Abstract
Objective: To identify genes potentially associated with cervical intraepithelial neoplasia (CIN) progression through bioinformatic approaches and clinicopathological verification. Methods: mRNA expression microarray data related to CIN progression were screened from the Gene Expression Omnibus (GEO) database and re-analyzed using bioinformatics approaches. Tissue microarray immunohistochemistry was conducted to assess the significant identified genes in CIN, cervical cancer, and normal tissues. Results: Biological annotation and text mining showed that 14 differentially expressed genes were directly or indirectly related to CIN progression. The expression of 5 up-regulated differentially expressed genes, namely, CCND2, TGFBR2, PRKCB, SH3KBP1 and WNT2B, was examined by tissue microarray immunohistochemistry, with the known CIN progression genes P16 and Ki-67 as the internal reference. Expression of TGFBR2, SH3KBP1, and WNT2B were not detected in CIN and cervical carcinoma, whereas no significant difference in the expression rate of PRKCB was detected (P > 0.05). CCND2, P16, and Ki-67 expression showed a gradual increasing trend in normal, CIN, and cervical cancer. Conclusions: 14 differentially expressed genes were associated with CIN progression, as indicated by the microarray data analysis results. CCND2 may be a new marker for the prediction of CIN progression in addition to P16 and Ki-67.
Keywords: Cervical intraepithelial neoplasia, progression, expression profiling microarray, gene, bioinformatics, immunohistochemistry
Introduction
Cervical cancer is the second most common female malignancy, with poor prognosis and the fourth highest mortality rate in the world [1]. Cervical intraepithelial neoplasia (CIN) is a precursor lesion for cervical cancer, classified histologically as low-grade CIN (CIN1) or high-grade CIN (CIN2 and CIN3). According to laboratory surveys from the College of American Pathologists (CAP), more than 1 million women develop CIN1 each year, and 500,000 are diagnosed with high-grade CIN (CIN2 and CIN3) [2]. Approximately 30% of the patients with high-grade CIN could progress to cervical cancer if left untreated [3,4], and another 20-40% of those patients will show spontaneous regression [5-7]. The existing pathologic histological methods are still unable to identify the CIN cases that might progress or spontaneously regress; therefore, the identification of CIN progression markers is necessary for the early prediction and diagnosis of cervical cancer to avoid overtreatment and adverse pregnancy outcomes due to cervical conization for those wishing to retain fertility.
In recent years, data mining has been performed at the molecular level using bioinformatic approaches such as sequence alignment, biochip data extraction, visual mapping, biological data clustering, pathway analyses, statistical analyses, and promoter prediction, and these approaches guide laboratory experiments and promote more rapid genomic annotation [8], providing novel research ideas for studying the molecular pathogenesis of various diseases. Therefore, bioinformatics approaches are a feasible and potentially valuable approach to data mining and the prediction of gene function on the basis of many large databases and networks [8]. For example, Zou J [9] identified 38 differentially expressed genes and 9 microRNAs associated with ovarian cancer progression through the Gene Expression Omnibus (GEO) [10] database and comprehensive bioinformatic analyses. Liu X [11] identified 6 differentially expressed genes associated with ovarian cancer progression through the GEO database, including ALDH1A2. However, genes associated with CIN progression have never been identified using the GEO database. In this study, using mRNA expression profiles retrieved from the GEO database, we screened for differentially expressed genes that are closely associated with CIN progression and development. The correlation between the differentially expressed genes and CIN progression was examined through comprehensive bioinformatics analyses, including text mining, biological process annotation, biological pathway enrichment, and protein/gene interaction analysis. Based on the results, we identified genes likely to play key roles in the development of high-grade CIN, and analyzed the correlation between these genes, CIN, and cervical cancer based on clinical samples.
Materials and methods
Acquisition and analysis of datasets
Microarray data published prior to January 2016 detailing cervical intraepithelial neoplasia progression-related mRNA expression profiles were retrieved and downloaded from the National Center for Biotechnology Information (NCBI) GEO database (http://www. ncbi.nlm nih.gov/geo). Queries were conducted using “cervical intraepithelial neoplasia” as a keyword. The search was restricted to studies of expression profiling by array and the species to Homo sapiens. We downloaded the mRNA expression microarray datasets GSE63514 [12] and GSE51993 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE51993).
Differentially expressed genes were identified using the GEO2R tool through the GEO database. GEO2R [13] is an R programming language-based dataset analysis tool that was launched in 2012, which is useful to compare two sets of samples under the same experimental conditions to screen differentially expressed genes. In the present study, differentially expressed genes associated with CIN progression were screened using a p-value less than 0.05 and a fold change of at least 2 (> 2-fold change) as the threshold.
Bioinformatics analyses
Intersection analysis of multiple genes was performed using Venn Diagram [14] (http://bioinformatics.psb.ugent.be/webtools/Venn/). Pathway enrichment of the biologically common differentially expressed genes was performed using the DAVID online tool [15-17] (https://david.ncifcrf.gov/home.jsp). Annotation of biologic processes between genes and CIN progression was performed using the Coremine Medical online database (http://www.coremine.com/medical/) [18]. The protein/gene interaction analysis was performed using the GeneMANIA online tool (http://www.genemania.org/) [19].
Tissue samples and data collection
From the database for the Department of Pathology, Affiliated Tumor Hospital of Guangxi Medical University and Liuzhou People’s Hospital, we collected pathology reports of 130 women who underwent both liquid-based cervical cytology and HPV DNA load tests from January 2013 and June 2015. Samples from 68 patients with CIN were obtained after cervical conization. Cancer tissues were taken after radical hysterectomy from 33 early squamous cervical cancer patients without treatment before surgery. Normal cervical tissues were obtained after hysterectomy from 29 uterine leiomyoma patients. The CIN and cervical cancer patients in the group were followed up for an average of 12 ± 3.4 months. All of the hematoxylin and eosin-stained slides from the individual paraffin blocks were reviewed and confirmed by 2 different pathologists. The tissue sample size should be adequate to construct tissue microarray (TMA) blocks. All patients underwent preoperative detection of human papillomavirus (HPV) and cervical cytology. HPV detection was performed using the hybrid capture method, with a relative light unit/positive control critical value (RLU/CO) ≥ 1.0 indicating a positive test. Cytology diagnosis of HPV was performed using the 2001 version of the TBS the classification system. The CIN naming system and WHO diagnostic criteria were used for pathologic diagnosis.
TMA construction and immunohistochemistry
The TMA blocks were established using a previously described method [20]. Briefly, hematoxylin and eosin-stained slides from all paraffin-embedded tissue blocks were marked by a pathologist to locate representative areas for TMA samples. Cylinders (2 mm in diameter) were punched from the blocks corresponding to the marked area, and 4-um-thick sections were placed into the tissue microarray paraffin blocks, containing on average 25 to 30 punch biopsies. Immunohistochemistry reactions were performed as follows. Briefly, the TMA slides were deparaffinized, sequentially dehydrated, pretreated in 3% hydrogen peroxide, boiled with citrate antigen retrieval solution, and incubated in 1% BSA to inhibit nonspecific antibody binding. Then, the TMA slides were incubated in a humidified chamber at 4°C overnight with the primary antibodies. The slides were incubated with a biotinylated antibody, followed by streptavidin-peroxidase for 30 min. Subsequently, the expression staining was visualized by exposing the slides to a diaminobenzidine solution for 1 min. Hematoxylin counterstaining was applied the slides for 10 min, followed by washing with water, dehydration in graded ethanol solutions, clearing with xylene, and mounting.
Immunostaining was assessed by two independent pathologists. CCND2 immunostaining was scored as follows: [21] 0, no staining or staining in < 5% of the cells; 1, staining in ≥ 5 to < 25% of the cells; 2, staining in ≥ 25 to < 50% of the cells; 3, staining in ≥ 50% to < 75% of the cells; and 4, staining in ≥ 75% of the cells. Scores of 0 and 1 were considered ‘negative’, whereas scores ≥ 2 were regarded as ‘positive’. Positive p16INK4a histology staining was defined as strong and diffuse staining of the basal and para-basal squamous cell compartment in at least the lower third of the epithelial thickness in well-oriented sections [22]. The Ki-67 labeling index was graded into low and high indices at a cut-off value of 20% [23].
Statistical analyses
All statistical analyses were conducted using the SPSS statistical software (version 19.0, SPSS, Chicago, IL, USA). The Chi-square test and Fisher’s exact test were applied to compare the expression results in the cervical tissues. Univariate and multivariate analyses of clinical factors affecting the expression of CCND2 were examined using Chi-square tests and logistic regression analyses, respectively. Associations between gene expression and hrHPV were examined using the Chi-square test. Receiver operating characteristic (ROC) curve and logistic regression analyses were introduced to evaluate the diagnostic capabilities of the identified genes. All tests were performed using a P value of 0.05 for significance.
Results
Screening of differentially expressed genes and pathway enrichment
In the present study, the mRNA expression profiling datasets screened from the GEO database were GSE63514 [12] and GSE51993. The data of GSE63514 included 14 CIN1 lesions, 22 CIN2 lesions, 40 CIN3 lesions, and 28 cancer specimens, whereas the GSE51993 data comprised 7 normal (HPV-), 9 CIN1 lesions (HPV+) and 8 CIN3 lesions (HPV+) (Table 1). The CIN and normal samples served as the experimental group and control group, respectively. By using adjusted P value < 0.05 and a fold change > 2 as the threshold, the GSE63514 and GSE51993 datasets were screened using the GEO2R tool to reveal the differentially expressed genes between the experimental and control groups. A total of 999 up-regulated differentially expressed genes and 858 down-regulated differentially expressed genes were identified from the GSE51993 dataset, and 5,028 up-regulated differentially expressed genes and 4,303 down-regulated differentially expressed genes from the GSE63514 dataset. The up-regulated and down-regulated differential expression genes in the two datasets were intersected using the Venn Diagram tool, and 144 commonly up-regulated genes and 219 commonly down-regulated genes were found to be significantly differentially expressed in the two datasets (Figure 1). Biological signaling pathway enrichment was then performed on the above 144 up-regulated and 219 down-regulated genes using DAVID to identify the biological significance of the common differentially expressed genes in the regulation of CIN progression at the unitary level. The pathway enrichment identified 3 significant signaling pathways, the Wnt, endocytosis, and Vibrio cholerae infection pathways, according to their P values. Fourteen differentially expressed genes were found among the 3 pathways that exhibited an identical expression pattern trend between the 2 datasets (Table 2). Among them, only SH3KBP1 has been shown to be highly expressed in cervical cancer[24]; none of the other genes have been previously reported.
Table 1.
Gene expression microarray datasets related to CIN progression
Figure 1.
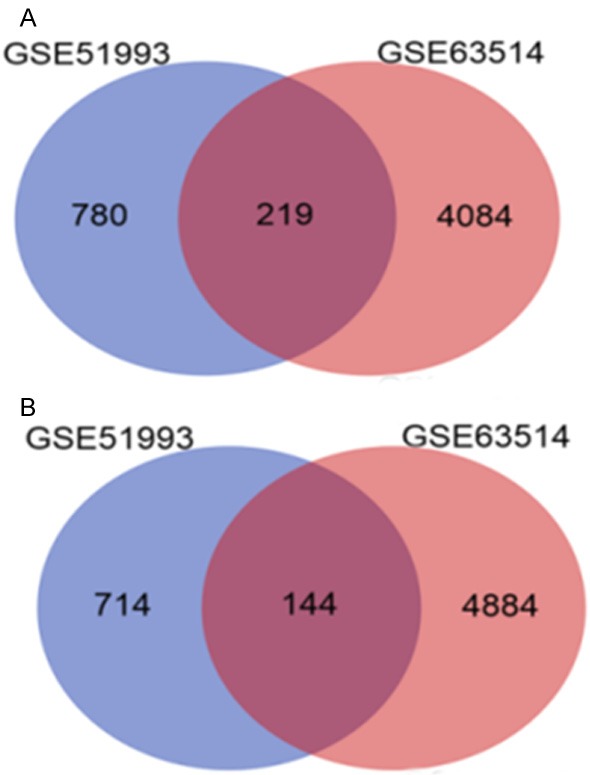
Analysis of the common differentially expressed genes in the two microarray datasets of GSE63514 and GSE51993 by using the Venn diagram tool (A, 219 common down-regulated genes; B, 144 common up-regulated genes).
Table 2.
The 14 differentially expressed genes that exhibited exactly the same expression pattern trends
| Gene name | Signal pathway | LogFC* | Expression pattern | Literature related to CIN progression | |
|---|---|---|---|---|---|
|
| |||||
| GSE63514 | GSE51993 | ||||
| CCND2 | Wnt | 0.0446 | 0.504157 | Upregulated | - |
| NFATC1 | Wnt | 0.879 | 0.168807 | Upregulated | - |
| PRKCB | Wnt | 0.847 | 0.462753 | Upregulated | - |
| PPP2R5D | Wnt | 0.488 | 0.266376 | Upregulated | - |
| WNT2B | Wnt | 0.847 | 0.511759 | Upregulated | - |
| SH3KBP1 | Endocytosis | 0.578 | 0.429254 | Upregulated | Highly expressed in cervical cancer [24] |
| HSPA6 | Endocytosis | 1.18 | 0.435959 | Upregulated | - |
| PIKFYVE | Endocytosis | 0.266 | 0.126757 | Upregulated | - |
| RABEP1 | Endocytosis | 0.483 | 0.224578 | Upregulated | - |
| TGFBR2 | Endocytosis | 0.0341 | 0.153118 | Upregulated | - |
| KDELR1 | Vibrio cholerae infection | -0.212 | -0.23526 | Downregulated | - |
| MUC2 | Vibrio cholerae infection | -0.0647 | -0.2518 | Downregulated | - |
| KCNQ1 | Vibrio cholerae infection | -0.156 | -0.21546 | Downregulated | - |
| TJP2 | Vibrio cholerae infection | -0.26 | -0.49813 | Downregulated | - |
A positive logFC value shows that the gene is upregulated, and a negative logFC value indicates that the gene is downregulated.
- No reports have been reported or retrieved.
Bioinformatics analysis of the correlation between the differentially expressed genes and CIN progression
To reveal the biologic significance of the 14 commonly differentially expressed genes in the regulation of CIN progression at the unitary level, biological process annotation and text mining were performed on the above-described genes using the COREMINE tool. A co-occurrence analysis of the literature was conducted using the gene names and “Cervical Intraepithelial Neoplasia”, “Progression”, and “Recurrence” as keywords. The results revealed that all of the commonly differentially expressed genes were present in the text-mining networks, except PPP2R5D and PIKFYVE. Among the 12 present genes, 3 genes (CCND2, TGFBR2 and MUC2) were directly related to CIN progression. In addition to significant correlations with “Cervical Intraepithelial Neoplasia”, “Progression”, and “Recurrence”, the genes were also significantly associated with each other, with the relationship between TGFBR2 and WNT2B presented as an example in Figure 2.
Figure 2.
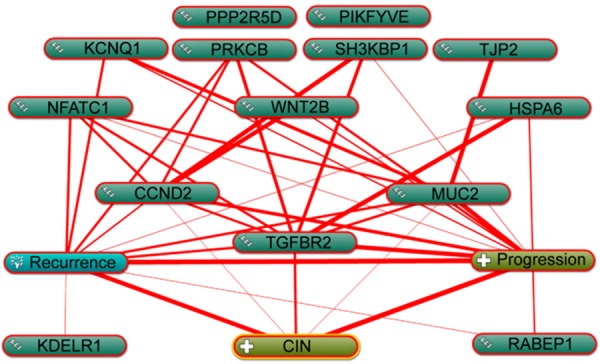
Function prediction and analysis of the 14 identified genes related to progression and recurrence using the Coremine tool. The thickest red line indicates the strongest association.
Protein/gene interaction analysis
An advanced search using the keywords ‘Cervical Intraepithelial Neoplasia’ and ‘Progression’ was performed in the PubMed database, and 24 protein-coding genes were found to be significantly correlated with CIN progression, including CDKN2A [25], AQP8 [26], CADM1 [27], CCL2 [28], CTNNB1 [23], EGFR [29], ERBB2 [29], FHIT [30], IL17A [31], IMP3 [32], MAPK1 [33], MKI67 [34], MTDH [35], MYC [30], PHGDH [36], PIK3CA [33], PRKCI [37], PTGS2 [29], STK11 [33], TP53 [33], TP53BP1 [38], TP63 [39], FOXO1 [40] and ITCH [41]. The relationships between the 14 potentially differentially expressed genes and 24 previously reported protein genes were analyzed using GeneMANIA (http://www.genemania.org/), a web-based tool for the rapid and accurate prediction of gene function based on multiple networks derived from different genomic/proteomic data sources [19]. The protein/gene interactions in the network include pathway, co-localization, co-expression, physical interaction, genetic interaction, and shared protein domains. As shown in Figure 3, all of the 14 screened genes interacted with the 24 reported genes either directly or indirectly, indicating that these genes may be functionally relevant. Accordingly, the screened genes may also be associated with CIN progression. Particularly, CCND2 was found to interact directly with 16 CIN progression relevant genes including CDKN2A, CADM1, CCL2, CTNNB1, ERBB2, IMP3, MYC, PHGDH, PIK3CA, PRKCI, PTGS2, TP53, TP53BP1, TP63, FOXO1 and ITCH. Moreover, the 14 screened genes also interacted with each other through various means. For example, CCND2 directly interacted with PRKCB, SH3KBP1, TGFBR2 and KDELR1; TGFBR2 directly interacted with CCND2 and 7 other genes; PRKCB directly interacted with CCND2 and 6 other genes; and SH3KBP1 directly interacted with CCND2 and 5 other genes.
Figure 3.
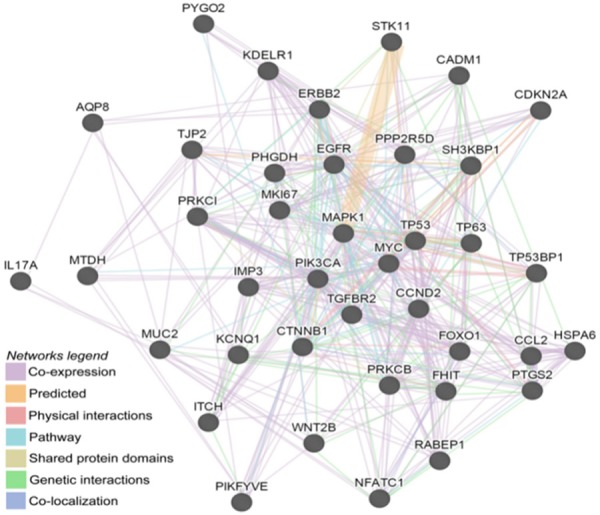
Protein/gene interaction network between the 24 known genes and the 14 screened genes generated using the GeneMANIA tool. The types of interactions between proteins/genes are indicated in the network legend of the figure.
Expression of different genes in cervical lesions and the analysis of clinical factors
The expression of 5 up-regulated differentially expressed genes, including CCND2, TGFBR2, PRKCB, SH3KBP1 and WNT2B, which were significantly associated in the above text-mining networks and protein/gene interaction analysis, was examined by tissue microarray immunohistochemistry, with the known CIN progression genes P16 and Ki-67 as the internal reference. Each primary antibody is summarized in Table 3. The positive expression rates of CCND2 in normal, CIN, and cervical cancer tissues were significantly different at 14%, 29% and 64%, respectively, and their expression increased gradually with the increase in cervical lesion grade (Table 4 and Figure 4). The positive expression rates of P16 and Ki-67 in normal, CIN and cervical cancer tissues were also significantly different (P < 0.05) at 17% and 69%, 91% and 21%, and 57% and 79%, respectively. The increase in cervical lesions also indicated a gradual increase in the trend (Table 4 and Figure 4). There was no significant difference in the expression rate of PRKCB (P > 0.05), whereas WNT2B, SH3KBP1 and TGFBR2 were not detected in the CIN and cervical cancer tissues. The results did not change significantly after CIN was subdivided into the CIN1, CIN 2, and CIN 3 subgroups and compared with the normal and cervical cancer groups (Table 5).
Table 3.
Antibodies used for immunohistochemical staining
| Antibodies | Type | Dilution | Source |
|---|---|---|---|
| CCND2 | Rabbit Monoclonal Antibody | 1:50 | Santa Cruz Biotechnology (sc-754) |
| PRKCB | Rabbit Monoclonal Antibody | 1:200 | Abcam (ab18512) |
| WNT2B | Rabbit Polyclonal Antibody | 1:200 | Santa Cruz Biotechnology (sc-134152) |
| SH3KBP1 | Rabbit Monoclonal Antibody | 1:200 | Abcam (ab151574) |
| TGFBR2 | Mouse Monoclonal Antibody | 1:200 | Abcam (ab78419) |
| P16 | Mouse Monoclonal Antibody | 1:100 | Cell Signaling technology (#2407) |
| Ki-67 | Rabbit Monoclonal Antibody | 1:400 | Cell Signaling technology (#9027) |
Table 4.
Expression of different genes in normal, CIN, and cervical cancer tissues
| Gene | Normal (n = 29) | CIN (n = 68) | Cervical cancer (n = 33) | X2 | P |
|---|---|---|---|---|---|
| CCND2 | 18.649 | 0.000 | |||
| Negative | 25 (86%) | 48 (71%) | 12 (36%) | ||
| Positive | 4 (14%) | 20 (29%) | 21 (64%) | ||
| PRKCB | 4.577 | 0.101 | |||
| Negative | 24 (83%) | 46 (68%) | 19 (58%) | ||
| Positive | 5 (17%) | 22 (32%) | 14 (42%) | ||
| P16 | 68.63 | 0.000 | |||
| Negative | 24 (83%) | 21 (31%) | 3 (9%) | ||
| Positive | 5 (17%) | 47 (69%) | 30 (91%) | ||
| Ki-67 | 19.54 | 0.000 | |||
| Negative | 23 (79%) | 29 (43%) | 7 (21%) | ||
| Positive | 6 (21%) | 39 (57%) | 26 (79%) |
Figure 4.
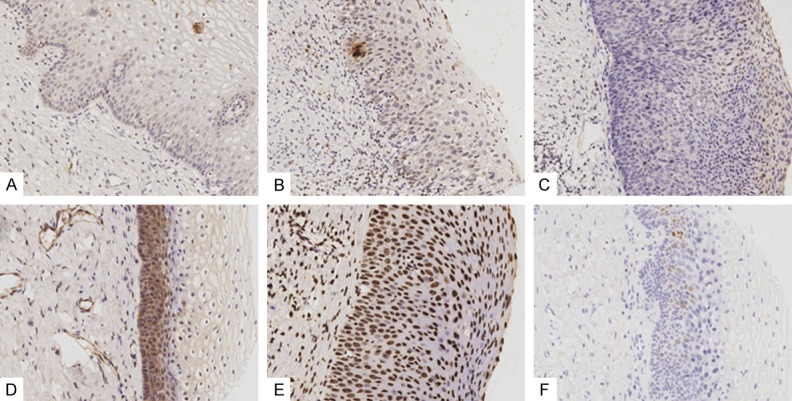
Representative positive examples of immunohistochemistry of identified genes on CIN tissues. A. CCND2 (CIN1); B. CCND2 (CIN2); C. CCND2 (CIN3); D. PRKCB (CIN1); E. P16 (CIN3); F. Ki-67 (CIN2). Immunohistochemistry, DAB, brown stain, magnification × 20.
Table 5.
The expression of each gene in normal, CIN1, CIN2, CIN3 and cervical cancer tissues
| Gene | Normal (n = 29) | CIN 1 (n = 14) | CIN 2 (n = 23) | CIN 3 (n = 31) | Cervical cancer (n = 33) | P |
|---|---|---|---|---|---|---|
| CCND2 | 0.000 | |||||
| Negative | 25 (86%) | 12 (86%) | 19 (83%) | 17 (55%) | 12 (36%) | |
| Positive | 4 (14%) | 2 (14%) | 4 (17%) | 14 (45%) | 21 (64%) | |
| PRKCB | 0.30 | |||||
| Negative | 24 (83%) | 10 (71%) | 16 (70%) | 20 (65%) | 19 (58%) | |
| Positive | 5 (17%) | 4 (29%) | 7 (30%) | 11 (35%) | 14 (42%) | |
| P16 | 0.00 | |||||
| Negative | 24 (83%) | 6 (43%) | 7 (30%) | 8 (26%) | 3 (9%) | |
| Positive | 5 (17%) | 8 (57%) | 16 (70%) | 23 (74%) | 30 (91%) | |
| Ki-67 | 0.00 | |||||
| Negative | 23 (79%) | 6 (57%) | 8 (35%) | 15 (48%) | 7 (21%) | |
| Positive | 6 (21%) | 8 (43%) | 15 (65%) | 16 (52%) | 26 (79%) |
A clinical analysis of CCND2 expression was performed based on the immunohistochemical protein expression and clinical data. The univariate analysis showed that high expression of CCND2 was positively correlated with high-grade CIN (P < 0.05) but not significantly associated with age, age at first sexual intercourse, number of sexual partners, contraceptive methods, number of pregnancies, number of births, P16 level and Ki-67 level (P > 0.05). Logistic regression analysis that included CCND2 as a dependent variable indicated a significant difference in the CIN grade, with an odds ratio (OR) of 6.847 (P < 0.05) (Table 6).
Table 6.
Logistic regression analysis of factors affecting CCND2 expression
| CCND2 (+) (n = 20) | CCND2 (-) (n = 48) | OR | P | |
|---|---|---|---|---|
| Age | 0.797 | 0.760 | ||
| ≥ 40 | 13 | 34 | ||
| < 40 | 7 | 14 | ||
| Primary Sex Age | 0.514 | 0.332 | ||
| < 19 | 11 | 20 | ||
| ≥ 20 | 9 | 28 | ||
| Sexual Partners | 0.005 | 0.945 | ||
| 1 | 14 | 34 | ||
| ≥ 2 | 6 | 14 | ||
| Contraceptive | 0.137 | 0.712 | ||
| Yes | 17 | 39 | ||
| No | 3 | 9 | ||
| Pregnancy Times | 0.021 | 0.884 | ||
| ≥ 3 | 4 | 7 | ||
| < 3 | 16 | 31 | ||
| Birth Times | 1.511 | 0.219 | ||
| ≥ 3 | 7 | 10 | ||
| <3 | 13 | 38 | ||
| hrHPV | 3.898 | 0.048 | ||
| Negative | 3 | 19 | ||
| Positive | 17 | 29 | ||
| P16 | 1.103 | 0.294 | ||
| Negative | 8 | 13 | ||
| Positive | 12 | 35 | ||
| Ki-67 | 0.677 | 0.410 | ||
| Negative | 7 | 22 | ||
| Positive | 13 | 26 | ||
| CIN Grade | 6.847 | 0.033 | ||
| I | 2 | 12 | ||
| II | 4 | 19 | ||
| III | 14 | 17 |
CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus.
The correlation between CCND2 expression and hrHPV infection was analyzed. The results indicated that CCND2 was significantly associated with hrHPV infection (P < 0.05). The logistic regression analysis that included CCND2 as the dependent variable showed that hrHPV was statistically significant, with an OR of 3.898 (P < 0.05) (Table 6).
The relation between hrHPV and the expression of genes in CIN and cervical cancer tissues was analyzed. The expression of CCND2, P16 and Ki-67 in hrHPV-positive CIN tissues was significantly higher than that in hrHPV-negative CIN tissues (P < 0.05), but not in cervical cancer tissues (P > 0.05) (Table 7). These results indicated that the expression of the three genes in CIN increased significantly with hrHPV infection, suggesting that during the progression of CIN to cervical cancer, the expression of CCND2, P16 and Ki-67 were closely associated with hrHPV infection.
Table 7.
Relationship between the expression of each gene and hrHPV infection
| CIN | P value | Cervical cancer | P value | |||
|---|---|---|---|---|---|---|
|
|
|
|||||
| hrHPV (-) | hrHPV (+) | hrHPV (-) | hrHPV (+) | |||
| CCND2 | 0.048 | 0.610 | ||||
| Negative | 19 | 29 | 2 | 10 | ||
| Positive | 3 | 17 | 2 | 19 | ||
| P16 | 0.018 | 0.330 | ||||
| Negative | 11 | 10 | 1 | 3 | ||
| Positive | 11 | 36 | 3 | 26 | ||
| Ki-67 | 0.016 | 0.190 | ||||
| Negative | 14 | 15 | 2 | 5 | ||
| Positive | 8 | 31 | 2 | 24 | ||
CIN, cervical intraepithelial neoplasia; HPV, human papillomavirus.
Combined detection of genes in CIN diagnosis
The area under the curve (AUC) of CCND2 for the diagnosis of CIN was 0.58 (P > 0.05), and for P16 and Ki-67 it was 0.76 and 0.68, respectively, suggesting that CCND2 was less valuable for the diagnosis of CIN than P16 and Ki-67. The sensitivities of CCND2, P16 and Ki-67 in the diagnosis of CIN were 29.4%, 69.1% and 57.4%, respectively, and the specificities were 86.2%, 82.8% and 79.3%, respectively (Table 8).
Table 8.
Sensitivity, specificity, and accuracy of p16, Ki-67, and CCND2 in CIN diagnosis
| Gene | Sensitivity (%) | Specificity (%) | Positive predictive values | Negative predictive values | AUC (95% CI) | P |
|---|---|---|---|---|---|---|
| CCND2 | 29.4 | 86.2 | 83.3 | 34.2 | 0.58 (0.47-0.68) | 0.0684 |
| P16 | 69.1 | 82.8 | 90.4 | 53.3 | 0.76 (0.66-0.84) | < 0.0001 |
| Ki-67 | 57.4 | 79.3 | 86.7 | 44.2 | 0.68 (0.58-0.77) | 0.0002 |
Using normal and CIN as the dependent variables, CCND2, Ki-67, P16, age, hrHPV status, age at first sexual intercourse, number of sexual partners, contraceptive methods, number of pregnancies and number of births were selected by the logistic diagnostic model. The results showed that only Ki-67, P16 and hrHPV status were statistically significant (P < 0.05), and the OR values were 10.34, 15.49 and 13.70, respectively, indicating that the diagnostic value of Ki-67, P16 and hrHPV-positive status were 10.34, 15.49 and 13.70 times greater than those of hrHPV-negative status, respectively (Table 9).
Table 9.
Result of CIN diagnostic logistic regression model
| Factor | B | S.E. | Wals | P | OR | 95% CI |
|---|---|---|---|---|---|---|
| ki67 | 2.34 | 0.76 | 9.51 | 0.002 | 10.33595 | 2.34-45.60 |
| P16 | 2.74 | 0.75 | 13.52 | 0.000 | 15.49167 | 3.60-66.75 |
| hrHPV | 2.62 | 0.74 | 12.37 | 0.000 | 13.70814 | 3.19-58.97 |
| Constant | -8.748 |
Results of the follow-up
Sixty-eight CIN patients in the group were followed up for an average of 12 ± 3.4 months. The results indicated that 2 patients had recurrence, and their pathology before surgery was CIN3. The immunohistochemical analysis showed that these two patients were P16 and Ki-67 positive, with one patient being CCND2 positive. Moreover, recurrence was not detected in 33 of the cervical cancer patients during the follow-up.
Discussion
CIN is a precursor lesion of cervical cancer. The occurrence and progression of CIN is a complex process with multi-step and multi-factor participation. At present, many of the mechanisms of CIN have not been illuminated at the molecular level. Accurate predictions of the progression rates of CIN lesions are limited due to a lack of sensitive markers for neoplastic progression. Therefore, it is very important to further explore the regulatory mechanism of CIN progression at the molecular level. To our knowledge, this study is the first to use GEO profile data mining to identify the signaling pathways and key genes related to the regulation of CIN progression. In this study, using integrated bioinformatics and clinical sample data analyses, three signaling pathways, including the Wnt, endocytosis, and Vibrio cholerae infection pathways, were identified to be significantly associated with CIN progression, and CCND2, which is part of the Wnt pathway, may be a potential key gene associated with CIN progression. This study has a certain role at the molecular level in further studies of CIN progression. CCND2 is expected to be a new molecular marker for CIN progression, in addition to P16 and Ki-67, to guide clinicians to stratify CIN therapy.
The reanalysis and reuse of public data has a potentially great effect in scientific research [42]. The GEO database is a publically available international repository containing high-throughput microarray and second-generation sequencing functional genomic datasets that are submitted by the research community. By 2012, the GEO database comprised more than 32,000 public series submitted directly by 13,000 laboratories, hosting 800,000 samples derived from > 1,600 organisms [13]. The datasets presented to the GEO database often comply with journal or grant directives that require data to be made publicly available in a MIAME-supportive [43] database. As a result, the GEO database has supporting data and links to nearly 20,000 published manuscripts [13]. In this study, through the rigorous screening of qualified GEO database microarray data, two sets of CIN-related mRNA expression profiles (Table 1) were chosen to analyze the pathways and genes involved in CIN progression, further revealing the molecular mechanism of CIN progression.
Based on the above data mining and bioinformatics analyses, this study identified three potential CIN progression-associated signaling pathways, including the Wnt, endocytosis and Vibrio cholerae infection pathways, which is, to some extent, consistent with previous research results. For example, endocytosis is involved in the signal transduction of a variety of cellular physiological activities, including cell growth and differentiation, cell chemotaxis, and immune responses [44]. Dynamin 2, the core gene in the endocytosis pathway, was proved to be a helpful biomarker in grading CIN lesions and is a candidate biomarker for the detection of low-grade CIN with high sensitivity [45]. Combined with the endocytosis pathway gene SH3KBP1, which is highly expressed in cervical cancer [24], the results suggest that the endocytosis signaling pathway may also play an important role in CIN progression. Recent research has shown that the Wnt signaling pathway is significantly associated with the promotion of cervical cancer cell proliferation and cell migration/invasion [46,47], as well as the progression of cervical cancer by promoting the epithelial-mesenchymal transition [48], which has also been suggested in other tumors [49-51]. Given the close relationship between CIN and cervical cancer, it has been proposed that alterations in the Wnt pathway are involved in CIN progression, and studies of CIN progression based on this signaling pathway may provide a new solution for blocking CIN progression. Although there are no reports on the relationship between the Vibrio cholera infection pathway and CIN, cholera toxin (CT) as a carrier/adjuvant may enhance human T-cell responses to HPV oncoprotein in vitro using dendritic cells (DCs) as antigen-presenting cells [52]. Based on the notion that hrHPV may be responsible for both cervical carcinoma and CIN, the Vibrio cholera infection pathway may also be involved with cervical lesions.
In terms of the 14 differentially expressed genes in the three signaling pathways, SH3KBP1 was found to be highly expressed in cervical cancer tissues [24], but the other 13 genes were not reported in CIN and cervical cancer. Moreover, through biological annotation, it was found that some of the differentially expressed genes were directly associated with CIN progression. Although not all of the genes were directly related, most of them were associated with tumor progression and recurrence. In addition, these genes weresignificantly related to each other, suggesting that the 14 identified genes may play an important role in the progression of CIN. Three genes, CCND2, TGFBR2 and MUC2, which were directly associated with CIN progression, may be directly involved in the regulation of CIN progression. Based on Gene MANIA, it was found that all of the selected genes not only interacted with the 24 known CIN progression genes but also interacted with each other in a variety of ways, further supporting that these screened genes may be functionally associated with and participate in the regulation of CIN progression at the unitary level.
Of the 14 differentially expressed genes, CCND2 and TGFBR2 may have the most significant associations with CIN progression. First, only three genes were identified as having a direct association with CIN progression by biological annotation, and these included CCND2 and TGFBR2. Second, in the complete protein/gene interaction network, comprising 24 known genes, CCND2 and TGFBR2 exhibited the greatest numbers of interactions, interacting with 16 and 12 of the 24 known genes, respectively. These known genes have been reported to be associated with the progression of CIN, suggesting that CCND2 and TGFBR2 may be more closely related to CIN progression. Moreover, CCND2 directly interacted with 4 of the screened differentially expressed genes, and TGFBR2 also had a direct interaction with 8 of the screened differentially expressed genes in the protein/gene network. For the above reasons, CCND2 and TGFBR2 may be important candidate genes for CIN progression regulation. However, the results need to be confirmed by further studies.
To verify the relationship between the differentially expressed genes and CIN progression, this study conducted immunohistochemical detection using a tissue microarray. The selected genes for immunohistochemical analysis included the significant genes from the bioinformatics analysis and the reported known genes P16 and Ki-67. Results indicated that the up-regulation of CCND2 was positively correlated with CIN grade, and that hrHPV-positive infection status also increased with CIN grade. For this reason, it was speculated that CCND2 may play a role in CIN progression to cervical cancer. CCND2 (cyclin D2) is one of the cyclin family members that regulates the cell cycle. CCND2 activates cell cycle-dependent kinase 4 (CDK4) and CDK6 in the cell cycle, forms a complex with CDK4 and CDK6 as a regulatory subunit, and plays a key role in regulating cell cycle transition from the G1 phase to S phase [53]. Previous studies have confirmed that CCND2 is highly expressed in tumors, including ovarian cancer and testicular cancer, and CCND2 knockout inhibits cancer cell proliferation [54]. A recent report indicated that mutations in the vicinity of the CCND2 gene were closely related to the occurrence of colorectal cancer [55]. Du et al. [56] found that CCND2 down-regulated the cell cycle G1/S transition, which inhibited the proliferation of HeLa cells. However, to date, no studies have investigated the relation between CCND2 and CIN. Our study showed that the expression of CCND2 was significantly associated with CIN grade and hrHPV infection status, indicating that CCND2 may be a new marker for the early diagnosis of CIN progression.
Four significantly up-regulated genes in the bioinformatics analysis, PRKCB, WNT2B, SH3KBP1 and TGFBR2, were not expressed or differentially expressed in cervical cancer tissues. The reason may be due to the low specificity of the high-throughput database and the differences between the screening and validation methods. Therefore, other methods, such as RT-PCR and western blotting, should be employed for functional verification.
The tumor suppressor protein p16 may be one of the most studied markers in CIN progression. P16 is the only biomarker currently used clinically for CIN diagnosis and has been recommended by the College of American Pathologists and the American Society for Colposcopy and Cervical Pathology (ASCCP) [57]. However, P16 has not been applied clinically for the diagnosis of CIN progression and prognosis. Several studies have found that the expression of P16 is associated with the progression of cervical lesions [25,58,59]. Our study also indicated that P16 was significantly associated with CIN progression, and P16 may be a marker of CIN progression.
Recently, the expression of the proliferating antigen gene Ki-67 was found to increase gradually in CIN1, CIN2, CIN3, and squamous cell carcinoma by immunohistochemistry [60], which is consistent with the results of our study. Based on our analysis of the differences in Ki-67 expression in normal, CIN, and cervical cancer tissues, Ki-67 may predict both low-grade and high-grade CIN progression. However, some scholars have found that Ki-67 was irrelevant for evaluating CIN progression [61], indicating that more studies to prove its usefulness as a progression marker are needed.
CCND2, Ki-67, and P16 were positively correlated with the hrHPV infection status in CIN and cervical cancer tissues, suggesting that these three genes are not only associated with CIN progression but are also synergistic in their carcinogenic effects with hrHPV. However, correlation analysis showed that CCND2 was not associated with Ki-67 and P16, suggesting that the mechanism of action of CCND2 and HPV is not related to that of Ki-67, P16, and HPV. Therefore, the specific mechanism of the involvement of these three genes in CIN progression needs to be further studied.
Most cases of CIN can be accurately diagnosed through cervical biopsy specimens using hematoxylin and eosin (HE) staining. However, when the lesions are also combined with immature metaplasia, inflammatory epithelial hyperplasia and epithelial atrophy, the diagnosis of CIN is difficult using only HE staining. Therefore, as a means of screening cervical lesions, P16, Ki-67 and other immunohistochemical markers have been gradually incorporated to compensate for the inadequacy of simple HE staining. However, there are still a small number of high-grade CIN cases that do not express the above genes [62], which was confirmed in our study. The ROC curve analysis showed that CCND2 alone was not valuable for the diagnosis of CIN, but when combined, the sensitivity for CIN diagnosis increased. This indicates the need for multiple immunohistochemical assays to avoid clinically over-treating false positive cases and inadequately treating false negative cases.
Several limitations of this study should be considered. First, this study was only cross-sectional design, not a prospective study that can follow up patients with CIN for progression analysis; therefore, there might have been confounders that were not recognized or controlled. The small sample size may have affected the results. Finally, hrHPV-negativity of the CIN samples might have led to a bias in the effect.
Conclusions
In this study, differentially expressed genes were screened using bioinformatics approaches and the GEO database, and using pathway enrichment, biological annotation and protein/gene interaction analyses, three pathways with 14 differentially expressed genes related to CIN progression were identified. The Wnt signaling pathway may be an important pathway in the development of CIN, and CCND2 and TGFBR2 may be important candidate genes associated with the regulation of CIN progression. The study of CIN progression based on signaling pathways and genes may provide new solutions to block the progression of CIN. The molecular marker CCND2, which was verified by subsequent tissue microarray immunohistochemistry, may provide an important basis for further studies and clinical applications (Figure 5). The next steps will be to increase the number of cases, extend the follow-up time, and utilize other experimental methods to study the molecular mechanism of CCND2 up-regulation associated with CIN progression.
Figure 5.
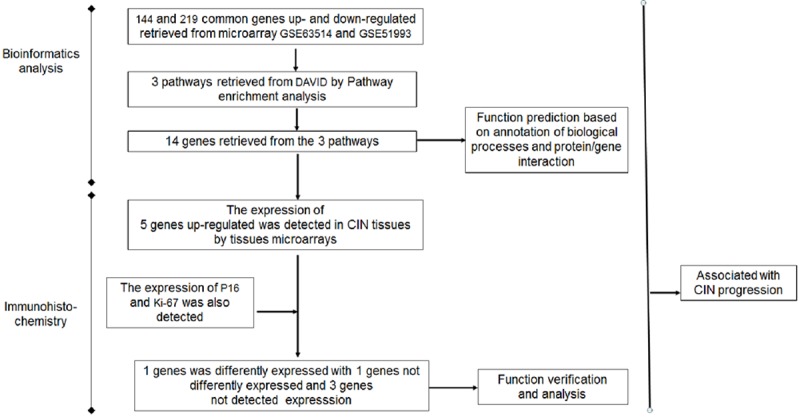
Procedure for identification of biological processes and genes associated with CIN progression.
Acknowledgements
The study was funded by the Public Welfare Special Funds of National Health and Family Planning Commission of China (201402010) and Key Laboratory of High-Incidence-Tumor Prevention & Treatment of Guangxi Medical University (GXK2001606).
Disclosure of conflict of interest
None.
References
- 1.Ramondetta L. What is the appropriate approach to treating women with incurable cervical cancer? J Natl Compr Canc Netw. 2013;11:348–55. doi: 10.6004/jnccn.2013.0044. [DOI] [PubMed] [Google Scholar]
- 2.Jones BA, Davey DD. Quality management in gynecologic cytology using interlaboratory comparison. Arch Pathol Lab Med. 2000;124:672–81. doi: 10.5858/2000-124-0672-QMIGCU. [DOI] [PubMed] [Google Scholar]
- 3.McCredie MR, Sharples KJ, Paul C, Baranyai J, Medley G, Jones RW, Skegg DC. Natural history of cervical neoplasia and risk of invasive cancer in women with cervical intraepithelial neoplasia 3: a retrospective cohort study. Lancet Oncol. 2008;9:425–34. doi: 10.1016/S1470-2045(08)70103-7. [DOI] [PubMed] [Google Scholar]
- 4.Peto J, Gilham C, Deacon J, Taylor C, Evans C, Binns W, Haywood M, Elanko N, Coleman D, Yule R, Desai M. Cervical HPV infection and neoplasia in a large population-based prospective study: the Manchester cohort. Br J Cancer. 2004;91:942–53. doi: 10.1038/sj.bjc.6602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munk AC, Gudlaugsson E, Ovestad IT, Lovslett K, Fiane B, Hidle B, Kruse AJ, Skaland I, Janssen EA, Baak JP. Interaction of epithelial biomarkers, local immune response and condom use in cervical intraepithelial neoplasia 2-3 regression. Gynecol Oncol. 2012;127:489–94. doi: 10.1016/j.ygyno.2012.09.010. [DOI] [PubMed] [Google Scholar]
- 6.Ovestad IT, Gudlaugsson E, Skaland I, Malpica A, Munk AC, Janssen EA, Baak JP. The impact of epithelial biomarkers, local immune response and human papillomavirus genotype in the regression of cervical intraepithelial neoplasia grades 2-3. J Clin Pathol. 2011;64:303–7. doi: 10.1136/jcp.2010.083626. [DOI] [PubMed] [Google Scholar]
- 7.Grimm C, Polterauer S, Natter C, Rahhal J, Hefler L, Tempfer CB, Heinze G, Stary G, Reinthaller A, Speiser P. Treatment of cervical intraepithelial neoplasia with topical imiquimod: a randomized controlled trial. Obstet Gynecol. 2012;120:152–9. doi: 10.1097/AOG.0b013e31825bc6e8. [DOI] [PubMed] [Google Scholar]
- 8.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zou J, Yin F, Wang Q, Zhang W, Li L. Analysis of microarray-identified genes and microRNAs associated with drug resistance in ovarian cancer. Int J Clin Exp Pathol. 2015;8:6847–58. [PMC free article] [PubMed] [Google Scholar]
- 10.Davis S, Meltzer PS. GEOquery: a bridge between the Gene Expression Omnibus (GEO) and bioconductor. Bioinformatics. 2007;23:1846–7. doi: 10.1093/bioinformatics/btm254. [DOI] [PubMed] [Google Scholar]
- 11.Liu X, Gao Y, Zhao B, Li X, Lu Y, Zhang J, Li D, Li L, Yin F. Discovery of microarray-identified genes associated with ovarian cancer progression. Int J Oncol. 2015;46:2467–78. doi: 10.3892/ijo.2015.2971. [DOI] [PubMed] [Google Scholar]
- 12.den Boon JA, Pyeon D, Wang SS, Horswill M, Schiffman M, Sherman M, Zuna RE, Wang Z, Hewitt SM, Pearson R, Schott M, Chung L, He Q, Lambert P, Walker J, Newton MA, Wentzensen N, Ahlquist P. Molecular transitions from papillomavirus infection to cervical precancer and cancer: role of stromal estrogen receptor signaling. Proc Natl Acad Sci U S A. 2015;112:E3255–64. doi: 10.1073/pnas.1509322112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, Boutros PC. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics. 2011;12:35. doi: 10.1186/1471-2105-12-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis G Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 18.de Leeuw N, Dijkhuizen T, Hehir-Kwa JY, Carter NP, Feuk L, Firth HV, Kuhn RM, Ledbetter DH, Martin CL, van Ravenswaaij-Arts CM, Scherer SW, Shams S, Van Vooren S, Sijmons R, Swertz M, Hastings R. Diagnostic interpretation of array data using public databases and internet sources. Hum Mutat. 2012;33:930–40. doi: 10.1002/humu.22049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mostafavi S, Ray D, Warde-Farley D, Grouios C, Morris Q. GeneMANIA: a real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008;9(Suppl 1):S4. doi: 10.1186/gb-2008-9-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie D, Sham JS, Zeng WF, Lin HL, Che LH, Wu HX, Wen JM, Fang Y, Hu L, Guan XY. Heterogeneous expression and association of beta-catenin, p16 and c-myc in multistage colorectal tumorigenesis and progression detected by tissue microarray. Int J Cancer. 2003;107:896–902. doi: 10.1002/ijc.11514. [DOI] [PubMed] [Google Scholar]
- 21.Georgieva J, Sinha P, Schadendorf D. Expression of cyclins and cyclin dependent kinases in human benign and malignant melanocytic lesions. J Clin Pathol. 2001;54:229–35. doi: 10.1136/jcp.54.3.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergeron C, Ronco G, Reuschenbach M, Wentzensen N, Arbyn M, Stoler M, von Knebel Doeberitz M. The clinical impact of using p16 (INK4a) immunochemistry in cervical histopathology and cytology: an update of recent developments. Int J Cancer. 2015;136:2741–51. doi: 10.1002/ijc.28900. [DOI] [PubMed] [Google Scholar]
- 23.Chang MS, Oh S, Jung EJ, Park JH, Jeon HW, Lee TS, Kim JH, Choi E, Byeon SJ, Park IA. High-risk human papillomavirus load and biomarkers in cervical intraepithelial neoplasia and cancer. APMIS. 2014;122:427–36. doi: 10.1111/apm.12163. [DOI] [PubMed] [Google Scholar]
- 24.Ma Y, Ye F, Xie X, Zhou C, Lu W. Significance of PTPRZ1 and CIN85 expression in cervical carcinoma. Arch Gynecol Obstet. 2011;284:699–704. doi: 10.1007/s00404-010-1693-9. [DOI] [PubMed] [Google Scholar]
- 25.Liao GD, Sellors JW, Sun HK, Zhang X, Bao YP, Jeronimo J, Chen W, Zhao FH, Song Y, Cao Z, Zhang SK, Xi MR, Qiao YL. p16INK4A immunohistochemical staining and predictive value for progression of cervical intraepithelial neoplasia grade 1: a prospective study in China. Int J Cancer. 2014;134:1715–24. doi: 10.1002/ijc.28485. [DOI] [PubMed] [Google Scholar]
- 26.Chang H, Shi Y, Tuokan T, Chen R, Wang X. Expression of aquaporin 8 and phosphorylation of Erk1/2 in cervical epithelial carcinogenesis: correlation with clinicopathological parameters. Int J Clin Exp Pathol. 2014;7:3928–37. [PMC free article] [PubMed] [Google Scholar]
- 27.Mazumder Indra D, Mitra S, Roy A, Mondal RK, Basu PS, Roychoudhury S, Chakravarty R, Panda CK. Alterations of ATM and CADM1 in chromosomal 11q22.3-23.2 region are associated with the development of invasive cervical carcinoma. Hum Genet. 2011;130:735–48. doi: 10.1007/s00439-011-1015-8. [DOI] [PubMed] [Google Scholar]
- 28.Iwata T, Fujii T, Morii K, Saito M, Sugiyama J, Nishio H, Morisada T, Tanaka K, Yaguchi T, Kawakami Y, Aoki D. Cytokine profile in cervical mucosa of Japanese patients with cervical intraepithelial neoplasia. Int J Clin Oncol. 2015;20:126–33. doi: 10.1007/s10147-014-0680-8. [DOI] [PubMed] [Google Scholar]
- 29.Fukazawa EM, Baiocchi G, Soares FA, Kumagai LY, Faloppa CC, Badiglian-Filho L, Coelho FR, Goncalves WJ, Costa RL, Goes JC. Cox-2, EGFR, and ERBB-2 expression in cervical intraepithelial neoplasia and cervical cancer using an automated imaging system. Int J Gynecol Pathol. 2014;33:225–34. doi: 10.1097/PGP.0b013e318290405a. [DOI] [PubMed] [Google Scholar]
- 30.Samir R, Asplund A, Tot T, Pekar G, Hellberg D. High-risk HPV infection and CIN grade correlates to the expression of c-myc, CD4+, FHIT, E-cadherin, Ki-67, and p16INK4a. J Low Genit Tract Dis. 2011;15:280–6. doi: 10.1097/LGT.0b013e318215170c. [DOI] [PubMed] [Google Scholar]
- 31.Miranda LN, Reginaldo FP, Souza DM, Soares CP, Silva TG, Rocha KB, Jatoba CA, Donadi EA, Andrade JM, Goncalves AK, Crispim JC. Greater expression of the human leukocyte antigen-G (HLA-G) and interleukin-17 (IL-17) in cervical intraepithelial neoplasia: analytical cross-sectional study. Sao Paulo Med J. 2015;133:336–42. doi: 10.1590/1516-3180.2013.7170009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Del Gobbo A, Bonoldi E, Cribiu FM, Franceschetti I, Matinato C, Fiori S, Gianelli U, Bosari S. Insulin-like growth factor II mRNA binding protein 3 (IMP3) expression in cervical intraepithelial neoplasia and its relationship with HIV-infection status. Sex Health. 2014;12:22–4. doi: 10.1071/SH13144. [DOI] [PubMed] [Google Scholar]
- 33.Jung SH, Choi YJ, Kim MS, Baek IP, Lee SH, Lee AW, Hur SY, Kim TM, Lee SH, Chung YJ. Progression of naive intraepithelial neoplasia genome to aggressive squamous cell carcinoma genome of uterine cervix. Oncotarget. 2015;6:4385–93. doi: 10.18632/oncotarget.2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruse AJ, Baak JP, Janssen EA, Bol MG, Kjellevold KH, Fianne B, Lovslett K, Bergh J. Low- and high-risk CIN 1 and 2 lesions: prospective predictive value of grade, HPV, and Ki-67 immuno-quantitative variables. J Pathol. 2003;199:462–70. doi: 10.1002/path.1316. [DOI] [PubMed] [Google Scholar]
- 35.Huang K, Li LA, Meng Y, You Y, Fu X, Song L. High expression of astrocyte elevated gene-1 (AEG-1) is associated with progression of cervical intraepithelial neoplasia and unfavorable prognosis in cervical cancer. World J Surg Oncol. 2013;11:297. doi: 10.1186/1477-7819-11-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rotondo JC, Bosi S, Bassi C, Ferracin M, Lanza G, Gafa R, Magri E, Selvatici R, Torresani S, Marci R, Garutti P, Negrini M, Tognon M, Martini F. Gene expression changes in progression of cervical neoplasia revealed by microarray analysis of cervical neoplastic keratinocytes. J Cell Physiol. 2015;230:806–12. doi: 10.1002/jcp.24808. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima T, Asai-Sato M, Akimoto K, Nagashima Y, Taguri M, Sasaki K, Nakaya MA, Asano R, Tokinaga A, Kiyono T, Hirahara F, Ohno S, Miyagi E. Aberrant expression of the cell polarity regulator aPKClambda/iota is associated with disease progression in cervical intraepithelial neoplasia (CIN): a possible marker for predicting CIN prognosis. Int J Gynecol Pathol. 2016;35:106–17. doi: 10.1097/PGP.0000000000000228. [DOI] [PubMed] [Google Scholar]
- 38.Matsuda K, Miura S, Kurashige T, Suzuki K, Kondo H, Ihara M, Nakajima H, Masuzaki H, Nakashima M. Significance of p53-binding protein 1 nuclear foci in uterine cervical lesions: endogenous DNA double strand breaks and genomic instability during carcinogenesis. Histopathology. 2011;59:441–51. doi: 10.1111/j.1365-2559.2011.03963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhu D, Jiang XH, Jiang YH, Ding WC, Zhang CL, Shen H, Wang XL, Ma D, Hu Z, Wang H. Amplification and overexpression of TP63 and MYC as biomarkers for transition of cervical intraepithelial neoplasia to cervical cancer. Int J Gynecol Cancer. 2014;24:643–8. doi: 10.1097/IGC.0000000000000122. [DOI] [PubMed] [Google Scholar]
- 40.Zhang B, Gui LS, Zhao XL, Zhu LL, Li QW. FOXO1 is a tumor suppressor in cervical cancer. Genet Mol Res. 2015;14:6605–16. doi: 10.4238/2015.June.18.3. [DOI] [PubMed] [Google Scholar]
- 41.Zhou Y, Tao F, Cheng Y, Xu F, Yao F, Feng D, Miao L, Xiao W, Ling B. Up-regulation of ITCH is associated with down-regulation of LATS1 during tumorigenesis and progression of cervical squamous cell carcinoma. Clin Invest Med. 2014;37:E384–94. doi: 10.25011/cim.v37i6.22243. [DOI] [PubMed] [Google Scholar]
- 42.Rung J, Brazma A. Reuse of public genome-wide gene expression data. Nat Rev Genet. 2013;14:89–99. doi: 10.1038/nrg3394. [DOI] [PubMed] [Google Scholar]
- 43.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, Gaasterland T, Glenisson P, Holstege FC, Kim IF, Markowitz V, Matese JC, Parkinson H, Robinson A, Sarkans U, Schulze-Kremer S, Stewart J, Taylor R, Vilo J, Vingron M. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet. 2001;29:365–71. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 44.McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–33. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- 45.Lee YY, Song SY, Do IG, Kim TJ, Kim BG, Lee JW, Bae DS. Dynamin 2 expression as a biomarker in grading of cervical intraepithelial neoplasia. Eur J Obstet Gynecol Reprod Biol. 2012;164:180–4. doi: 10.1016/j.ejogrb.2012.05.029. [DOI] [PubMed] [Google Scholar]
- 46.Wei H, Wang N, Zhang Y, Wang S, Pang X, Zhang S. Wnt-11 overexpression promoting the invasion of cervical cancer cells. Tumour Biol. 2016;37:11789–98. doi: 10.1007/s13277-016-4953-x. [DOI] [PubMed] [Google Scholar]
- 47.Ramos-Solano M, Alvarez-Zavala M, Garcia-Castro B, Jave-Suarez LF, Aguilar-Lemarroy A. [Wnt signalling pathway and cervical cancer] . Rev Med Inst Mex Seguro Soc. 2015;53:S218–24. [PubMed] [Google Scholar]
- 48.Song E, Yu W, Xiong X, Kuang X, Ai Y, Xiong X. Astrocyte elevated gene-1 promotes progression of cervical squamous cell carcinoma by inducing epithelial-mesenchymal transition via Wnt signaling. Int J Gynecol Cancer. 2015;25:345–55. doi: 10.1097/IGC.0000000000000381. [DOI] [PubMed] [Google Scholar]
- 49.Nojima M, Suzuki H, Toyota M, Watanabe Y, Maruyama R, Sasaki S, Sasaki Y, Mita H, Nishikawa N, Yamaguchi K, Hirata K, Itoh F, Tokino T, Mori M, Imai K, Shinomura Y. Frequent epigenetic inactivation of SFRP genes and constitutive activation of Wnt signaling in gastric cancer. Oncogene. 2007;26:4699–713. doi: 10.1038/sj.onc.1210259. [DOI] [PubMed] [Google Scholar]
- 50.Jain S, Singhal S, Lee P, Xu R. Molecular genetics of hepatocellular neoplasia. Am J Transl Res. 2010;2:105–18. [PMC free article] [PubMed] [Google Scholar]
- 51.Deka J, Wiedemann N, Anderle P, Murphy-Seiler F, Bultinck J, Eyckerman S, Stehle JC, Andre S, Vilain N, Zilian O, Robine S, Delorenzi M, Basler K, Aguet M. Bcl9/Bcl9l are critical for Wnt-mediated regulation of stem cell traits in colon epithelium and adenocarcinomas. Cancer Res. 2010;70:6619–28. doi: 10.1158/0008-5472.CAN-10-0148. [DOI] [PubMed] [Google Scholar]
- 52.Nurkkala M, Wassen L, Nordstrom I, Gustavsson I, Slavica L, Josefsson A, Eriksson K. Conjugation of HPV16 E7 to cholera toxin enhances the HPV-specific T-cell recall responses to pulsed dendritic cells in vitro in women with cervical dysplasia. Vaccine. 2010;28:5828–36. doi: 10.1016/j.vaccine.2010.06.068. [DOI] [PubMed] [Google Scholar]
- 53.Musgrove EA, Caldon CE, Barraclough J, Stone A, Sutherland RL. Cyclin D as a therapeutic target in cancer. Nat Rev Cancer. 2011;11:558–72. doi: 10.1038/nrc3090. [DOI] [PubMed] [Google Scholar]
- 54.Ortega S, Malumbres M, Barbacid M. Cyclin D-dependent kinases, INK4 inhibitors and cancer. Biochim Biophys Acta. 2002;1602:73–87. doi: 10.1016/s0304-419x(02)00037-9. [DOI] [PubMed] [Google Scholar]
- 55.Jia WH, Zhang B, Matsuo K, Shin A, Xiang YB, Jee SH, Kim DH, Ren Z, Cai Q, Long J, Shi J, Wen W, Yang G, Delahanty RJ, Ji BT, Pan ZZ, Matsuda F, Gao YT, Oh JH, Ahn YO, Park EJ, Li HL, Park JW, Jo J, Jeong JY, Hosono S, Casey G, Peters U, Shu XO, Zeng YX, Zheng W. Genome-wide association analyses in East Asians identify new susceptibility loci for colorectal cancer. Nat Genet. 2013;45:191–6. doi: 10.1038/ng.2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Du X, Lin LI, Zhang L, Jiang J. microRNA-195 inhibits the proliferation, migration and invasion of cervical cancer cells via the inhibition of CCND2 and MYB expression. Oncol Lett. 2015;10:2639–43. doi: 10.3892/ol.2015.3541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Darragh TM, Colgan TJ, Thomas Cox J, Heller DS, Henry MR, Luff RD, McCalmont T, Nayar R, Palefsky JM, Stoler MH, Wilkinson EJ, Zaino RJ, Wilbur DC Members of the LAST Project Work Groups. The lower anogenital squamous terminology standardization project for HPV-associated lesions: background and consensus recommendations from the college of American pathologists and the American society for colposcopy and cervical pathology. Int J Gynecol Pathol. 2013;32:76–115. doi: 10.1097/PGP.0b013e31826916c7. [DOI] [PubMed] [Google Scholar]
- 58.Tsoumpou I, Arbyn M, Kyrgiou M, Wentzensen N, Koliopoulos G, Martin-Hirsch P, Malamou-Mitsi V, Paraskevaidis E. p16(INK4a) immunostaining in cytological and histological specimens from the uterine cervix: a systematic review and meta-analysis. Cancer Treat Rev. 2009;35:210–20. doi: 10.1016/j.ctrv.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Omori M, Hashi A, Nakazawa K, Yuminamochi T, Yamane T, Hirata S, Katoh R, Hoshi K. Estimation of prognoses for cervical intraepithelial neoplasia 2 by p16INK4a immunoexpression and high-risk HPV in situ hybridization signal types. Am J Clin Pathol. 2007;128:208–17. doi: 10.1309/0UP5PJK9RYF7BPHM. [DOI] [PubMed] [Google Scholar]
- 60.Zhou WQ, Sheng QY, Sheng YH, Hou WJ, Xu GX, Wu YM, Lu H. Expressions of survivin, P16(INK4a), COX-2, and Ki-67 in cervical cancer progression reveal the potential clinical application. Eur J Gynaecol Oncol. 2015;36:62–8. [PubMed] [Google Scholar]
- 61.Baak JP, Kruse AJ, Garland SM, Skaland I, Janssen EA, Tabrizi S, Fagerheim S, Robboy S, Nilsen ST. Combined p53 and retinoblastoma protein detection identifies persistent and regressive cervical high-grade squamous intraepithelial lesions. Am J Surg Pathol. 2005;29:1062–6. [PubMed] [Google Scholar]
- 62.Bergeron C, Ordi J, Schmidt D, Trunk MJ, Keller T, Ridder R. Conjunctive p16INK4a testing significantly increases accuracy in diagnosing high-grade cervical intraepithelial neoplasia. Am J Clin Pathol. 2010;133:395–406. doi: 10.1309/AJCPXSVCDZ3D5MZM. [DOI] [PubMed] [Google Scholar]


