Abstract
Motility is important for virulence, biofilm formation, and the environmental adaptation of many bacteria. Vibrio parahaemolyticus (V. parahaemolyticus) contains two flagellar systems that are responsible for motility, and are tightly regulated by transcription regulators and sigma factors. In this study, we identified a novel transcription factor, VPA1701, which regulates the swarming motility of V. parahaemolyticus. The VPA1701 deletion mutant (ΔVPA1701) eliminated the swarming motility on the surface of BHI agar plates and reduced colonization in infant rabbits. RNA-seq assays, confirmed by qRT-PCR, indicated that VPA1701 regulated the expression of lateral flagellar cluster genes. Further analyses revealed that VPA1701 directly binds to the promoter region of the flgBCDEFGHIJKL cluster to regulate the expression of lateral flagellar genes. CalR was originally identified as a repressor for the swarming motility of V. parahaemolyticus, and it was inhibited by calcium. In this study, we found that VPA1701 could inhibit the expression of the calR gene to increase the swarming motility of V. parahaemolyticus. Calcium downregulated the expression of calR, indicating that calcium could increase swarming motility of ΔVPA1701 by inhibiting calR. Thus, this study illustrates how the transcription factor VPA1701 regulates the expression of lateral flagellar genes and calR to control the swarming motility of V. parahaemolyticus.
Keywords: Vibrio parahaemolyticus, VPA1701, CalR, lateral flagellar system, swarming motility
1. Introduction
V. parahaemolyticus is a foodborne pathogen sometimes found in contaminated seafood, which can cause acute-gastroenteritis and hemorrhagic sepsis in humans [1,2]. The major virulence factors of V. parahaemolyticus are hemolysin (thermostable direct hemolysin [TDH], TDH-related hemolysin [TRH]), type III secretion systems (T3SS1 and T3SS2), type VI secretion systems (T6SS1 and T6SS2), biofilm formation, motility, protease production and iron uptake system [3,4,5,6,7,8]. The pathogenesis processes of V. parahaemolyticus include motility, adhesion, invasion, proliferation, production of toxins in vivo, and the damage to cells and tissues [9,10]. Motility plays an essential role in the infection of V. parahaemolyticus in the host, and the protease production can also benefit host invasion. The V. parahaemolyticus contains two flagellar systems: polar flagellum and lateral flagellum. The polar flagellum is encoded by the polar flagellar genes responsible for the swimming motility, and the lateral flagellum is responsible for the swarming motility encoded by lateral flagellar genes [11]. A previous study also indicated that V. parahaemolyticus lacking flagellum could not adhere to either normal or degenerating tissue cells [12]. Therefore, the flagellum may help bacteria to adhere to the surface of cells and promote the biofilm formation that contributes to the pathogenicity of bacteria [13,14].
Motility provides a more favorable living condition for microorganisms, and 60% of bacteria contain the flagellar systems responsible for motility. These flagellar systems can be divided into three types: the polar flagellar system (Vibrio cholerae), the lateral flagellar system (Salmonella enterica), and the dual flagellar systems (V. parahaemolyticus, Vibrio alginolyticus, and Aeromonas spp) [14,15]. In V. parahaemolyticus, the dual flagellar systems play an essential role in complex environments, the sheathed polar flagellar system powered by MotAB and MotXY sodium pumps propels the bacterium in a liquid environment, while the lateral flagellar system motor remains to be identified [16,17]. The polar flagellar system is encoded by more than 60 genes located in chromosome I and regulated by FlaK, and the lateral flagellar system is encoded by 38 genes located in chromosome II and regulated by LafK. Additionally, the transcriptional regulator LafK may compensate for the function of FlaK to enable swimming ability in some bacteria [18].
The expression of flagellar genes is hierarchical, with specific transcription regulators and sigma factors controlling the early, middle, and late gene expression. The flagellar genes and specific regulators are similar in vibriosis [19]. The expression of polar flagellum genes is modulated by the master regulator FlaK, which is a σ54-dependent regulator and activates the expression of class II flagellar genes. Subsequently, the regulators FlaM and σ54 initiate the expression of class III genes. The expression of class II and class III genes is in the middle of the hierarchy. Lastly, the expression of class IV genes is directly activated by a σ28-dependent regulator [20]. In the expression of the lateral flagellar system, the master regulator LafK activates the class II genes, including flgAMN, flgBCDEFGHIJ, fliDSTKLAmotAB, and motYlafKfliEFGHI, early in the hierarchy. Then, the expression of class III genes, including flgKL, lafA, fliDSTKLAmotAB, and flgMN, are activated by σ28, which is encoded by fliA [14,20,21]. Thus, the expression of flagellar genes is tightly regulated by the specific regulators and sigma factors in a spatiotemporal-dependent way.
The motility is affected by various cellular processes, such as the quorum sensing system, type III secretion system (T3SS), signaling molecules, and transcription regulators. The quorum sensing system plays an essential role in the regulation of motility in V. alginolyticus, V. harveyi, and V. parahaemolyticus [22,23,24]. In V. parahaemolyticus, the T3SS1 transcription regulator ExsA inhibits the swarming motility by decreasing the expression of lafK, the master regulator of the lateral flagellar system [25]. The second messenger c-di-GMP directly binds to the flagellar master regulator FlrA and inhibits the motility of Vibrio cholerae [26]. Glucose regulates the synthesis of flagellum by inhibiting the polar localization of FapA in V. vulnificus [27]. Furthermore, a global transcriptional regulator, CalR, downregulates the expression of laf genes to inhibit the swarming motility of V. parahaemolyticus [25]. Additionally, ToxR acts as an activator of both swarming and swimming motility in V. parahaemolyticus [28].
The objective of this study was to assess the role of VPA1701 in the regulation of swarming motility in V. parahaemolyticus. A previous study used transposon-insertion sequencing (TIS) to identify 230 genes that contributed to the colonization of V. parahaemolyticus in infant rabbit small intestines [29]. In this study, we selected 22 of those genes to construct the deletion mutant strains and found that the colonization and swarming motility of ΔVPA1701 had significantly decreased compared to the wild type. Furthermore, the VPA1701 directly bound to the promoter region of the flagellar genes to regulate the expression of lateral flagellar genes. Lastly, VPA1701 inhibited the expression of calR to control the swarming motility of V. parahaemolyticus.
2. Results and Discussion
2.1. Essential Roles of VPA1701 in the Colonization and Swarming Motility of V. parahaemolyticus
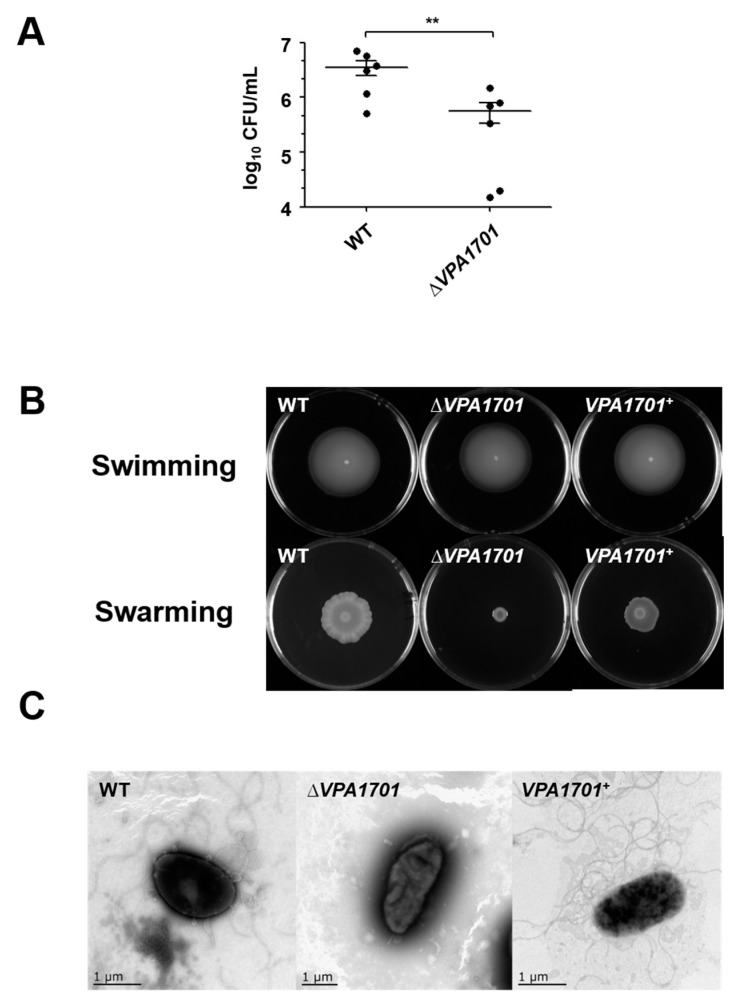
VPA1701 has been identified as a transcriptional regulatory protein in the GntR family that contains a conserved helix-turn-helix (HTH) for DNA binding at the N-terminus and an effector binding domain involved in oligomerization at the C-terminus [30]. BlastP search against GenBank indicated that V. parahaemolyticus VPA1701 shared 98.86%, 98.48%, 92.72%, and 89.27% identities to the homologous proteins in Vibrio harveyi, Vibrio campbellii, Vibrio vulnificus, and Vibrio cholerae, respectively (Supplementary Figure S1). The previous study used TIS to demonstrate that VPA1701 was involved in the colonization of V. parahaemolyticus in the intestines of infant rabbits [29]. In this study, we constructed the VPA1701 deletion mutant and complemented strains to confirm the function of VPA1701 in V. parahaemolyticus. Infant rabbits received a dose of the bacteria, delivered orogastrically, and observations showed that the colonization of V. parahaemolyticus was significantly reduced by ∆VPA1701 compared to the WT (Figure 1A).
Figure 1.
Essential roles of VPA1701 in the in vivo colonization and motility of V. parahaemolyticus. (A) The infant rabbit received WT and ∆VPA1701 orogastrically, and the infant rabbit was killed to obtain the colonization data. The results are shown as the mean ± SD (n = 6); ** p < 0.05. (B) V. parahaemolyticus WT, ∆VPA1701, and VPA1701+ were cultured in semi-solid plates (LB with 0.3% Agar) at 37 °C and solid plates (BHI with 1.5% Agar) at 30 °C to observe the swimming and swarming ability separately. (C) The generation of the lateral flagellum was observed by TEM of WT, ∆VPA1701, and VPA1701+.
During colonization and infection, the bacteria have evolved several mechanisms to control the expression of genes to enable them to adapt to different environments. This colonization ability was essential for the infection of V. parahaemolyticus, and the previous study has found that motility was important for the colonization and infection of V. parahaemolyticus and Vibrio coralliilyticus [9,31]. The quorum sensing regulator SwrT could inhibit a GntR family protein SwrZ to control the expression of laf genes and regulate the swarming motility of V. parahaemolyticus [32]. Therefore, the motility of WT, ∆VPA1701, and complemented strains were measured, and the results showed that there was no significant difference in swimming motility between WT and ∆VPA1701 (Figure 1B). However, the swarming motility of ∆VPA1701 was significantly decreased compared to the WT, and the swarming motility of the complemented strain recovered to the level of the WT (Figure 1B). Furthermore, the TEM results indicated that the lateral flagellum of ∆VPA1701 was eliminated, while the WT and VPA1701+ were surrounded by a significant amount of lateral flagellum (Figure 1C). These results indicated that VPA1701 protein regulated the swarming motility and colonization of V. parahaemolyticus.
2.2. The Global Transcriptional Analysis of VPA1701 in V. parahaemolyticus
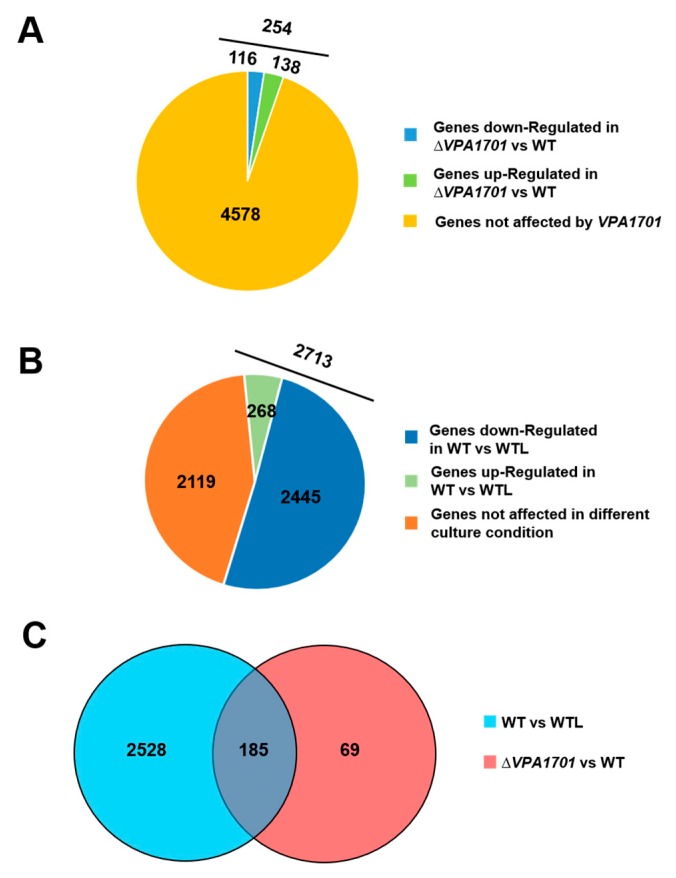
The previous experiment showed that the VPA1701 protein regulated the swarming motility of V. parahaemolyticus, and RNA-seq identified the genes regulated by VPA1701 in the BHI agar plate. The VPA1701 protein regulated 254 genes, which 116 genes were downregulated and 138 genes were upregulated (log2FC ≥ 2 or log2FC ≤ −2, p < 0.001) in the ΔVPA1701 strain compared to the WT (Figure 2A). Swarming motility was analyzed in the agar plate, and the RNA-seq was also used to identify the different gene expression between the BHI agar plate and liquid medium. The results indicated that 2713 genes were regulated in the wild type cultured in agar plates (WT) compared to the wild type cultured in liquid medium (WTL) (log2FC ≥ 2 or log2FC ≤ −2, p < 0.001). Resultantly, 268 genes were upregulated, and 2445 genes were downregulated (Figure 2B), indicating that several genes were downregulated in the BHI agar plate. To identify the co-regulon of VPA1701 and the WT cultured in the BHI agar plates, we identified 185 overlapping genes between the regulons of VPA1701 and wild type cultured in the BHI agar plates (Figure 2C), indicating that these genes may be associated with the motility of V. parahaemolyticus.
Figure 2.
RNA-seq reveals the genes regulated by VPA1701 in the BHI agar plate. (A) The number of genes regulated by VPA1701. (B) The number of genes regulated by the WT cultured in BHI plate with 1.5% agar compared to the wild type cultured in the liquid (WTL). (C) Venn diagrams perform the number of genes co-regulated by VPA1701 and the BHI agar plate condition.
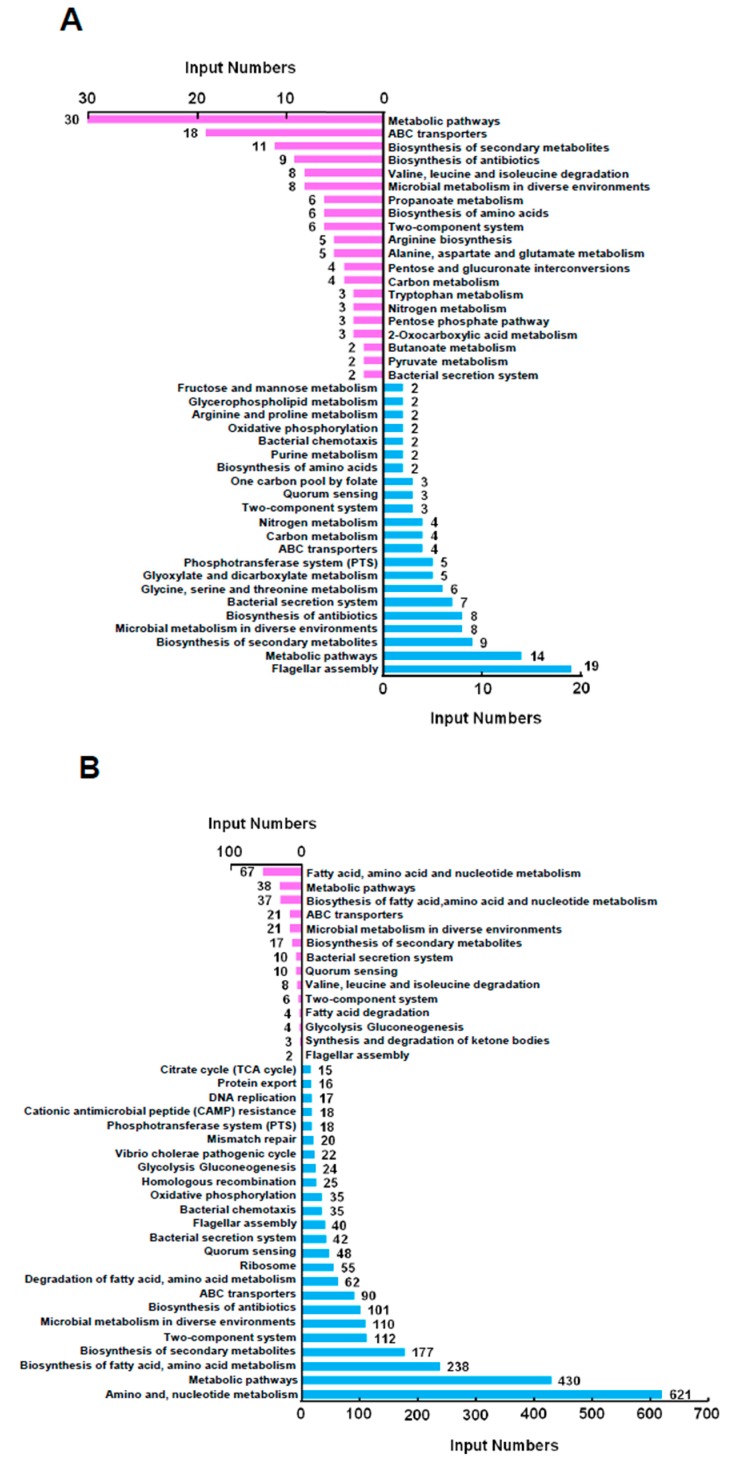
Based on the RNA-seq data, we performed a KEGG analysis to elucidate further the pathways regulated by VPA1701 (Figure 3A). We found that the metabolic pathways and flagellar assembly genes were altered in the ΔVPA1701 strain. Results showed that 44 genes were involved in the metabolic pathways (30 genes were upregulated and 14 genes were downregulated), and 22 genes were involved in the ABC transporters (18 genes were upregulated and four genes were downregulated). Moreover, two-component system pathways contained three downregulated genes and six upregulated genes in the ΔVPA1701 strain. Nineteen genes of the lateral flagellar system were downregulated in the ΔVPA1701 compared to the WT, indicating that the regulation of swarming motility by VPA1701 was enabled by the expression of lateral flagellar genes in V. parahaemolyticus.
Figure 3.
KEGG pathway analysis of the genes regulated by VPA1701 and the BHI agar plate condition. (A) Analysis of the gene expression in ΔVPA1701 vs. WT. (B) Analysis of the gene expression in WT vs. WTL. The pink indicates the upregulated genes and the blue indicates the downregulated genes.
Moreover, we analyzed the KEGG pathway of the genes regulated by the WT cultured in the BHI agar plates compared to the liquid medium (Figure 3B). We found that 688 genes were involved in fatty acid, amino acid, and nucleotide metabolism pathways (67 genes were upregulated, and 621 genes were downregulated). A total of 118 genes in the two-component system were regulated in the BHI agar plate (112 genes were downregulated and six were genes upregulated). Moreover, ten genes were downregulated, and 42 genes were upregulated in the bacterial secretion system pathway. The type II secretion system and type III secretion system were downregulated, while the type VI secretion system was upregulated. Additionally, 40 polar flagellar genes that were responsible for swimming motility were downregulated in the BHI agar plate. Two lateral flagellar genes, VPA1548 (lafK, the transcriptional activator) and VPA1551 (fliS, a flagellar assembly gene), were upregulated. These two genes were downregulated in the ΔVPA1701 strain, indicating that VPA1701 may regulate the expression of the lateral flagellar genes responsible for the swarming motility of V. parahaemolyticus.
2.3. Transcriptional Levels of the Lateral Flagellar Genes in ΔVPA1701
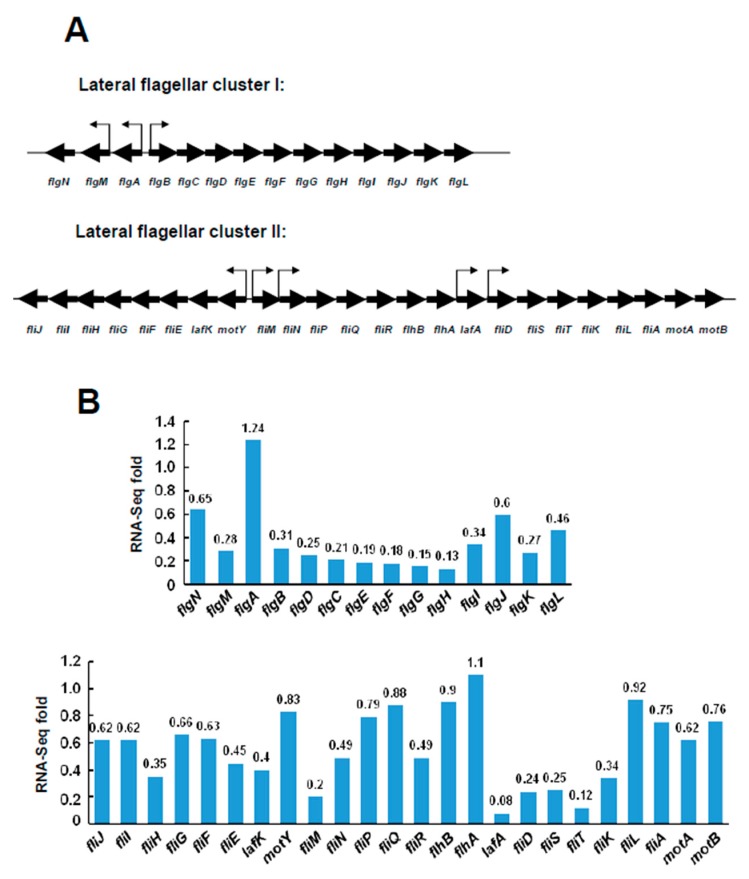
RNA-seq analysis indicated that the lateral flagellar genes were downregulated in the ΔVPA1701 strain. Then, 38 genes of the lateral flagellar system were divided into two clusters, with the lateral flagellar cluster I containing 14 genes (flgNM, flgA, and flgBCDEFGHIJKL, from VPA0261 to VPA0274) and the lateral flagellar cluster II containing 24 genes (fliJIHGFElafKmotY, fliM, fliNPQRflhBA, lafA, and fliDSTKLAmotAB, from VPA1532 to VPA1557). The arrows in Figure 4A indicate the predicted promoter regions of the flagellar gene clusters. The flagellar gene clusters were downregulated by VPA1701, as shown in Figure 4B. Almost all of the genes located in the lateral flagellar cluster I were downregulated in ΔVPA1701 compared to the WT, indicating that the expression of flgBCDEFGHIJKL was regulated by VPA1701 (Figure 4B). Furthermore, the lafA, fliD, fliS, fliT, and fliK genes of the lateral flagellar cluster II were significantly decreased in ΔVPA1701, suggesting that these genes were also regulated by VPA1701 (Figure 4B). Combined results of the RNA-seq analysis demonstrated that the VPA1701 protein as a transcriptional activator that regulated the expression of the lateral flagellar genes in V. parahaemolyticus.
Figure 4.
The transcription levels of lateral flagellar genes in ΔVPA1701. (A) Genetic map of the lateral flagellar cluster I and cluster II genes in V. parahaemolyticus. The black arrows represent the position of the predicted promoters. (B) Transcription levels of the two lateral flagellar genes in ∆VPA1701 compared to the WT indicated by RNA-seq.
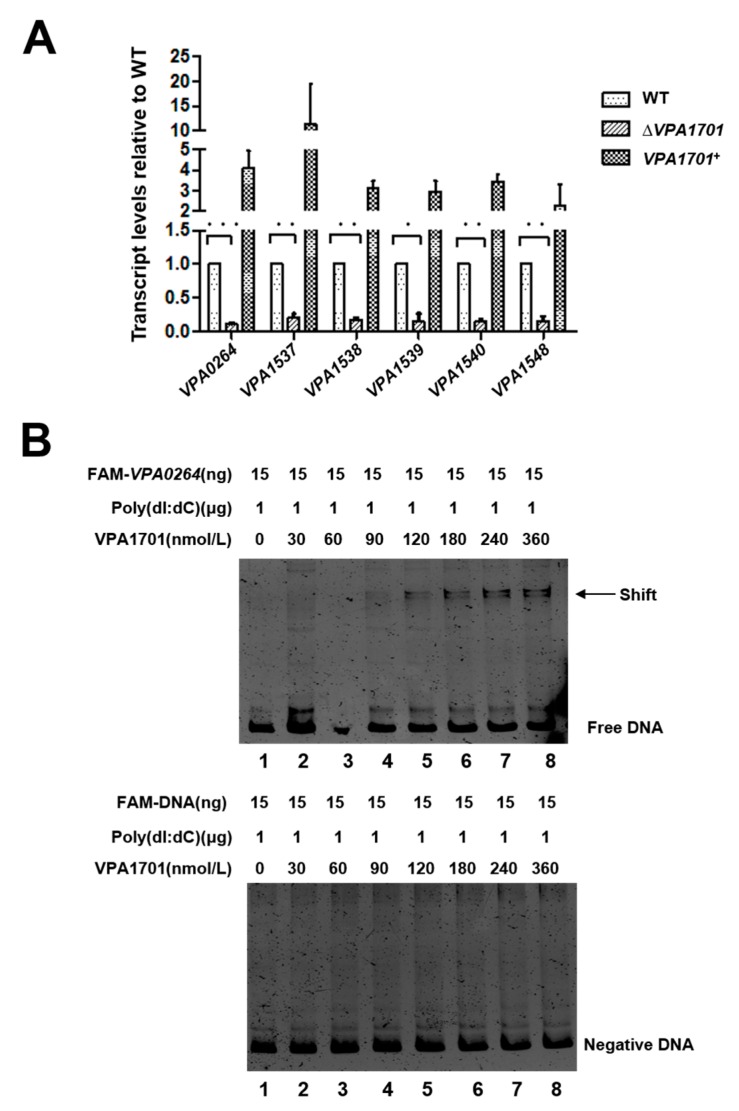
2.4. VPA1701 Directly Binds the Promoter of VPA0264 to Activate the Expression of Lateral Flagellar Genes
The RNA-seq data showed that VPA1701 significantly downregulated the lateral flagellar cluster I genes and the VPA1548-VPA1553 genes of the lateral flagellar cluster II. Thus, we selected VPA0264, located in the lateral flagellar cluster I, and VPA1537, VPA1538, VPA1539, VPA1540, and VPA1548, located in the lateral flagellar cluster II to confirm the regulation of the lateral flagellar genes by VPA170 using qRT-PCR. The expression of these six genes was significantly decreased in ΔVPA1701, and the complemented strain had the same transcription level as the WT (Figure 5A). Next, we used EMSA to observe the binding of VPA1701 to the promoter of lateral flagellar genes in vitro. The VPA1701 protein bound to the promoter region of VPA0264 in a concentration-dependent manner with high concentration of a non-specific competitor. The gyrB probe that was used as a negative control remained unbound with the highest concentration of VPA1701 protein (Figure 5B). However, the VPA1701 could not bind to the other promoters of lateral flagellar genes (Supplementary Figure S2). In conclusion, VPA1701 protein binds directly to the promoter region of flgBCDEFGHIJKL to activate the expression of the lateral flagellar system that controls the swarming motility of V. parahaemolyticus.
Figure 5.
VPA1701 directly bound to the promoter of VPA0264. (A) qRT-PCR analysis of the transcription level of lateral flagellar genes (VPA0264, VPA1537, VPA1538, VPA1539, VPA1540, and VPA1548) cultured in BHI agar plates at 30 °C. The data are presented as mean ± SD (n = 3); *: p < 0.01; **: p < 0.05; ***: p < 0.001. (B) EMSAs were performed with purified VPA1701 protein, and the promoter region of the VPA0264 was analyzed. 15 ng of FAM-labeled probes were added to the EMSA reaction and mixed with non-specific competitor DNA (poly (dI:dC)).
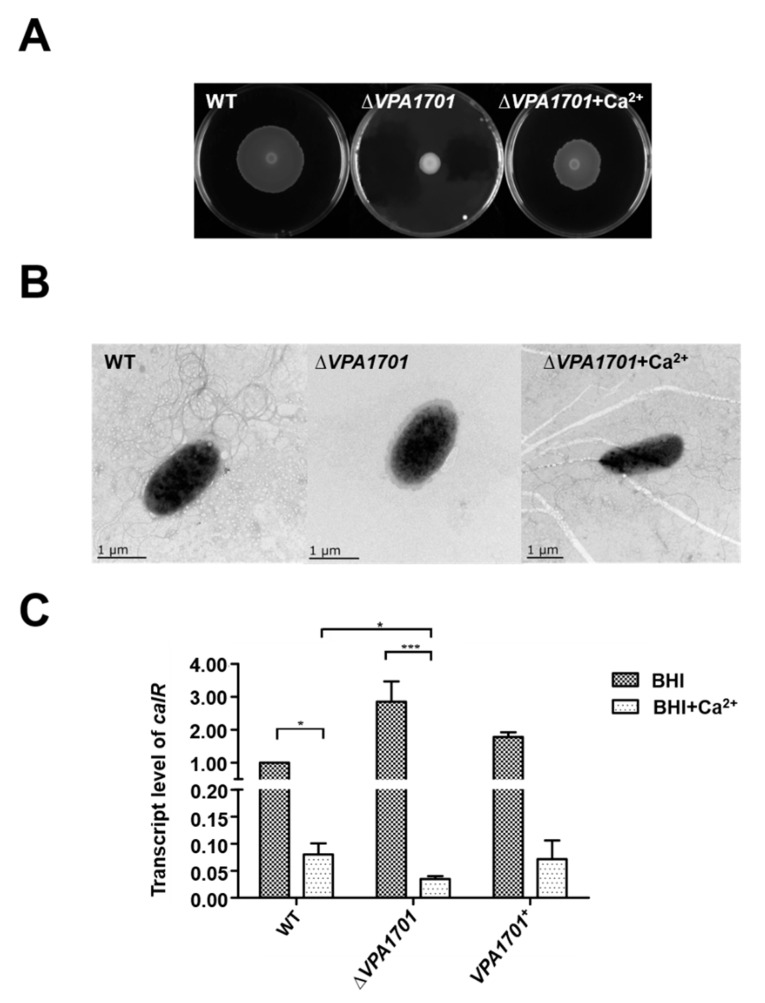
2.5. Calcium Restored the Swarming Ability of ∆VPA1701
CalR is a LysR-type transcriptional regulator that was inhibited by the Calcium in V. parahaemolyticus [25]. CalR was originally identified as a repressor of the swarming motility, TDH2, and T3SS1, and previous studies have demonstrated that the biofilm formation, hemolytic activity and T6SS2 also could be regulated by CalR in V. parahaemolyticus [25,33,34,35]. When the ∆VPA1701 strain was cultured in the BHI agar plates with 5 mM calcium, the swarming motility of ∆VPA1701 was recovered, but the swarming level was lower than the WT (Figure 6A). This may have been due to the decreased expression of lateral flagellar genes in the absence of VPA1701. TEM analysis further indicated that calcium restored the generation of the lateral flagellum of ∆VPA1701 (Figure 6B). In addition, we used qRT-PCR to detect the expression level of calR in WT, ∆VPA1701, and VPA1701+ in the BHI agar plates, with and without 5 mM calcium. The transcription level of calR significantly increased in ∆VPA1701 compared to the WT without calcium, and it decreased by 50% with 5 mM calcium (Figure 6C), indicating that the inhibition of calcium to calR had a greater influence on the expression of calR than VPA1701. Furthermore, the expression of calR was significantly decreased in ∆VPA1701 with 5 mM calcium compared to the ∆VPA1701 mutant strain without calcium (Figure 6C). Together, these results demonstrated that VPA1701 inhibited the expression of calR to regulate the swarming motility of V. parahaemolyticus, and calcium compensated for the inhibition of calR by VPA1701 and restored the swarming motility of ∆VPA1701.
Figure 6.
Calcium restores the swarming motility of ∆VPA1701. (A) The swarming motility analysis of WT and ∆VPA1701 in the BHI agar plates, with and without 5 mM calcium. (B) The generation of lateral flagella in the WT and ∆VPA1701 cultured in the BHI agar plates, with and without 5 mM, calcium was observed by TEM. (C) The transcription levels of calR in WT and ∆VPA1701 cultured in the BHI agar plates, with and without 5mM calcium, was assessed using qRT-PCR. The data are presented as mean ± SD (n = 3); *: p < 0.01; **: p < 0.05; ***: p < 0.001.
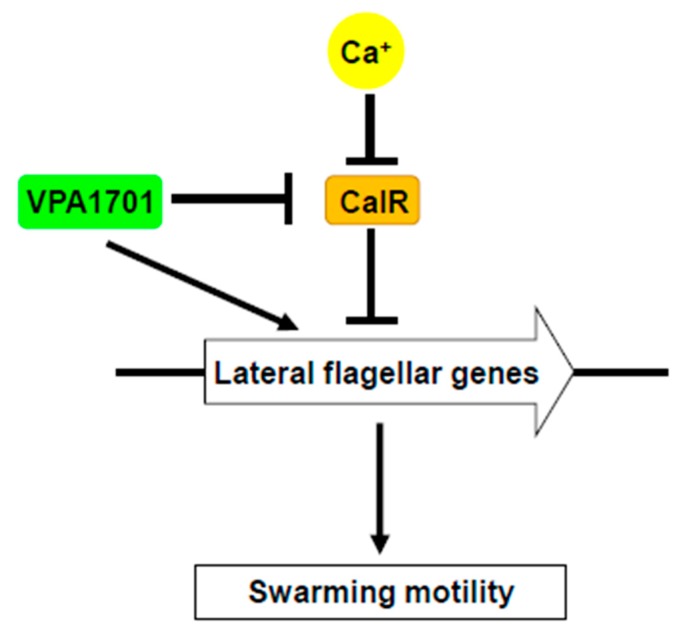
Based on these results, we propose a model for the regulation of the swarming motility by VPA1701 in V. parahaemolyticus (Figure 7). VPA1701 is a transcriptional regulator in the GntR family that specifically binds to the flgBCDEFGHIJKL promoter region to regulate the expression of lateral flagellar genes that are responsible for the swarming motility. Moreover, VPA1701 also inhibits the transcription of calR and promotes the expression of lateral flagellar genes in V. parahaemolyticus. In the presence of calcium, the expression of calR is inhibited, and the swarming motility of ∆VPA1701 is restored. These data facilitate an improved understanding of the regulatory networks of the transcription factor VPA1701 contributing to the swarming motility and colonization of V. parahaemolyticus.
Figure 7.
The regulatory network of VPA1701 for swarming motility in V. parahaemolyticus VPA1701 signaling increases the expression of lateral flagellar genes and inhibits the transcription of the calR gene to control the swarming motility of V. parahaemolyticus. Calcium and VPA1701 inhibit the expression of calR and de-repression of CalR to the lateral flagellar genes to control the swarming motility.
3. Materials and Methods
3.1. Bacterial Strains, Plasmids and Culture Conditions
V. parahaemolyticus RIMD 2210633 wild-type (WT) and its deviants (mutants and complements), as well as E. coli, were grown in Luria-Bertani (LB) broth at 37 °C, shaken at 200 rpm/min. The swarming motility analysis of V. parahaemolyticus strains were cultured in Brain Heart Infusion (BHI) with 1.5% agar. When necessary, suitable antibiotics were added: carbenicillin (Carb, 50 μg/mL), chloramphenicol (Cm, 25 μg/mL), and kanamycin (Km, 50 μg/mL). Detailed information on the bacterial strains, plasmids, and primers used in this study is listed in Supplementary Tables S1 and S2.
3.2. Construction of the VPA1701 Mutant and Complemented Strains
An in-frame deletion mutant of V. parahaemolyticus RIMD 2210633 VPA1701 was constructed as previously described in Reference [36]. The upstream fragment (643 bp) and downstream fragment (764 bp) of VPA1701 were amplified by PCR with specific primers (Supplementary Table S2) and connected by overlap PCR. The overlap fragments (1407 bp) were cloned into the suicide vector pDM4 with Sac I/Xho I sites and then transformed into E. coli DH5α λpir and E. coli SM10 λpir. The recombinant plasmid VPA1701::pDM4 was transformed into the wild type by conjugation, and the conjugant was plated in LB agar with Carb and Cm. Then, the second cross-over recombination was placed in LB agar containing 10% sucrose. The mutant strain (∆VPA1701) was verified by PCR (VPA1701-out/in-F/R) and sequencing.
To construct the VPA1701 complemented strain, the RBS and ORF regions of VPA1701 were amplified by PCR with specific primers (VPA1701-F/R) and then cloned into the pMMB207 plasmid with Sal I/Sac I sites. Then, the recombinant plasmid (VPA1701::pMMB207) was transformed into E. coli SM10 λpir and conjugated to the ∆VPA1701 strain. The conjugants were placed in LB agar containing Carb and Cm and confirmed by PCR (pMMB207-F/R) and sequencing. When necessary, 0.1 mM IPTG was added to induce the expression of VPA1701. The complemented strain was defined as VPA1701+.
3.3. Motility Analysis
The swimming and swarming motility assays were set up as previously described in Reference [37]. WT, ∆VPA1701, and VPA1701+ strains were cultured overnight and diluted to 1:100 in new LB broth. Then, 2 μL of diluted cultures (OD600 = 1.0) were spotted on the LB plates containing 0.3% agar at 37 °C for 12 h and BHI plates containing 1.5% agar at 30 °C for 24 h to investigate the swimming and swarming motility separately. All the experiments were conducted three times.
3.4. Transmission Electron Microscope (TEM) of the Lateral Flagellar
The WT, ∆VPA1701, and VPA1701+ strains were incubated in BHI agar for 15 h, washed gently with 0.01 M PBS, and then placed into separate 2 mL tubes. Subsequently, 5 μL of bacterial suspensions were dripped onto the copper grid with a supporting membrane and they were left for 5 min until the mesh was dry enough to cover. Then, 5% uranyl acetate was used to cover the dry samples for 30 min in a dry environment and observed by TEM (JEM 2100, Tokyo, Japan).
3.5. Quantification Real-Time Reverse Transcription PCR (qRT-PCR)
The V. parahaemolyticus WT, ∆VPA1701, and VPA1701+ strains were cultured in BHI agar plates for 15 h, and then the cell cultures were harvested from the plates. The total RNA was extracted using the Bacterial Total RNA Extraction Kit (Shangon Biotech, Shanghai, China) and treated with RNase-free DNase I to remove the contamination of genomic DNA. Then, 1 μg of total RNA was used to generate the first strand of cDNA by the reverse transcriptase (Takara, Tsuruga, Japan). The qRT-PCR was performed by the specific primers listed in the Supplementary Table S2, using an ABI StepOnePlus Real-Time PCR System with SYBR® Premix Ex TaqTM (Takara, Tsuruga, Japan). The transcription levels were normalized to the gyrB in each sample using the 2−ΔΔCt method, as described in Reference [38].
3.6. RNA-Seq Analysis
The V. parahaemolyticus WT and ∆VPA1701 strains were cultured in BHI agar plates for 15 h, the wild type was cultured in LB broth for 9 h, and then the bacterial cells were collected. Total RNA was extracted using the Bacterial Total RNA Extraction Kit (Shangon Biotech, Shanghai, China). RNA samples were digested with DNase I (Promega, Madison, WI, USA) to eliminate genomic DNA contamination. Three parallel RNA samples of each strain were sequenced using the illumina HiSeq (GENEWIZ, Suzhou, China). The subsequent procedures and statistical analysis were conducted as previously described in Reference [39].
3.7. Overexpression and Purification of VPA1701 Protein
The ORF region of the VPA1701 was amplified by PCR with specific primers (Supplementary Table S2) and then cloned into the pET30a plasmid. The recombinant plasmid (VPA1701::pET30a) was transformed into E. coli BL21(DE3), cultured in LB plates containing Carb to select the positive clone with the primers (pET30a-F/R), and then confirmed by sequencing.
The bacteria VPA1701::pET30a/BL21 was cultured overnight, diluted to 1% in 200 mL of fresh LB broth, and IPTG was added to induce the expression of protein until the OD600 was between 0.4–0.6. Then this was cultured at 22 °C for 12 h, shaken at 120 rpm/min. The cell cultures were collected, washed, and resuspended. The Ni2+ column purified the His-tagged VPA1701 protein. The VPA1701 protein was verified by 12% SDS-PAGE, and the concentration was determined using a nucleic acid analyzer (Nano-200, Shanghai, China).
3.8. Electrophoretic Mobility Shift Assay (EMSA)
The EMSA analysis was performed as previously described in Reference [38]. The DNA probes were amplified by the specific primers using a FAM fluorescent label (Supplementary Table S2) and purified using a DNA Gel Purification Kit. The concentration of the DNA probes was measured using the nucleic acid analyzer. The EMSA reaction (20 μL) was mixed with 15 ng of DNA probes, 1 μg of Poly-dIdC, and 4 μL of 5× Binding buffer (10 mM NaCl, 0.1 mM DTT, 0.1 mM EDTA, 10 mM Tris, pH 7.4), thereby increasing the amount of VPA1701-his protein. Then it was incubated at 25 °C for 30 min, where 2 μL of 10× EMSA/Gel shift loading buffer was added to each sample, and it was separated by a 6% native-PAGE gel in 0.5× TBE buffer (54 mM Tris-base, 27.5 mM Boric acid, 4.12 mM EDTA) for 2 h with 100 V. The gel was scanned using a Typhoon FLA 9500 (GE healthcare, Uppsala, Sweden). The gyrB promoter was used as a negative control.
3.9. Infant Rabbit Model of V. parahaemolyticus Infection
Infant rabbits were used to observe the intestinal colonization of V. parahaemolyticus [40,41]. The overnight cultured WT and ∆VPA1701 were placed in fresh LB medium at 30 °C for 18 h. Then, 2 mL of each sample was collected and washed using PBS three times, and then resuspended with a 4 ml 2.5% NaHCO3 solution for each sample. Three-day-old rabbits received 100 μL of bacterial suspension, delivered orogastrically, and they were observed for diarrhea and death for 36 h. Then the rabbits were euthanized and dissected, and 0.1 g of intestinal tissue was removed, smashed, and suspended with 1 mL of PBS. Then this was diluted to 10−1–10−6 and plated in LB agar containing Carb to determine the colonization of V. parahaemolyticus. All animal experiments of this study were approved by the institutional administrative committee and ethics committee of laboratory animals (Approval No. SYXK 2016-0019) and conducted following the guidelines approved by the Animal Welfare and Ethics Committees of Yangzhou University.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/8/4/235/s1. Table S1: Bacterial strains and plasmids used in this study. Table S2: Primers used in this study. Figure S1: Multiple amino acid sequence alignments of VPA1701 proteins in Vibrios. The amino acid sequence alignments of VPA1701 protein among V. parahaemolyticus and the homology proteins in Vibrio harveyi, Vibrio campbellii, Vibrio vulnificus and Vibrio cholerae. The identical and highly conserved amino acid sequence were indicated by red rectangular frames. Figure S2: No specific binding of VPA1701 and the promoter regions of VPA1548 and VPA1551. EMSA assay using purified VPA1701 protein and VPA1548 and VPA1551 probes with the increasing amount of VPA1701 protein.
Author Contributions
Conceptualization, D.G. and X.J.; Methodology, D.G., H.M., Y.L. and H.G.; Softwore, D.G.; Validation, H.M.; Formal Analysis, Y.L. and H.G.; Investigation, H.M.; Resources, X.J.; Data Curation, D.G.; Writing—Original Draft preparation, D.G. and H.M.; Writing—Review & Editing, D.G. and X.J. Visualization, H.M.; Supervision, X.J.; Project Adminstration, X.J.; Funding Acquisition, D.G.
Funding
This work was supported by the National Natural Science Foundation of China (31700122) and China Postdoctoral Science Foundation (2018M632388).
Conflicts of Interest
The authors declare no conflict of interest. The funding sponsors had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript, and in the decision to publish the results.
References
- 1.Nair G.B., Ramamurthy T., Bhattacharya S.K., Dutta B., Takeda Y., Sack D.A. Global dissemination of Vibrio parahaemolyticus serotype O3:K6 and its serovariants. Clin. Microbiol. Rev. 2007;20:39–48. doi: 10.1128/CMR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Austin B. Vibrios as causal agents of zoonoses. Vet. Microbiol. 2010;140:310–317. doi: 10.1016/j.vetmic.2009.03.015. [DOI] [PubMed] [Google Scholar]
- 3.Makino K., Oshima K., Kurokawa K., Yokoyama K., Uda T., Tagomori K., Iijima Y., Najima M., Nakano M., Yamashita A., et al. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of Vibrio cholerae. Lancet. 2003;361:743–749. doi: 10.1016/S0140-6736(03)12659-1. [DOI] [PubMed] [Google Scholar]
- 4.Salomon D., Gonzalez H., Updegraff B.L., Orth K. Vibrio parahaemolyticus type VI secretion system 1 is activated in marine conditions to target bacteria, and is differentially regulated from system 2. PLoS ONE. 2013;8:e61086. doi: 10.1371/journal.pone.0061086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han N., Mizan M.F.R., Jahid I.K., Ha S.D. Biofilm formation by Vibrio parahaemolyticus on food and food contact surfaces increases with rise in temperature. Food Control. 2016;70:161–166. doi: 10.1016/j.foodcont.2016.05.054. [DOI] [Google Scholar]
- 6.Li L., Meng H., Gu D., Li Y., Jia M. Molecular mechanisms of Vibrio parahaemolyticus pathogenesis. Microbiol. Res. 2019;222:43–51. doi: 10.1016/j.micres.2019.03.003. [DOI] [PubMed] [Google Scholar]
- 7.Salamone M., Nicosia A., Bennici C., Quatrini P., Catania V., Mazzola S., Ghersi G., Cuttitta A. Comprehensive Analysis of a Vibrio parahaemolyticus Strain Extracellular Serine Protease VpSP37. PLoS ONE. 2015;10:e0126349. doi: 10.1371/journal.pone.0126349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salamone M., Nicosia A., Ghersi G., Tagliavia M. Vibrio Proteases for Biomedical Applications: Modulating the Proteolytic Secretome of V. alginolyticus and V. parahaemolyticus for Improved Enzymes Production. Microorganisms. 2019;7:387. doi: 10.3390/microorganisms7100387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merrell B.R., Walker R.I., Joseph S.W. In vitro and in vivo pathologic effects of Vibrio parahaemolyticus on human epithelial cells. Can. J. Microbiol. 1984;30:381–388. doi: 10.1139/m84-056. [DOI] [PubMed] [Google Scholar]
- 10.O’Boyle N., Houeix B., Kilcoyne M., Joshi L., Boyd A. The MSHA pilus of Vibrio parahaemolyticus has lectin functionality and enables TTSS-mediated pathogenicity. Int. J. Med. Microbiol. 2013;303:563–573. doi: 10.1016/j.ijmm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 11.McCarter L., Silverman M. Surface-induced swarmer cell differentiation of Vibrio parahaemolyticus. Mol. Microbiol. 1990;4:1057–1062. doi: 10.1111/j.1365-2958.1990.tb00678.x. [DOI] [PubMed] [Google Scholar]
- 12.Kearns D.B. A field guide to bacterial swarming motility. Nat. Rev. Microbiol. 2010;8:634–644. doi: 10.1038/nrmicro2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Murray T.S., Ledizet M., Kazmierczak B. Swarming motility, secretion of type 3 effectors and biofilm formation phenotypes exhibited within a large cohort of Pseudomonas aeruginosa clinical isolates. J. Med. Microbiol. 2010;59:511–520. doi: 10.1099/jmm.0.017715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Merino S., Shaw J.G., Tomás J.M. Bacterial lateral flagella: An inducible flagellar system. FEMS Microbiol. Lett. 2006;263:127–135. doi: 10.1111/j.1574-6968.2006.00403.x. [DOI] [PubMed] [Google Scholar]
- 15.Inaba S., Nishigaki T., Takekawa N., Kojima S., Homma M. Localization and domain characterization of the SflA regulator of flagellar formation in Vibrio alginolyticus. Genes Cells. 2017;22:619–627. doi: 10.1111/gtc.12501. [DOI] [PubMed] [Google Scholar]
- 16.McCarter L.L. Polar Flagellar Motility of the Vibrionaceae. Microbiol. Mol. Biol. Rev. 2001;65:445–462. doi: 10.1128/MMBR.65.3.445-462.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boles B.R., McCarter L.L. Insertional Inactivation of Genes Encoding Components of the Sodium-Type Flagellar Motor and Switch of Vibrio parahaemolyticus. J. Bacteriol. 2000;182:1035–1045. doi: 10.1128/JB.182.4.1035-1045.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim Y.K., McCarter L.L. Cross-Regulation in Vibrio parahaemolyticus: Compensatory Activation of Polar Flagellar Genes by the Lateral Flagellar Regulator LafK. J. Bacteriol. 2004;186:4014–4018. doi: 10.1128/JB.186.12.4014-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim Y.K., McCarter L.L. Analysis of the polar flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 2000;182:3693–3704. doi: 10.1128/JB.182.13.3693-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stewart B.J., McCarter L.L. Lateral flagellar gene system of Vibrio parahaemolyticus. J. Bacteriol. 2003;185:4508–4518. doi: 10.1128/JB.185.15.4508-4518.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shinoda S., Okamoto K. Formation and function of Vibrio parahaemolyticus lateral flagella. J. Bacteriol. 1977;129:1266–1271. doi: 10.1128/jb.129.3.1266-1271.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tian Y., Wang Q., Liu Q., Ma Y., Cao X., Guan L., Zhang Y. Involvement of LuxS in the regulation of motility and flagella biogenesis in Vibrio alginolyticus. Biosci. Biotechnol. Biochem. 2008;72:1063–1071. doi: 10.1271/bbb.70812. [DOI] [PubMed] [Google Scholar]
- 23.Henares B.M., Xu Y., Boon E.M. A Nitric Oxide-Responsive Quorum Sensing Circuit in Vibrio harveyi Regulates Flagella Production and Biofilm Formation. Int. J. Mol. Sci. 2013;14:16473–16484. doi: 10.3390/ijms140816473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu R., Tang H., Qiu Y., Yang W., Yang H., Zhou D., Huang X., Hu L., Zhang Y. Quorum sensing regulates the transcription of lateral flagellar genes in Vibrio parahaemolyticus. Future Microbiol. 2019;14:1043–1053. doi: 10.2217/fmb-2019-0048. [DOI] [PubMed] [Google Scholar]
- 25.Gode-Potratz C.J., Chodur D.M., McCarter L.L. Calcium and Iron Regulate Swarming and Type III Secretion in Vibrio parahaemolyticus. J. Bacteriol. 2010;192:6025–6038. doi: 10.1128/JB.00654-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Srivastava D., Hsieh M.L., Khataokar A., Neiditch M.B., Waters C.M. Cyclic di-GMP inhibits Vibrio cholerae motility by repressing induction of transcription and inducing extracellular polysaccharide production. Mol. Microbiol. 2013;90:1262–1276. doi: 10.1111/mmi.12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park S., Park Y.H., Lee C.R., Kim Y.R., Seok Y.J. Glucose induces delocalization of a flagellar biosynthesis protein from the flagellated pole. Mol. Microbiol. 2016;101:795–808. doi: 10.1111/mmi.13424. [DOI] [PubMed] [Google Scholar]
- 28.Chen L., Qiu Y., Tang H., Hu L.F., Yang W.H., Zhu X.J., Huang X.X., Wang T., Zhang Y.Q. ToxR Is Required for Biofilm Formation and Motility of Vibrio Parahaemolyticus. Biomed. Environ. Sci. 2018;31:848–850. doi: 10.3967/bes2018.112. [DOI] [PubMed] [Google Scholar]
- 29.Hubbard T.P., Chao M.C., Abel S., Blondel C.J., Abel Zur Wiesch P., Zhou X., Davis B.M., Waldor M.K. Genetic analysis of Vibrio parahaemolyticus intestinal colonization. Proc. Natl. Acad. Sci. USA. 2016;113:6283–6288. doi: 10.1073/pnas.1601718113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haydon D.J., Guest J.R. A new family of bacterial regulatory proteins. FEMS Microbiol. Lett. 1991;63:291–295. doi: 10.1111/j.1574-6968.1991.tb04544.x. [DOI] [PubMed] [Google Scholar]
- 31.Ushijima B., Häse C.C. Influence of Chemotaxis and Swimming Patterns on the Virulence of the Coral Pathogen Vibrio coralliilyticus. J. Bacteriol. 2018;200:e00791-17. doi: 10.1128/JB.00791-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaques S., McCarter L.L. Three new regulators of swarming in Vibrio parahaemolyticus. J. Bacteriol. 2006;88:2625–2635. doi: 10.1128/JB.188.7.2625-2635.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Osei-Adjei G., Zhang Y., Gao H., Yang W., Zhou D., Huang X., Yang H., Zhang Y. CalR is required for the expression of T6SS2 and the adhesion of Vibrio parahaemolyticus to HeLa cells. Arch. Microbiol. 2017;199:931–938. doi: 10.1007/s00203-017-1361-6. [DOI] [PubMed] [Google Scholar]
- 34.Gao H., Zhang L., Osei-Adjei G., Yang W., Zhou D., Huang X., Yang H., Yin Z., Zhang Y. Transcriptional regulation of cpsQ-mfpABC and mfpABC by CalR in Vibrio parahaemolyticus. Microbiologyopen. 2017;6:e470. doi: 10.1002/mbo3.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y., Zhang Y., Gao H., Zhang L., Yin Z., Huang X., Zhou D., Yang H., Yang W., Wang L. Vibrio parahaemolyticus CalR down regulates the thermostable direct hemolysin (TDH) gene transcription and thereby inhibits hemolytic activity. Gene. 2017;613:39–44. doi: 10.1016/j.gene.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Zhou X., Konkel M.E., Call D.R. Vp1659 is a Vibrio parahaemolyticus type III secretion system 1 protein that contributes to translocation of effector proteins needed to induce cytolysis, autophagy, and disruption of actin structure in HeLa cells. J. Bacteriol. 2010;192:3491–3502. doi: 10.1128/JB.01493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whitaker W.B., Richards G.P., Boyd E.F. Loss of Sigma Factor RpoN Increases Intestinal Colonization of Vibrio parahaemolyticus in an Adult Mouse Model. Infect. Immun. 2014;82:544–556. doi: 10.1128/IAI.01210-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gu D., Guo M., Yang M., Zhang Y., Zhou X., Wang Q. A σE-Mediated Temperature Gauge Controls a Switch from LuxR-Mediated Virulence Gene Expression to Thermal Stress Adaptation in Vibrio alginolyticus. PLoS Pathog. 2016;12:e1005645. doi: 10.1371/journal.ppat.1005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu Y., Zhao L., Yang M., Yin K., Zhou X., Leung K.Y., Liu Q., Zhang Y., Wang Q. Transcriptomic dissection of the horizontally acquired response regulator EsrB reveals its global regulatory roles in the physiological adaptation and activation of T3SS and the cognate effector repertoire in Edwardsiella piscicida during infection toward turbot. Virulence. 2017;8:1355–1377. doi: 10.1080/21505594.2017.1323157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Abel S., Waldor M.K. Infant Rabbit Model for Diarrheal Diseases. Curr. Protoc. Microbiol. 2015;38:6A.6.1-15. doi: 10.1002/9780471729259.mc06a06s38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ritchie J.M., Rui H., Zhou X., Iida T., Kodoma T., Ito S., Davis B.M., Bronson R.T., Waldor M.K. Inflammation and Disintegration of Intestinal Villi in an Experimental Model for Vibrio parahaemolyticus-Induced Diarrhea. PLoS Pathog. 2012;8:e1002593. doi: 10.1371/journal.ppat.1002593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.