Key Points
PCNSL has a unique molecular profile distinct from that of systemic DLBCL.
BCL6 rearrangements are associated with a poor prognosis in PCNSL.
Abstract
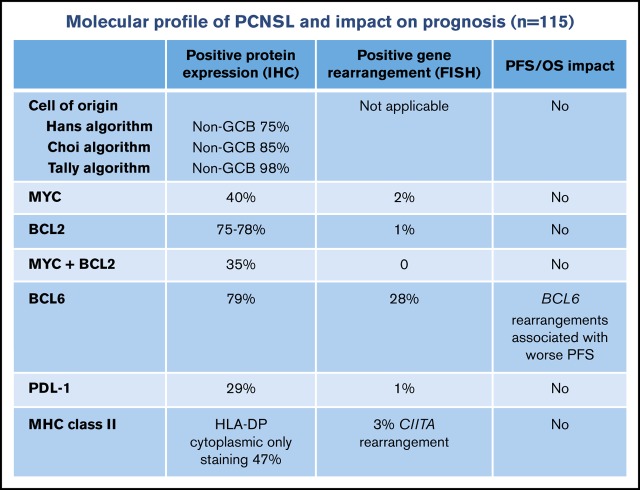
The objective of this study was to evaluate the distribution and prognostic impact of a broad range of molecular attributes in a large cohort of immunocompetent patients with primary central nervous system lymphoma (PCNSL) by using tissue microarray. Patients diagnosed with PCNSL were initially identified in the BC Cancer Lymphoid Cancer clinical and pathology databases. Tissue microarrays were constructed by using archival formalin-fixed paraffin-embedded diagnostic biopsy tissue. Immunohistochemistry and fluorescent in situ hybridization studies were performed. A total of 115 patients with PCNSL with diffuse large B-cell lymphoma (DLBCL) histology were identified. The majority of cases (≥75%) had a non–germinal center B-cell phenotype according to immunohistochemistry algorithms, but cell of origin did not affect progression-free or overall survival. MYC (40%), BCL2 (75%), and programmed death-ligand 1 (29%) protein expression were common, but their corresponding gene rearrangements were rare (≤1% each), suggesting that alternate mechanisms were driving expression. There were no dual rearrangements involving MYC and BCL2. Only 22% of cases had membranous expression of major histocompatibility complex class II, suggesting a mechanism for escape from immune surveillance. Epstein-Barr virus–encoded RNA was positive in 1 immunocompetent patient. BCL6 protein expression (77%) and BCL6 rearrangements (31%) were frequent; the latter was the only factor associated with a poor prognosis in the overall cohort and in the subgroup of 52 patients treated with high-dose methotrexate–based regimens. This large population-based study shows that prominent molecular features of PCNSL are unique and different from those of systemic DLBCL. These results may better inform drug development in PCNSL.
Visual Abstract
Introduction
Primary diffuse large B-cell lymphoma (DLBCL) of the central nervous system (CNS), also known as primary CNS lymphoma (PCNSL), is an aggressive non-Hodgkin lymphoma that exclusively involves the CNS, including brain parenchyma, leptomeninges, or intraocular regions. Several retrospective studies performed over the past decade suggest that the biology of PCNSL is unique and different from that of systemic DLBCL.1-3 However, the pathogenesis of PCNSL remains poorly understood, in part due to its relative rarity but also because CNS biopsies are often stereotactic needle biopsies, small surgical biopsies, or obtained after a course of corticosteroids and may therefore not yield sufficient material for analysis.
Analyzing a broad range of molecular abnormalities in a large cohort of uniformly treated patients is necessary to understand the biology of PCNSL. From a prognostic standpoint, phenotypic and genotypic factors associated with outcomes in systemic DLBCL such as cell of origin (COO) or aberrations in MYC/BCL2/BCL6 may not necessarily be applicable to PCNSL. From a treatment perspective, molecular profiling of PCNSL could help select patients for specific therapies, especially in the era of noncytotoxic novel agents.4 The objective of the current study was to evaluate the distribution and prognostic impact of a broad range of molecular attributes in a large cohort of unselected immunocompetent patients with newly diagnosed PCNSL by using tissue microarray (TMA).
Materials and methods
Patient identification
Patients with a brain biopsy result showing a B-cell non-Hodgkin lymphoma between 1998 and 2010 were initially identified in the BC Cancer Centre for Lymphoid Cancer clinical and pathology databases. Archival formalin-fixed paraffin-embedded diagnostic biopsy tissue was retrieved, and TMAs were constructed. All brain biopsy samples were centrally reviewed by a BC Cancer hematopathologist at the time of TMA construction if a central review for clinical purposes had not been performed previously. Central pathology review reports and medical records were subsequently examined to verify the diagnosis of PCNSL with DLBCL morphology and to obtain clinical and treatment data before inclusion in the current analysis. Patients without PCNSL, including those with non-DLBCL morphology and secondary CNS relapse of systemic DLBCL, were excluded. HIV-positive patients were also excluded.
The majority of patients underwent contrast-enhanced computed tomography and/or magnetic resonance imaging of the head, chest, abdomen, and pelvis. Deep brain lesions were defined as those localized to the periventricular region, corpus callosum, basal ganglia, brainstem, or cerebellum.5 Ocular slit-lamp examinations and cerebrospinal fluid analyses were obtained when feasible; positron emission tomography scans were not routinely performed, however.
Treatment
During the study period, intravenous methotrexate-based chemotherapy regimens were recommended for patients with adequate renal function and otherwise good performance status. The MIDVAP regimen was used between 1988 and 1999, and it included methotrexate 1 g/m2 together with doxorubicin, vincristine, procarbazine, dexamethasone, and whole-brain radiotherapy (WBRT) with 35 Gy in 20 fractions.6 Single-agent high-dose methotrexate (HDMTX) 8 g/m2 followed by leucovorin rescue was introduced in January 2000, and the addition of intravenous rituximab 375 mg/m2 together with the first 4 doses of HDMTX was introduced in 2006. Treatment modality was switched to WBRT in cases of significant methotrexate toxicity or insufficient response.7,8 Consolidative high-dose chemotherapy and autologous stem cell transplantation were not routinely recommended during the study period. Patients who were not considered candidates for intravenous chemotherapy were treated with WBRT alone or best supportive care (BSC), the latter category including corticosteroids.
TMAs, immunohistochemistry, and in situ hybridization
TMAs were constructed by using duplicate 0.6-mm diameter cores from diagnostic formalin-fixed paraffin-embedded tissue biopsies of CNS tissue. Immunohistochemistry (IHC) was performed on 4-μm TMA sections using routine protocols on a fully automated Ventana BenchMark XT system (Ventana Medical Systems, Tucson, AZ). IHC staining was evaluated independently by 2 to 3 hematopathologists (K.L.T., G.W.S., and M.A.M.), and discrepancies were resolved by 1 to 2 hematopathologists (G.W.S. and R.D.G.).
COO was determined by estimating protein expression in malignant cells using IHC in 10% increments for CD10 (Novocastra 56C6; ≥30% expression positive), BCL6 (Novocastra LN22; ≥30% expression positive), MUM1 (Dako MUM1p; ≥30% expression positive for the Hans algorithm or ≥80% for the Choi algorithm), FOXP1 (Fischer JC12; ≥ 80% expression positive), LMO2 (Santa Cruz 1A9-1; ≥30% expression positive), and GCET1 (Abcam RMA341; ≥80% expression positive). COO was then assigned by using 3 different IHC algorithms: Hans, Choi, and Tally.9-11
Additional IHC included MYC (Epitomics Y69; ≥40% expression positive), BCL2 (Dako 124 and Epitomics E17; ≥50% expression positive), and programmed death-ligand 1 (PD-L1) (Dako 22C3; ≥1% expression positive) on tumor cells. Expression of the HLA DPQR antigens of major histocompatibility complex (MHC) II by IHC (Dako CR3/43; ≥10% expression positive) was performed as previously described. The pattern of staining was further described as restricted to cytoplasm and/or cell membrane.12,13
To determine Epstein-Barr virus infection status of the large lymphoid cells, in situ hybridization for Epstein-Barr virus–encoded RNA (EBER; Ventana Medical Systems) was performed on 4-μm TMA sections using routine protocols on a fully automated Ventana Benchmark XT system. Staining in ≥80% of malignant cells was considered positive.
Fluorescence in situ hybridization
Fluorescence in situ hybridization (FISH) was performed as previously described.14 Separate break-apart assays were performed for MYC, BCL2, and BCL6 using the LSI MYC, BCL2, and BCL6 dual color break-apart commercial probes, respectively (Abbott Molecular, Chicago, IL). PDL1/2 (9p24.1 locus) and CIITA (16p13 locus) FISH were performed as previously described using in-house bacterial artificial chromosome probes (Spectrum Green and Orange).15 FISH signal patterns in 200 interphase nuclei were independently scored (S.B.-N. and K.L.T.) using an Olympus BX61 microscope (40×) and ARIOL software (version 3.4; Genetix, San Jose, CA). A positive rearrangement was defined as a break-apart identified in >5% of nuclei with split green/red signals or the loss of one or more red or green signals. In situations in which >20% of nuclei had ≥3 fusion signals, this was further categorized as gain (3 or 4 fusion signals) or amplification (≥5 fusion signals). All other signal findings were regarded as negative.
Statistical analysis
Group comparisons were performed with Fisher’s exact test. Progression-free survival (PFS) was defined as the time from diagnosis to relapse/progression or death from any cause. Patients who were switched to WBRT for persistent disease upon completion of HDMTX were coded as having a progression event. Overall survival (OS) was defined as the time from diagnosis to death from any cause. PFS and OS were estimated by using the Kaplan-Meier method, and group comparisons were made with the log-rank test. Hazard ratios (HRs) and 95% confidence intervals (CIs) were estimated in univariate and multivariate Cox proportional hazards models. Variables with P < .1 in univariate analysis were entered into the multivariate models, which used a backward likelihood ratio selection method, and results with P < .05 were considered statistically significant. All statistical analyses were performed with SPSS version 14.0 (IBM Corporation, Armonk, NY). The study was approved by the UBC/BC Cancer Research Ethics Board.
Results
Patient characteristics
A total of 157 cases were initially identified and included in the TMAs. However, 42 were excluded after review of central pathology and medical records: failure to obtain sufficient tumor content for the TMA and/or failure of IHC/FISH testing (n = 14), secondary CNS relapse of systemic DLBCL (n = 10), HIV infection (n = 10), PCNSL with non-DLBCL histology (n = 4), and unavailable clinical/treatment data (n = 4). A total of 115 patients were therefore included in the current analysis, and characteristics are shown in Table 1.
Table 1.
Patient and disease characteristics
| Characteristic | Entire cohort (N = 115), n (%) or n/N (%) | HDMTX subgroup (n = 52), n (%) or n/N (%) |
|---|---|---|
| Patient characteristics | ||
| Age >60 y | 81 (70) | 29 (56) |
| Male sex | 68 (59) | 32 (62) |
| Performance status ≥2 | 89 (77) | 32 (62) |
| Elevated lactate dehydrogenase | 24/83 (29) | 14/51 (27) |
| Ocular involvement | 5 (4) | 5 (10) |
| Deep brain lesions | 67/111 (60) | 31/50 (62) |
| ≥2 brain lesions | 46/111 (41) | 24/50 (48) |
| Largest mass ≥4 cm | 61/109 (56) | 31 (60) |
| Extra-CNS disease | 0 (0) | 0 (0) |
| Frontline treatment | ||
| BSC | 23 (20) | |
| WBRT | 40 (35) | |
| HDMTX-based chemotherapy | 52 (45) | |
| HDMTX regimen | 41 (79) | |
| MIDVAP regimen | 11 (21) | |
| Rituximab | 11 (21) | |
| Radiation | 18 (35) | |
| Autologous SCT | 0 (0) | |
| IHC for cell of origin | ||
| CD10 positive | 4/110 (4) | 1/50 (2) |
| BCL6 positive | 85/108 (79) | 34/49 (69) |
| MUM1 positive* | 33/109 (30) | 14/50 (28) |
| FOXP1 positive | 84/111 (76) | 34/50 (68) |
| LMO2 positive | 34/106 (32) | 14/47 (30) |
| GCET1 positive | 5/114 (4) | 2/51 (4) |
| Cell of origin algorithms | ||
| Non-GCB (Hans algorithm) | 82/109 (75) | 38/50 (76) |
| Non-GCB (Choi algorithm) | 93/109 (85) | 41/50 (82) |
| Non-GCB (Tally algorithm) | 108/110 (98) | 50/50 (100) |
| IHC for other proteins | ||
| MYC positive | 37/93 (40) | 16/38 (42) |
| BCL2 positive | ||
| 124 antibody | 69/92 (75) | 31/39 (79) |
| E17 antibody | 87/111 (78) | 40/50 (80) |
| MYC + BCL2 positive | ||
| 124 antibody (BCL2) | 30/88 (35) | 13/37 (35) |
| E17 antibody (BCL2) | 32/92 (35) | 13/38 (34) |
| PD-L1 positive | 31/107 (29) | 13/46 (28) |
| HLA-DP positive | ||
| Cytoplasmic staining only | 53/112 (47) | 28/51 (55) |
| Membranous staining† | 25/112 (22) | 8/51 (16) |
| EBER positive | 1/108 (1) | 0 (0) |
SCT, stem cell transplantation.
Cutoff (Hans algorithm) of ≥30%.
All membranous positive cases also had cytoplasmic staining.
Primary treatment modalities included HDMTX-containing regimens in 52 (45%) patients, WBRT alone in 40 (35%) patients, and BSC in 23 (20%) patients. Among the 52 patients treated with chemotherapy, 11 (21%) received MIDVAP and 41 (79%) received HDMTX. Rituximab was also given to 11 (21%) of 41 patients treated with HDMTX. All patients treated with MIDVAP received WBRT as part of the protocol, and 7 patients treated with HDMTX were switched to WBRT due to inability to tolerate methotrexate. The median dose of WBRT in these 7 patients plus the 40 patients treated with WBRT (as a single modality) was 35 Gy (range, 20-50 Gy) administered over a median of 20 days (range, 5-25 days). None of the patients in this study received consolidative high-dose chemotherapy and autologous stem cell transplantation.
IHC, EBER, and FISH results
The majority of tumors had a non–germinal center B-cell (GCB) phenotype as determined by the Hans (75%), Choi (85%), and Tally (98%) algorithms. There was 74% agreement between algorithms in the assignment of COO. Table 1 shows the expression of individual proteins used to assign COO, although 5% to 6% of patients were not assigned a COO due to failure of ≥1 of the IHC stains.
Protein expression for MYC was positive in 40% of patients, BCL2 in 75% to 78% of patients, and dual expression of MYC/BCL2 was observed in 35% of cases. All cases with dual MYC/BCL2 expression had a non-GCB phenotype according to all 3 algorithms, except for 1 case assigned a GCB phenotype according to the Hans algorithm. Overall, MYC (n = 2) and BCL2 (n = 1) rearrangements were uncommon (Table 2). There were no dual rearrangements involving MYC and BCL2, although one case with an MYC rearrangement also had a concurrent BCL6 rearrangement.
Table 2.
FISH results in the entire cohort
| Gene | FISH result, n/N (%) | |||||
|---|---|---|---|---|---|---|
| Failed | Informative | |||||
| Normal | Break-apart | Gain | Amplification | Deletion | ||
| MYC | 7/115 (6) | 94/108 (87) | 2/108 (2) | 14/108 (13) | 0 (0) | 0 (0) |
| BCL2 | 5/115 (4) | 81/110 (74) | 1/110 (1) | 28/110 (25) | 1/110 (1) | 0 (0) |
| BCL6 | 6/115 (6) | 94/109 (86) | 30/109 (28) | 11/109 (10) | 2/109 (1) | 1/109 (1) |
| PD-L1/2 | 14/115 (12) | 84/101 (83) | 1/101 (1) | 13/101 (13) | 3/101 (3) | 1/101 (1) |
| CIITA | 10/115 (9) | 92/105 (88) | 3/105 (3) | 12/105 (11) | 1/105 (1) | 0 (0) |
BCL6 rearrangements were present in 28% of patients; however, a much higher proportion exhibited BCL6 protein expression (79%). BCL6 IHC was positive in 27 (93%) of 29 patients with BCL6 rearrangements and was negative in 17 (23%) of 73 patients without BCL6 rearrangements, for a 43% concordance rate (supplemental Table 1). Table 2 shows BCL6 copy number alterations that included 1 deletion, 2 amplifications, 11 gains (5 also had break-apart), and 94 normal. Supplemental Table 1 also shows that BCL2 and LMO2 protein expression (but not BCL6 protein expression) was the only factor associated with the presence of BCL6 rearrangements.
PD-L1 protein expression was positive in 31 (29%) of 107 patients, whereas only 1 patient had a PD-L1 rearrangement. MHC class II protein expression was positive in 78 (70%) of 112 patients, with staining restricted to the cytoplasm (47%) or cell membrane (22%). All cases with membranous staining also had cytoplasmic staining. CIITA rearrangements were identified in 3 patients. EBER was positive in 1 case (confirmed HIV-negative).
Outcomes and prognostic factors
With a median follow-up of 8 years (range, 8 months to 16 years) in living patients, the 5-year PFS and OS estimates were 11% and 24%, respectively. Treatment modality (HDMTX vs BSC: HR, 0.09 [95% CI, 0.05-0.18]; WBRT vs BSC: HR, 0.16 [95% CI, 0.09-0.30], P < .001) as well as the presence of BCL6 rearrangements (HR, 1.68 [95% CI, 1.08-2.60], P = .021) were the only variables significantly associated with worse PFS in univariate analysis, as shown in supplemental Table 2. Age >60 years (HR, 1.78 [95% CI, 1.13-2.81], P = .013), poor performance status (HR, 2.27 [95% CI, 1.35-3.84], P = .002), and treatment modality (HDMTX vs BSC: HR, 0.06 [95% CI, 0.03-0.12]; WBRT vs BSC: HR, 0.18 [95% CI, 0.10-0.32], P < .001) were significantly associated with worse OS. The presence of BCL6 rearrangements was not associated with OS (HR, 1.28 [95% CI, 0.81-2.01], P = .286). The 1 patient with dual MYC and BCL6 rearrangements progressed after 2 cycles of single-agent HDMTX and died shortly after a course of palliative WBRT.
None of the other clinical and pathologic variables, including COO, MYC/BCL2, and BCL6 protein expression, were associated with PFS or OS. Figure 1 displays PFS and OS according to treatment, and Figure 2 presents PFS and OS according to the presence or absence of BCL6 rearrangements. Table 3 shows that in multivariate analysis, only the presence of BCL6 rearrangements remained significantly associated with PFS, whereas only treatment modality remained significantly associated with OS.
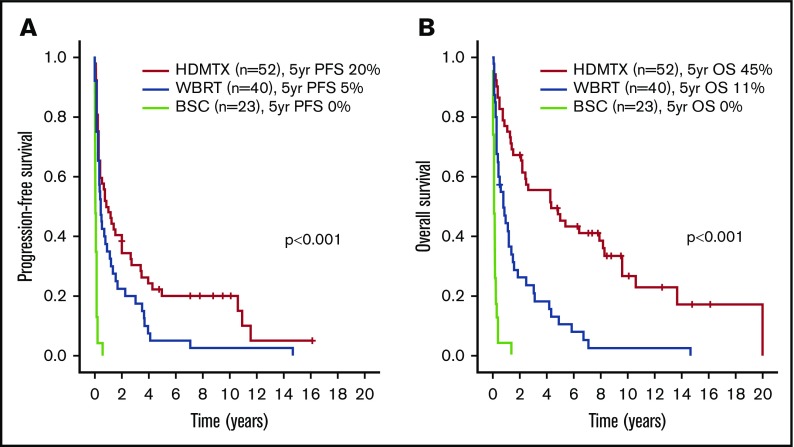
Figure 1.
Outcomes according to frontline treatment in the entire cohort. PFS (A) and OS (B).
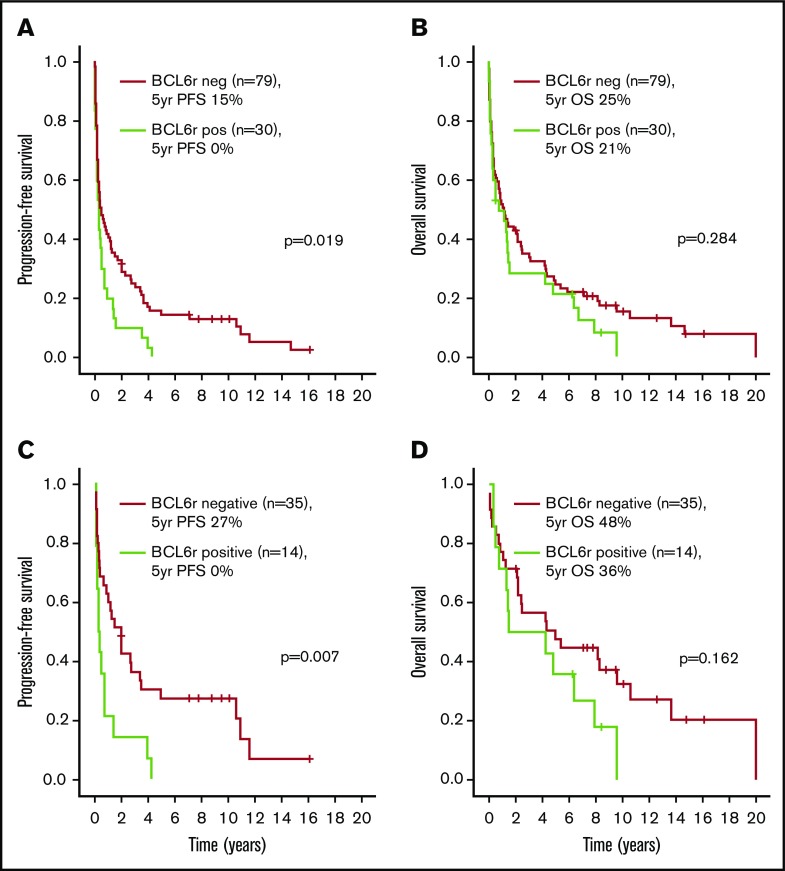
Figure 2.
Outcomes according to the presence of BCL6 rearrangements (BCL6r). PFS (A) and OS (B) in the entire cohort. PFS (C) and OS (D) in the HDMTX subgroup.
Table 3.
Multivariate analyses
| Cohorts and variables | PFS, HR (95% CI) | OS, HR (95% CI) |
|---|---|---|
| Entire cohort | n = 79 | n = 109 |
| Age (>60 y vs ≤60 y) | Not included | 1.49 (0.93-2.40), P = .099 |
| Performance status (2-4 vs 0-1) | Not included | 1.17 (0.67-2.06), P = .587 |
| LDH (elevated vs normal) | 1.61 (0.96-2.70), P = .071 | Not included |
| Treatment (HDMTX vs others) | 0.69 (0.43-1.11), P = .125 | 0.30 (0.19-0.46), P < .001 |
| BCL6 FISH (positive vs negative) | 2.06 (1.22-3.49), P = .007 | 1.55 (0.98-2.46), P = .064 |
| HDMTX only | n = 48 | n = 45 |
| Sex (male vs female) | Not included | 2.59 (1.16-5.81), P = .021 |
| LDH (elevated vs normal) | 1.76 (0.89-3.48), P = .104 | Not included |
| LMO2 IHC (positive vs negative) | Not included | 1.33 (0.61-2.92), P = .479 |
| BCL6 FISH (positive vs negative) | 2.62 (1.33-5.17), P = .006 | 1.12 (0.46-2.71), P = .806 |
LDH, lactate dehydrogenase.
Characteristics and outcomes of patients treated with HDMTX
A subgroup analysis was performed in the 52 patients treated with HDMTX-based regimens. Patient and disease characteristics are summarized in Table 1. With a median follow-up of 8.5 years (range, 2-16 years) in living patients, the 5-year PFS and OS were 20% and 45%, respectively, as shown in Figure 1. Supplemental Table 2 and Table 3 show that the presence of BCL6 rearrangements was the only variable associated with worse PFS in univariate (HR, 2.46 [95% CI, 1.26-4.84], P = .009) (Figure 2C-D) and multivariate analyses. Male sex was the only variable associated with worse OS in univariate (HR, 2.52, [95% CI, 1.15-5.49], P = .020) and multivariate analyses. The same analysis was repeated in the 40 patients treated with WBRT alone, and no prognostic factors, including BCL6 rearrangements, were identified (data not shown).
Discussion
In this large population-based cohort, we performed a broad analysis of the biology and prognosis of PCNSL with DLBCL morphology using TMAs and found a distinct molecular profile that is very different from what has been reported for systemic DLBCL.1-3,16,17 Consistent with other reports, the majority of cases had a non-GCB phenotype according to the IHC algorithms, but COO did not affect PFS or OS.18-23 Protein overexpression (MYC, BCL2, BCL6, and PD-L1) seemed to be independent of their corresponding gene rearrangements. With the exception of BCL6 rearrangements, FISH results (MYC, BCL2, PD-L1, and CIITA rearrangements) were not associated with PFS or OS partly because they were extremely uncommon (<5%).
Even though MYC and BLC2 protein expression was common (40% and 75%-80%, respectively), their corresponding gene rearrangements were extremely uncommon (≤3%, including no cases with dual MYC and BCL2 translocations), suggesting that alternate mechanisms were driving protein expression in PCNSL.19,20,24-28 MYC protein overexpression may be related to the high proliferative activity of PCNSL or activation of the NF-κB pathway, but the precise mechanisms remain unknown and cannot be further clarified by our data.24,25,29 Dual expression of MYC and BCL2 was strongly associated with a non-GCB phenotype, similar to systemic DLBCL.17,30,31 MYC and/or BCL2 overexpression was not associated with outcomes in our study, although other retrospective series using different methodologies and patient bases have reported a negative association.28,32
BCL6 rearrangements were frequent (∼1 in 3 patients), which is consistent with previous retrospective series in which the prevalence of BCL6 rearrangements (mostly chromosomal translocations) ranged between 10% and 40%.1,24-26,33 BCL6 rearrangements were the only factor associated with a poor PFS on multivariate analysis in the overall cohort and in the subgroup of patients treated with HDMTX-based regimens. From a biologic standpoint, BCL6 is a proto-oncogene encoding a transcriptional repressor necessary for normal germinal center formation. Translocations between BCL6 and various immunoglobulin and non-immunoglobulin partner genes deregulate this transcriptional repressor and prevent BCL6 from binding to its own promoter, leading to abnormal B-cell development.33,34 Our FISH analyses used a single probe, and therefore we could not evaluate for specific translocation partners or discrete chromosomal abnormalities such as deletions of the long arm of chromosome 6 [Del(6)(q22)].25 This aspect of PCNSL lymphomagenesis may also partially explain its non-GCB phenotype.
Conversely, BCL6 protein expression was very frequent (∼80%) and was strongly correlated with the presence of BCL6 rearrangements, although it did not play a prognostic role. The presence of BCL6 protein expression has been associated with variable outcomes (favorable, unfavorable, or no association) in various retrospective series of patients with PCNSL.21,22,32,33,35,36 These studies, including ours, have used different methods to test for BCL6 expression and have evaluated outcomes in various cohorts with dissimilar therapies and distributions of prognostic factors. As a germinal center marker in a disease with largely a non-GCB phenotype, it remains unknown whether BCL6 protein expression plays an independent, substantial role in PCNSL.
Unlike certain forms of systemic DLBCL such as primary mediastinal large B-cell lymphoma, we found that CIITA rearrangements were very rare (3 of 105).37 Using a targeted DNA sequencing approach, Chapuy et al only found 1 of 24 cases with a CIITA rearrangement.2 We also found that only 22% of cases had membranous expression of MHC class II that typifies antigen-presenting cells, including B cells. Our group previously showed that a cytoplasmic-only pattern (but not membranous) conferred a worse prognosis in systemic DLBCL.13 We did not find a similar association, raising the hypothesis that the reduced membranous expression in PCNSL plays an important role in the biology of PCNSL, likely related to mechanisms of escape from immune surveillance.
Similarly, PD-L1 expression was common, but PD-L1/PD-L2 translocations were very uncommon (1 of 101).19 Chapuy et al identified PD-L1/PD-L2 chromosomal rearrangements in 3 of 24 patients through genomic sequencing but also noted frequent 9p24.1/PD-L1/PD-L2 copy gains (28 of 42 patients in a separate extension cohort) using ligand-specific quantitative polymerase chain reaction. The latter may provide a more robust mechanism for PD-L1 upregulation.2 We found that 13 of 101 cases had PD-L1/PD-L2 copy gains, but these were not associated with PD-L1 protein expression or prognosis (data not shown). Altogether, these data suggest that alternate mechanisms lead to PD-L1/PD-L2 deregulation, again highlighting the role of immune evasion in PCNSL.
To the best of our knowledge, this analysis is the largest published cohort of PCNSL testing a broad range of molecules with IHC, ISH, and FISH in a TMA. Our study captured >80% of the denominator of patients with PCNSL diagnosed in British Columbia between 1998 and 2010 and is therefore reflective of results in a general population with excellent long-term follow-up. However, our cohort was enriched with patients expected to have a poor prognosis (70% aged >60 years, 78% with performance status >1, and 56% not eligible for HDMTX-based regimens) and treated in an era in which relatively obsolete treatments were used. Outcomes in the 52 patients treated with HDMTX-based therapies remained poor (5-year PFS, 20%; 5-year OS, 45%), which may have limited our ability to detect any other relevant prognostic factors. It is unknown whether results may have been different in the subgroup of patients receiving HDMTX-based therapies plus additional strategies such as other drugs that cross the blood–brain barrier, immediate WBRT consolidation, etoposide/cytarabine consolidation, or consolidation with autologous stem cell transplantation.38-40
In conclusion, we found that PCNSL has a distinct biology and prognosis. Overall outcomes in this cohort were poor, reflecting a general population base and an earlier treatment era, which may have abrogated the prognostic impact of other factors. This methodology should be repeated in larger cohorts of patients with PCNSL receiving contemporary treatments known to be associated with improved outcomes. Other molecules relevant to PCNSL such as MYD88, CD79b, ETV6, PIM-1, CARD11, and EZH2 should be examined.1,2,41,42 Other methods such as whole-genome/exome sequencing and peripheral blood micro-RNA could provide additional prognostic information.43,44 Our results may better inform drug development in PCNSL, including those targeting the B-cell receptor pathway, immune evasion, and the tumor microenvironment.4,39,45-48
Supplementary Material
The full-text version of this article contains a data supplement.
Acknowledgments
The authors acknowledge the generous donation from Ami and Bella Haasz as well as Dianne Bentz to partially cover publication costs.
This study was supported in part by the Terry Fox Foundation Strategic Health Research Training Program in Cancer Research at Canadian Institutes of Health Research grant TGT-53912 (K.L.T. and G.W.S.). This study was also supported in part by research funding from the Terry Fox Research Institute (Overcoming treatment failure in lymphoid cancers), the Michael Smith Foundation for Health Research (Genetic mechanisms underlying immune privilege in malignant lymphomas), and the BC Cancer Centre for Lymphoid Cancer.
Authorship
Contribution: D.V., K.L.T., R.D.G., and G.W.S. designed the research; D.V., K.L.T., S.B.-N., M.A.M., R.D.G., and G.W.S. performed the research and analyzed data; and all authors contributed data and reviewed and approved the final manuscript.
Conflict-of-interest disclosure: D.V., T.N.S., J.M.C., L.H.S., K.J.S., and D.W.S. have received research funding from Roche to support the BC Cancer Centre for Lymphoid Cancer database. D.V. reports honoraria/advisory board from Roche, Celgene, Seattle Genetics, Lundbeck, Janssen, AstraZeneca, Gilead, and AbbVie, outside the submitted work. C.S. reports research funding from Bristol-Myers Squibb and Tioma; consultancy from Juno Therapeutics, Roche, and Seattle Genetics; and patents and royalties from NanoString, outside of the submitted work. J.M.C. reports research funding from Janssen, Genentech, Merck, Bayer, Roche, Lilly, Seattle Genetics, Takeda, Amgen, and Cephalon; and patents and royalties from NanoString, outside of the submitted work. L.H.S. reports honoraria/advisory boards from Roche/Genentech, AbbVie, Amgen, Apobiologix, Celgene, Gilead, Janssen, Kite, Karyopharm, Lundbeck, Merck, Seattle Genetics, Teva, and TG Therapeutics; and research funding from Roche/Genentech, outside the submitted work. K.J.S. reports honoraria/advisory board from BMS, Merck, Novartis, Verastem, AbbVie, Servier, and Seattle Genetics, outside the submitted work. D.W.S. reports grants from Janssen; research contracts from Roche/Genentech and NanoString; and personal fees from Celgene, outside the submitted work. R.D.G. reports patents and royalties from NanoString, outside the submitted work. The remaining authors declare no competing financial interests.
Correspondence: Diego Villa, BC Cancer–Vancouver Cancer Centre, 600 West 10th Ave, Vancouver, BC V5Z 4E6, Canada; e-mail: dvilla@bccancer.bc.ca.
References
- 1.Braggio E, Van Wier S, Ojha J, et al. Genome-wide analysis uncovers novel recurrent alterations in primary central nervous system lymphomas. Clin Cancer Res. 2015;21(17):3986-3994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapuy B, Roemer MG, Stewart C, et al. Targetable genetic features of primary testicular and primary central nervous system lymphomas. Blood. 2016;127(7):869-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kagoya Y, Nannya Y, Nakamura F, Kurokawa M. Gene expression profiles of central nervous system lymphoma predict poor survival in patients with diffuse large B-cell lymphoma. Br J Haematol. 2014;166(5):794-797. [DOI] [PubMed] [Google Scholar]
- 4.Illerhaus G, Schorb E, Kasenda B. Novel agents for primary central nervous system lymphoma: evidence and perspectives. Blood. 2018;132(7):681-688. [DOI] [PubMed] [Google Scholar]
- 5.Ferreri AJ, Blay JY, Reni M, et al. Prognostic scoring system for primary CNS lymphomas: the International Extranodal Lymphoma Study Group experience. J Clin Oncol. 2003;21(2):266-272. [DOI] [PubMed] [Google Scholar]
- 6.Shenkier TN, Voss N, Chhanabhai M, et al. The treatment of primary central nervous system lymphoma in 122 immunocompetent patients: a population-based study of successively treated cohorts from the British Colombia Cancer Agency. Cancer. 2005;103(5):1008-1017. [DOI] [PubMed] [Google Scholar]
- 7.Kansara R, Shenkier TN, Connors JM, et al. Rituximab with high-dose methotrexate in primary central nervous system lymphoma. Am J Hematol. 2015;90(12):1149-1154. [DOI] [PubMed] [Google Scholar]
- 8.Biccler JL, Savage KJ, Brown PDN, et al. Risk of death, relapse or progression, and loss of life expectancy at different progression-free survival milestones in primary central nervous system lymphoma. Leuk Lymphoma. 2019;60(10):2516-2523. [DOI] [PubMed] [Google Scholar]
- 9.Choi WW, Weisenburger DD, Greiner TC, et al. A new immunostain algorithm classifies diffuse large B-cell lymphoma into molecular subtypes with high accuracy. Clin Cancer Res. 2009;15(17):5494-5502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hans CP, Weisenburger DD, Greiner TC, et al. Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103(1):275-282. [DOI] [PubMed] [Google Scholar]
- 11.Meyer PN, Fu K, Greiner TC, et al. Immunohistochemical methods for predicting cell of origin and survival in patients with diffuse large B-cell lymphoma treated with rituximab. J Clin Oncol. 2011;29(2):200-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rimsza LM, Farinha P, Fuchs DA, Masoudi H, Connors JM, Gascoyne RD. HLA-DR protein status predicts survival in patients with diffuse large B-cell lymphoma treated on the MACOP-B chemotherapy regimen. Leuk Lymphoma. 2007;48(3):542-546. [DOI] [PubMed] [Google Scholar]
- 13.Kendrick S, Rimsza LM, Scott DW, et al. Aberrant cytoplasmic expression of MHCII confers worse progression free survival in diffuse large B-cell lymphoma. Virchows Arch. 2017;470(1):113-117. [DOI] [PubMed] [Google Scholar]
- 14.Chin SF, Daigo Y, Huang HE, et al. A simple and reliable pretreatment protocol facilitates fluorescent in situ hybridisation on tissue microarrays of paraffin wax embedded tumour samples. Mol Pathol. 2003;56(5):275-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steidl C, Shah SP, Woolcock BW, et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nature. 2011;471(7338):377-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz R, Wright GW, Huang DW, et al. Genetics and pathogenesis of diffuse large B-cell lymphoma. N Engl J Med. 2018;378(15):1396-1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Scott DW, Mottok A, Ennishi D, et al. Prognostic significance of diffuse large B-cell lymphoma cell of origin determined by digital gene expression in formalin-fixed paraffin-embedded tissue biopsies. J Clin Oncol. 2015;33(26):2848-2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Camilleri-Broët S, Crinière E, Broët P, et al. A uniform activated B-cell-like immunophenotype might explain the poor prognosis of primary central nervous system lymphomas: analysis of 83 cases. Blood. 2006;107(1):190-196. [DOI] [PubMed] [Google Scholar]
- 19.Four M, Cacheux V, Tempier A, et al. PD1 and PDL1 expression in primary central nervous system diffuse large B-cell lymphoma are frequent and expression of PD1 predicts poor survival. Hematol Oncol. 2017;35(4):487-496. [DOI] [PubMed] [Google Scholar]
- 20.Gill KZ, Iwamoto F, Allen A, et al. MYC protein expression in primary diffuse large B-cell lymphoma of the central nervous system. PLoS One. 2014;9(12):e114398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreher S, Jöhrens K, Strehlow F, et al. Prognostic impact of B-cell lymphoma 6 in primary CNS lymphoma. Neuro-oncol. 2015;17(7):1016-1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Momota H, Narita Y, Maeshima AM, et al. Prognostic value of immunohistochemical profile and response to high-dose methotrexate therapy in primary CNS lymphoma. J Neurooncol. 2010;98(3):341-348. [DOI] [PubMed] [Google Scholar]
- 23.Raoux D, Duband S, Forest F, et al. Primary central nervous system lymphoma: immunohistochemical profile and prognostic significance. Neuropathology. 2010;30(3):232-240. [DOI] [PubMed] [Google Scholar]
- 24.Brunn A, Nagel I, Montesinos-Rongen M, et al. Frequent triple-hit expression of MYC, BCL2, and BCL6 in primary lymphoma of the central nervous system and absence of a favorable MYC(low)BCL2 (low) subgroup may underlie the inferior prognosis as compared to systemic diffuse large B cell lymphomas. Acta Neuropathol. 2013;126(4):603-605. [DOI] [PubMed] [Google Scholar]
- 25.Cady FM, O’Neill BP, Law ME, et al. Del(6)(q22) and BCL6 rearrangements in primary CNS lymphoma are indicators of an aggressive clinical course. J Clin Oncol. 2008;26(29):4814-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montesinos-Rongen M, Zühlke-Jenisch R, Gesk S, et al. Interphase cytogenetic analysis of lymphoma-associated chromosomal breakpoints in primary diffuse large B-cell lymphomas of the central nervous system. J Neuropathol Exp Neurol. 2002;61(10):926-933. [DOI] [PubMed] [Google Scholar]
- 27.Nosrati A, Monabati A, Sadeghipour A, Radmanesh F, Safaei A, Movahedinia S. MYC, BCL2, and BCL6 rearrangements in primary central nervous system lymphoma of large B cell type. Ann Hematol. 2019;98(1):169-173. [DOI] [PubMed] [Google Scholar]
- 28.Son SM, Ha SY, Yoo HY, et al. Prognostic impact of MYC protein expression in central nervous system diffuse large B-cell lymphoma: comparison with MYC rearrangement and MYC mRNA expression. Mod Pathol. 2017;30(1):4-14. [DOI] [PubMed] [Google Scholar]
- 29.Deckert M, Montesinos-Rongen M, Brunn A, Siebert R. Systems biology of primary CNS lymphoma: from genetic aberrations to modeling in mice. Acta Neuropathol. 2014;127(2):175-188. [DOI] [PubMed] [Google Scholar]
- 30.Hu S, Xu-Monette ZY, Tzankov A, et al. MYC/BCL2 protein coexpression contributes to the inferior survival of activated B-cell subtype of diffuse large B-cell lymphoma and demonstrates high-risk gene expression signatures: a report from The International DLBCL Rituximab-CHOP Consortium Program. Blood. 2013;121(20):4021-4031, quiz 4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Staiger AM, Ziepert M, Horn H, et al. ; German High-Grade Lymphoma Study Group . Clinical impact of the cell-of-origin classification and the MYC/BCL2 dual expresser status in diffuse large B-cell lymphoma treated within prospective clinical trials of the German High-Grade Non-Hodgkin’s Lymphoma Study Group. J Clin Oncol. 2017;35(22):2515-2526. [DOI] [PubMed] [Google Scholar]
- 32.Kim S, Nam SJ, Kwon D, et al. MYC and BCL2 overexpression is associated with a higher class of Memorial Sloan-Kettering Cancer Center prognostic model and poor clinical outcome in primary diffuse large B-cell lymphoma of the central nervous system. BMC Cancer. 2016;16:363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwindt H, Akasaka T, Zühlke-Jenisch R, et al. Chromosomal translocations fusing the BCL6 gene to different partner loci are recurrent in primary central nervous system lymphoma and may be associated with aberrant somatic hypermutation or defective class switch recombination. J Neuropathol Exp Neurol. 2006;65(8):776-782. [DOI] [PubMed] [Google Scholar]
- 34.Pasqualucci L, Migliazza A, Basso K, Houldsworth J, Chaganti RS, Dalla-Favera R. Mutations of the BCL6 proto-oncogene disrupt its negative autoregulation in diffuse large B-cell lymphoma. Blood. 2003;101(8):2914-2923. [DOI] [PubMed] [Google Scholar]
- 35.Lin CH, Kuo KT, Chuang SS, et al. Comparison of the expression and prognostic significance of differentiation markers between diffuse large B-cell lymphoma of central nervous system origin and peripheral nodal origin. Clin Cancer Res. 2006;12(4):1152-1156. [DOI] [PubMed] [Google Scholar]
- 36.Lossos C, Bayraktar S, Weinzierl E, et al. LMO2 and BCL6 are associated with improved survival in primary central nervous system lymphoma. Br J Haematol. 2014;165(5):640-648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mottok A, Woolcock B, Chan FC, et al. Genomic alterations in CIITA are frequent in primary mediastinal large B cell lymphoma and are associated with diminished MHC class II expression. Cell Reports. 2015;13(7):1418-1431. [DOI] [PubMed] [Google Scholar]
- 38.Omuro A, Correa DD, DeAngelis LM, et al. R-MPV followed by high-dose chemotherapy with TBC and autologous stem-cell transplant for newly diagnosed primary CNS lymphoma. Blood. 2015;125(9):1403-1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rubenstein JL, Geng H, Fraser EJ, et al. Phase 1 investigation of lenalidomide/rituximab plus outcomes of lenalidomide maintenance in relapsed CNS lymphoma. Blood Adv. 2018;2(13):1595-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ferreri AJM, Cwynarski K, Pulczynski E, et al. ; International Extranodal Lymphoma Study Group (IELSG) . Whole-brain radiotherapy or autologous stem-cell transplantation as consolidation strategies after high-dose methotrexate-based chemoimmunotherapy in patients with primary CNS lymphoma: results of the second randomisation of the International Extranodal Lymphoma Study Group-32 phase 2 trial. Lancet Haematol. 2017;4(11):e510-e523. [DOI] [PubMed] [Google Scholar]
- 41.Bruno A, Boisselier B, Labreche K, et al. Mutational analysis of primary central nervous system lymphoma. Oncotarget. 2014;5(13):5065-5075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakamura T, Tateishi K, Niwa T, et al. Recurrent mutations of CD79B and MYD88 are the hallmark of primary central nervous system lymphomas. Neuropathol Appl Neurobiol. 2016;42(3):279-290. [DOI] [PubMed] [Google Scholar]
- 43.Zhou Y, Liu W, Xu Z, et al. Analysis of genomic alteration in primary central nervous system lymphoma and the expression of some related genes. Neoplasia. 2018;20(10):1059-1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth P, Keller A, Hoheisel JD, et al. Differentially regulated miRNAs as prognostic biomarkers in the blood of primary CNS lymphoma patients. Eur J Cancer. 2015;51(3):382-390. [DOI] [PubMed] [Google Scholar]
- 45.Lionakis MS, Dunleavy K, Roschewski M, et al. Inhibition of B cell receptor signaling by ibrutinib in primary CNS lymphoma. Cancer Cell. 2017;31(6):833-843.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grommes C, Tang SS, Wolfe J, et al. Phase 1b trial of an ibrutinib-based combination therapy in recurrent/refractory CNS lymphoma. Blood. 2019;133(5):436-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nayak L, Iwamoto FM, LaCasce A, et al. PD-1 blockade with nivolumab in relapsed/refractory primary central nervous system and testicular lymphoma. Blood. 2017;129(23):3071-3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tun HW, Johnston PB, DeAngelis LM, et al. Phase 1 study of pomalidomide and dexamethasone for relapsed/refractory primary CNS or vitreoretinal lymphoma. Blood. 2018;132(21):2240-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.