Abstract
The aberrant expression of microRNAs (miRNAs) underlies a series of human diseases, including ovarian cancers. In our previous study, we found that miR-129-1-3p and miR-129-2-3p levels were significantly decreased in serous ovarian cancer via a microarray and quantitative PCR. In this study, we investigated the pathological role of miR-129-3p in an ovarian cancer cell line, SKOV3 cells. The results demonstrated that miR-129-3p overexpression distinctively inhibited the proliferation, migration, and invasion of ovarian cancer cells. The regulator of cell cycling, BZW1 (basic leucine zipper and W2 domains 1), was validated as a novel direct target of miR-129-3p. Specifically, miR-129-3p bound directly to the 3’ untranslated region of BZW1 and suppressed its expression. Our results indicate that miR-129-3p serves as a tumor suppressor by targeting BZW1 in ovarian cancer cells and highlight that the restoration of miR-129-3p might be a novel therapeutic strategy for ovarian cancer.
Keywords: miR-129-3p, serous ovarian cancer, BZW1
Introduction
Ovarian cancer is one of the leading causes of cancer-associated deaths among women worldwide and is the leading cause of death among gynecologic malignancies [1,2]. Ovarian cancer mainly includes four major histologic subtypes, namely, serous, mucinous, endometrioid, and clear cell, with serous being the most common [3,4]. Over the last few decades, the 5-year survival rate for ovarian cancer patients has substantially improved due to the progression in combined chemotherapy and biotherapy strategies, but the overall cure rate remains extremely poor relative to other female reproductive tract malignancies [5-7]. Therefore, elucidation of the mechanisms underlying the occurrence and development of ovarian cancer is crucial to uncovering new potential targets to treat this highly lethal disease.
MicroRNA (miRNA) is a class of endogenous small non-coding RNAs that post-transcriptionally regulates gene expression through partly binding to complementary sequences in the 3’ untranslated regions (3’UTR) of target mRNAs [8-10]. The aberrant expression of miRNAs underlies a spectrum of human diseases, including ovarian cancers [11-15]. Importantly, it is well documented that normalizing or correcting the aberrant miRNAs could return a cell or organ from a pathological state to its normal phenotype [16,17]. In our previous study, we identified the deregulated miRNAs in serous ovarian cancer via a microarray and quantitative PCR (qPCR), and found that miR-129-1-3p and miR-129-2-3p levels were significantly decreased in serous ovarian cancer compared with the normal fimbriated distal end of the fallopian tube [18]. In this study, we investigated the pathological role of miR-129-3p in an ovarian cancer cell line, SKOV3 cells. The results demonstrated that miR-129-3p overexpression distinctively inhibited the proliferation, migration, and invasion of ovarian cancer cells. Moreover, we found that basic leucine zipper and W2 domains 1 (BZW1) was a direct target of miR-129-3p.
Materials and methods
Cell culture and transfection
The human serous ovarian cancer cell line, SKOV3, was purchased from the Shanghai Institute of Biochemistry and Cell Biology at the Chinese Academy of Sciences. The cells were cultured with McCoy’s 5A Medium (Gibco) containing 15% fetal bovine serum (FBS) (Gibco), 50 IU/mL penicillin, and 50 IU/mL streptomycin at 37°C in a humidified atmosphere containing 5% CO2. The cells were transfected with miR-129-1-3p or miR-129-2-3p mimics or a negative control (NC) miRNA (Applied Biosystems) using the HiPerFect Reagent (Qiagen) according to the manufacturer’s protocols.
Total RNA extraction and qPCR
Total RNA was extracted from the cells using the Trizol reagent (Invitrogen) according to the manufacturer’s protocol. The concentration and purity of the total RNA samples were measured using the NanoDROP 2000 (Gene Company Limited, CA, USA). Next, cDNA was generated using the miScript II Reverse Transcription Kit (Qiagen) according to the manufacturer’s protocol. qPCR was performed using the miScript SYBR Green PCR Kit (Qiagen) on the ABI7500 Fast Real-time qPCR system (Applied Biosystems, USA). U6 snRNA or GAPDH was used as an internal control, and the fold change was calculated by the 2-ΔΔCt method [19]. The sequences of primers used in this study were as follows: BZW1 forward primer 5’-CCCTCAGTAAAGCAAACAGGA-3’ and reverse primer 5’-AACCCAACATTTCCAGCAAC-3’; GAPDH forward primer 5’-GCACCGTCAAGGCTGAGAAC-3’ and reverse primer 5’-TGGTGAAGACGCCAGTGGA-3’.
Scratch assay
SKOV3 cells were seeded at a concentration of 5 × 104 cells per well in a 96-well plate and transfected with miRNA mimics or NC for 24 h. The cells were cultured until they formed a monolayer that occupied 100% of the surface area. Next, a linear wound was made by scratching the monolayer with a toothpick. Then, the medium was changed to a medium containing 1% FBS (Gibco). Images were captured and the gap size was measured. Then the cells were cultured for another 24 h, and images were captured and the gap size was measured. The experiment was repeated three times.
MTT assay and cell cycle analysis
Transfected SKOV3 cells were seeded into 96-well plates at a density of 5000 cells per well in a final volume of 200 μL, and cultured for 24, 48, 76 h. MTT (20 μL of 5 mg/mL) was added to each well for 4 h at 37°C, followed by the removal of the culture medium and the addition of 150 μL DMSO (Sigma). The optical density was measured at a wavelength of 490 nm using a MRX II absorbance reader (DYNEX Technologies, Chantilly, Virginia, USA). For cell cycle analysis, transfected SKOV3 cells were harvested and fixed in 70% ice-cold ethanol at 4°C for 24 h. The cells were subsequently incubated with 20 μg/mL propidium iodide (Sigma) for 30 min at 4°C, and the cell cycle distribution was analyzed using flow cytometry (FACScan, BD Biosciences; San Diego, CA, USA). The experiment was repeated three times.
Cell migration assay
The assay was performed using a Transwell insert chamber. Transfected SKOV3 cells (5 × 104 per mL) were seeded on the top chamber with a serum-free medium, while McCoy’s 5A complete medium containing 20% FBS was used as the chemoattractant in the lower chamber. Following the incubation of the cells for 24 h, the cells located on the top surface of the insert were removed, while those on the bottom surface were fixed with methanol for 30 min and stained with methyl violet for 15 min. Cell counting was performed under an inverted microscope (Olympus, Tokyo, Japan) to determine their relative numbers. Ten random visual fields were observed. The experiment was repeated three times.
Cell invasion assay
The assay was performed using a transwell insert chamber coated with Matrigel (BD Biosciences). The concentration of cells seeded was 10 × 104/mL. The remaining procedures were the same as those described for the cell migration assay. The experiment was repeated three times.
Western blotting
Transfected SKOV3 cells were harvested, and total cell protein was extracted on ice using a RIPA lysis buffer in the presence of freshly added protease inhibitors (Thermo), then quantified by the BCA method (Pierce). A total of 50 μg protein extract per lane was loaded onto a 10% SDS-polyacrylamide gel and transferred to a PVDF membrane (Pierce). Nonspecific binding was blocked with 5% skim milk in PBS. The membrane was incubated with mouse anti-BZW1 antibody (Abcam, ab85090) overnight at 4°C. HRP Conjugated Rabbit Anti-Goat IgG (Beijing Zhong Shan, Pv6000) was used as secondary antibody, and a mouse anti-GAPDH antibody (BBI, D19539-0100) was used as an internal standard. The immuno-reactive bands were visualized with an infrared image processing system to analyze the molecular weight and optical density.
3’UTR luciferase reporter assay
The human wild-type (WT) or mutant (MUT) 3’UTRs of BZW1 containing the predicted miR-129-3p binding sites were synthesized (South Gene Technology, China) and cloned into the pGL3.0-control vectors (Promega). SKOV3 cells were seeded in 24-well plates, then transfected with 50 nM miRNA or a NC (Applied Biosystems) and co-transfected with 100 ng of pGL3 containing the WT or MUT 3’UTR of BZW1, using HiPerFect reagent (Invitrogen) according to the manufacturer’s instructions. pRL-TK vectors (5 μg per well) were co-transfected as endogenous controls for luciferase activity. After 48 h, cells were lysed, and luciferase activities were measured using a dual-luciferase assay kit (Promega).
Statistical analysis
The statistical analysis was performed using SPSS software 13.0 (SPSS, Chicago, USA). All of the data are presented as the mean ± SD. The significance of difference between two groups was identified using a Student’s t-test. Multiple comparisons were performed by one-way ANOVA and followed by a Bonferroni post-test for comparison between two groups. P values less than 0.05 were considered significant.
Results
MiR-129-3p inhibited the proliferation of ovarian cancer cells
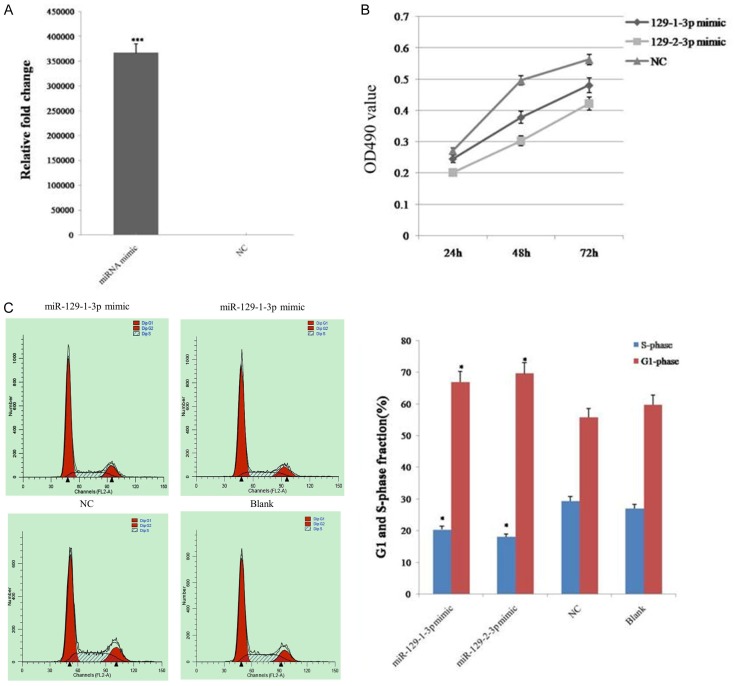
To investigate whether miR-129-1-3p and miR-129-2-3p play a role in tumorigenesis, an ovarian cancer cell line, SKOV3, was transfected with miRNA mimics, and the miRNA expression level was verified by qPCR (Figure 1A). The MTT assay revealed that miR-129-1-3p and miR-129-2-3p overexpression induced a marked inhibition of the growth rate of SKOV3 cells, compared to the negative control (NC) (Figure 1B). In addition, flow cytometry analysis indicated that miR-129-1-3p and miR-129-2-3p overexpression resulted in more cells being arrested at the G0-G1 phase and fewer cells accumulating in the S phase, compared to the corresponding controls (Figure 1C). These results suggested that miR-129-1-3p and miR-129-2-3p inhibited the proliferation of ovarian cancer cells.
Figure 1.
The proliferation of ovarian cancer cells was inhibited after the over-expression of miR-129-3p. A. Expression level of miR-129-1-3p in Skov3 ovarian cancer cells after transfection of miRNA mimics detected by qPCR. B. Determination of cell viability with the MTT assay. C. Determination of the cell cycle using flow cytometry. Data are expressed as the mean ± sd. from three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; compared with NC group.
MiR-129-3p inhibited the migration of ovarian cancer cell
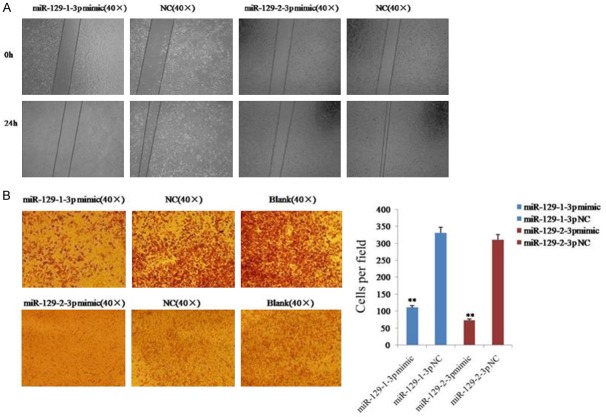
The effect of miR-129-1-3p and miR-129-2-3p on the migratory ability of SKOV3 cells was investigated by a scratch assay and a transwell migration assay. The result of the scratch assay showed that the cells from the miR-129-1-3p or miR-129-2-3p mimics-transfected group had an obviously weaker migratory or wound healing ability than the cells in the NC group (Figure 2A). Additionally, the result of the transwell migration assay revealed that 110 ± 9 cells passed through the membrane in the miR-129-1-3p mimic group, while 330 ± 10 cells passed through the membrane in the NC group (Figure 2B). Similarly, 73 ± 17 cells from the miR-129-2-3p mimic group migrated through the membrane, while 310 ± 10 cells passed through the membrane in the NC group (Figure 2B). These results indicated that miR-129-1-3p and miR-129-2-3p inhibited the migration of ovarian cancer cells.
Figure 2.
Migration of ovarian cancer cells was inhibited after the over-expression of miR-129-3p. A. Determination of cell migration with the scratch assay. B. Analysis of cell migration using a transwell migration assay. Data are expressed as the mean ± sd. from three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; compared with NC group.
MiR-129-3p inhibited the invasion of ovarian cancer cells
The effect of miR-129-1-3p and miR-129-2-3p on cell invasion was determined using the transwell assay with Matrigel. The result revealed that 71 ± 10 cells invaded the membrane in the miR-129-1-3p mimic group, while 174 ± 16 cells invaded the membrane in the NC group (Figure 3). Similarly, 102 ± 15 cells from the miR-129-2-3p mimic group invaded the membrane, while 332 ± 53 cells invaded the membrane in the NC group (Figure 2B). These results indicated that miR-129-1-3p and miR-129-2-3p inhibited the invasion of ovarian cancer cells.
Figure 3.
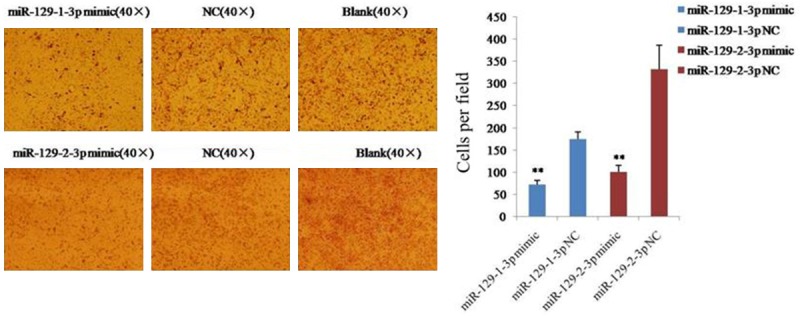
Determination of cell invasive ability using a transwell insert chamber coated with Matrigel. Data are expressed as the mean ± sd. from three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; compared with NC group.
BZW1 was a direct target of miR-129-3p
The TargetScan database shows that BZW1 is a potential target of miR-129-3p, and the sequence of the putative target site of miR-129-3p in the 3’UTR of BZW1 mRNA is highly conservative among species (Figure 4A). To confirm the prediction, we first detected the mRNA levels of this gene in SKOV3 cells after transfection with miR-129-1-3p or miR-129-2-3p mimics by qPCR. Our data showed that BZW1 mRNA levels in cancer cells were distinctly decreased after the elevation of miR-129-2-3p instead of miR-129-1-3p (Figure 4B). In addition, we examined the protein level of BZW1 in SKOV3 cells after the overexpression of miR-129-1-3p or miR-129-2-3p by western blotting. The result revealed that the protein level of BZW1 was obviously lower in both miR-129-1-3p and miR-129-2-3p transfected cells (Figure 4C). Finally, we analyzed the activity of the putative target site of miR-129-3p in the 3’UTR of BZW1 mRNA. To this end, we generated a reporter construct that contains the firefly luciferase gene fused to the putative target site. This construct was transiently transfected into SKOV3 cells together with miR-129-1-3p or miR-129-2-3p or a NC miRNA. We observed a marked reduction in luciferase activity in cells transfected with miR-129-1-3p but not miR-129-2-3p (Figure 4D). In contrast, mutation of 5 nt in the miR-129-3p seed sequence led to a complete abrogation of reporter inhibition (Figure 4D). Collectively, these data indicated that BZW1 was a direct target of miR-129-1-3p and miR-129-2-3p in SKOV3 cells.
Figure 4.
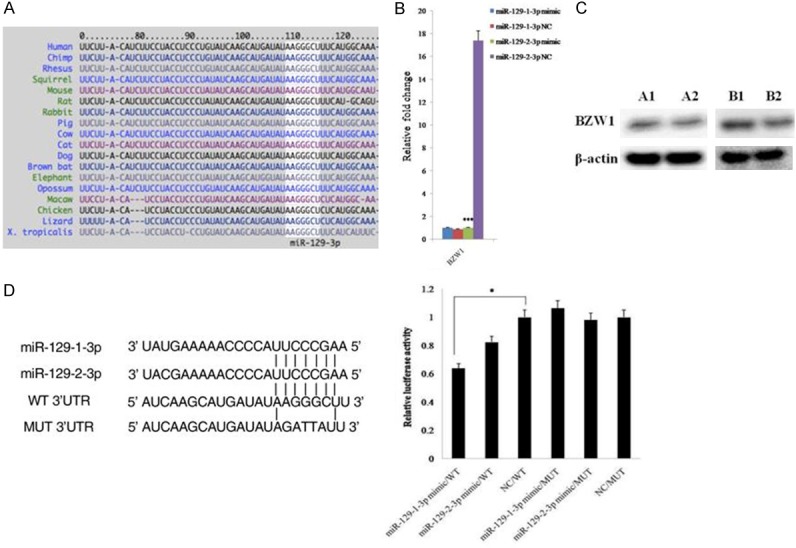
Validation of the relationship between miR-129-3p and BZW1. (A) The position of the miR-129-3p target site in BZW1 3’UTR and its conservation among species. The figure is from the TargetScan website (www.targetscan.org). (B, C) Expression of BZW1 in cancer cells after transfection with miR-129-1-3p or miR-129-2-3p detected by qPCR (B) and western blotting (C). A1 is the NC group, and A2 is the miR-129-1-3p mimic group; B1 is the NC group, and B2 is the miR-129-2-3p mimic group. (D) Luciferase reporter assays for SKOV3 cells transfected with pRL-TK vectors carrying BZW1 wild type (WT) or mutant (MUT) 3’UTR in the absence or presence of miR-129-3p mimics. Data are expressed as the mean ± sd. from three independent experiments. *, P < 0.05; **, P < 0.01; and ***, P < 0.001; compared with NC group.
Discussion
MiR-129-1-3p and miR-129-2-3p are a miRNA family, which derive from different miRNA precursors but share the same seed sequence. The mir-129-1 gene is located in a fragile site region of the human genome near FRA7H on chromosome 7q32, where it is frequently deleted in many cancers. The mir-129-2 gene is located in a CpG island on 11p11.2. Therefore, miR-129 is believed to be involved in the regulation of tumorigenesis [20]. Actually, accumulating evidence has shown that miR-129-3p is a tumor suppressor. Reduced expression of miR-129 has been reported in several types of tumor cell lines, as well as primary tumor tissues. Kang et al. reported that miR-129-2 suppresses the proliferation and migration of esophageal carcinoma cells [21]; Yu et al. showed that miR-129 family members play an important role in regulating cell proliferation in gastric cancer cells [22], and gastric juice miR-129-1-3p and miR-129-2-3p are potential biomarkers for gastric cancer screening [23]. In addition, reduced miR-129 expression levels have been found to be involved in numerous types of human cancer, such as hepatocellular carcinoma, bladder cancer, endometrial cancer and colorectal carcinoma [24-27]. In this study, we further proved that miR-129-3p functions as a tumor suppressor in serous ovarian cancer cells.
BZW1, also known as KIAA0005 or BZAP45, is a member of the bZIP super-family of transcription factors, which can activate histone H4 gene transcription and serve as a co-regulator of other transcription factors for the control of cell cycling. BZW1 has been reported to promote the growth of salivary mucoepidermoid carcinoma [28]. A bioinformatics analysis shows that BZW1 is a potential target of miR-129-3p, and the target site in BZW1 mRNA is highly conservative among species. Importantly, our data demonstrated that BZW1 is a new target of miR-129-3p. Thus, we speculate that miR-129-3p regulates the growth of ovarian cancer cells by directly targeting BZW1. However, it is worth pointing out that other target genes, such as CDK6 and SOX4 [25,29], which were validated in previous studies, might also be involved in regulating this process.
Ovarian cancer is a “silent killer” for women worldwide. Despite ongoing efforts to develop an effective treatment strategy, the overall cure rate remains approximately 30% [2]. Therefore, it is crucial to uncover new potential targets to treat this highly lethal disease. MiRNAs have been identified as crucial regulators of gene expression, and their deregulation is a hallmark of human diseases. Therefore, targeting these regulators can offer a novel miRNA-directed therapeutic approach. Accumulating evidence shows that miRNAs are involved in the occurrence and development of ovarian cancer, and targeting aberrant miRNAs might be a novel therapeutic approach for ovarian cancer [11-15]. In this study, we found that miR-129-3p was down-regulated in ovarian cancer, and the over-expression of miR-129-3p inhibited the proliferation, migration, and invasion of ovarian cancer cells, which indicates that miR-129-3p is a potential target to treat ovarian cancer. Further study using in vivo approaches is needed to validate these phenomena.
In summary, our study confirmed the reduced expression of miR-129-1-3p and miR-129-2-3p in ovarian cancer, and for the first time reported them as potential tumor suppressors in ovarian cancer, which inhibit the proliferation, migration, and invasion of ovarian cancer cells by targeting BZW1. The restoration of miR-129-3p might be a novel therapeutic strategy for preventing the occurrence and progression of ovarian cancer.
Acknowledgements
This work was supported by the Shaanxi Province Natural Science Basic Research Project (no. 2014K11-01-01-25) and the National Basic Research Program (973 program) of China (no. 2015CB553703).
Disclosure of conflict of interest
None.
References
- 1.Holschneider CH, Berek JS. Ovarian cancer: epidemiology, biology, and prognostic factors. Semin Surg Oncol. 2000;19:3–10. doi: 10.1002/1098-2388(200007/08)19:1<3::aid-ssu2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 2.Bast RC Jr, Hennessy B, Mills GB. The biology of ovarian cancer: new opportunities for translation. Nat Rev Cancer. 2009;9:415–28. doi: 10.1038/nrc2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Muggia F, Kosloff R. Investigational agents for epithelial ovarian cancer. Expert Rev Anticancer Ther. 2005;5:855–868. doi: 10.1586/14737140.5.5.855. [DOI] [PubMed] [Google Scholar]
- 4.Hensley ML. Epithelial ovarian cancer. Curr Treat Options Oncol. 2002;3:131–141. doi: 10.1007/s11864-002-0059-3. [DOI] [PubMed] [Google Scholar]
- 5.Fu S, Kavanagh JJ, Hu W, Bast RC Jr. Clinical application of oxaliplatin in epithelial ovarian cancer. Int J Gynecol Cancer. 2006;16:1717–1732. doi: 10.1111/j.1525-1438.2006.00654.x. [DOI] [PubMed] [Google Scholar]
- 6.Markman M, Bookman MA. Second-line treatment of ovarian cancer. Oncologist. 2000;5:26–35. doi: 10.1634/theoncologist.5-1-26. [DOI] [PubMed] [Google Scholar]
- 7.Modesitt SC, Jazaeri AA. Recurrent epithelial ovarian cancer: pharmacotherapy and novel therapeutics. Expert Opin Pharmacother. 2007;8:2293–2305. doi: 10.1517/14656566.8.14.2293. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011;12:861–874. doi: 10.1038/nrg3074. [DOI] [PubMed] [Google Scholar]
- 11.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Ménard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 12.Taylor DD, Gercel-Taylor C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol Oncol. 2008;110:13–21. doi: 10.1016/j.ygyno.2008.04.033. [DOI] [PubMed] [Google Scholar]
- 13.Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Bützow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu X, Macdonald DM, Huettner PC, Feng Z, El Naqa IM, Schwarz JK, Mutch DG, Grigsby PW, Powell SN, Wang X. A miR-200 microRNA cluster as prognostic marker in advanced ovarian cancer. Gynecol Oncol. 2009;114:457–464. doi: 10.1016/j.ygyno.2009.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Yang N, Kaur S, Volinia S, Greshock J, Lassus H, Hasegawa K, Liang S, Leminen A, Deng S, Smith L, Johnstone CN, Chen XM, Liu CG, Huang Q, Katsaros D, Calin GA, Weber BL, Bützow R, Croce CM, Coukos G, Zhang L. MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res. 2008;68:10307–10314. doi: 10.1158/0008-5472.CAN-08-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wurdinger T, Costa FF. Molecular therapy in the microRNA era. Pharmacogenomics J. 2007;7:297–304. doi: 10.1038/sj.tpj.6500429. [DOI] [PubMed] [Google Scholar]
- 17.He X, Xie J, Zhang D, Su Q, Sai X, Bai R, Chen C, Luo X, Gao G, Pan W. Recombinant adeno-associated virus-mediated inhibition of microRNA-21 protects mice against the lethal schistosome infection by repressing both IL-13 and transforming growth factor beta 1 pathways. Hepatology. 2015;61:2008–2017. doi: 10.1002/hep.27671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y, Yao L, Liu F, Hong J, Chen L, Zhang B, Zhang W. Characterization of microRNA expression in serous ovarian carcinoma. Int J Mol Med. 2014;34:491–498. doi: 10.3892/ijmm.2014.1813. [DOI] [PubMed] [Google Scholar]
- 19.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 20.Calin GA, Sevignani C, Dumitru CD, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M, Croce CM. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kang M, Li Y, Liu W, Wang R, Tang A, Hao H, Liu Z, Ou H. miR-129-2 suppresses proliferation and migration of esophageal carcinoma cells through downregulation of SOX4 expression. Int J Mol Med. 2013;32:51–58. doi: 10.3892/ijmm.2013.1384. [DOI] [PubMed] [Google Scholar]
- 22.Yu X, Song H, Xia T, Han S, Xiao B, Luo L, Xi Y, Guo J. Growth inhibitory effects of three miR-129 family members on gastric cancer. Gene. 2013;532:87–93. doi: 10.1016/j.gene.2013.09.048. [DOI] [PubMed] [Google Scholar]
- 23.Yu X, Luo L, Wu Y, Yu X, Liu Y, Yu X, Zhao X, Zhang X, Cui L, Ye G, Le Y, Guo J. Gastric juice miR-129 as a potential biomarker for screening gastric cancer. Med Oncol. 2013;30:365. doi: 10.1007/s12032-012-0365-y. [DOI] [PubMed] [Google Scholar]
- 24.Lu CY, Lin KY, Tien MT, Wu CT, Uen YH, Tseng TL. Frequent DNA methylation of MiR-129-2 and its potential clinical implication in hepatocellular carcinoma. Genes Chromosomes Cancer. 2013;52:636–643. doi: 10.1002/gcc.22059. [DOI] [PubMed] [Google Scholar]
- 25.Huang YW, Liu JC, Deatherage DE, Luo J, Mutch DG, Goodfellow PJ, Miller DS, Huang TH. Epigenetic repression of microRNA-129-2 leads to overexpression of SOX4 oncogene in endometrial cancer. Cancer Res. 2009;69:9038–9046. doi: 10.1158/0008-5472.CAN-09-1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dyrskjøt L, Ostenfeld MS, Bramsen JB, Silahtaroglu AN, Lamy P, Ramanathan R, Fristrup N, Jensen JL, Andersen CL, Zieger K, Kauppinen S, Ulhøi BP, Kjems J, Borre M, Orntoft TF. Genomic profiling of microRNAs in bladder cancer: miR-129 is associated with poor outcome and promotes cell death in vitro. Cancer Res. 2009;69:4851–4860. doi: 10.1158/0008-5472.CAN-08-4043. [DOI] [PubMed] [Google Scholar]
- 27.Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, Prosper F, Garcia-Foncillas J. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- 28.Li S, Chai Z, Li Y, Liu D, Bai Z, Li Y, Situ Z. BZW1, a novel proliferation regulator that promotes growth of salivary muocepodermoid carcinoma. Cancer Lett. 2009;284:86–94. doi: 10.1016/j.canlet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 29.Wu J, Qian J, Li C, Kwok L, Cheng F, Liu P, Perdomo C, Kotton D, Vaziri C, Anderlind C, Spira A, Cardoso WV, Lü J. miR-129 regulates cell proliferation by downregulating Cdk6 expression. Cell Cycle. 2010;9:1809–1818. doi: 10.4161/cc.9.9.11535. [DOI] [PubMed] [Google Scholar]