential targets of miR-210 in pancreatic cancer cells. Hepatobiliary Pancreat Dis Int. 2012;11:319–324. doi: 10.1016/s1499-3872(12)60168-4. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, Sun H, Dai H, Walsh R, Imakura M, Schelter J, Burchard J, Dai X, Chang AN, Diaz RL. MicroRNA miR-210 modulates cellular response to hypoxia through the MYC antagonist MNT. Cell Cycle. 2009;8:2756–2768. doi: 10.4161/cc.8.17.9387. [DOI] [PubMed] [Google Scholar]
- 28.Nan Z, Shaoyou L, Yaning G, Junyu R, Xixi T, Jian D. Effect of miRNA-210 on proliferation, migration and invasion of human breast cancer cells. Chinese Journal of Cancer Biotherapy. 2013;20:289–294. [Google Scholar]
- 29.Kai AK, Lo RC, Lee JM, Wong JC, Ng IO. Hypoxia-inducible miR-210 is involved in hypoxia-induced cell migration and invasion in human hepatocellular carcinoma. AACR. 2012 [Google Scholar]
- 30.Zhang C, Tian W, Meng L, Qu L, Shou C. PRL-3 promotes gastric cancer migration and invasion through a NF-κB-HIF-1α-miR-210 axis. J Mol Med (Berl) 2016;94:401–415. doi: 10.1007/s00109-015-1350-7. [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y, Yan J, Wang L, Dai H, Li N, Hu W, Cai H. HIF-1α promotes breast cancer cell MCF-7 proliferation and invasion through regulating miR-210. Cancer Biother Radiopharm. 2017;32:297–301. doi: 10.1089/cbr.2017.2270. [DOI] [PubMed] [Google Scholar]
- 32.Tran MN, Choi W, Navai N, Wszolek MF, Lee IC, Siefker-Radtke AO, McConkey DJ. Abstract LB-472: p63 inhibits epithelial-mesenchymal transition by promoting the expression of miR205 in human bladder cancer cells. Cancer Res. 2012;72:472. [Google Scholar]


