Abstract
Introduction
The expression of MiR-135b-5p was up-regulated while Krüppel-like factor 4 (KLF4) expression was extremely low in human gastric carcinoma (GC) tissues. This study aimed to explore the role of miR-135b-5p in GC cells and its influence on various cell capacity and viability by targeting KLF4.
Material and methods
The dual-luciferase reporter assay was first performed and the target relationship between miR-135b-5p and KLF4 was confirmed. Then three GC cell lines and the human normal gastric epithelial cell line (GES1) were analyzed for the expression level of miR-135b-5p and KLF4 mRNA by RT-qPCR. The BGC-823 GC cell line was chosen for subsequent assays.
Results
The expression of miR-135b-5p and KLF4 was manipulated via transfection. The changes of proliferation, invasion, migration, viability, cycle and apoptosis of GC cells were evaluated by MTS, colony formation assay, transwell assay, wound healing assay and flow cytometry assay, respectively. Overexpression of MiR-135b-5p enhanced viability, proliferation, invasion and migration of GC cells, increased cell viability and reduced cell apoptosis. Replenishing of KLF4 functioned oppositely.
Conclusions
The inhibitory effects of ectopic KLF4 could be attenuated by co-transfection of miR-135b-5p. Collective data suggested that miR-135b-5p has a tumor-promoting role in GC cells via downregulating KLF4. Hence, inhibition of miR-135b-5p could be valuable for treatment of gastric cancer.
Keywords: gastric cancer, Krüppel-like factor 4, MiR-135b-5p, BGC-823
Introduction
A large number of people in the world suffer from gastric cancer (GC), which is characterized by high mortality [1]. As reported, approximately 723,000 deaths occurred in 2012 and this figure accounted for 8.8% of all cancer-related deaths in 2012 [2]. The incidence of GC varies from region to region. Eastern Asia exhibits the highest incidence of GC whereas Northern Africa exhibits the lowest incidence of GC [1]. Several exogenous risk factors for GC have been confirmed: dietary habits, smoking, alcohol consumption, obesity, aging, viruses and bacteria such as Helicobacter pylori [3]. Besides environmental and behavioral factors, a variety of tumor-related genes have also been reported in GC, including both oncogenes and tumor suppressive genes [4]. Surgery is so far the most effective approach for GC and it is usually carried out with chemotherapy or radiotherapy [5]. The 5-year survival rate decreases from 60% to 2% when tumor recurrence, invasion or metastasis occurs [5]. Since at least 15% of GC patients experience tumor metastasis and surgical procedures may fail to improve the survival status of GC patients, studies are conducted to discover alternative therapies that are able to ameliorate this issue [6, 7].
MicroRNAs (miRNAs) are endogenous small non-coding RNA molecules with a length of 18–24 nucleotides and they modulate gene expression as posttranscriptional regulators [8, 9]. MicroRNAs regulate gene expression by controlling mRNA stability, degradation, and gene translation. The discovery of miRNAs was traced back to the early 1990s [10]; however, they were not recognized as biological regulators until the early 2000s [11]. Since many miRNAs are deregulated in various types of cancers, researchers suspected that miRNAs may act as tumor suppressors or oncogenes [12–14]. Several miRNAs are aberrantly expressed in GC tissues [15, 16] and they are likely to be used as biomarkers for GC screening and diagnosis [17]. For example, microRNA-429 functions as a tumor suppressor of GC cells by targeting FSCN1 [18] while microRNA-16a functions as an oncogene in GC, stimulating proliferation and metastasis in vitro and in vivo [19]. Therefore, ongoing research in this area is worthwhile.
MiR-135b-5p is a microRNA which has been suggested to be associated with cancer cell proliferation and migration since overexpression of miR-135b-5p was often discovered in cancer cells [20]. It has also been reported that miR-135b-5p can directly target several genes, including MEF2C [21] and DISC1 [22]. Krüppel-like factor 4 (KLF4) is predicted as an another target of miR-135b-5p and it is characterized as a pluripotency factor [23]. For example, KLF4 is able to regulate cell cycle progression and differentiation, which are important for the corresponding life cycles of some cancer viruses such as HPV [24]. KLF4 is suppressed by miR-10b and downregulation of KLF4 facilitates the proliferation and metastases of osteosarcoma cells [25]. However, in a subset of melanoma, ectopic expression of KLF4 promotes melanoma cell growth and reduces cell apoptosis, suggesting that depletion of KLF4 could help to control melanoma growth [26]. In our research, we confirmed the low expression of KLF4 in GC tissues compared with adjacent tissues. Hence, we attempted to uncover the regulatory relationship between miR-135b-5p and KLF4, which could provide a profound understanding of the corresponding mechanism of GC carcinogenesis.
In this study, sufficient evidence indicated that ectopic miR-135b-5p negatively regulated KLF4 expression, which also contributed to the proliferation, migration and invasion of GC cells. Therefore, downregulation of miR-135b-5p could be a novel strategy for GC clinical treatment.
Material and methods
Patients and tissue specimens
Sixty (33 male and 27 female patients, average age 45) gastric cancer specimens were obtained from patients who were hospitalized in the Department of Gastrointestinal Colorectal and Anal Surgery of China-Japan Union Hospital of Jilin University. Written consent forms were signed by all the patients and this study was approved by the hospital ethics committee (IRB number: CJH5413-16-05-15). All patients were first diagnosed with gastric cancer and treated without radiotherapy or chemotherapy. Pathology was graded in accordance with World Health Organization criteria. All specimens consisted of gastric cancer tissues and adjacent non-tumorous tissues (distance to GC tissues > 5 cm).
Cell culture
Human gastric carcinoma cell lines BGC-823, MKN-28, SGC-7901 and the human normal gastric epithelial cell line (GES1) (BNBio, Beijing, China) were cultured in RPMI 1640 medium with 10% FBS, 100 U/ml penicillin and 100 U/ml streptomycin under standard culture conditions, 2–3 passages.
Cell transfection
MiR-135b-5p mimics and KLF4 cDNA were synthesized by Molbase biological reagent company. After amplification by PCR, KLF4 cDNA was inserted into the pcDNA3.1 plasmid. BGC-823 cells were first seeded in six-well plates and when cell growth reached 70–80% confluence, transfection was performed with the help of Lipofectamine 2000 reagents (Invitrogen, Carlsbad, CA, USA). Cells were harvested at 48 h after transfection. Transfection efficiency was confirmed using RT-qPCR and Western blot.
QRT-PCR
Total RNA was isolated with Trizol reagent (Thermo Fisher Scientific, MA, USA) and was reverse-transcribed into cDNA before RT-qPCR analysis. The miRNA and mRNA expression levels were normalized against U6 and GAPDH respectively and quantified using the 2–ΔΔCt method. The primers are listed in Table I.
Table I.
Primer sequences for RT-PCR
| cDNA | Forward | Reverse |
|---|---|---|
| U6 | 5’-CTCGCTTCGGCAGCACA-3’ | 5’-AACGCTTCACGAATTTGCGT-3’ |
| MiR-135b-5p | 5’-ACACTCCAGCTGGGUAUGGCUUUUCAUUCCU-3’ | |
| GADPH | 5’-TGAAGGTCGGAGTCAACGG-3’ | 5’-CCTGGAAGATGGTGATGGG-3’ |
| KLF4 | 5’- CGGGCTGATGGGCAAGTT-3’ | 5’-GGGCAGGAAGGATGGGTAA-3’ |
Western blots
Proteins were extracted using RIPA lysis (Beyotime, Shanghai, China) and the protein concentrations were quantified with BCA Kit (Sigma-Aldrich, St. Louis, MO, USA). Proteins were transferred from SDS-PAGE gel to PVDF membranes, which were then blocked with 5% nonfat milk and subsequently incubated with KLF4 and GAPDH antibodies (Abcam, Cambridge, MA, USA) at 4°C overnight. Then the membranes were incubated with secondary antibodies (Abcam) for 1 h at room temperature, followed by analysis with enhanced chemiluminescence (ECL) Western blotting detection reagents (GE Healthcare, Little Chalfont, Buckinghamshire, UK).
Dual-luciferase reporter assay
According to TargetScan (http://www.targetscan.org/), KLF4 is one of miR-135b-5p’s targets. Sequences containing the miR-124-3p target region of EDNRB 3′UTR and its mutation (Sangon, Shanghai, China) were inserted into pmirGLO reporting vector (Promega, USA). Then, pmirGLO recombinant vector was co-transfected with miR-124-3p mimics, inhibitor and control oligo in HEK293T cells. Firefly and Renilla luciferase activities were detected consecutively at 48 h after co-transfection.
MTS assay
BGC-823 cells of different groups were seeded in a 96-well plate at a density of 1 × 103 per well. The absorptions of cells was measured at various time point (0, 12, 24, 36, 48, 60, 72 h) using a Cell Titer 96 AQueous One Solution Cell Proliferation Assay (MTS) (Promega, Beijing, China). The cell absorbance at OD 570 nm was measured after staining.
Colony formation assay
Transfected BGC-823 cells were seeded in six-well plates (500 cells per well) and incubated in a standard environment for 2 weeks. Then colonies were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet. Five random fields were photographed and the number of colonies was counted.
Transwell assay
Transwell assays were performed using 8 μm-pore transwell chambers (Corning company) coated with Matrigel (BD Bioscience). 500 μl of medium containing 10% FBS (the attractant) was added into lower chambers, and 100 μl serum-free medium into upper chambers. The transfected BGC-823 cells were plated in the upper chambers. After 48-hour incubation, the cells on the bottom surface of the upper chambers were cleaned with a cotton swab. Invading cells on another surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 30 min. Five random fields under a microscope were photographed and then the cell numbers were counted.
Wound healing assay
At 48 h after transfection, 8 × 105 cells were cultured into 35 mm2 culture dishes and 200 μl sterile pipette tips were used to scratch the culture surface. The wound closure was measured to determine the migration of cells. Photographs were taken at 0 and 24 h. Each experiment was repeated 3 times.
Flow cytometry
For cell cycle analysis, transfected cells were stained with PI using the Cycletest PLUS DNA Reagent Kit (BD Biosciences). The FITC Annexin V Apoptosis Detection Kit (BD Biosciences, Bedford, MA, USA) was used for apoptosis analysis according to the protocol of the manufacturer. Afterwards, a flow cytometer (BD FACS Aria III, San Jose, CA, USA) was applied to count cells in different status.
Statistical analysis
The results were presented as mean ± SD. Normally distributed measurement data were compared with the t test or analysis of variance (ANOVA); otherwise data were compared with the nonparametric rank-sum test. A p-value less than 0.01 or 0.05 was considered to indicate a statistically significant difference.
Results
Expression levels of miR-135b-5p and KLF4 in GC tissues and para-carcinoma normal tissues, GC cells and normal cells
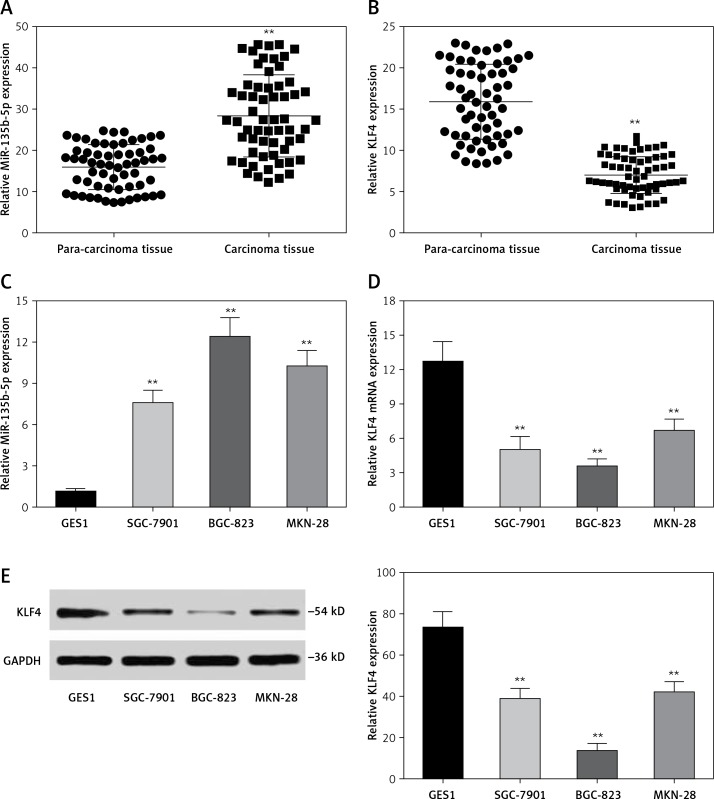
We analyzed the expression level of miR-135b-5p and KLF4 in GC tissues (n = 60) and corresponding para-carcinoma normal tissues using RT-qPCR. The results indicated that miR-135b-5p expression levels in GC tissues were significantly higher than that in para-carcinoma tissues (p < 0.05, Figure 1 A), whereas the KLF4 expression levels in GC tissues were significantly lower than in para-carcinoma tissues (p < 0.05, Figure 1 B). Meanwhile, the results of RT-qPCR showed that miR-135b-5p expression levels in BGC-823, MKN-28 and SGC-7901 GC cell lines were significantly higher compared with that in the GES1 cell line (p < 0.05, Figure 1 C), and miR-135b-5p expression levels in BGC-823 were the most significant. Both RT-qPCR results and western blot results indicated that the KLF4 expression in GC cell lines was remarkably suppressed (Figures 1 D, E).
Figure 1.
MiR-135b-5p was up-regulated in GC tissues and cell lines, KLF4 was down-regulated in GC tissues and cell lines. A, B – Expression level of miR-135b-5p and KLF4 mRNA in GC tissues (n = 60) and para-carcinoma tissues using RT-qPCR. C, D – Expression level of miR-135b-5p and KLF4 mRNA in GC cell lines and normal cell line using RT-qPCR. E – Protein level of KLF4 in GC cell lines and normal cell line detected using western blot
**p < 0.05.
MiR-135b-5p directly targets KLF4 3′UTR
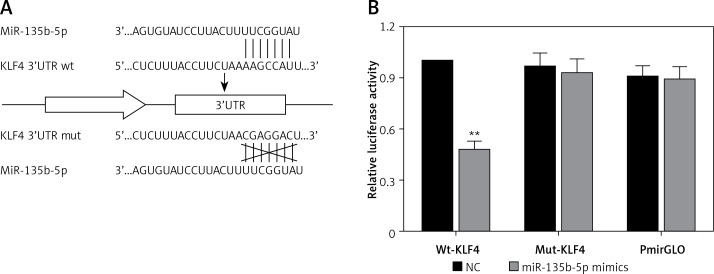
To further investigate the association between miR-135b-5p and KLF4, we first found the potential miR-135b-5p targeting site in the KLF4 3′UTR region using the TargetScan database (Figure 2 A). Wild-type and mutated 3′UTR were synthesized and harbored in the pmirGLO reporting plasmid. Co-transfection was performed in HEK 293T cells and dual luciferase activity was detected at 48 h after transfection. Compared with the control group, luciferase activities of cells co-transfected with miR-135b-5p mimics and KLF4 wild-type 3′UTR-pmirGLO plasmid were remarkably decreased in HEK293T cells (p < 0.05), whereas luciferase activities of cells co-transfected with miR-135b-5p mimics and mutated KLF4 3′UTR pmirGLO plasmid showed no significant differences compared with the control group (Figure 2 B), which demonstrated that KLF4 3′UTR was the target of miR-135b-5p.
Figure 2.
MiR-135b-5p targets KLF4 3′UTR. A – The target relation of miR-135b-5p and KLF4 3′UTR, and the mutated sites in KLF4 3′UTR. B – Luciferase activity of cells co-transfected with miR-135b-5p mimics and the wild type or mutated 3′UTR of KLF4 was detected by luciferase assay
All data are presented as mean ± SD; **p < 0.05.
MiR-135b-5p inhibits expression of KLF4
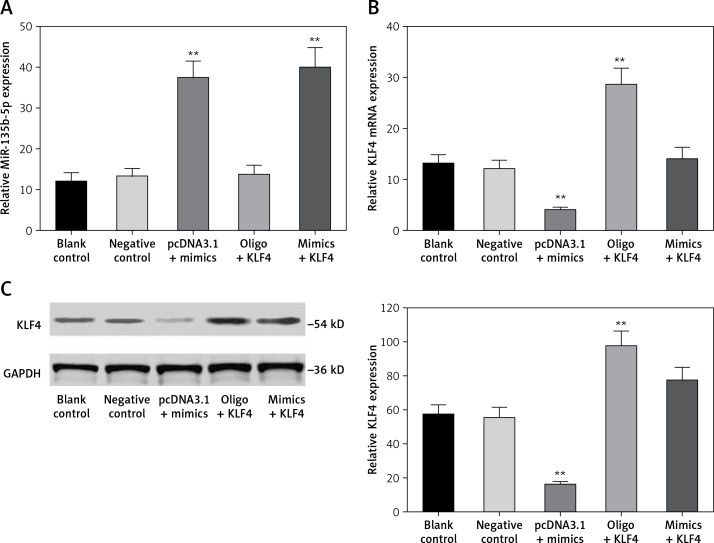
In order to investigate the regulatory relationship between miR-135b-5p and KLF4, we first manipulated the expression of miR-135b-5p and KLF4 in BGC-823 cells via transfection. The cells were divided into the following five groups: the blank control group treated with no reagents, the negative control group transfected with pcDNA3.1 vector and miR mimics control, the pcDNA3.1 + mimics group transfected with pcDNA3.1 vector and miR-135b-5p mimics, the oligo + KLF4 group transfected with miR mimics control and pcDNA3.1-KLF4 cDNA, and the mimics + KLF4 group transfected with miR-135b-5p mimics and pcDNA3.1-KLF4 cDNA. QRT-PCR and western blot were applied to determine the expression of miR-135b-5p and KLF4 and confirm the transfection efficiency. MiR-135b-5p expression significantly increased in the pcDNA3.1 + mimics group and mimics + KLF4 group (p < 0.05, Figure 3 A). There was no significant difference in miR-135b-5p expression among the blank control, negative control and oligo + KLF4 group, indicating that KLF4 has no effect on miR-135b-5p expression. However, both mRNA expression and protein level of KLF4 were repressed in the pcDNA3.1 + mimics group and overexpressed in the oligo + KLF4 group compared to the blank control group (p < 0.05, Figures 3 B, C). Intriguingly, the upregulation of KLF4 by pcDNA3.1-KLF4 cDNA was attenuated by co-transfection of miR-135b-5p in BGC-823 cells. Collective data indicated that miR-135b-5p could directly target KLF4 and suppress its expression.
Figure 3.
MiR-135b-5p inhibits KLF4 expression. A, B – At 48 h after transfection of BGC-823 cells, RT-qPCR was used to analyze the expression level of miR-135b-5p and KLF4 mRNA in blank control group, negative control group, pcDNA3.1 + mimics group, oligo + KLF4 group and mimics + KLF4 group. C – Expression level of KLF4 in all groups was detected using western blot
All data are presented as mean ± SD; **p < 0.05.
MiR-135b-5p modulates biological functions of BGC-823 cells via inhibiting KLF4
Since the regulatory relationship between miR-135b-5p and KLF4 has been confirmed, the effects of the miR-135b-5p/KLF4 axis on gastric cancer cell behaviors which may explain the aberrant expression of miR-135b-5p and KLF4 in gastric cancer tissues and cells drew our attention. Hence, after manipulating miR-135b-5p and KLF4 expression, we accessed the proliferation, migration, invasion, cell cycle and cell apoptosis of transfected cells.
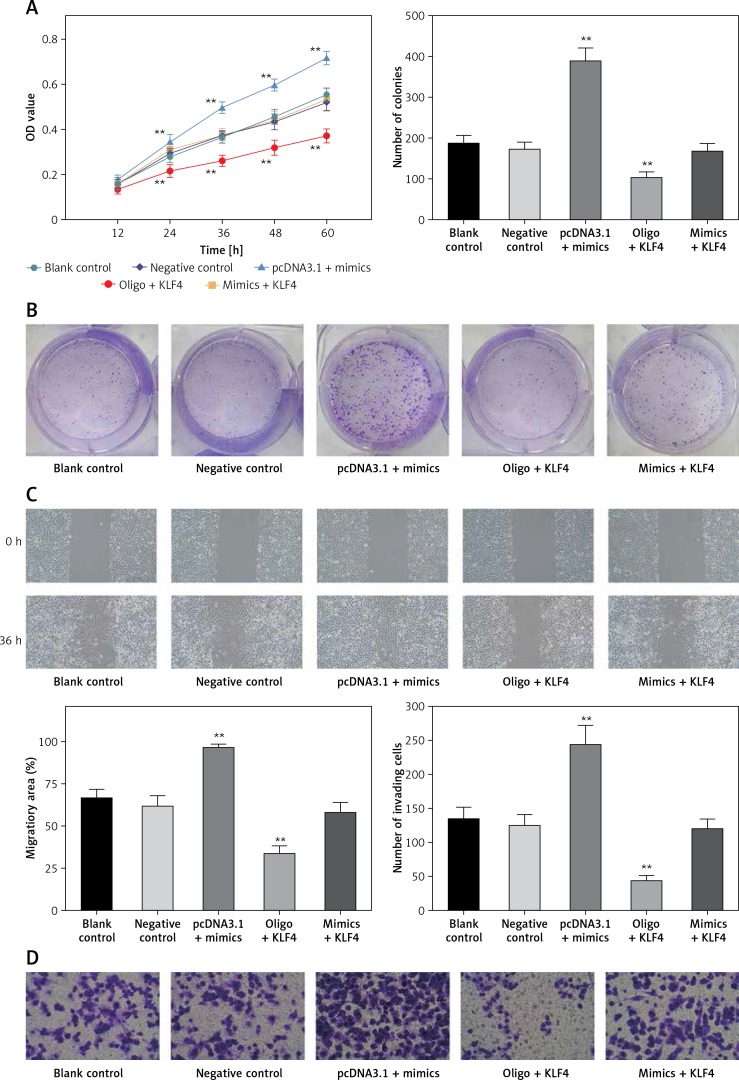
MTS assays suggested that there were no significant differences among blank control, negative control and mimics + KLF4 group. In the 24–72 h period, BGC-823 cells viability significantly increased in the miR-135b-5p high-expressed group and decreased in the KLF4 overexpressed group (p < 0.05, Figure 4 A). Expectedly, overexpression of miR-135b-5p and KLF4 in the mimics + KLF4 group showed a result of antagonism. Similar results were obtained via colony formation assay. At 48 h after transfection, the colony number of different groups was counted (Figure 4 B). Overexpression of miR-135b-5p facilitated colony formation compared to the blank control group while upregulation of KLF4 efficiently repressed the colony number (p < 0.05). The repression was impaired by co-transfection of miR-135b-5p and there was no significant difference in colony number among blank control, negative control and mimics + KLF4 group. All those observations suggested that miR-135b-5p promotes the proliferation of BGC-823 cells via repressing KLF4.
Figure 4.
MiR-135b-5p expression and KLF4 regulated expression affects cell proliferation, migration, invasion capacity of BGC-823 cells. A – MTT assays detected cell viability. B – Colony formation assays analyzed cell proliferation capacity. C – Wound healing assays analyzed cell migration capacity. D – Transwell assays analyzed cell invasion capacity
All data are presented as mean ± SD; **p < 0.05.
Transwell assays and wound healing assays indicated that there were no significant differences between the blank control group and the negative control group (p > 0.05, Figures 4 C, D). Compared with the blank control group, the number of invading cells of the miR-135b-5p high-expressed group increased significantly (p < 0.05), which meant cell invasion capacity was enhanced; the wound closure in pcDNA3.1 + mimics was also larger, suggesting an enhanced cell migratory capacity. However, the number of invading cells of the KLF4 high-expressed group decreased significantly, and the wound closure was smaller (p < 0.05). Still, the weakened cell invading and migratory capacity could be repaired by co-transfection of miR-135b-5p mimics.
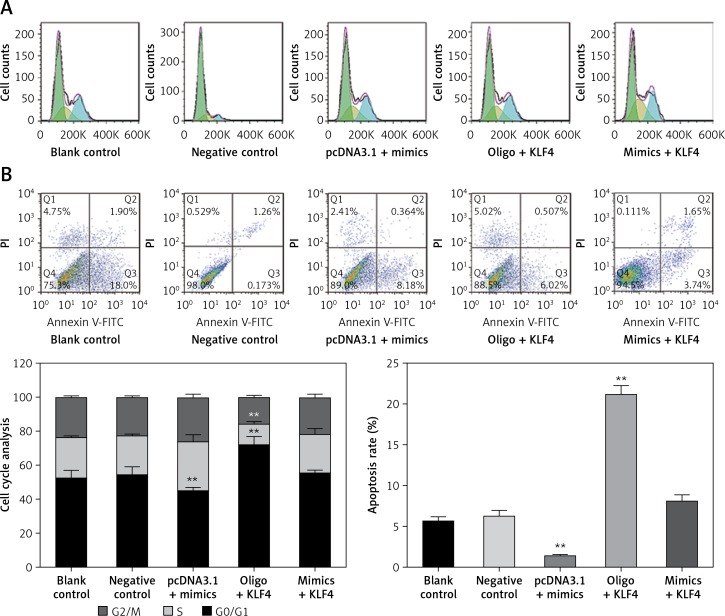
To further study the changes of cell cycle and cell apoptosis rate, flow cytometry and specialized staining were applied. Compared with the control group, the proportion of BGC-823 cells in G0/G1 phase decreased significantly, the proportion of cells in S and G2/Mphases increased significantly, and the cell apoptosis rate decreased significantly in the miR-135b-5p mimics group (p < 0.05, Figures 5 A, B). In the oligo + KLF4 group, the cell proportion of G0/G1 phase increased significantly, the cell proportion of S and G2/Mphases decreased significantly, and the apoptosis rate increased (p < 0.05). Hence, miR-135b-5p could efficiently promote cell mitosis and suppress cell apoptosis while KLF4 plays an opposite role. Then in the mimics + KLF4 group, the inhibitory effects of KLF4 were attenuated by miR-135b-5p. Taken together, the evidence proved that miR-135b-5p promotes cell proliferation, migration, invasion, and mitosis, and reduces the cell apoptosis rate of BGC-823 cells via downregulating KLF4.
Figure 5.
MiR-135b-5p expression and KLF4 regulated expression affects cell cycle and apoptosis of BGC-823 cells. A – After PI staining, flow cytometry was used to analyze cell cycle of transfected cells. B – After PI and Annexin V-FITC staining, flow cytometry was used to count cell apoptosis rate
All data are presented as mean ± SD; **p < 0.05.
Discussion
Accumulating evidence has proven that many miRNAs are altered in gastric cancer cells and lead to the deregulation of various genes, indicating their roles as either oncogenes or tumor suppressors. For instance, miR-524-5p suppressed the growth and invasion of GC cells [27]. MiR-216b was found downregulated in human GC cells, which could inhibit proliferation and cell cycle progression by targeting HDACB [28]. Also, miR-429 functioned as a tumor suppressor in GC cells, downregulating the oncogene FSCN1 [29]. On the other hand, there were studies revealing that miR-532-5p could promote cell growth, migration and invasion as an oncogenic miRNA by directly targeting RUNX3 [30]. In addition, miR-935 was markedly upregulated in GC tissues and cells, and facilitated cell proliferation through targeting SOX7 [31]. These studies showed that the dysregulation of miRNAs may play different roles in GC, working together to regulate the metabolism in cancer cells. In the present study, we found that miR-135b-5p was overexpressed in GC cells, compared with the GES1 cell line. Our experiment results first demonstrated that the overexpression of miR-135b-5p could significantly enhance the proliferation, migration, invasion and mitosis of GC cells. Apoptosis was also suppressed by miR-135b-5p, indicating a tumor-promoting role of miR-135b-5p in gastric cancer cells.
MiR-135b-5p has been found to modulate gene expression in various cancers. For instance, by targeting MEF2C, miR-135b-5p could promote cell proliferation and migration in atherosclerosis [32]; Wang et al. reported that miR-135b-5p could regulate the level of DISC1 mRNA, and they also found that DISC1 regulation was allele specific, i.e. miR-135b-5p only bound to the major allele (A), not to the minor allele (G) in a neuropsychiatric disorder [33]. However, a limited number of studies have been carried out to investigate the interaction of miR-135b and its target genes in gastric cancer progression. Only those by Gao et al. and Liu et al. demonstrated that miR-135b and miR-29c were promising tissue-specific biomarkers of gastric carcinogenesis [34, 35]. Tseng et al. found that miR-135b-5p was upregulated and could be modulated by interference with β-catenin in gastric cancer [36]. The interplay between miR-135b-5p and KLF4 possibly played an important role in gastric cancer development, as suggested by our study described herein.
KLF4 is a critical cellular transcription factor which is able to regulate cell cycle progression and differentiation [37]. It is also known as the gut-enriched Kruppel-like factor, engaging in gastrointestinal epithelial cell proliferation and differentiation, and is found downregulated in gastrointestinal cancers [38]. KLF4 could also promote ghrelin expression by binding to the ghrelin promoter [39]. Althubiti et al. have suggested that the downregulation of KLF4 was related to diffuse-type gastric cancer [40]. Furthermore, it was shown that KLF4 negatively regulates epithelial-mesenchymal transition (EMT) in GI cells, indicating its role in cancer cells as a tumor suppressor gene [41]. Dopamine was also shown to hinder the proliferation of GC cells by upregulating KLF4, which is an inhibitor of the cell cycle, by reducing AKT phosphorylation [42]. Thus, the dysregulation of KLF4 is assumed to be closely related to GC pathogenesis. To be specific, the low level of KLF4 is speculated to lead to the progression of gastric cancers. In line with former studies, we observed low-level expression of KLF4 in GC cells, compared with GES1 cell lines, and replenishment of KLF4 could significantly inhibit the proliferation, migration and invasion and induce G0/G1 arrest and cell apoptosis of BGC-823 cells, indicating that KLF4 had a tumor suppressive effect in GC cells. However, the limitations of this study should be taken into consideration. For example, KLF4 is not the only target of miR-135b-5p, and similarly miR-135b-5p is not the only miRNA which regulates KLF4. Thus because of the complicated regulation network, our study just contributed to the comprehensiveness of the gastric cancer mechanism.
In conclusion, we have proved that miR-135b-5p is overexpressed in GC cells while KLF4 expression is inhibited, supplementing the previous studies. Furthermore, the overexpression of miR-135b-5p can promote the proliferation, migration as well as invasion of GC cells, suggesting that miR-135b-5p may serve as an onco-miRNA. More importantly, we have demonstrated that there is a negative relationship between miR-135b-5p and KLF4, which has never been studied before, showing the possibility that the knockout of miR-135b-5p in GC tumor cells may serve as a therapeutic strategy by overexpressing anti-oncogenes such as KLF4. However, further in vivo studies need to be carried out to investigate the influence of the miR-135b-5p/KLF4 axis on gastric cancer.
Acknowledgments
This study was partly supported by the Industry Technology Research and Development Project of Jilin Province of China (2013C014-2), the Scientific and Technological Development Project of Jilin Province of China (20140204028YY), and the Industry Technology Research and Development Project of Jilin Province of China (2014Y083).
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–8. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380:2095–128. doi: 10.1016/S0140-6736(12)61728-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang F, Meng W, Wang B, Qiao L. Helicobacter pylori-induced gastric inflammation and gastric cancer. Cancer Lett. 2014;345:196–202. doi: 10.1016/j.canlet.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 4.Yasui W, Oue N, Kuniyasu H, Ito R, Tahara E, Yokozaki H. Molecular diagnosis of gastric cancer: present and future. Gastric Cancer. 2001;4:113–21. doi: 10.1007/pl00011733. [DOI] [PubMed] [Google Scholar]
- 5.Global Burden of Disease Cancer Collaboration; Fitzmaurice C, Allen C, Barber RM, et al. Global, regional, and national cancer incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 cancer groups, 1990 to 2015: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2016;3:524–48. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhong D, Ran J, Zhang X, et al. Syntenin is expressed in human gliomas and may correlate with tumor migration. Arch Med Sci. 2015;11:1303–7. doi: 10.5114/aoms.2015.49212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen W, Zheng R, Zhang S, Zhao P, Zeng H, Zou X. Report of cancer incidence and mortality in China, 2010. Ann Transl Med. 2014;2:61. doi: 10.3978/j.issn.2305-5839.2014.04.05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hao K, Luk JM, Lee NP, et al. Predicting prognosis in hepatocellular carcinoma after curative surgery with common clinicopathologic parameters. BMC Cancer. 2009;9:389. doi: 10.1186/1471-2407-9-389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barr M., Jr The long arms of anencephaly: a refutation. Birth Defects Res A Clin Mol Teratol. 2009;85:710–4. doi: 10.1002/bdra.20591. [DOI] [PubMed] [Google Scholar]
- 10.Ji J, Shi J, Budhu A, et al. MicroRNA expression, survival, and response to interferon in liver cancer. N Engl J Med. 2009;361:1437–47. doi: 10.1056/NEJMoa0901282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Overwater A, Kessels K, Elias SG, et al. Endoscopic resection of high-risk T1 colorectal carcinoma prior to surgical resection has no adverse effect on long-term outcomes. Gut. 2018;67:284–90. doi: 10.1136/gutjnl-2015-310961. [DOI] [PubMed] [Google Scholar]
- 12.Zhao JJ, Chen PJ, Duan RQ, Li KJ, Wang YZ, Li Y. miR-630 functions as a tumor oncogene in renal cell carcinoma. Arch Med Sci. 2016;12:473–8. doi: 10.5114/aoms.2016.59918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Royo H, Cavaille J. Non-coding RNAs in imprinted gene clusters. Biol Cell. 2008;100:149–66. doi: 10.1042/BC20070126. [DOI] [PubMed] [Google Scholar]
- 14.Martianov I, Ramadass A, Serra Barros A, Chow N, Akoulitchev A. Repression of the human dihydrofolate reductase gene by a non-coding interfering transcript. Nature. 2007;445:666–70. doi: 10.1038/nature05519. [DOI] [PubMed] [Google Scholar]
- 15.Schmitz KM, Mayer C, Postepska A, Grummt I. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–9. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong R, Zhang EB, Yin DD, et al. Long noncoding RNA PVT1 indicates a poor prognosis of gastric cancer and promotes cell proliferation through epigenetically regulating p15 and p16. Mol Cancer. 2015;14:82. doi: 10.1186/s12943-015-0355-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen ZZ, Huang L, Wu YH, Zhai WJ, Zhu PP, Gao YF. LncSox4 promotes the self-renewal of liver tumour-initiating cells through Stat3-mediated Sox4 expression. Nat Commun. 2016;7:12598. doi: 10.1038/ncomms12598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gong C, Maquat LE. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3’ UTRs via Alu elements. Nature. 2011;470:284–8. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu TP, Liu XX, Xia R, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–61. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 20.Tsang WP, Ng EK, Ng SS, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010;31:350–8. doi: 10.1093/carcin/bgp181. [DOI] [PubMed] [Google Scholar]
- 21.Zhu M, Chen Q, Liu X, et al. lncRNA H19/miR-675 axis represses prostate cancer metastasis by targeting TGFBI. FEBS J. 2014;281:3766–75. doi: 10.1111/febs.12902. [DOI] [PubMed] [Google Scholar]
- 22.Li X, Wang H, Yao B, Xu W, Chen J, Zhou X. lncRNA H19/miR-675 axis regulates cardiomyocyte apoptosis by targeting VDAC1 in diabetic cardiomyopathy. Sci Rep. 2016;6:36340. doi: 10.1038/srep36340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lv J, Fan HX, Zhao XP, et al. Long non-coding RNA unigene 56159 promotes epithelial-mesenchymal transition by acting as a ceRNA of miR-140-5p in hepatocellular carcinoma cells. Cancer Lett. 2016;382:166–75. doi: 10.1016/j.canlet.2016.08.029. [DOI] [PubMed] [Google Scholar]
- 24.Wu T, Qu L, He G, et al. Regulation of laryngeal squamous cell cancer progression by the lncRNA H19/miR-148a-3p/DNMT1 axis. Oncotarget. 2016;7:11553–66. doi: 10.18632/oncotarget.7270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Wang B, Chen LQ, et al. miR-10b promotes invasion by targeting KLF4 in osteosarcoma cells. Biomed Pharmacother. 2016;84:947–53. doi: 10.1016/j.biopha.2016.09.108. [DOI] [PubMed] [Google Scholar]
- 26.Riverso M, Montagnani V, Stecca B. KLF4 is regulated by RAS/RAF/MEK/ERK signaling through E2F1 and promotes melanoma cell growth. Oncogene. 2017;36:3322–33. doi: 10.1038/onc.2016.481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun M, Nie F, Wang Y, et al. LncRNA HOXA11-AS promotes proliferation and invasion of gastric cancer by scaffolding the chromatin modification factors PRC2, LSD1, and DNMT1. Cancer Res. 2016;76:6299–310. doi: 10.1158/0008-5472.CAN-16-0356. [DOI] [PubMed] [Google Scholar]
- 28.Xu Z, Wang X, Shen X, Luo C, Wu P, Zeng H. One-stage lumbopelvic fixation in the treatment of lumbosacral junction tuberculosis. Eur Spine J. 2015;24:1800–5. doi: 10.1007/s00586-015-3863-8. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell JS, Li N, Weinhold N, et al. Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Nat Commun. 2016;7:12050. doi: 10.1038/ncomms12050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan L, Sun M, Liu GJ, et al. Long noncoding RNA PVT1 promotes non-small cell lung cancer cell proliferation through epigenetically regulating LATS2 expression. Mol Cancer Ther. 2016;15:1082–94. doi: 10.1158/1535-7163.MCT-15-0707. [DOI] [PubMed] [Google Scholar]
- 31.Xu MD, Wang Y, Weng W, et al. A positive feedback loop of lncRNA-PVT1 and FOXM1 facilitates gastric cancer growth and invasion. Clin Cancer Res. 2017;23:2071–80. doi: 10.1158/1078-0432.CCR-16-0742. [DOI] [PubMed] [Google Scholar]
- 32.Zheng J, Yu F, Dong P, et al. Long non-coding RNA PVT1 activates hepatic stellate cells through competitively binding microRNA-152. Oncotarget. 2016;7:62886–97. doi: 10.18632/oncotarget.11709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, Yuan JH, Wang SB, et al. Oncofetal long noncoding RNA PVT1 promotes proliferation and stem cell-like property of hepatocellular carcinoma cells by stabilizing NOP2. Hepatology. 2014;60:1278–90. doi: 10.1002/hep.27239. [DOI] [PubMed] [Google Scholar]
- 34.Gao SB, Xu B, Ding LH, et al. The functional and mechanistic relatedness of EZH2 and menin in hepatocellular carcinoma. J Hepatol. 2014;61:832–9. doi: 10.1016/j.jhep.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 35.Liu H, Liu Y, Liu W, Zhang W, Xu J. EZH2-mediated loss of miR-622 determines CXCR4 activation in hepatocellular carcinoma. Nat Commun. 2015;6:8494. doi: 10.1038/ncomms9494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tseng YY, Moriarity BS, Gong W, et al. PVT1 dependence in cancer with MYC copy-number increase. Nature. 2014;512:82–6. doi: 10.1038/nature13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo W, Zhu X, Liu W, et al. MYC associated zinc finger protein promotes the invasion and metastasis of hepatocellular carcinoma by inducing epithelial mesenchymal transition. Oncotarget. 2016;7:86420–32. doi: 10.18632/oncotarget.13416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461–8. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–14. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Althubiti M, Rada M, Samuel J, et al. BTK Modulates p53 activity to enhance apoptotic and senescent responses. Cancer Res. 2016;76:5405–14. doi: 10.1158/0008-5472.CAN-16-0690. [DOI] [PubMed] [Google Scholar]
- 41.Gong B, Wang Z, Zhang M, et al. Disturbed P53-MDM2 feedback loop contributes to thoracic aortic dissection formation and may be a result of TRIM-25 overexpression. Ann Vasc Surg. 2017;40:243–51. doi: 10.1016/j.avsg.2016.07.091. [DOI] [PubMed] [Google Scholar]
- 42.Harris SL, Levine AJ. The p53 pathway: positive and negative feedback loops. Oncogene. 2005;24:2899–908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]