Abstract
During carcinogenesis, growth, proliferation, invasion and metastasis, increasing evidence shows that autophagy and endoplasmic reticulum stress (ER stress) are regulated in nasopharyngeal carcinoma, a finding drawing more attention from physicians and scientists. As one of the carbon-based nano-materials, graphene oxide (GO) has been extensively used for its advantages, such as biocompatibility, an ultrahigh surface to volume ratio, abundant surface groups, and a special photothermal effect. The present study is designed to explore the effects of GO on autophagy and ER stress in nasopharyngeal carcinoma cells. Our findings will provide scientific bases for the clinical application of GO and the development of new analogues. GO inhibits the proliferation of HONE1 cells, promotes their apoptosis in a concentration-dependent manner and enhances the expression of the ER stress chaperone GRP78 in HONE1 cells. These results suggest that GO could affect HONE1 cells through the autophagic and ER stress pathways. Thus, GO inhibits the proliferation of nasopharyngeal carcinoma cells via the induction of cytotoxic autophagy. In addition, ER stress is also activated as an adaptive response, so blocking ER stress may enhance the sensitivity of nasopharyngeal carcinoma cells to GO.
Keywords: Graphene oxide, endoplasmic reticulum stress, autophagy, nasopharyngeal carcinoma cells, apoptosis
Introduction
With more rapid urbanization, an increasing aged population, and higher economic development, the mortality and morbidity rates of nasopharyngeal carcinoma have sharply increased globally during recent decades. More progress has been made in understanding nasopharyngeal carcinoma oncology, and the efficacy and prognosis of nasopharyngeal carcinoma have also been improved by in-depth clinical practice and study. The treatment of nasopharyngeal carcinoma with radiotherapy has been effectively applied, but its practice has been limited by chemotherapy resistance, adverse effects caused by anticancer drugs, and the impaired quality of life caused by surgical resection [1,2]. Although the mechanisms of nasopharyngeal carcinoma are not fully elucidated, studies have provided data to support the hypothesis that autophagy and ER stress are associated with the occurrence and development of nasopharyngeal carcinoma, and more attention has been paid to it by physicians and scientists. Therefore, it is urgent to find a drug with better sensitivity and specificity to resist nasopharyngeal carcinoma.
As one of the carbon-based nano-materials, graphene oxide (GO) has been extensively used for its advantages, such as biocompatibility, an ultrahigh surface to volume ratio, abundant surface groups, and a special photothermal effect [3-6]. Accumulating evidence has shown that GO plays a pivotal role in the treatment of nasopharyngeal carcinoma [7-9]. Meanwhile, several studies have found that autophagy and ER stress is critically involved in the growth of tumor cells [10-12]. Moreover, evidence is lacking which shows the relationship between GO through autophagy and endoplasmic reticulum stress and the pathogenesis of nasopharyngeal carcinoma [13,14]. Accordingly, we hypothesized that GO may have potential therapeutic effects in nasopharyngeal carcinoma.
Therefore, the aim of this study was to evaluate whether GO alleviates the growth of nasopharyngeal carcinoma by regulating autophagy and ER stress.
Materials and methods
Chemical and biological reagents
HONE1 was purchased from the Otwo Biotech (Shenzhen) Inc. (Shenzhen, China). GO was purchased from the Nanjing Xianfeng Nano Material Technology Co., Ltd. (Nanjing, China), (catalogue No: 100683). CCK8 was obtained from Japan Tongren Company. GRP78 antibodies, LC3B antibodies, β-actin antibodies, and rabbit anti-IgG antibodies were purchased from Cell Signaling Technology, Inc. (Danvers, MA, USA). 4-PBA was purchased from Sigma (St. Louis, MO, USA). The total RNA was extracted from lung samples with TRIzol (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. cDNA was prepared using oligo (dT) primers (PrimeScript™RT reagent Kit, TAKALA, Kusatsu, Japan). Quantitative RT-PCR was performed in duplicate with SYBR Green I (SYBR®Premix Ex Taq™, TAKALA) using an Applied Biosystems 7500 analyzer (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Cell culture
The HONE1 cells were cultured in 1 mL DMEM containing 10% fetal bovine serum (FBS), 100 U/ml penicillin and 100 U/ml streptomycin at 37°C in 5% CO2 incubator. The cells in mid-log phase were used in the experiments.
Cell viability assay
Cell Viability was determined using a CCK8 assay. The HONE1 cells were seeded in 96-well flat bottom microtiter microplates at a density of 6.0 × 103 cells per well, the cells were cultured for 24 h, and then they were treated with 25, 50, 75, and 100 μg/ml GO at room temperature for 6 h, respectively. The control group and zero adjustment well were also set up. Next, the cells were incubated with 10 μL of CCK8 labeling reagent for 6 h. The absorbance value per well at 450 nm was read using an automatic multiwell spectrophotometer. All the CCK8 assays were performed in triplicate. Cell viability was determined according to the following formula: (experimental absorbance value-blank absorbance value/control absorbance value-blank absorbance value) × 100%.
Morphology observation
The HONE1 cells were treated with 0, 50 and 100 μg/ml GO for 6 h. After being washed with PBS, the cell morphology was observed with an inverted microscope.
Western blotting detection for GRP78 and LC3B
As in the previously described method, the expression levels of GRP78 and LC3B in the HONE1 cells were detected with western blotting [15]. Briefly, the cell sample was centrifuged at 12000 r/min for 20 min at 4°C, the supernatant was collected, and the sample protein concentration was measured with a nanodrop ultra-micro sample spectrophotometer. About 80 μL of each protein sample was mixed with 20 uL of 5× sodiumdodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer (Baomanbi, Shanghai, China). The mixture was boiled for 5 minutes and then cooled for another 5 minutes at room temperature. For the western blot assay, 80 μg of total protein per lane was separated by 12% SDS-PAGE and then transferred to polyvinylidene difluoride (PVDF) membranes (0.22 μm; Millipore Corp., Billerica, MA, USA). The PVDF membranes were blocked with 5% skim milk diluted in Tri-buffered saline (TBS) for 1 h at room temperature and then incubated with antibodies for rabbit anti-GRP78 (1:1,000 dilution; Cell Signaling Technology, Inc. Danvers, MA, USA), or Rabbit monoclonal anti-caspase-3 and anti-light chain 3 (LC3B) (1:1,000 dilution; Cell Signaling Technology, Inc. Danvers, MA, USA), and β-actin (1:1,000 dilution; Cell Signaling Technology, Inc. Danvers, MA, USA) and diluted in primary antibody dilution buffer overnight at 4°C according to the manufacturers’ instructions. After washing with TBST (TBS containing 0.05% Tween), the membranes were incubated with rabbit anti-IgG antibodies (1:5,000; Cell Signaling Technology, Inc. Danvers, MA, USA) diluted in a secondary antibody dilution buffer. After washing as above, the signals of each sample were measured using a sweep membrane apparatus (LI-COR). β-actin was used to assess the loading and transfer quality. Quantification was performed using Odyssey software.
Quantitative real-time polymerase chain reaction (PCR) analysis
For quantifying, the total RNA was extracted from lung samples with TRIzol (Invitrogen, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. cDNA was prepared using oligo (dT) primers (PrimeScript™RT reagent Kit, TAKALA, Kusatsu, Japan). Quantitative RT-PCR was performed in duplicate with SYBR Green I (SYBR®Premix Ex Taq™, TAKALA) using an Applied Biosystems 7500 analyzer (Thermo Fisher Scientific) according to the manufacturer’s instructions. DNA was amplified under the following conditions: denaturation at 95°C for 30 s, extension at 95°C for 5 s, 60°C for 34 s, and the samples were amplified for 40 cycles. β-actin was used as an internal control, and levels of each gene were normalized to β-actin expression using the ΔΔCt-method. The identity of the amplified products was examined using agarose gel electrophoresis and melt curve analysis.
Indirect immunofluorescence assay
LC3B was detected by an indirect immunofluorescence assay (IIF). HONE1 cells were seeded to coverslips in the 6-well plates and treated with GO, 4-PBA, 4-PBA+GO or PBS (control) for 6 h. After PBS washes, the cells were fixed with 4% paraformaldehyde and permeabilized as described. Briefly, they were washed with PBS, followed by 0.1% Triton X-100 for 10 min and washed again with PBS. After being blocked with 1% BSA for 1 h, they were incubated with the primary antibody (1:100 dilution) overnight at 4°C. After being washed with PBST, the HONE1 cells were incubated with the secondary antibody (1:500 dilution) for 1 h at room temperature in the dark. After they were washed, the cells were visualized with a fluorescence inverted microscope and the images were captured. A LUX multifunction enzyme labeling instrument was used for the quantitative detection of the fluorescence intensity (RFU).
Transmission electron microscope (TEM)
The autophagosome was observed by transmission electron microscope (TEM). The HONE1 cells were collected and centrifuged, and then they were increased with 2.5% glutaraldehyde for the night at 4°C. After being washed, the cells were fixed with osmium acid for 2 h. After embedding, the sections were stained and observed by electron microscopy.
Statistical analysis
Statistical analysis of data was performed using the SPSS statistical software (version 19.0, SPSS IBM, Chicago, IL, USA) and GraphPad Prism version 5.0 (GraphPad Software, San Diego, CA, USA). The data are presented as means ± standard deviation (SD). Statistical comparisons of the data were performed using one-way ANOVA. P-values of less than 0.05 were considered to indicate statistically significant differences.
Results
Effects of GO on hematopoietic cell proliferation
Cell viability was determined using a CCK8 assay. The HONE1 cells were treated with 25, 50, 75, and 100 μg/ml of GO for 6 h. Cell viability from each experimental group is shown in Figure 1. The results show that with an increase in GO concentration, cell viability gradually decreases, in a dose-dependent manner. Moreover, compared with the GO group, the 4-PBA group showed markedly increased cell viability.
Figure 1.
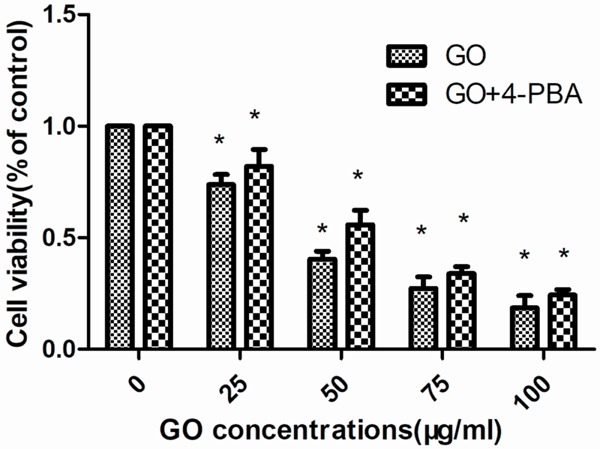
Cell viability was determined using a CCK8 assay. The HONE1 cells were treated with 25, 50, 75 and 100 μg/ml GO for 6 h. Cell viability from each experimental group is shown. Data are presented as the mean ± standard deviation (SD). The results found that with the increase of GO concentration, cell viability gradually decreased, in a dose-dependent manner. Moreover, compared with the GO group, the 4-PBA group showed markedly increased cell viability. P<0.05 indicates a statistically significant difference.
Effects of GO on cell morphology
The HONE1 cells were treated with 0, 50, and 100 μg/ml GO for 6 h, and cell morphology was shown in Figure 2. Compared with the control group, some dead cells were observed in the GO group; however, most of the HONE1 cells were necrotic, and the morphology of the cells was changed in the 100 μg/ml GO group.
Figure 2.
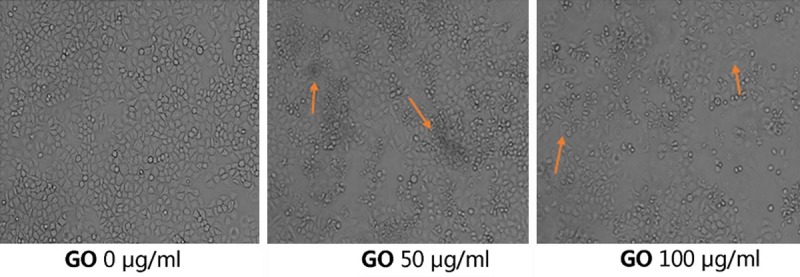
The HONE1 cells were treated with 0, 50, and 100 μg/ml GO for 6 h and the cell morphology is shown. Compared with the control group, some dead cells were observed in the GO group; however, most HONE1 cells were necrotic, and the morphology of the cells was changed in the 100 μg/ml GO group.
GO effects the protein expression of GRP78 and LC3B
The contents of the endoplasmic reticulum marker protein GRP78 and the autophagy protein LC3B were detected by western blot, as shown in Figure 3A. The results showed that with the increase of GO concentration, the content of GRP78 and LC3II increased gradually. After adding 4-PBA, the content of GRP78 and LC3II decreased relatively, suggesting that GO can induce endoplasmic reticulum stress and autophagy in both cells, and the blockade of ER stress response can reduce the conversion of LC3I to LC3II, indicating that ER stress is involved in GO-induced autophagy in HONE1 cells.
Figure 3.
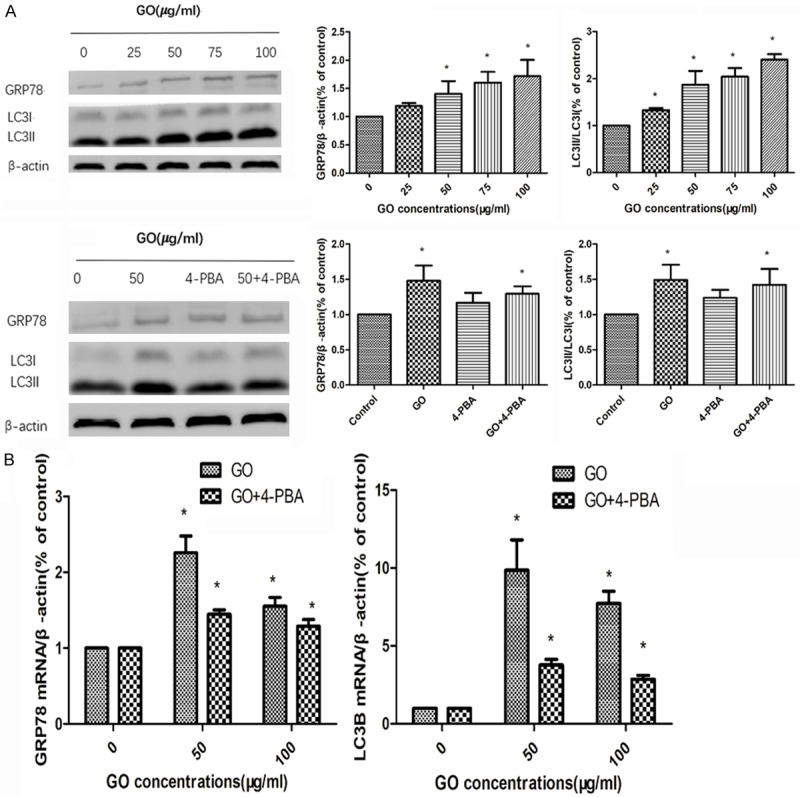
To evaluate the effects of GO on the endoplasmic reticulum and autophagy, the protein and mRNA expression of GRP78 and LC3B were analyzed by western blotting and quantitative real-time PCR, respectively (A. The protein expression of GRP78 and LC3B were analyzed by western blotting; B. The mRNA expression of GRP78 and LC3B were analyzed by RT-PCR). Data are presented as the mean ± SD, P<0.05 indicates a statistically significant difference.
GO effects the mRNA expression of GRP78 and LC3B
To evaluate the effects of GO on the endoplasmic reticulum and autophagy, the mRNA expression of GRP78 and LC3B were analyzed by quantitative real-time PCR. As shown in Figure 3B, compared with the control group, GO-administrated cells in the GO group increased the mRNA expression of GRP78 and LC3B. Conversely, 4-PBA treatment decreased the mRNA expression of GRP78 and LC3B compared to the GO group. The above results demonstrated that endoplasmic reticulum stress was involved in GO induced autophagy in the HONE1 cells.
GO treatment regulates autophagy
HONE1 cells were treated with GO (50 μg/ml) for 6 h, and the results are shown in Figure 4. Compared with the control group, the cells in the GO group showed an increased expression of LC3B, and nuclear morphologic changes were observed. These findings revealed that GO could induce autophagy.
Figure 4.
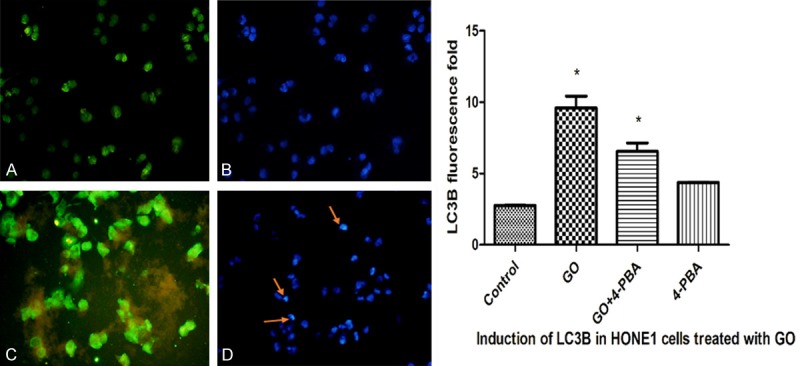
HONE1 cells were treated with GO (50 μg/ml) for 6 h, and the results are shown in the following figure (A. Cytoplasm in the control group; B. Cell nuclei in the control group; C. Cytoplasm in the GO group; D. Cell nuclei in the GO group).
GO stimulates HONE1 cells to produce autophagosomes
By means of transmission electron microscopy, the gold standard for autophagy is intracellular autophagosomes. 50 μg/ml of GO and an equal amount of FBS-free medium were co-cultured with HONE1 cells for 6h as a negative control and subjected to a series of treatments and observed by transmission electron microscopy. As shown in Figure 5, the GO-treated cells showed a large number of autophagosomes with a bilayer membrane vesicular structure.
Figure 5.
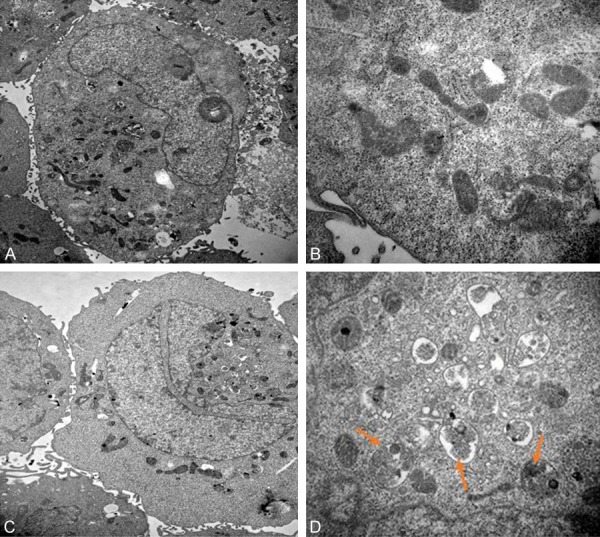
GO stimulates HONE1 cells to produce autophagosome. 50 μg/ml of GO and an equal amount of FBS-free medium were co-cultured with HONE1 cells for 6 h as a negative control and subjected to a series of treatments and observed by transmission electron microscopy. The GO-treated cells showed a large number of autophagosomes with a bilayer membrane vesicular structure. (A. TEM: 10000× in control group; B. TEM: 40000× in control group; C. TEM: 10000× in GO group; D. TEM: 40000× cell nuclei in GO group. The red arrow refers to the autophagosome).
Discussion
Owing to its large surface area and surface-active properties, previously published studies have shown that GO plays a crucial role in cancer therapy, which has the following functions and merits: its drug-loading capacity and photothermal therapy [16-18]. Although there have been many studies on GO-associated cancer, the underlying mechanism is still not conclusively known. However, studies have demonstrated that autophagy and ER stress play an important role in the development and outcome of nasopharyngeal carcinoma [19-21].
As reported, ER stress occurs when newly-synthesized, unfolded or misfolded proteins selectively accumulate in the endoplasmic reticulum when its function has been disturbed by differing pathological conditions. Furthermore, the previous studies have confirmed that ER stress can mediate the expression of PERK, IRE1/X-box binding protein-1 and then in turn, activate transcription factor-6 (ATF6) signaling pathways as protective measures, resulting in general translational attenuation. However, excessive or aberrant ER stress can result in cell injury or death by the activation of apoptotic pathways [22-24]. Meanwhile, as an inherent molecular chaperone in the endoplasmic reticulum, glucose-regulated protein 78 (GRP78) is a marker of ER stress [25]. Therefore, studies have provided data to support the hypothesis that the expression of GRP78 is associated with the occurrence and development of ER stress. In this study, our results demonstrated that GO significantly inhibited the expression of GRP78 in HONE1 cells in a dose-dependent manner. Therefore, these findings indicated that GO promoted the apoptosis of nasopharyngeal carcinoma cells through ER stress.
Interestingly, recent evidence suggests that autophagy is a double-edged sword in tumorigenesis and metastasis [26,27]. Extensive autophagy or inappropriate activation of autophagy may result in autophagic cell death. As a marker protein, LC3II can determine the mechanism of autophagy [28]. In our study, we demonstrated that the expression of LC3II was markedly shifted in a dose- and time-independent manner by western blotting and RT-PCR assay. Furthermore, compared with the control group, the GO-administrated HONE1 cells in the GO group increased the protein expression of LC3II. Conversely, 4-PBA treatment decreased the protein expression of LC3II compared to the GO group. The RT-PCR result is consistent with the western blotting assay. The above results demonstrated that endoplasmic reticulum stress was involved in GO induced autophagy in HONE1 cells.
In addition, the findings of the present study show that GO inhibited cell proliferation of HONE1 cells in a dose- and time-independent manner. Consistent with previous studies, an indirect immunofluorescence assay and a transmission electron microscope demonstrated that GO could indicate autophagy. The above result is consistent with previous studies [16].
Taken together, our results demonstrated that autophagy and ER stress were shown to be involved in the mechanism of nasopharyngeal carcinoma. GO could induce autophagy and ER stress and suppress cell proliferation, which provides a new avenue for the treatment of nasopharyngeal carcinoma. Nevertheless, there are of course some limitations to our present study. Further studies are needed to elucidate its possible mechanism.
Acknowledgements
This study was supported by grants from the National Natural Science Foundation of China (no: 81560309), the Open Issues of The Guangxi Talent Highland for Emergency and Rescue Medicine, Guangxi Colleges and Universities, the Key Laboratory of Emergency Medicine Research (ref: GXJZ201503) and the Guangxi Natural Science Foundation of China (no: 2017GXNSFBA198109).
Disclosure of conflict of interest
None.
References
- 1.Ali AS, Al-Shraim M, Al-Hakami AM, Jones IM. Epstein-barr virus: clinical and epidemiological revisits and genetic basis of oncogenesis. Open Virol J. 2015;9:7–28. doi: 10.2174/1874357901509010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chua MLK, Wee JTS, Hui EP, Chan ATC. Nasopharyngeal carcinoma. Lancet. 2016;387:1012–1024. doi: 10.1016/S0140-6736(15)00055-0. [DOI] [PubMed] [Google Scholar]
- 3.Qu Y, He F, Yu C, Liang X, Liang D, Ma L, Zhang Q, Lv J, Wu J. Advances on graphene-based nanomaterials for biomedical applications. Mater Sci Eng C Mater Biol Appl. 2018;90:764–780. doi: 10.1016/j.msec.2018.05.018. [DOI] [PubMed] [Google Scholar]
- 4.Wang H, Chen Q, Zhou S. Carbon-based hybrid nanogels: a synergistic nanoplatform for combined biosensing, bioimaging, and responsive drug delivery. Chem Soc Rev. 2018;47:4198–4232. doi: 10.1039/c7cs00399d. [DOI] [PubMed] [Google Scholar]
- 5.Singh DP, Herrera CE, Singh B, Singh S, Singh RK, Kumar R. Graphene oxide: an efficient material and recent approach for biotechnological and biomedical applications. Mater Sci Eng C Mater Biol Appl. 2018;86:173–197. doi: 10.1016/j.msec.2018.01.004. [DOI] [PubMed] [Google Scholar]
- 6.Zhao Q, Lin Y, Han N, Li X, Geng H, Wang X, Cui Y, Wang S. Mesoporous carbon nanomaterials in drug delivery and biomedical application. Drug Deliv. 2017;24:94–107. doi: 10.1080/10717544.2017.1399300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ma K, Fu D, Liu Y, Rui Dai, Yu D, Guo Z, Cui C, Wang L, Xu J, Mao C. Cancer cell targeting, controlled drug release and intracellular fate of biomimetic membrane-encapsulated drug-loaded nano-graphene oxide nanohybrids. J Mater Chem B. 2018;6:5080–5090. doi: 10.1039/C8TB00804C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arya BD, Mittal S, Joshi P, Pandey AK, Ramirez-Vick JE, Singh SP. Graphene oxide-chloroquine nanoconjugate induce necroptotic death in A549 cancer cells through autophagy modulation. Nanomedicine (Lond) 2018;13:2261–2282. doi: 10.2217/nnm-2018-0086. [DOI] [PubMed] [Google Scholar]
- 9.Assali A, Akhavan O, Mottaghitalab F, Adeli M, Dinarvand R, Razzazan S, Arefian E, Soleimani M, Atyabi F. Cationic graphene oxide nanoplatform mediates miR-101 delivery to promote apoptosis by regulating autophagy and stress. Int J Nanomedicine. 2018;13:5865–5886. doi: 10.2147/IJN.S162647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tang Z, Zhao L, Yang Z, Liu Z, Gu J, Bai B, Liu J, Xu J, Yang H. Mechanisms of oxidative stress, apoptosis, and autophagy involved in graphene oxide nanomaterial anti-osteosarcoma effect. Int J Nanomedicine. 2018;13:2907–2919. doi: 10.2147/IJN.S159388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan YG, Gurunathan S. Combination of graphene oxide-silver nanoparticle nanocomposites and cisplatin enhances apoptosis and autophagy in human cervical cancer cells. Int J Nanomedicine. 2017;12:6537–6558. doi: 10.2147/IJN.S125281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu T, Liang H, Liu L, Gong Y, Ding Y, Liao G, Cao Y. Influence of pristine and hydrophobic ZnO nanoparticles on cytotoxicity and endoplasmic reticulum (ER) stress-autophagy-apoptosis gene expression in A549-macrophage co-culture. Ecotoxicol Environ Saf. 2018;167:188–195. doi: 10.1016/j.ecoenv.2018.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Park HB, Kim MJ, Jung BD, Lee S, Park CK, Yang BK, Cheong HT. Effect of endoplasmic reticulum (ER) stress inhibitor treatment during parthenogenetic activation on the apoptosis and in vitro development of parthenogenetic porcine embryos. Dev Reprod. 2018;22:235–244. doi: 10.12717/DR.2018.22.3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ou L, Lin S, Song B, Liu J, Lai R, Shao L. The mechanisms of graphene-based materials-induced programmed cell death: a review of apoptosis, autophagy, and programmed necrosis. Int J Nanomedicine. 2017;12:6633–6646. doi: 10.2147/IJN.S140526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu W, Xie F, Zhang Y, Wang XX, Xia LL, Wu XL, Gao Z. A novel regulatory function for miR-217 targetedly suppressing fibronectin expression in keloid fibrogenesis. Int J Clin Exp Pathol. 2018;11:1866–1877. [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang C, Wang LM. Inhibition of autophagy attenuated curcumol-induced apoptosis in MG-63 human osteosarcoma cells via Janus kinase signaling pathway. Oncol Lett. 2017;14:6387–6394. doi: 10.3892/ol.2017.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hasanzade Z, Raissi H. Assessment of the chitosan-functionalized graphene oxide as a carrier for loading Thioguanine, an antitumor drug and effect of urea on adsorption process: combination of DFT computational and molecular dynamics simulation studies. J Biomol Struct Dyn. 2018;27:1–38. doi: 10.1080/07391102.2018.1496140. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Dong J, Zhang T, Peng Q. Graphene-based nanomaterials and their potentials in advanced drug delivery and cancer therapy. J Control Release. 2018;286:64–73. doi: 10.1016/j.jconrel.2018.07.034. [DOI] [PubMed] [Google Scholar]
- 19.Fang J, Liu Y, Chen Y, Ouyang D, Yang G, Yu T. Graphene quantum dots-gated hollow mesoporous carbon nanoplatform for targeting drug delivery and synergistic chemo-photothermal therapy. Int J Nanomedicine. 2018;13:5991–6007. doi: 10.2147/IJN.S175934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim MH, Jeung IC, Jeong J, Yoon SJ, Lee SH, Park J, Kang YS, Lee H, Park YJ, Lee HG, Lee SJ, Han BS, Song NW. Graphene oxide induces apoptotic cell death in endothelial cells by activating autophagy via calcium-dependent phosphorylation of c-Jun N-terminal kinases. Acta Biomater. 2016;46:191–203. doi: 10.1016/j.actbio.2016.09.018. [DOI] [PubMed] [Google Scholar]
- 21.Jiang JH, Pi J, Jin H, Cai JY. Functional graphene oxide as cancer-targeted drug delivery system to selectively induce oesophageal cancer cell apoptosis. Artif Cells Nanomed Biotechnol. 2018;5:1–11. doi: 10.1080/21691401.2018.1492418. [DOI] [PubMed] [Google Scholar]
- 22.Bhat TA, Chaudhary AK, Kumar S, O’Malley J, Inigo JR, Kumar R, Yadav N, Chandra D. Endoplasmic reticulum-mediated unfolded protein response and mitochondrial apoptosis in cancer. Biochim Biophys Acta Rev Cancer. 2017;1867:58–66. doi: 10.1016/j.bbcan.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oakes SA. Endoplasmic reticulum proteostasis: a key checkpoint in cancer. Am J Physiol Cell Physiol. 2017;312:C93–C102. doi: 10.1152/ajpcell.00266.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Axten JM. Protein kinase R(PKR)-like endoplasmic reticulum kinase (PERK) inhibitors: a patent review (2010-2015) Expert Opin Ther Pat. 2017;27:37–48. doi: 10.1080/13543776.2017.1238072. [DOI] [PubMed] [Google Scholar]
- 25.Yin Y, Sun G, Li E, Kiselyov K, Sun D. ER stress and impaired autophagy flux in neuronal degeneration and brain injury. Ageing Res Rev. 2017;34:3–14. doi: 10.1016/j.arr.2016.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin P, Wei P, Zhang Y, Lin J, Sha R, Hu Y, Zhang J, Zhou W, Yao H, Ren L, Yang JY, Liu Y, Wen L. Autophagy-mediated clearance of ubiquitinated mutant huntingtin by graphene oxide. Nanoscale. 2016;8:18740–18750. doi: 10.1039/c6nr07255k. [DOI] [PubMed] [Google Scholar]
- 27.Kouroku Y, Fujita E, Tanida I, Ueno T, Isoai A, Kumagai H, Ogawa S, Kaufman RJ, Kominami E, Momoi T. ER stress (PERK/eIF2alpha phosphorylation) mediates the polyglutamine-induced LC3 conversion, an essential step for autophagy formation. Cell Death Differ. 2007;14:230–9. doi: 10.1038/sj.cdd.4401984. [DOI] [PubMed] [Google Scholar]
- 28.Zhang T, Qi Y, Liao M, Xu M, Bower KA, Frank JA, Shen HM, Luo J, Shi X, Chen G. Autophagy is a cell self-protective mechanism against arsenic-induced cell transformation. Toxicol Sci. 2012;130:298–308. doi: 10.1093/toxsci/kfs240. [DOI] [PubMed] [Google Scholar]


