Abstract
A few new papers report that mutations in some genes belonging to the group 3 of plant sulfate transporter family result in low phytic acid phenotypes, drawing novel strategies and approaches for engineering the low-phytate trait in cereal grains. Here, we shortly review the current knowledge on phosphorus/sulfur interplay and sulfate transport regulation in plants, to critically discuss some hypotheses that could help in unveiling the physiological links between sulfate transport and phosphorus accumulation in seeds.
Keywords: sulfate transporters, phytic acid, sulfur, phosphorous
1. Background
Phytic acid (PA)—the major phosphorus (P) store in seeds—cannot be digested by humans and monogastric animals who lack the digestive enzyme phytase. For this reason, almost 90% of phytate consumed by humans is excreted, contributing to eutrophication of rivers, lakes, and oceans [1]. Furthermore, high levels of PA largely prevent the absorption of essential metals in the intestine, thus reducing further the nutritional value of the seeds [2,3].
In the last decades, several approaches have been proposed to solve the seed PA-related problems, including the engineering of crops for high phytase activity in seeds, or the selection of suitable low phytic acid (lpa) genotypes for crop breeding [4]. Today, numerous lpa genotypes have been identified and studied in several major crops, including maize, barley, wheat, rice, soybean, and common bean, reveling several mutations and alleles that could be potentially useful for breeding. However, a large part of the lpa phenotypes is caused by mutations in genes involved in PA biosynthesis or compartmentalization and often results in undesirable pleiotropic effects on yield-related traits and agronomic performances, since PA and inositol phosphates play pivotal roles in a plethora of developmental and signaling processes [4,5]. As a result, the use of these genetic resources to engineer seed PA content has proven to be challenging. Most recent advances in this research topic revealed that mutations in some members of the sulfate transporter gene family might result in lpa phenotypes. Unfortunately, little data are available to explain such effects fully or to develop new strategies for engineering seed PA content. Trying to fill this gap, here, we shortly review the current knowledge on plant sulfate transporters, trying to provide a glimpse into the complex and, in many respects, unexpected connections among the regulatory layers of sulfur (S) and P homeostasis in plants.
2. Sulfate Transporters: A Short Overview
S is an essential nutrient for plants. It is found in the amino acids cysteine and methionine, which are essential components of proteins and peptides, in vitamins and cofactors, and in a plethora of secondary compounds. S plays important and critical roles in a wide variety of cellular processes involved in plant development and response to environmental changes [6,7,8,9].
Sulfate (SO42−) ions in the rhizosphere are the major source of S for plants. They are absorbed by roots and then allocated to different sinks by mean of specific sulfate transporters (SULTRs). The oxidized S atom in SO42− is then reduced and assimilated into cysteine, before entering other metabolic pathways, or directly used for sulfation reactions [9,10,11]. SULTRs are classified as H+/SO42− co-transporters, are integrated into membranes by 12 membrane-spanning domains, and contain a carboxyl-terminal region, named STAS (Sulfate Transporter/AntiSigma-factor), which is thought to be critical for both activity and stability of the transporters, as well as for their interaction with other proteins [9,12,13,14].
A multigene family encodes plant SULTRs. In the best-characterized species—Arabidopsis thaliana and, to a lesser extent, rice (Oryza sativa L.)—12 SULTR genes have been reported [14,15]. SULTRs can be divided into four functional groups or subfamilies, according to their amino acid sequences. The members of each group have specialized functions for SO42− uptake and distribution within the cells and among plant organs, as indicated by their different tissue and subcellular localization, and regulation pathways.
Group 1 of the family encodes high-affinity SULTRs. Two members of this group, SULTR1;1 and SULTR1;2, are mainly expressed in the outermost cell layers of the root (root hairs, epidermis, and cortex), where they largely contribute in determining the rate of SO42− uptake. Arabidopsis sultr1;1sultr1;2 double-knockout lines are severely impaired in growth and unable to take up SO42− at low external concentrations [16,17,18,19]. Although these transporters seem to share the same function, they are differently regulated to fulfill the plant demand for S-containing compounds under different SO42− availabilities or soil conditions. In the currently accepted model, SULTR1;2 is thought to be the major component of the SO42− uptake system under normal S supply, whereas SULTR1;1 should play a most significant role under S deficiency or during other stresses [16,17,20,21,22].
Sulfate ions absorbed by root are translocated to shoot throughout the xylem and then distributed to different sink organs and tissues. It has been proposed that SULTR2;1, a low-affinity SULTR expressed in pericycle and xylem parenchyma, may play a pivotal role in controlling the amount of SO42− available to be loaded into the xylem, by acting as a scavenger reabsorbing the excess of the anion in the apoplastic space inside the root stele. Under S starvation, the increase in the transcript level of SULTR2;1 could help in maintaining adequate fluxes of SO42− directed to the xylem [16,23]. It is important to note that a local expression of SULTR2;1 has also been observed in the xylem parenchyma and phloem cells of the leaves, and that it is not possible to rule out that SULTR2;1 transcript is also expressed below detection levels in the phloem companion cells of the root [16,24].
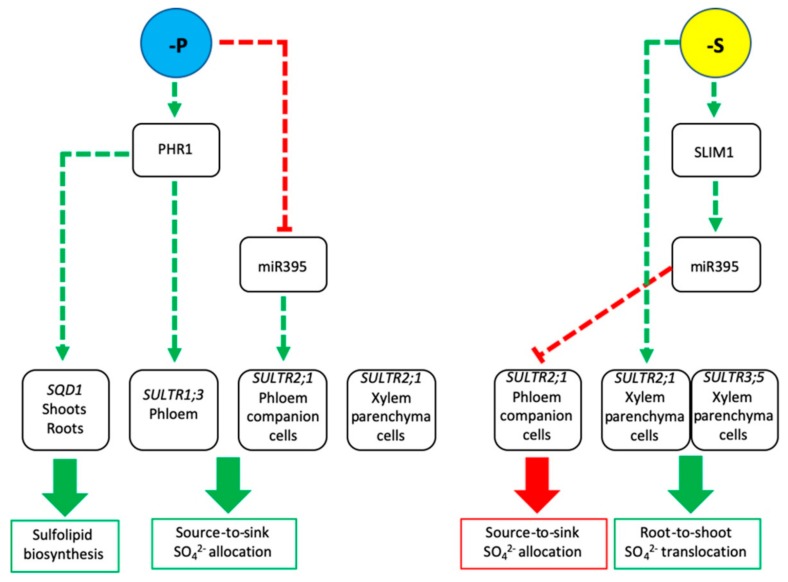
An interesting regulatory circuit controls SO42− translocation and partitioning at the post-transcriptional level (Figure 1). The SULTR2;1 mRNA is targeted and degraded by the miRNA-395 (miR395), which accumulates under S deficiency mainly in the companion cells of the phloem of both root and shoot [24]. The induction of miR395 is, in turn, activated by SLIM1/EIL3 (SULFUR LIMITATION 1/ETHYLENE-INSENSITIVE3-LIKE3), a major regulator gene belonging to the EIL family transcription factors, which controls the expression of several S-responsive genes [25,26]. The mechanism by which miR395 controls SULTR2;1 transcript level is not conventional, since the accumulation of both miR395 and SULTR2;1 mRNA is induced under S starvation. However, the non-overlapping spatial expression domains of the two transcripts allows miR395 to restrict the expression of SULTR2;1 to the xylem parenchyma cells of the root, thus inhibiting long-distance SO42− transport to sink tissues via the phloem and facilitating, at the same time, xylem SO42− translocation to the leaves [24,25].
Figure 1.
Main regulatory circuits controlling SO42− distribution in response to P or S status. Under S deficiency, the induction of SULTR2;1, in xylem parenchyma cells, and miR395, in phloem companion cells, enhances root-to-shoot SO42− translocation. In this condition, the co-expression of SULTR3,5 could help the activity of SULTR2;1 in reabsorbing the excess of SO42− in the apoplastic space of the root. Under P deficiency, an extra regulatory circuit involving PHR1 allows changes in SO42− to support sulfolipids biosynthesis.
Another low-affinity SULTR belonging to group 2, SULTR2;2, seems to be involved in controlling the source-to-sink distribution of SO42− inside the plant. Localization analyses indicate that SULTR2;2 may play a role in the transport of SO42− via root phloem, as well as in the distribution of the anion from leaf vasculature to the leaf palisade and mesophyll, which are thought to be the primary sites for SO42− assimilation [16]. Finally, long-distance transport of SO42− from source to sink organs could also involve SULTR1;3, a high-affinity SULTR of group 1, as indicated by the peculiar expression of this transporter in sieve elements and companion cells of the phloem [27].
Inside the cells, SO42− is further transported into the vacuole and chloroplast/plastid, where it is compartmentalized as S store or reduced and assimilated into cysteine for further metabolic processes, respectively. To date, tonoplast proteins mediating vacuolar SO42− influx have not been identified. On the other hand, SULTR4;1 and SULTR4;2 are known to be involved in downloading SO42− from the vacuoles under S limiting conditions [28].
Recently, all five members of group 3 have been indicated as redundantly involved in SO42− uptake across the chloroplast envelope membrane [29,30]. However, these observations do not appear to be conclusive, since several other functions could be postulated for these transporters on the base of observations that are crucial for our dissertation about the hypothetical links between SULTRs and lpa phenotypes. It is important to note that if, on the one hand, reasonable uncertainties about the capacity of both SULTR1s and SULTR2s to selectively move SO42− do not exist, on the other, no direct evidence has been provided about the actual SO42− transport activity of most of the SULTR3 subfamily members [14]. A few papers indeed indicate that both substrate preference and subcellular localization of some SULTR3s could be different than expected.
Kataoka et al. [31] reported that SULTR3;5 is expressed in the root vasculature of Arabidopsis—showing the same expression domain of the low-affinity SULTR2;1—and subcellular localizes on the plasma membrane. The heterologous expression of SULTR3;5 in yeasts defective for SO42− uptake shows that this protein does not transport SO42− itself, whereas it enhances the SO42− uptake capacity of SULTR2;1 when co-expressed in the same yeast mutant. These results, along with the observation that the Arabidopsis sultr3;5 mutant retains more SO42− in the root under S starvation, strongly suggest that SULTR3;5 may have the function to help SULTR2;1 in retrieval apoplastic SO42−, contributing in this way to root-to-shoot SO42− translocation (Figure 1).
SULTR3;4 from rice and Arabidopsis have been recently indicated as SULTR-like phosphorus distribution transporters (SPDTs) playing essential roles in controlling the allocation of phosphate to grains and developing tissues, respectively [32,33]. Tissue-specific expression analyses show that SULTR3;4/SPDT of rice is expressed in the xylem region of both enlarged- and diffuse-vascular bundles of nodes [32]. The Arabidopsis ortholog gene shows a more complex expression pattern, since it is mainly expressed in the fascicular cambium between the xylem and phloem and in the interfascicular cambium of lower stem, as well as in the cambial zone of the leaf petiole, rosette basal region, hypocotyl, and in the parenchyma cells of both xylem and phloem surrounding the cambial zone [33]. Moreover, the SULTR3;4/SPDTs are localized at the plasma membrane, show proton-dependent transport activities for phosphate, do not transport SO42−, and are up-regulated by phosphate deficiency but not under S starvation [32,33]. Mutations in OsSULTR3;4/SPDT alter the distribution of P in rice plants, decreasing both total P (−20%) and phytate (−30%) in the brown de-husked grains, without affecting yield, seed germination, and seedling vigor.
Another member of group 3, SULTR3;3, has been indicated as implicated in PA accumulation in barley and rice grains. Zhao et al. [34] recently reported that disruptions in rice SULTR3;3 gene are the casual events of two interesting allelic mutations, previously described as lpa-MH86-1 and Os-lpa-Z9B-1, since they produce grains with a reduced concentration of both PA and total P [35]. Tissue-specific expression analyses reveal that OsSULTR3;3 is expressed in the vascular bundles of shoots, leaves, flowers, and seeds, but not in the roots. This protein seems to be localized in the endoplasmic reticulum, when expressed in onion epidermal cells, and it does not show any transport activity for both SO42− and phosphate when heterologously expressed in yeast mutant strains defective for SO42− or phosphate uptake, or Xenopus oocytes. However—as underlined by Zhao et al. [34]—the lack of transport activity for SO42− or phosphate in heterologous systems does not necessarily mean that OsSULTR3;3 does not have a role in SO42− of phosphate transport, since its activity may depend on other proteins or post-translational modifications not present in non-plant hosts. Moreover, OsSULTR3;3 mutations affect the concentrations of total P and phosphate of both root and shoot—which result higher in the mutants than in the wild type—but also reduce the concentrations of SO42− in the same organs. Finally, transcriptional analyses performed on developing grains reveal that OsSULTR3;3 disruptions are associated with significant changes in the transcript level of genes involved in S and P homeostasis, suggesting a possible role of this gene in the cross-talk between the two nutrients [34]. Interestingly, a single base pair substitution in the last exon of an ortholog gene of OsSULTR3;3 (designed as HvST) has also been identified as the causal event for the low phytic acid phenotype of the lpa1-1 barley mutants [36].
Taken as a whole, these findings strongly indicate that expression domains and subcellular localizations, as well as substrate preferences of the SULTR3 subfamily members are variable, and may depend on plant species, development stage, or experimental approaches used to study their functions. Further efforts will be necessary to understand better whether this variability could play a role in the regulation of SO42− fluxes under different environmental conditions, also concerning the level of other essential mineral nutrients.
3. Sulfur and Phosphorous Interplay
Similar to S, P is also an essential macronutrient for plants. P is found as phosphate ester in the majority of the molecular constituents essential for plant cell functions, including nucleic acids, proteins, phospholipids, sugars, ATP, and NADPH. Important aspects related to P acquisition and homeostasis in plants have been recently reviewed elsewhere [37,38,39]. Here, we mainly focus our attention on S and P interplay by analyzing specific aspects related to SO42− transport and distribution inside the plants.
Although it is clear that S or P deficiencies have diverse phenotypic effects on plant growth, development, and productivity, intriguing interconnected responses to the internal levels of these two nutrients have been described at metabolic and transcriptional levels, suggesting the existence of coordination between S and P homeostasis. Rouached [40] pointed out that deficiency or surplus of only one of the two nutrients often results in changes in the expression levels of genes specifically involved in controlling the homeostasis of the other nutrient and underlined as comparable molecular mechanisms regulate both SO42− and phosphate transport in plants.
At the metabolic level, one of the most evident relationships between S and P is linked to membrane composition. It is known that cells can replace sulfolipids by phospholipids under S starvation, as well as they are able to replace phospholipids by sulfolipids and/or galactolipids under P starvation [41,42,43,44,45,46]. In Arabidopsis, the synthesis of sulfolipids is catalyzed by two enzymes, SQD1 and SQD2, whose expressions are increased by P starvation [42,43]. Although lipid shifts could be interpreted as adaptive mechanisms for plant survival under different nutrient availabilities, the physiological and biochemical consequences of phospholipids-sulfolipids substitutions on plant membrane functions are still unclear. Reprogramming membrane compositions under nutrient deficiency could have profound impacts on both S and P availability for plant metabolism. Moreover, recent studies have shown that the lipid environment and lipid-protein interactions may have crucial roles in modulating the functions as well as the conformational dynamics of membrane transporters [47].
Unfortunately, our basic knowledge about the interactions between P metabolism and SO42− transport is limited. A few papers show that P deficiency or perturbations in P metabolism may impact the SO42− allocation inside the plants. It has been reported that SO42− concentration increases in roots and decreases in shoots of Arabidopsis as a consequence of reduced phosphate availabilities in the growing medium [48]. Transcriptional analysis of the main SULTR genes implicated in long-distance SO42− transport reveals that, under P starvation, the transcript of AtSULTR1;3 accumulates in both roots and shoots, whereas that of AtSULTR2;1 weakly accumulates only in the roots. In the same conditions, AtSQD1 transcript increases in both roots and shoots, indicating that adaptive modulations of SULTRs controlling the inter-organ distribution of SO42− are required for the replacement of phospholipid by sulfolipids induced by P starvation [48]. Most of these responses seem to be dependent on PHR1 (PHOSPHATE RESPONSE1), a gene encoding a protein belonging to the MYB-CC family transcription factors involved in the activation of several phosphate starvation-induced genes (PSI) [38,49]. PHR1 binds to an imperfect palindromic motif, named P1BS, which is prevalent in the promoter of the PSI genes [49,50]. Interestingly cis-regulatory motifs for PHR1-dependent gene activation are also present in the promoters of both AtSULTR1;3 and AtSQD1 genes, whose expressions are coherently reduced in the Arabidopsis phr1 mutant grown under P starvation [48,51,52]. Interestingly, other evidence indicates PHR1 as the convergent point for the cross-talk between P and other essential nutrients, such as zinc and iron [53,54]. AtSULTR2;1, is up-regulated by P starvation in a PHR1-independent manner, since AtSULTR2;1 transcript further accumulates in phr1 P deficient plants [48]. In this context, the observation that the expression of miR395—the microRNA that mainly controls the spatial expression of SULTR2;1 in vascular tissues—is suppressed under P deficiency, allows us to speculate about the existence of an extra regulatory circuit which controls the inter-organ distribution of SO42− under P starvation [55]. In this circuit (Figure 1): (i) the suppression of miR395 should allow SULTR2;1 to control root-to-shoot SO42− translocation via the xylem route, as well as the source-to-sink SO42− re-allocation via the phloem; (ii) PHR1 activates the expression of SULTR1;3 increasing further the capacity of the plants to move SO42− from source to sink tissues. Unfortunately, no other evidence is available to support this extra regulatory circuit further and to fully appreciate its possible physiological impact on S metabolism in P deficient plants. Finally, the observation that Arabidopsis lines engineered for low PA content show alterations in SO42− distribution and changes in expression of some SULTRs suggests the existence of another level of complexity in the cross-talk between S and P, which directly involves PA [56].
4. SULTRs as Novel Elements in the lpa Network
As mentioned above, genetic lesions in some genes putatively involved in sulfate transport result in lpa phenotypes in rice and barley [32,34,36]. Interestingly, all the mutations described thus far affect putative SULTR genes belonging to the SULTR3 subfamily, which includes elements whose functions are still objects of debate. For a detailed description of the SULTR3/lpa alleles, readers are referred to Cominelli et al. [57].
Differently from other SULTRs, whose capability to transport SO42− has mostly been proven using yeast mutants as heterologous expression systems, the function of the SULTR3s as SO42− transporters has only been hypothesized on the base of their sequence homologies with other SULTRs. Moreover, species-specific differences could explain the variability observed for the subcellular membrane localization of SULTR3 subfamily members.
Mineral nutrients required for plant growth are absorbed by the roots from the soil solution and then released to the xylem to be translocated to different tissues together with the transpiration flow. However, transpiration cannot be considered as the sole driving force for the root-to-shoot movement of nutrients, since developing organs such as new leaves and seeds are not photosynthetically active. Recently, nodes of gramineous plants have been identified as the main actors controlling nutrient delivery to developing tissues in a transpiration-independent way [58,59]. Several rice transporters involved in the intervascular transfer of nutrients from enlarged vascular bundles to diffuse vascular bundles of nodes seem to be essential to ensure this process [59]. Among these, OsSULTR3;4/SPDT has been indicated as pivotal in controlling phosphate delivery to developing tissues since it shows a proton-dependent transport activity for phosphate (but not for SO42−), and it is highly expressed in the node 1 of rice at the reproductive stage [32]. Moreover, OsSULTR3;4/SPDT knockout mutants reduce P allocation to new leaves and grains, raveling the essential role of this transporter in switching phosphate toward developing leaves and grains.
The recent finding that the Arabidopsis SULTR3;4 ortholog gene also controls xylem-to-phloem phosphate transfer, strongly suggests that sequence homology of SULTR3;4 with other SULTRs does not necessarily indicate that they share the same function [33]. All these observations not only show SULTR3;4s as phosphate transporters rather than as SO42− transporters, but may also explain the role of these proteins in the lpa network.
If, on the one hand, the recent description of SULTR3;4/SPDTs as phosphate transporters seems to leave no room for doubt, on the other, assessment of the role of SULTR3;3s on P allocation still appears challenging. Rice SULTR3;3 is mainly expressed in vascular tissues and does not show any transport activity for SO42− and phosphate [34]. Further studies are thus needed to uncover its function and subcellular localization. However, the lack of specific transport activity for SO42− has also been indicated for the Arabidopsis SULTR3;5, which has been described as an essential component of the SO42− transport system that facilitates the root-to-shoot SO42− translocation in the vasculature [31]. Although the mechanisms controlling SO42− allocation in rice are still known, it is possible to speculate that also OsSULT3;3 could have a role in SO42− partitioning among organs, by helping the activity of some other vascular transporter, or in SO42− transport into the chloroplast, as recently suggested for its ortholog in Arabidopsis [30]. Rice sultr3;3 mutants show significant alterations in S and P homeostasis, as indicated by the reduced concentration of SO42− in both shoots and roots, as well as by the accumulation of transcripts of several S- and P-responsive genes in developing grains. Disruption in OsSULTR3;3 also affects the concentrations of various grain metabolites not directly involved in PA biosynthesis. In particular, the reduced level of cysteine, along with the accumulation of its precursor serine, seems to indicate an insufficient supply of S during seed differentiation. Interestingly, reduced levels of cysteine have also been observed in the chloroplasts isolated from different Arabidopsis sultr3 mutants [30]. Thus, the alterations in S homeostasis could be interpreted as the primary physiological event that reduces the accumulation of P in the grains of sultr3;3 mutants. Finally, since total P and phosphate concentrations in root and shoot are higher in mutants than in the wild type, we may further speculate about the existence of mechanisms that somehow limit the systemic mobility of P in the plant. The analysis of the membrane lipid composition could provide in the next future a possible explanation for this phenomenon since substitution of sulfolipids by phospholipids caused by an insufficient S supply could increase the amount of P immobilized within cell membranes.
5. Conclusions and Perspectives
The implication of SULTRs in seed P accumulation not only provides novel opportunities to design routes for the breading of new lpa varieties in important cereal crops but also reveals the existence of a complex network of interactions between S and P homeostasis. The recent finding showing the involvement of SULTR3;4/SPTDs in delivering phosphate, and not sulfate, to developing tissues opens new questions about the nature of the other members of the SULTR3 subfamily [32,33]. Further investigations aimed at determining their substrate preference between sulfate and phosphate are then essential to unveil the actual role of these transporters in the control of nutrient homeostasis. In this context, the recent study of Cao et al. [29], suggesting that all the Arabidopsis SULTR3 homologs may redundantly mediate sulfate import into the chloroplast, needs to be carefully reconsidered since chloroplasts isolated from the sultr3 quintuple mutant retain about 50% of the sulfate uptake capacity of the wild type. Redundancy versus diversity will be the novel challenge to face.
Acknowledgments
The music of Gustav Mahler’s V Symphony inspired this work. We would like to thank Maurizio Cocucci and Silvia Morgutti for their precious support during the writing of the manuscript.
Author Contributions
G.A.S. and F.F.N. equally contributed in discussing this opinion. F.F.N. wrote the manuscript. G.A.S. and F.F.N. revised and approved the final version of the opinion.
Funding
This work received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bohn L., Meyer A.S., Rasmussen S.K. Phytate: Impact on environment and human nutrition. A challenge for molecular breeding. J. Zhejiang Univ. Sci. B. 2008;9:165–191. doi: 10.1631/jzus.B0710640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raboy V. Seeds for a better future: “Low phytate”, grains help to overcome malnutrition and reduce pollution. Trends Plant Sci. 2001;6:458–462. doi: 10.1016/S1360-1385(01)02104-5. [DOI] [PubMed] [Google Scholar]
- 3.Schlemmer U., Frølich W., Prieto R.M., Grases F. Phytate in foods and significance for humans: Food sources, intake, processing, bioavailability, protective role and analysis. Mol. Nutr. Food Res. 2009;53:S330–S375. doi: 10.1002/mnfr.200900099. [DOI] [PubMed] [Google Scholar]
- 4.Raboy V. Approaches and challenges to engineering seed phytate and total phosphorus. Plant Sci. 2009;177:281–296. doi: 10.1016/j.plantsci.2009.06.012. [DOI] [Google Scholar]
- 5.Sparvoli F., Cominelli E. Seed biofortification and phytic acid reduction: A conflict of interest for the plant? Plants. 2015;4:728–755. doi: 10.3390/plants4040728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Leustek T., Martin M.N., Bick J.A., Davies J.P. Pathways and regulation of sulfur metabolism revealed through molecular and genetic studies. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2000;51:141–165. doi: 10.1146/annurev.arplant.51.1.141. [DOI] [PubMed] [Google Scholar]
- 7.Saito K. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiol. 2004;136:2443–2450. doi: 10.1104/pp.104.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nocito F.F., Lancilli C., Giacomini B., Sacchi G.A. Sulfur metabolism and cadmium stress in higher plants. Plant Stress. 2007;1:142–156. [Google Scholar]
- 9.Takahashi H., Kopriva S., Giordano M., Saito K., Hell R. Sulfur assimilation in photosynthetic organisms: Molecular functions and regulations of transporters and assimilatory enzyme. Annu. Rev. Plant Biol. 2011;62:157–184. doi: 10.1146/annurev-arplant-042110-103921. [DOI] [PubMed] [Google Scholar]
- 10.Mugford S.G., Lee B.R., Koprivova A., Matthewman C., Kopriva S. Control of sulfur partitioning between primary and secondary metabolism. Plant J. 2011;65:96–105. doi: 10.1111/j.1365-313X.2010.04410.x. [DOI] [PubMed] [Google Scholar]
- 11.Koprivova A., Kopriva S. Sulfation pathways in plants. Chem. Biol. Interact. 2016;259:23–30. doi: 10.1016/j.cbi.2016.05.021. [DOI] [PubMed] [Google Scholar]
- 12.Shibagaki N., Grossman A.R. The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J. Biol. Chem. 2006;281:22964–22973. doi: 10.1074/jbc.M603462200. [DOI] [PubMed] [Google Scholar]
- 13.Shibagaki N., Grossman A.R. Binding of cysteine synthase to the STAS domain of sulfate transporter and its regulatory consequences. J. Biol. Chem. 2010;285:25094–25102. doi: 10.1074/jbc.M110.126888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi H. Sulfate transport systems in plants: Functional diversity and molecular mechanisms underlying regulatory coordination. J. Exp. Bot. 2019;70:4075–4087. doi: 10.1093/jxb/erz132. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S., Asif M.H., Chakrabarty D., Tripathi R.D., Trivedi P.K. Differential expression and alternative splicing of rice sulphate transporter family members regulate sulphur status during plant growth, development and stress conditions. Funct. Integr. Genom. 2011;11:259–273. doi: 10.1007/s10142-010-0207-y. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi H., Watanabe-Takahashi A., Smith F.W., Blake-Kalff M., Hawkesford M.J., Saito K. The roles of three functional sulphate transporters involved in uptake and translocation of sulphate in Arabidopsis thaliana. Plant J. 2000;23:171–182. doi: 10.1046/j.1365-313x.2000.00768.x. [DOI] [PubMed] [Google Scholar]
- 17.Yoshimoto N., Takahashi H., Smith F.W., Yamaya T., Saito K. Two distinct high-affinity sulfate transporters with different inducibilities mediate uptake of sulfate in Arabidopsis roots. Plant J. 2002;29:465–473. doi: 10.1046/j.0960-7412.2001.01231.x. [DOI] [PubMed] [Google Scholar]
- 18.Yoshimoto N., Inoue E., Watanabe-Takahashi A., Saito K., Takahashi H. Posttranscriptional regulation of high-affinity sulfate transporters in Arabidopsis by sulfur nutrition. Plant Physiol. 2007;145:378–388. doi: 10.1104/pp.107.105742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barberon M., Berthomieu P., Clairotte M., Shibagaki N., Davidian J.C., Gosti F. Unequal functional redundancy between the two Arabidopsis thaliana high-affinity sulphate transporters SULTR1;1 and SULTR1;2. New Phytol. 2008;180:608–619. doi: 10.1111/j.1469-8137.2008.02604.x. [DOI] [PubMed] [Google Scholar]
- 20.El Kassis E., Cathala N., Rouached H., Fourcroy P., Berthomieu P., Terry N., Davidian J.C. Characterization of a selenate-resistant Arabidopsis mutant. Root growth as a potential target for selenate toxicity. Plant Physiol. 2007;143:1231–1241. doi: 10.1104/pp.106.091462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rouached H., Wirtz M., Alary R., Hell R., Arpat A.B., Davidian J.C., Fourcroy P., Berthomieu P. Differential regulation of the expression of two high-affinity sulfate transporters, SULTR1.1 and SULTR1.2, in Arabidopsis. Plant Physiol. 2008;147:897–911. doi: 10.1104/pp.108.118612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferri A., Lancilli C., Maghrebi M., Lucchini G., Sacchi G.A., Nocito F.F. The sulfate supply maximizing Arabidopsis shoot growth is higher under long- than short-term exposure to cadmium. Front. Plant Sci. 2017;8:854. doi: 10.3389/fpls.2017.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maruyama-Nakashita A., Watanabe-Takahashi A., Inoue E., Yamaya T., Saito K., Takahashi H. Sulfur-responsive elements in the 3’-nontranscribed intergenic region are essential for the induction of SULFATE TRANSPORTER 2;1 gene expression in Arabidopsis roots under sulfur deficiency. Plant Cell. 2015;27:1279–1296. doi: 10.1105/tpc.114.134908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawashima C.G., Yoshimoto N., Maruyama-Nakashita A., Tsuchiya Y.N., Saito K., Takahashi H., Dalmay T. Sulphur starvation induces the expression of microRNA-395 and one of its target genes but in different cell types. Plant J. 2009;57:313–321. doi: 10.1111/j.1365-313X.2008.03690.x. [DOI] [PubMed] [Google Scholar]
- 25.Kawashima C.G., Matthewman C.A., Huang S., Lee B.R., Yoshimoto N., Koprivova A., Rubio-Somoza I., Todesco M., Rathjen T., Saito K., et al. Interplay of SLIM1 and miR395 in the regulation of sulfate assimilation in Arabidopsis. Plant J. 2011;66:863–876. doi: 10.1111/j.1365-313X.2011.04547.x. [DOI] [PubMed] [Google Scholar]
- 26.Maruyama-Nakashita A., Nakamura Y., Tohge T., Saito K., Takahashi H. Arabidopsis SLIM1 is a central transcriptional regulator of plant sulfur response and metabolism. Plant Cell. 2006;18:3235–3251. doi: 10.1105/tpc.106.046458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshimoto N., Inoue E., Saito K., Yamaya T., Takahashi H. Phloem-localizing sulfate transporter, Sultr1;3, mediates re-distribution of sulfur from source to sink organs in Arabidopsis. Plant Physiol. 2003;131:1511–1517. doi: 10.1104/pp.014712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kataoka T., Watanabe-Takahashi A., Hayashi N., Ohnishi M., Mimura T., Buchner P., Hawkesford M.J., Yamaya T., Takahashi H. Vacuolar sulfate transporters are essential determinants controlling internal distribution of sulfate in Arabidopsis. Plant Cell. 2004;16:2693–2704. doi: 10.1105/tpc.104.023960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cao M.J., Wang Z., Wirtz M., Hell R., Oliver D.J., Xiang C.B. SULTR3;1 is a chloroplast-localized sulfate transporter in Arabidopsis thaliana. Plant J. 2013;73:607–616. doi: 10.1111/tpj.12059. [DOI] [PubMed] [Google Scholar]
- 30.Chen Z., Zhao P.X., Miao Z.Q., Qi G.F., Wang Z., Yuan Y., Ahmad N., Cao M.J., Hell R., Wirtz M., et al. SULTR3s function in chloroplast sulfate uptake and affect ABA biosynthesis and the stress response. Plant Physiol. 2019;180:593–604. doi: 10.1104/pp.18.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kataoka T., Hayashi N., Yamaya T., Takahashi H. Root-to-shoot transport of sulfate in Arabidopsis. Evidence for the role of SULTR3;5 as a component of low-affinity sulfate transport system in the root vasculature. Plant Physiol. 2004;136:4198–4204. doi: 10.1104/pp.104.045625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yamaji N., Takemoto Y., Miyaji T., Mitani-Ueno N., Yoshida K.T., Ma J.F. Reducing phosphorus accumulation in rice grains with an impaired transporter in the node. Nature. 2017;541:92–95. doi: 10.1038/nature20610. [DOI] [PubMed] [Google Scholar]
- 33.Ding G., Lei G.J., Yamaji N., Yokosho K., Mitani-Ueno N., Huang S., Ma J.F. Vascular cambium-localized AtSPDT mediates xylem-to-phloem transfer of phosphorus for its preferential distribution in Arabidopsis. Mol. Plant. 2019 doi: 10.1016/j.molp.2019.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Zhao H., Frank T., Tan Y., Zhou C., Mabnoune M., Arpat A.B., Cui H., Huang J., He Z., Poirier Y., et al. Disruption of OsSULTR3;3 reduces phytate and phosphorus concentrations and alters the metabolite profile in rice grains. New Phytol. 2016;211:926–939. doi: 10.1111/nph.13969. [DOI] [PubMed] [Google Scholar]
- 35.Liu Q.L., Xu X.H., Ren X.L., Fu H.W., Wu D.X., Shu Q.Y. Generation and characterization of low phytic acid germplasm in rice (Oryza sativa L.) Theor. Appl. Genet. 2007;114:803–814. doi: 10.1007/s00122-006-0478-9. [DOI] [PubMed] [Google Scholar]
- 36.Ye H., Zhang X.Q., Broughton S., Westcott S., Wu D., Lance R., Li C. A nonsense mutation in a putative sulphate transporter gene results in low phytic acid in barley. Funct. Integr. Genom. 2011;11:103–110. doi: 10.1007/s10142-011-0209-4. [DOI] [PubMed] [Google Scholar]
- 37.Wang F., Deng M., Xu J., Zhu X., Mao C. Molecular mechanisms of phosphate transport and signaling in higher plants. Semin. Cell Dev. Biol. 2018;74:114–122. doi: 10.1016/j.semcdb.2017.06.013. [DOI] [PubMed] [Google Scholar]
- 38.Ham B.K., Chen J., Yan Y., Lucas W.J. Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 2018;49:1–9. doi: 10.1016/j.copbio.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Veneklaas E.J., Lambers H., Bragg J., Finnegan P.M., Lovelock S.E., Plaxton W.C., Price C.A., Scheible W.R., Shane M.W., White P.J., et al. Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 2012;195:306–320. doi: 10.1111/j.1469-8137.2012.04190.x. [DOI] [PubMed] [Google Scholar]
- 40.Rouached H. Multilevel coordination of phosphate and sulfate homeostasis in plants. Plant Signal. Behav. 2011;6:952–955. doi: 10.4161/psb.6.7.15318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Härtel H., Essigmann B., Lokstein H., Hoffmann-Benning S., Peters-Kotting M., Benning C. The phospholipid-deficient pho1 mutant of Arabidopsis thaliana is affected in the organization, but not in the light acclimation, of the thylakoid membrane. Biomembranes. 1998;1415:205–218. doi: 10.1016/S0005-2736(98)00197-7. [DOI] [PubMed] [Google Scholar]
- 42.Essigmann B., Guler S., Narang R.A., Linke D., Benning C. Phosphate availability affects the thylakoid lipid composition and the expression of SQD1, a gene required for sulfolipid biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA. 1998;95:1950–1955. doi: 10.1073/pnas.95.4.1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu B., Xu C., Benning C. Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc. Natl. Acad. Sci. USA. 2002;99:5732–5737. doi: 10.1073/pnas.082696499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sugimoto K., Sato N., Tsuzuki M. Utilization of a chloroplast membrane sulfolipid as a major internal sulfur source for protein synthesis in the early phase of sulfur starvation in Chlamydomonas reinhardtii. FEBS Lett. 2007;581:4519–4522. doi: 10.1016/j.febslet.2007.08.035. [DOI] [PubMed] [Google Scholar]
- 45.Tjellström H., Andersson M.X., Larsson K.E., Sandelius A.S. Membrane phospholipids as a phosphate reserve: The dynamic nature of phospholipid-to-digalactosyl diacylglycerol exchange in higher plants. Plant Cell Environ. 2008;31:1388–1398. doi: 10.1111/j.1365-3040.2008.01851.x. [DOI] [PubMed] [Google Scholar]
- 46.Sugimoto K., Tsuzuki M., Sato N. Regulation of synthesis and degradation of a sulfolipid under sulfur-starved conditions and its physiological significance in Chlamydomonas reinhardtii. New Phytol. 2010;185:676–686. doi: 10.1111/j.1469-8137.2009.03115.x. [DOI] [PubMed] [Google Scholar]
- 47.Martens C., Shekhar M., Borysik A.J., Lau A.M., Reading E., Tajkhorshid E., Booth P.J., Politis A. Direct protein-lipid interactions shape the conformational landscape of secondary transporters. Nat. Commun. 2018;9:4151. doi: 10.1038/s41467-018-06704-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rouached H., Secco D., Arpat B., Poirier Y. The transcription factor PHR1 plays a key role in the regulation of sulfate shoot-to-root flux upon phosphate starvation in Arabidopsis. BMC Plant Biol. 2011;11:19. doi: 10.1186/1471-2229-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rubio V., Linhares F., Solano R., Martín A.C., Iglesias J., Leyva A., Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001;15:2122–2133. doi: 10.1101/gad.204401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bustos R., Castrillo G., Linhares F., Puga M.I., Rubio V., Pérez J., Solano R., Leyva A., Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010;6:e1001102. doi: 10.1371/journal.pgen.1001102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Franco-Zorrilla J.M., Gonzalez E., Bustos R., Linhares F., Leyva A., Paz-Ares J. The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 2004;55:285–293. doi: 10.1093/jxb/erh009. [DOI] [PubMed] [Google Scholar]
- 52.Stefanovic A., Ribot C., Rouached H., Wang Y., Chong J., Belbahri L., Delessert S., Poirier Y. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007;50:982–994. doi: 10.1111/j.1365-313X.2007.03108.x. [DOI] [PubMed] [Google Scholar]
- 53.Khan G.A., Bouraine S., Wege S., Li Y., de Carbonnel M., Berthomieu P., Poirier Y., Rouached H. Coordination between zinc and phosphate homeostasis involves the transcription factor PHR1, the phosphate exporter PHO1, and its homologue PHO1;H3 in Arabidopsis. J. Exp. Bot. 2014;65:871–884. doi: 10.1093/jxb/ert444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bournier M., Tissot N., Mari S., Boucherez J., Lacombe E., Briat J.F., Gaymard F. Arabidopsis ferritin 1 (AtFer1) gene regulation by the phosphate starvation response 1 (AtPHR1) transcription factor reveals a direct molecular link between iron and phosphate homeostasis. J. Biol. Chem. 2013;288:22670–22680. doi: 10.1074/jbc.M113.482281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hsieh L., Lin S., Shih A., Chen J., Lin W., Tseng C., Lin W.Y., Tseng C.Y., Li H.Y., Tzyy-Jen Chiou T.J. Uncovering small RNA-mediated responses to phosphate-deficiency in Arabidopsis by deep sequencing. Plant Physiol. 2009;151:2120–2132. doi: 10.1104/pp.109.147280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belgaroui N., Zaidi I., Farhat A., Chouayekh H., Bouain N., Chay S., Curie C., Mari S., Masmoudi K., Davidian J.C., et al. Over-expression of the bacterial phytase US417 in Arabidopsis reduces the concentration of phytic acid and reveals its involvement in the regulation of sulfate and phosphate homeostasis and signaling. Plant Cell Physiol. 2004;55:1912–1924. doi: 10.1093/pcp/pcu122. [DOI] [PubMed] [Google Scholar]
- 57.Cominelli E., Pilu R., Sparvoli F. Phytic acid, transporters and the generation of low phytic acid mutants in grain crops. Plants. 2019 doi: 10.3390/plants9010069. submitted for publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamaji N., Ma J.F. The node, a hub for mineral nutrient distribution in graminaceous plants. Trends Plant Sci. 2014;19:556–563. doi: 10.1016/j.tplants.2014.05.007. [DOI] [PubMed] [Google Scholar]
- 59.Yamaji N., Ma J.F. Node-controlled allocation of mineral elements in Poaceae. Curr. Opin. Plant Biol. 2017;39:18–24. doi: 10.1016/j.pbi.2017.05.002. [DOI] [PubMed] [Google Scholar]