Abstract
Interleukin-1 (IL-1) gene is known to be implicated in tuberculosis (TB). The present case-control study was designed to investigate whether IL-1 polymorphisms associated with TB susceptibility in a Chinese Tibetan cohort. 300 Tibetan tuberculosis patients and 300 healthy controls were recruited for 12 single-nucleotide polymorphisms (SNPs) genotyping of IL-1 gene via Sequenom MassARRAY analysis. The odds ratio (OR) and 95% confidence interval (CI) were analyzed by unconditional logistic regression to evaluate the effect of polymorphisms on the risk of tuberculosis. Among genotyped SNPs, a significant high risk between TB and SNP rs3783550 G/T, rs3783546 G/C, rs2856838 A/G, rs1609682 G/T, rs3783521 A/G genotype within IL-1α as well as rs1143623 G/G genotype within IL-1β was found based on multiple model analysis. In addition, IL-1α SNPs mapped in a 10 kb LD block with D’>0.98, suggesting that a significant linkage disequilibrium presence among these SNPs, and “TCATG” haplotype of IL-1α SNPs with a 1.47-fold risk for TB was observed (P = 0.007). In conclusion, our data shed new light on how IL-1 SNPs may contribute to tuberculosis susceptibility in the Chinese Tibetan population.
Keywords: IL-1, tuberculosis, gene polymorphisms, Chinese Tibetan population
Introduction
Tuberculosis (TB) is a chronic infectious disease caused by Mycobacterium tuberculosis (Mtb), and remains a global emergency of morbidity and mortality especially in Asia and Africa [1,2]. Approximately 2 million individuals die of TB and 9 million become infected annually based on the World Health Organization statistics [3]. The incidence of tuberculosis in China ranks as the top in the world [4], and the incidence of tuberculosis in Tibetans is high in China [5]. Conventional diagnosis of TB can be performed according to clinical manifestations, laboratory tests, and imaging studies.
The occurrence of TB at different rates among particular races, ethnicities, and families indicates a genetic predisposition to TB susceptibility [6]. Therefore, it is critical to understand if genetic variation is significantly related to more or less susceptibility to TB infection. Discovery of reliable genetic molecular markers are essential for early rapid diagnosis, improving therapeutic efficacy, and reducing drug toxicity. Several lines of evidence from genome-wide linkage studies and association-based candidate gene studies have defined a number of immune response genes for the development of active TB, including encoding pattern recognition receptors (TLR, CD14), C-type lectins, cytokines/chemokines, and their receptors (IFN-γ, TNF-α, IL-12, IL-10, MCP-1, MMP-1), major histocompatibility complex (MHC) molecules, vitamin D receptor (VDR), and proton-coupled divalent metal ion transporters (SLC11A1) [7,8]. Genetic variations in these genes have a diverse influence on the susceptibility to or protection against TB among particular families, ethnicities, and races.
IL-1 gene cluster, as a type of immune response gene, is located in a 430-kb region of DNA on the long arm of human chromosome 2 and codes some of pro-inflammatory cytokines, which might play a role in various neuropathologies involved in neuron inflammation. The largest IL-1 family of interleukins consist of IL-1α, IL-1β, and IL-1 receptor antagonist (IL-1RA). Among them, IL-1α and β are key players in the innate immune system, and also are endogenous pyrogens with similar activities to lipopolysaccharides (LPS), which are the major molecular components of the outer membrane of gram-negative bacteria [9,10]. Several lines evidence supporting an association between IL-1 innate immune gene associated with TB pathogenesis [6,11].
TB is a major public health problem among Tibetans, most of whom live in the Tibet Autonomous Region, and some groups reside in the Qinghai, Gansu, Sichuan, and Yunnan provinces of China. A study of genomic variations in Tibetan people suggested that a majority of the Tibetan gene pool may have diverged from that of the Han population around 3000 years ago [12]. To our best knowledge, there is no information between IL-1 gene polymorphisms and the susceptibility of tuberculosis in Chinese Tibetan population. Therefore, we performed this case-control study in order to overcome this limitation and provide useful information for TB treatment. A total of 12 SNPs were carefully selected for further genotyping based on 1000 Genomes Project (http://www.1000 genomes.org/) and dbSNP (https://www. ncbi.nlm.nih.gov/projects/SNP/) databases with MAFs>5% in Chinese Han population.
Materials and methods
Study participants
This case-control study was performed on 300 tuberculosis patients and 300 healthy individuals from Chinese Tibetan population. Tuberculosis cases were diagnosed according to the standard criteria of GB15987-1995 treated at the third Hospital of the Tibet Autonomous Region between September 2015 and July 2017. Control subjects without any clinical signs or tuberculosis history, and without any other severe diseases, were selected from normal Tibetans in physical examination. After signing the informed consent form, each subject was collected about 5 mL venous blood with EDTA and stored at -20°C until DNA extraction. This study was approved by the Clinical Research Ethics of Xizang Minzu University.
SNPs selection and genotyping
12 SNPs of the IL-1 gene were selected based on the databases of 1000 Genomes Project (http://www.1000genomes.org/) and dbSNP (https://www. ncbi.nlm.nih.gov/projects/SNP/) with MAFs>5% in Chinese Han population. Genomic DNA was extracted from peripheral blood samples by GoldMag-Mini Purification Kit (GoldMag Co. Ltd. Xi’an, People’s Republic of China) [13], and measurement of DNA concentration was performed by NanoDrop 2000 (Thermo Scientific, Waltham, MA, USA). Sequenom MassARRAY Assay Design 4.0 Software (Agena Bioscience Inc) was utilized to design a multiplexed SNP Mass EXTENDED assay [14]. SNP genotyping was carried out via the manufacturer’s protocol on the Sequenom MassARRAY RS1000 platform. Data management and analysis were performed using the Sequenom Typer 4.0 Software (Agena Bioscience Inc).
Statistical analysis
Microsoft Excel and SPSS 19.0 Software (SPSS Inc, Chicago, IL, USA) were used for statistical analyses. Deviation from the Hardy-Weinberg equilibrium (HWE) for each SNP was tested to measure the distribution of the polymorphism using a Chi-square test [15,16]. The P values obtained in our study were two-sided and P-value < 0.05 was considered significant. The odds ratio (OR) and 95% confidence interval (CI) were analyzed by unconditional logistic regression to evaluate the effect of polymorphisms on the risk of tuberculosis. Finally, software of haploview package (version 4.2) was adopted to evaluate the pairwise linkage disequilibrium (LD) among SNPs and the association between polymorphism loci and TB risk [17,18].
Results
The IL-1 polymorphism was analyzed in 300 TB patients and 300 unrelated healthy controls. Table 1 presented basic information for 12 genotyped SNPs with regard to their chromosomal position, allele, minor allele frequency, and HWE test results for all subjects. There was no deviation from Hardy-Weinberg equilibrium (HWE) for each SNP, suggesting good SNP genotyping quality. Almost all of SNPs except rs2853550, rs1143643, rs3136558, rs1143630 had strong association with TB risk (P < 0.05) based on allele frequencies Chi-square test.
Table 1.
Allele frequencies of candidates SNPs examined in IL-1 gene among cases and controls
| SNP | Gene | Chr | Position | Allele | Minor allele frequency | HWE P value | OR (95% CI) | P a | |
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| Case | Control | ||||||||
| rs3783550 | IL-1α | 2q13 | 113532885 | T/G | 0.410 | 0.333 | 0.7956 | 1.39 (1.10-1.76) | 0.021* |
| rs3783546 | IL-1α | 2q13 | 113534830 | C/G | 0.420 | 0.332 | 0.8963 | 1.46 (1.15-1.85) | 0.006* |
| rs2856838 | IL-1α | 2q13 | 113539972 | A/G | 0.297 | 0.240 | 0.6332 | 1.34 (1.03-1.73) | 0.040* |
| rs1609682 | IL-1α | 2q13 | 113540205 | T/G | 0.410 | 0.334 | 0.7957 | 1.38 (1.09-1.75) | 0.022* |
| rs3783521 | IL-1α | 2q13 | 113543577 | G/A | 0.430 | 0.333 | 0.7956 | 1.51 (1.19-1.91) | 0.002* |
| rs2853550 | IL-1β | 2q13 | 113587121 | A/G | 0.090 | 0.092 | 0.2923 | 0.98 (0.66-1.45) | 0.308 |
| rs1143643 | IL-1β | 2q13 | 113588302 | C/T | 0.527 | 0.470 | 0.7276 | 1.26 (1.00-1.58) | 0.128 |
| rs3136558 | IL-1β | 2q13 | 113591275 | G/A | 0.372 | 0.379 | 0.8057 | 0.97 (0.77-1.22) | 0.928 |
| rs1143630 | IL-1β | 2q13 | 113591655 | T/G | 0.162 | 0.155 | 0.5060 | 1.05 (0.77-1.43) | 0.936 |
| rs1143627 | IL-1β | 2q13 | 113594387 | G/A | 0.561 | 0.465 | 0.0470 | 1.47 (1.17-1.85) | < 0.001* |
| rs16944 | IL-1β | 2q13 | 113594867 | A/G | 0.561 | 0.455 | 0.0803 | 1.53 (1.22-1.92) | < 0.001* |
| rs1143623 | IL-1β | 2q13 | 113595829 | G/C | 0.487 | 0.373 | 0.0359 | 1.59 (1.26-2.00) | < 0.001* |
SNP: Single nucleotide polymorphism; OR = Odds ratio; 95% CI = 95% confidence interval; HWE: Hardy-Weinberg equilibrium.
P < 0.05 indicates statistical significance;
P values were calculated using Pearson Chi-square test/Fisher’s exact test.
Comparisons of the SNP genotypes and the risk of TB under the genetic model were listed in Table 2. Among genotyped SNPs, a positive association was found between significantly high TB risk and SNPs rs3783550 G/T (OR = 1.54; 95% CI = 1.11-2.54; P = 0.010), rs37483546 G/C (OR = 1.67; 95% CI = 1.20-2.34; P = 0.002), rs2856838 A/G (OR = 1.52; 95% CI = 1.10-2.10; P = 0.011), rs1609682 G/T (OR = 1.54; 95% CI = 1.10-2.14; P = 0.011), rs3783521 A/G (OR = 1.50; 95% CI = 1.28-2.52; P < 0.001) genotypes within IL-1α as well as rs1143623 G/G (OR = 2.56; 95% CI = 1.63-4.00; P < 1E-04) within IL-1β based on multiple models analysis.
Table 2.
Relationship between IL-1 SNPs and tuberculosis risk under multiple models of analysis
| SNP | Model | Genotype | Control | Case | OR (95% CI) | P a |
|---|---|---|---|---|---|---|
| rs3783550 | Codominant | G/G | 132 (44.0%) | 101 (33.8%) | 1.00 | 0.020* |
| G/T | 136 (45.3%) | 151 (50.5%) | 1.45 (1.03-2.05) | |||
| T/T | 32 (10.7%) | 47 (15.7%) | 1.92 (1.14-3.22) | |||
| Dominant | G/G | 132 (44.0%) | 101 (33.8%) | 1.00 | 0.010* | |
| G/T+T/T | 168 (56.0%) | 198 (66.2%) | 1.54(1.11-2.14) | |||
| Recessive | G/G+G/T | 268 (89.3%) | 252 (84.3%) | 1.00 | 0.067 | |
| T/T | 32 (10.7%) | 47 (15.7%) | 1.56 (0.97-2.53) | |||
| Overdominant | G/G+T/T | 164 (54.7%) | 148 (49.5%) | 1.00 | 0.210 | |
| G/T | 136 (45.3%) | 151 (50.5%) | 1.23 (0.89-1.70) | |||
| Log-additive | --- | --- | --- | 1.40 (1.10-1.79) | 0.006* | |
| rs3783546 | Codominant | G/G | 132 (44.3%) | 95 (32.2%) | 1.00 | 0.006* |
| G/C | 134 (45.0%) | 152 (51.5%) | 1.58 (1.11-2.24) | |||
| C/C | 32 (10.7%) | 48 (16.3%) | 2.08 (1.24-3.5) | |||
| Dominant | G/G | 132 (44.3%) | 95 (32.2%) | 1.00 | 0.002* | |
| G/C+C/C | 166 (55.7%) | 200 (67.8%) | 1.67 (1.20-2.34) | |||
| Recessive | G/G+G/C | 266 (89.3%) | 247 (83.7%) | 1.00 | 0.048* | |
| C/C | 32 (10.7%) | 48 (16.3%) | 1.62 (1.00-2.61) | |||
| Overdominant | G/G+C/C | 164 (55.0%) | 143 (48.5%) | 1.00 | 0.110 | |
| G/C | 134 (45.0%) | 152 (51.5%) | 1.30 (0.94-1.80) | |||
| Log-additive | --- | --- | --- | 1.48 (1.16-1.88) | 0.001* | |
| rs2856838 | Codominant | G/G | 170 (57.0%) | 140 (46.7%) | 1.00 | 0.039* |
| A/G | 113 (37.9%) | 142 (47.3%) | 1.53 (1.09-2.13) | |||
| A/A | 15 (5.0%) | 18 (6.0%) | 1.46 (0.71-3.00) | |||
| Dominant | G/G | 170 (57.0%) | 140 (46.7%) | 1.00 | 0.011* | |
| A/G+A/A | 128 (43.0%) | 160 (53.3%) | 1.52 (1.10-2.10) | |||
| Recessive | G/G+A/G | 283 (95.0%) | 282 (94.0%) | 1.00 | 0.600 | |
| A/A | 15 (5.0%) | 18 (6.0%) | 1.46 (0.71-3.00) | |||
| Overdominant | G/G+A/A | 185 (62.1%) | 158 (52.7%) | 1.00 | 0.020* | |
| A/G | 113 (37.9%) | 142 (47.3%) | 1.47 (1.06-2.04) | |||
| Log-additive | --- | --- | --- | 1.37 (1.05-1.80) | 0.020* | |
| rs1609682 | Codominant | G/G | 131 (44.0%) | 101 (33.8%) | 1.00 | 0.022* |
| G/T | 135 (45.3%) | 151 (50.5%) | 1.45 (1.02-2.06) | |||
| T/T | 32 (10.7%) | 47 (15.7%) | 1.91 (1.13-3.20) | |||
| Dominant | G/G | 131 (44.0%) | 101 (33.8%) | 1.00 | 0.011* | |
| G/T+T/T | 167 (56.0%) | 198 (66.2%) | 1.54 (1.10-2.14) | |||
| Recessive | G/G+G/T | 266 (89.3%) | 252 (84.3%) | 1.00 | 0.072 | |
| T/T | 32 (10.7%) | 47 (15.7%) | 1.55 (0.96-2.51) | |||
| Overdominant | G/G+T/T | 163 (54.7%) | 148 (49.5%) | 1.00 | 0.200 | |
| G/T | 135 (45.3%) | 151 (50.5%) | 1.23 (0.89-1.70) | |||
| Log-additive | --- | --- | --- | 1.40 (1.10-1.78) | 0.006* | |
| rs3783521 | Codominant | A/A | 132 (44.0%) | 89 (30.4%) | 1.00 | 0.002* |
| A/G | 136 (45.3%) | 156 (53.2%) | 1.70 (1.19-2.42) | |||
| G/G | 32 (10.7%) | 48 (16.4%) | 2.22 (1.32-3.75) | |||
| Dominant | A/A | 132 (44.0%) | 89 (30.4%) | 1.00 | < 0.001* | |
| A/G+G/G | 168 (56.0%) | 204 (69.6%) | 1.80 (1.28-2.52) | |||
| Recessive | A/A+A/G | 268 (89.3%) | 245 (83.6%) | 1.00 | 0.041 | |
| G/G | 32 (10.7%) | 48 (16.4%) | 1.64 (1.02-2.65) | |||
| Overdominant | A/A+G/G | 164 (54.7%) | 137 (46.8%) | 1.00 | 0.054 | |
| A/G | 136 (45.3%) | 156 (53.2%) | 1.37 (0.99-1.9) | |||
| Log-additive | --- | --- | --- | 1.55 (1.21-1.97) | < 0.001* | |
| rs2853550 | Codominant | G/G | 248 (82.9%) | 254 (84.7%) | 1.00 | 0.300 |
| A/G | 47 (15.7%) | 38 (12.7%) | 0.79 (0.50-1.25) | |||
| A/A | 4 (1.3%) | 8 (2.7%) | 1.95 (0.58-6.57) | |||
| Dominant | G/G | 248 (82.9%) | 254 (84.7%) | 1.00 | 0.570 | |
| A/G+A/A | 51 (17.1%) | 46 (15.3%) | 0.88 (0.57-1.36) | |||
| Recessive | G/G+A/G | 295 (98.7%) | 292 (97.3%) | 1.00 | 0.240 | |
| A/A | 4 (1.3%) | 8 (2.7%) | 2.02 (0.60-6.78) | |||
| Overdominant | G/G+A/A | 252 (84.3%) | 262 (87.3%) | 1.00 | 0.280 | |
| A/G | 47 (15.7%) | 38 (12.7%) | 0.78 (0.49-1.23) | |||
| Log-additive | --- | --- | --- | 0.98 (0.68-1.42) | 0.910 | |
| rs1143643 | Codominant | T/T | 82 (27.4%) | 67 (22.6%) | 1.00 | 0.130 |
| T/C | 153 (51.2%) | 146 (49.3%) | 1.17 (0.79-1.73) | |||
| C/C | 64 (21.4%) | 83 (28.0%) | 1.59 (1.00-2.51) | |||
| Dominant | T/T | 82 (27.4%) | 67 (22.6%) | 1.00 | 0.180 | |
| T/C+C/C | 217 (72.6%) | 229 (77.4%) | 1.29 (0.89-1.87) | |||
| Recessive | T/T+T/C | 235 (78.6%) | 213 (72.0%) | 1.00 | 0.060 | |
| C/C | 64 (21.4%) | 83 (28.0%) | 1.43 (0.98-2.08) | |||
| Overdominant | T/T+C/C | 146 (48.8%) | 150 (50.7%) | 1.00 | 0.650 | |
| T/C | 153 (51.2%) | 146 (49.3%) | 0.93 (0.67-1.28) | |||
| Log-additive | --- | --- | --- | 1.26 (1.00-1.58) | 0.048* | |
| rs3136558 | Codominant | A/A | 116 (38.9%) | 118 (39.3%) | 1.00 | 0.930 |
| A/G | 138 (46.3%) | 141 (47.0%) | 1.00 (0.71-1.42) | |||
| G/G | 44 (14.8%) | 41 (13.7%) | 0.92 (0.56-1.51) | |||
| Dominant | A/A | 116 (38.9%) | 118 (39.3%) | 1.00 | 0.920 | |
| A/G+G/G | 182 (61.1%) | 182 (60.7%) | 0.98 (0.71-1.37) | |||
| Recessive | A/A+A/G | 254 (85.2%) | 259 (86.3%) | 1.00 | 0.700 | |
| G/G | 44 (14.8%) | 41 (13.7%) | 0.91 (0.58-1.45) | |||
| Overdominant | A/A+G/G | 160 (53.7%) | 159 (53.0%) | 1.00 | 0.870 | |
| A/G | 138 (46.3%) | 141 (47.0%) | 1.03 (0.75-1.42) | |||
| Log-additive | --- | --- | --- | 0.97 (0.77-1.22) | 0.790 | |
| rs1143630 | Codominant | G/G | 212 (70.7%) | 208 (69.3%) | 1.00 | 0.940 |
| G/T | 83 (27.7%) | 87 (29.0%) | 1.07 (0.75-1.53) | |||
| T/T | 5 (1.7%) | 5 (1.7%) | 1.02 (0.29-3.57) | |||
| Dominant | G/G | 212 (70.7%) | 208 (69.3%) | 1.00 | 0.720 | |
| G/T+T/T | 88 (29.3%) | 92 (30.7%) | 1.07 (0.75-1.51) | |||
| Recessive | G/G+G/T | 295 (98.3%) | 295 (98.3%) | 1.00 | 1.000 | |
| T/T | 5 (1.7%) | 5 (1.7%) | 1.00 (0.29-3.49) | |||
| Overdominant | G/G+T/T | 217 (72.3%) | 213 (71.0%) | 1.00 | 0.720 | |
| G/T | 83 (27.7%) | 87 (29.0%) | 1.07 (0.75-1.52) | |||
| Log-additive | --- | --- | --- | 1.05 (0.77-1.45) | 0.740 | |
| rs1143627 | Codominant | G/G | 55 (18.5%) | 96 (32.3%) | 1.00 | 5E-04* |
| A/G | 166 (55.9%) | 141 (47.5%) | 0.49 (0.33-0.73) | |||
| A/A | 76 (25.6%) | 60 (20.2%) | 0.45 (0.28-0.73) | |||
| Dominant | G/G | 55 (18.5%) | 96 (32.3%) | 1.00 | 1E-04* | |
| A/G+A/A | 242 (81.5%) | 201 (67.7%) | 0.48 (0.33-0.7) | |||
| Recessive | G/G+A/G | 221 (74.4%) | 237 (79.8%) | 1.00 | 0.120 | |
| A/A | 76 (25.6%) | 60 (20.2%) | 0.74 (0.50-1.08) | |||
| Overdominant | G/G+A/A | 131 (44.1%) | 156 (52.5%) | 1.00 | 0.040* | |
| A/G | 166 (55.9%) | 141 (47.5%) | 0.71 (0.52-0.99) | |||
| Log-additive | --- | --- | --- | 0.67 (0.53-0.85) | 7E-04* | |
| rs16944 | Codominant | A/A | 54 (18.0%) | 95 (32.1%) | 1.00 | 2E-04* |
| A/G | 165 (55.0%) | 142 (48.0%) | 0.49 (0.33-0.73) | |||
| G/G | 81 (27.0%) | 59 (19.9%) | 0.41 (0.26-0.66) | |||
| Dominant | A/A | 54 (18.0%) | 95 (32.1%) | 1.00 | 1E-04* | |
| A/G+G/G | 246 (82.0%) | 201 (67.9%) | 0.46 (0.32-0.68) | |||
| Recessive | A/A+A/G | 219 (73.0%) | 237 (80.1%) | 1.00 | 0.042* | |
| G/G | 81 (27.0%) | 59 (19.9%) | 0.67 (0.46-0.99) | |||
| Overdominant | A/A+G/G | 135 (45.0%) | 154 (52.0%) | 1.00 | 0.086 | |
| A/G | 165 (55.0%) | 142 (48.0%) | 0.75 (0.55-1.04) | |||
| Log-additive | --- | --- | --- | 0.64 (0.51-0.81) | 2E-04* | |
| rs1143623 | Codominant | C/C | 109 (36.3%) | 80 (26.7%) | 1.00 | 1E-04* |
| G/C | 158 (52.7%) | 148 (49.3%) | 1.28 (0.89-1.84) | |||
| G/G | 33 (11.0%) | 72 (24.0%) | 2.97 (1.80-4.92) | |||
| Dominant | C/C | 109 (36.3%) | 80 (26.7%) | 1.00 | 0.011* | |
| G/C+G/G | 191 (63.7%) | 220 (73.3%) | 1.57 (1.11-2.22) | |||
| Recessive | C/C+G/C | 267 (89.0%) | 228 (76.0%) | 1.00 | < 1E-04* | |
| G/G | 33 (11.0%) | 72 (24.0%) | 2.56 (1.63-4.00) | |||
| Overdominant | C/C+G/G | 142 (47.3%) | 152 (50.7%) | 1.00 | 0.410 | |
| G/C | 158 (52.7%) | 148 (49.3%) | 0.88 (0.64-1.21) | |||
| Log-additive | --- | --- | --- | 1.63 (1.29-2.08) | < 1E-04* |
SNPs: Single nucleotide polymorphisms; OR = odds ratio; 95% CI = 95% confidence interval.
P < 0.05 indicates statistical significance;
p values were calculated by unconditional logistic regression.
The results for the association between the IL-1 haplotype and the risk of TB were listed in Table 3. Results revealed that five IL-1α SNPs (rs3783550, rs3783546, rs2856838, rs1609682, rs3783521) mapped in a 10 kb LD block with the D’>0.98 (Figure 1A), suggesting that a significant linkage disequilibrium presence among these SNPs. Furthermore, “TCATG” haplotype of IL-1α SNPs with a 1.47-fold risk for TB was observed (P = 0.007).
Table 3.
Haplotype frequencies and their association with tuberculosis risk
| Gene | SNPs | Haplotype | Frequency | OR (95% CI) | P a | |
|---|---|---|---|---|---|---|
|
| ||||||
| Case | Control | |||||
| IL-1α | rs3783550 | GGGGA | 0.575 | 0.667 | 1.00 | --- |
| rs3783546 | TCATG | 0.293 | 0.242 | 1.47 (1.11-1.94) | 0.007* | |
| rs2856838 | TCGTG | 0.113 | 0.091 | 1.42 (0.97-2.08) | 0.070 | |
| rs1609682 | ||||||
| rs3783521 | ||||||
| IL-1β | rs2853550 | GT | 0.474 | 0.530 | 1.00 | --- |
| rs1143643 | GC | 0.436 | 0.378 | 1.30 (1.02-1.67) | 0.034* | |
| AC | 0.090 | 0.092 | 1.10 (0.75-1.62) | 0.630 | ||
| IL-1β | rs1143630 | GAGG | 0.432 | 0.537 | 1.00 | --- |
| rs1143627 | GGAG | 0.324 | 0.220 | 1.86 (1.40-2.46) | < 0.0001* | |
| rs16944 | TGAG | 0.159 | 0.149 | 1.37 (0.96-1.95) | 0.081 | |
| rs1143623 | GGAC | 0.077 | 0.081 | 1.17 (0.76-1.80) | 0.480 | |
OR = odds ratio; 95% CI = 95% confidence interval.
P < 0.05 indicates statistical significance;
p vsalue were calculated by unconditional logistic regression.
Figure 1.
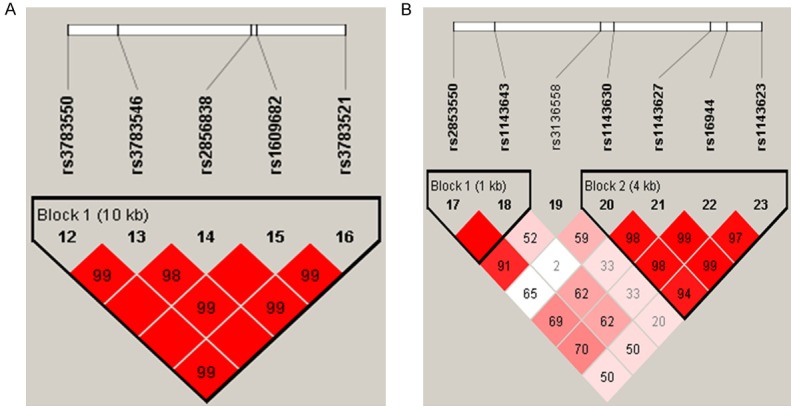
Haplotype block map for the SNPs in the IL-1 gene in our experiment. A. Hapotype block map for SNPs in the IL-1α gene. B. Hapotype block map for SNPs in the IL-1β gene.
Discussion
Current evidence suggests that the IL-1 cytokine family is a complex self-regulating system, and its members have been proposed as candidate genes in inflammatory disorders. In the present study, we aimed to investigate whether the IL-1 polymorphism is related to susceptibility to TB in Chinese Tibetan population. Among analyzed 12 SNPs in our study, we found a positive association between higher TB risk and IL-1α polymorphism (rs3783550, rs3783546, rs2856838, rs1609682, rs3783521) as well as rs1143623 within IL-1β. Furthermore, “TCATG” haplotype of IL-1α SNPs with a 1.47-fold risk for TB was observed (P = 0.007), suggesting that these SNPs may contribute to tuberculosis susceptibility in Chinese Tibetan population.
IL-1α is a well-characterized gene that encodes the IL-1α protein of the interleukin 1 cytokine family, and is involved in a wide range of inflammatory activities and immune responses [19]. Encouraged by several lines of evidence supporting a potential link between IL-1α polymorphism and various diseases risk. SNP rs3783550 located within or near the interleukin IL-1α is suggested to be related to endometriosis in Japanese samples [19], higher probability of preeclampsia developing in a Brazilians [20], and preterm prelabor rupture of membranes [21]. Two other SNPs (rs3783546, rs2856838) were associated with a measure of processing speed for cognitive ability [22], chronic rhinosinusitis with and without nasal polyposis, and decreased risk of malaria infection in northern Uganda [23], respectively. IL-1β exerts its primary proinflammatory effects by stimulating the formation of its main effector, IL-6, which drives the inflammation cascade [24]. IL-1β SNP rs1143623 is within a promoter GATA transcription factor family binding site. A correlation between rs1143623 and triglyceride and interleukin 6 metabolism [25], pathogenesis of schizophrenia [26], and higher IL-13 plasma levels in adults living with HIV/AIDS [27] were found in several novel studies.
In this study, a significantly high risk of TB in the presence of SNP rs3783550, rs3783546, rs2856838, rs1609682, rs3783521 within IL-1α as well as rs1143623 within IL-1β was found. To the best of our knowledge, IL-1α rs1609682 and rs3783521 have not previously been associated with TB or other diseases. In this study, rs1609682 and rs3783521 are in tight linkage disequilibrium with rs3783550, rs3783546 and rs2856838 mapped in a 10 kb LD block with the D’>0.98, suggesting that they may have similar functionality involved in TB risk. Importantly, with regard to the mechanism how these SNPs effect on TB susceptibility, one possibility is that they are likely to represent a genuine disease susceptibility locus involved in regulating NLRP3 or NF-κB signaling pathway, which enhanced the LPS-induced expression of inflammatory [28,29]. Alternatively, since IL-1α/β share common biological activities with the Toll/interleukin-1 receptor homology (TIR) domain, given their roles in inflammation response, it is not surprising to see specific polymorphisms of those loci contribute to the individual variability for TB susceptibility. However, these speculations need further investigation.
The results of this study also need to be considered in light of its limitations. Better support of our findings, requires replication in other samples. In addition, multiple testings are also considered. However, although we made many efforts to collect enough demographic and clinical information, there are still some unsatisfactory points.
In conclusion, IL-1 polymorphism, especially IL-1α variants, in our study were significantly associated with a higher risk of TB in the Chinese Tibetan population. Further investigation will be performed to determine how these relevant variants within IL-1 gene affect TB and determine the underlying mechanism via cell culture or knock-out mice models to provide a reliable theoretical basis for TB screening, early diagnosis and prognosis.
Acknowledgements
We would like to thank all the patients and individuals in this study for their participation. We are also very grateful for the assistance of the Clinical Research Ethics of Xizang Minzu University and the third Hospital of Tibet Autonomous Region who contributed blood samples and data for this study.
Disclosure of conflict of interest
None.
References
- 1.Orcau À, Caylà JA, Martínez JA. Present epidemiology of tuberculosis. Prevention and control programs. Enferm Infecc Microbiol Clin. 2011;29:2–7. doi: 10.1016/S0213-005X(11)70011-8. [DOI] [PubMed] [Google Scholar]
- 2.Takeda K, Akira S. Toll-like receptors in innate immunity. Int Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 3.Knechel NA. Tuberculosis: pathophysiology, clinical features, and diagnosis. Crit Care Nurse. 2009;29:34–43. doi: 10.4037/ccn2009968. [DOI] [PubMed] [Google Scholar]
- 4.Zumla A, George A, Sharma V, Herbert N Baroness Masham of Ilton. WHO’s 2013 global report on tuberculosis: successes, threats, and opportunities. Lancet. 2013;382:1765–7. doi: 10.1016/S0140-6736(13)62078-4. [DOI] [PubMed] [Google Scholar]
- 5.Wang LX, Cheng SM, Chen MT. The fifth sampling survey on tuberculosis epidemiology of China in 2010. Chinese Journal of Tuberculosi. 2012;8:485–508. [Google Scholar]
- 6.Mayer-Barber KD, Andrade BB, Oland SD, Amaral EP, Barber DL, Gonzales J, Derrick SC, Shi R, Kumar NP, Wei W, Yuan X, Zhang G, Cai Y, Babu S, Catalfamo M, Salazar AM, Via LE, Barry CE 3rd, Sher A. Host-directed therapy of tuberculosis based on interleukin-1 and type I interferon crosstalk. Nature. 2014;511:99–103. doi: 10.1038/nature13489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fol M, Druszczynska M, Wlodarczyk M, Ograczyk E, Rudnicka W. Immune response gene polymorphisms in tuberculosis. Acta Biochim Pol. 2015;62:633–40. doi: 10.18388/abp.2015_1130. [DOI] [PubMed] [Google Scholar]
- 8.Cliff JM, Kaufmann SH, McShane H, van Helden P, O’Garra A. The human immune response to tuberculosis and its treatment: a view from the blood. Immunol Rev. 2015;264:88–102. doi: 10.1111/imr.12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ainscough JS, Frank Gerberick G, Zahedi-Nejad M, Lopez-Castejon G, Brough D, Kimber I, Dearman RJ. Dendritic cell IL-1α and IL-1β are polyubiquitinated and degraded by the proteasome. J Biol Chem. 2014;289:35582. doi: 10.1074/jbc.M114.595686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kwak A, Lee Y, Kim H, Kim S. Intracellular interleukin (IL)-1 family cytokine processing enzyme. Arch Pharm Res. 2016;39:1556–64. doi: 10.1007/s12272-016-0855-0. [DOI] [PubMed] [Google Scholar]
- 11.Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun. 2012;80:3343–59. doi: 10.1128/IAI.00443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi X, Liang Y, Huerta-Sanchez E, Jin X, Cuo ZX, Pool JE, Xu X, Jiang H, Vinckenbosch N, Korneliussen TS, Zheng H, Liu T, He W, Li K, Luo R, Nie X, Wu H, Zhao M, Cao H, Zou J, Shan Y, Li S, Yang Q, Asan , Ni P, Tian G, Xu J, Liu X, Jiang T, Wu R, Zhou G, Tang M, Qin J, Wang T, Feng S, Li G, Huasang , Luosang J, Wang W, Chen F, Wang Y, Zheng X, Li Z, Bianba Z, Yang G, Wang X, Tang S, Gao G, Chen Y, Luo Z, Gusang L, Cao Z, Zhang Q, Ouyang W, Ren X, Liang H, Zheng H, Huang Y, Li J, Bolund L, Kristiansen K, Li Y, Zhang Y, Zhang X, Li R, Li S, Yang H, Nielsen R, Wang J, Wang J. Sequencing of 50 human exomes reveals adaptation to high altitude. Science. 2010;329:75–78. doi: 10.1126/science.1190371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geng TT, Xun XJ, Li S, Feng T, Wang LP, Jin TB, Hou P. Association of colorectal cancer susceptibility variants with esophageal cancer in a Chinese population. World J Gastroenterol. 2015;21:6898–6904. doi: 10.3748/wjg.v21.i22.6898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jin TB, Ren Y, Shi X, Jiri M, He N, Feng T, Yuan D, Kang L. Genetic variations in the CLNK gene and ZNF518B gene are associated with gout in case-control sample sets. Rheumatol Int. 2015;35:1141–1147. doi: 10.1007/s00296-015-3215-3. [DOI] [PubMed] [Google Scholar]
- 15.Gao L, Thakur A, Liang Y, Zhang S, Wang T, Chen T, Meng J, Wang L, Wu F, Jin T, Li X, Liu JJ, Chen C, Chen M. Polymorphisms in the TERT gene are associated with lung cancer risk in the Chinese Han population. Eur J Cancer Prev. 2014;23:497–501. doi: 10.1097/CEJ.0000000000000086. [DOI] [PubMed] [Google Scholar]
- 16.Su Q, Wang Y, Zhao J, Ma C, Wu T, Jin T, Xu J. Polymorphisms of PRLHR and HSPA12A and risk of gastric and colorectal cancer in the Chinese Han population. BMC Gastroenterol. 2015;15:107. doi: 10.1186/s12876-015-0336-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 18.Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci. Cell Res. 2005;15:97–98. doi: 10.1038/sj.cr.7290272. [DOI] [PubMed] [Google Scholar]
- 19.Sapkota Y, Low SK, Attia J, Gordon SD, Henders AK, Holliday EG, MacGregor S, Martin NG, McEvoy M, Morris AP, Takahashi A, Scott RJ, Kubo M, Zondervan KT, Montgomery GW, Nyholt DR. Association between endometriosis and the interleukin 1A (IL1A) locus. Hum Reprod. 2015;30:239–48. doi: 10.1093/humrep/deu267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Silva VR, Soardi FC, Tanaka SC, da Silva-Grecco RL, Paschoini MC, Balarin MA. Investigation of polymorphisms in pre-eclampsia related genes VEGF and IL1A. Arch Gynecol Obstet. 2015;291:1029–1035. doi: 10.1007/s00404-014-3503-2. [DOI] [PubMed] [Google Scholar]
- 21.Romero R, Friel LA, Velez Edwards DR, Kusanovic JP, Hassan SS, Mazaki-Tovi S, Vaisbuch E, Kim CJ, Erez O, Chaiworapongsa T, Pearce BD, Bartlett J, Salisbury BA, Anant MK, Vovis GF, Lee MS, Gomez R, Behnke E, Oyarzun E, Tromp G, Williams SM, Menon R. A genetic association study of maternal and fetal candidate genes that predispose to preterm prelabor rupture of membranes (PROM) Am J Obstet Gynecol. 2010;203:361, e1–361, e30. doi: 10.1016/j.ajog.2010.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marioni RE, Deary IJ, Murray GD, Fowkes FG, Price JF. Associations between polymorphisms in five inflammation-related genes and cognitive ability in older persons. Genes Brain Behav. 2010;9:348–352. doi: 10.1111/j.1601-183X.2010.00565.x. [DOI] [PubMed] [Google Scholar]
- 23.Legason ID, Pfeiffer RM, Udquim KI, Bergen AW, Gouveia MH, Kirimunda S, Otim I, Karlins E, Kerchan P, Nabalende H, Bayanjargal A, Emmanuel B, Kagwa P, Talisuna AO, Bhatia K, Yeager M, Biggar RJ, Ayers LW, Reynolds SJ, Goedert JJ, Ogwang MD, Fraumeni JF Jr, Prokunina-Olsson L, Mbulaiteye SM. Evaluating the causal link between malaria infection and endemic burkitt lymphoma in Northern Uganda: A Mendelian Randomization Study. Ebiomedicine. 2017;25:58–65. doi: 10.1016/j.ebiom.2017.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Woods A, Brull DJ, Humphries SE, Montgomery HE. Genetics of inflammation and risk of coronary artery disease: the central role of interleukin-6. Eur Heart J. 2000;21:1574–1583. doi: 10.1053/euhj.1999.2207. [DOI] [PubMed] [Google Scholar]
- 25.Delgado-Lista J, Garcia-Rios A, Perez-Martinez P, Solivera J, Yubero-Serrano EM, Fuentes F, Parnell LD, Shen J, Gomez P, Jimenez-Gomez Y, Gomez-Luna MJ, Marin C, Belisle SE, Rodriguez-Cantalejo F, Meydani SN, Ordovas JM, Perez-Jimenez F, Lopez-Miranda J. Interleukin 1B variant -1473G/C (rs1143623) influences triglyceride and interleukin 6 metabolism. J Clin Endocrinol Metab. 2011;96:E816–20. doi: 10.1210/jc.2010-2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kapelski P, Skibinska M, Maciukiewicz M, Pawlak J, Dmitrzak-Weglarz M, Szczepankiewicz A, Zaremba D, Twarowska-Hauser J. An association between functional polymorphisms of the interleukin 1 gene complex and schizophrenia using transmission disequilibrium test. Arch Immunol Ther Exp (Warsz) 2017;64:1–8. doi: 10.1007/s00005-016-0434-6. [DOI] [PubMed] [Google Scholar]
- 27.Gay CL, Zak RS, Lerdal A, Pullinger CR, Aouizerat BE, Lee KA. Cytokine polymorphisms and plasma levels are associated with sleep onset insomnia in adults living with HIV/AIDS. Brain Behav Immun. 2015;47:58–65. doi: 10.1016/j.bbi.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1β-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–25. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 29.Ito S, Hara Y, Kubota T. CARD8 is a negative regulator for NLRP3 inflammasome, but mutant NLRP3 in cryopyrin-associated periodic syndromes escapes the restriction. Arthritis Res Ther. 2014;16:R52. doi: 10.1186/ar4483. [DOI] [PMC free article] [PubMed] [Google Scholar]


