Abstract
The transition to reproduction is a crucial step in the life cycle of any organism. In Arabidopsis thaliana the establishment of reproductive growth can be divided into two phases: Firstly, cauline leaves with axillary meristems are formed and internode elongation begins. Secondly, lateral meristems develop into flowers with defined organs. Floral shoots are usually determinate and suppress the development of lateral shoots. Here, we describe a transposon insertion mutant in the Nossen accession with defects in floral development and growth. Most strikingly is the outgrowth of stems from the axillary bracts of the primary flower carrying secondary flowers. Therefore, we named this mutant flower-in-flower (fif). However, the transposon insertion in the annotated gene is not the cause for the fif phenotype. By means of classical and genome sequencing-based mapping, the mutation responsible for the fif phenotype was found to be in the LEAFY gene. The mutation, a G-to-A exchange in the second exon of LEAFY, creates a novel lfy allele and results in a cysteine-to-tyrosine exchange in the α1-helix of LEAFY’s DNA-binding domain. This exchange abolishes target DNA-binding, whereas subcellular localization and homomerization are not affected. To explain the strong fif phenotype against these molecular findings, several hypotheses are discussed.
Keywords: Arabidopsis thaliana, floral development, flower morphology, Ds transposon, classical/sequencing-based mapping, LEAFY, DNA-binding
1. Introduction
The development of flowers is indispensable for the reproductive success of angiosperm plants. During vegetative growth, the shoot apical meristem (SAM) develops leaves and/or branches, the latter with their own SAMs. After the switch to reproductive growth, the apical meristems give rise to flowers. Floral development differs crucially from vegetative shoot growth, as the flower possesses several types of organs of which the number, arrangement and morphology are species-specific. Furthermore, the development of lateral shoots is inhibited in flowers and floral shoots are determinate after the last reproductive organs have been initiated [1,2,3]. Thus, the coordination of complex molecular processes is necessary for successful floral development.
There has been significant progress in recent years towards understanding the molecular mechanisms underlying flower formation [4,5,6,7,8]. Central to this was the identification and cloning of the genes that initiate and maintain floral development in plant species, including Arabidopsis thaliana. The most intriguing discovery was the Arabidopsis loss-of-function mutants with structures that are intermediate between floral and vegetative shoots. The cloning of the corresponding genes revealed the existence of the master regulators required for the floral initiation process. To date, five regulatory master genes are known: LEAFY (LFY), APETALA1 (AP1), CAULIFLOWER (CAL), APETALA2 (AP2) and UNUSUAL FLORAL ORGANS (UFO) [3]. LFY and AP1 play a primary role in initiating the floral program, as the corresponding loss-of-function mutants do not generate shoots with floral characteristics and the ectopic expression of either gene induces precocious flower formation [9,10,11]. LFY, AP1 and CAL encode for transcription factors and are expressed predominantly in floral primordia [12,13,14].
During plant vegetative growth, LFY expression increases in newly formed leaves until a certain threshold is reached [15]. LFY then induces the expression of AP1/CAL genes by activation of the AP1/CAL promoters. Through their mutual transcriptional up-regulation, LFY and AP1/CAL cooperate to cause the floral transition [16,17]. Once the floral meristem is established, the floral initiation gene functions govern its spatial patterning by inducing the expression of the floral homeotic ABC genes, such as AP2, AP3, Pistillata (PI) and AGAMOUS (AG). The ABC gene functions in turn control the identity of the stereotypically arranged Arabidopsis floral organs [18,19].
In the course of our study of the influence of abiotic stress on flower symmetry, we searched for novel insertion mutants with defects in floral development or morphology in different Arabidopsis thaliana accessions. We focused on genes that had not yet been linked to flowering. A Ds transposon insertion mutant, which developed secondary inflorescences with partially aberrant flowers and had an aberrant growth phenotype, was identified in the No-0 accession. The wild type allele of the gene carrying the Ds transposon codes for a cystein/histidine-rich C1 domain protein [20,21]. However, a thorough genetic analysis revealed that the transposon-inserted allele is not the cause of the observed floral and developmental phenotype. Using classical mapping and mapping-by-sequencing, we eventually found a novel mutant allele of LFY to be responsible for the aberrant floral development, flower morphology and addressed the molecular reason for LFY malfunction.
2. Results
2.1. The Flower-in-Flower (fif) Transposon Insertion Line Displays a Novel Flower Phenotype
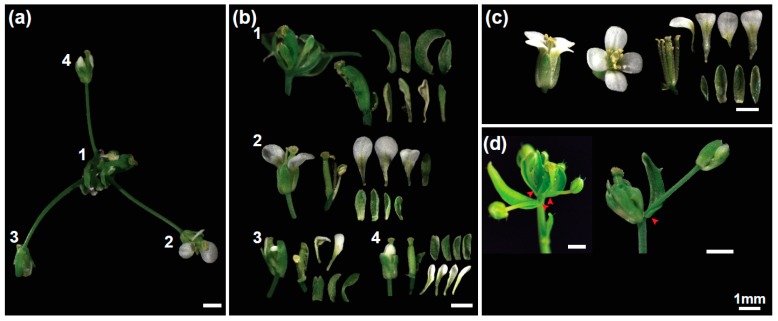
In order to identify novel Arabidopsis thaliana mutants with defects in flowering we screened the RIKEN Arabidopsis Phenome Information Database (RAPID; [22]). RAPID also covers a Ds transposon mutant collection in the Arabidopsis Nossen-0 (No-0) background [23,24]. We identified a transposon-tagged line (15-3794-1), which developed secondary inflorescences with partially aberrant flowers (Figure 1a). Because of this phenotype, we named this Arabidopsis mutant flower-in-flower (fif).
Figure 1.
Flower phenotype of the Arabidopsis thaliana (No-0) flower-in-flower (fif) mutant. (a) Overview over a representative fif mutant “inflorescence” displaying different flower types 1 to 4. (b) Floral organs of the primary fif flower (1) and different secondary fif flowers (2–4). (c) Flower of the wild type No-0 accession. (d) Primary flowers of the fif mutant with stems that outgrow from axillary bract meristems (red arrow heads) and carry secondary flowers. Size bar: 1 mm.
Wild type Arabidopsis flowers does not have bracts but consist of four concentric rings of four sepals, four petals, six stamens and two fused carpels (Figure 1c). In contrast, the primary flower of the fif mutant had bracts as well as sepals but the petals were incompletely developed or entirely missing (Figure 1b). In addition, there were either no stamens or the stamens displaying an aberrant development (Figure 1b). Furthermore, there were more than two carpels per flower, which were not or only partially overgrown and sterile. Most obvious, however, was the outgrowth of stems from the axillary meristems of the leaf-like sepal structures, which carried terminal secondary flowers (1d). A few secondary fif flowers showed a wild type-like phenotype and were fertile (Figure 1b,d).
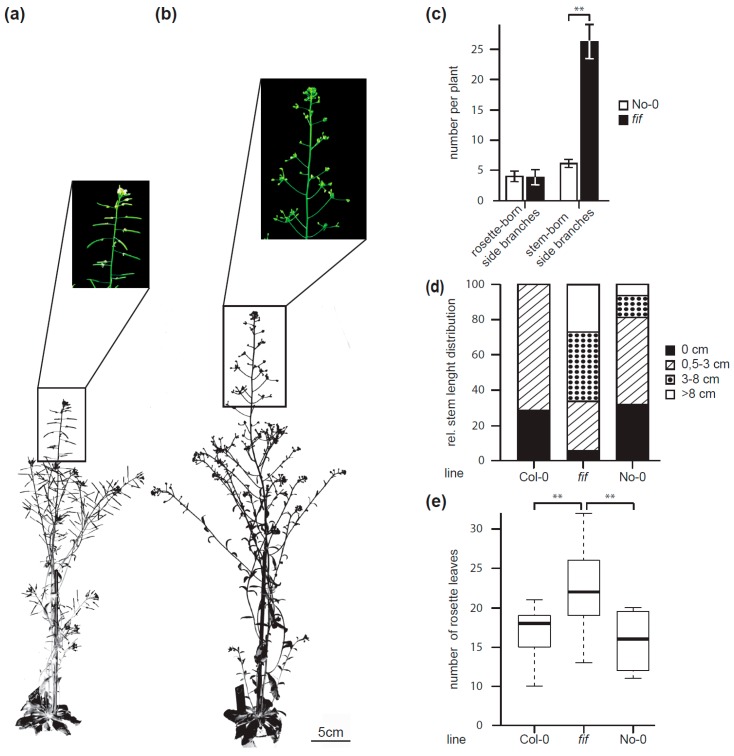
Furthermore, the fif mutant plant displayed an aberrant growth habitus compared to wild type No-0 (Figure 2a,b). The “bushy” appearance of the fif mutant was due to an enhanced number of stem-born side branches compared to wild type No-0, whereas the number of rosette-born side shoots was the same in fif and wild type No-0 plants (Figure 2c). Furthermore, the stem length of the fif mutant plants a month after sowing showed a different size distribution compared to wild type No-0 and Col-0 (Figure 2d). Whereas the majority of the stem length of the No-0 and Col-0 plants was at 0 cm and between 0.5 and 3.0 cm, the fif plants by majority exhibited stem lengths between 3 and 8 cm (Figure 2d). In addition, the fif mutant plants showed extended vegetative growth and, therefore, delayed flowering compared to wild type No-0 and Col-0 as indicated by the enhanced number of rosette leaves until flowering (Figure 2e).
Figure 2.
Growth habitus and flowering time of wild type No-0, wild type Col-0 and fif mutant plants. Overview over the growth habitus and magnification of the inflorescence of 6.5-weeks old wild type No-0 (a) and fif plants (b), grown side-by-side in the greenhouse. Size bar: 5.0 cm. (c) Number of rosette-born side branches and stem-born side branches of wild type No-0 (white bars) and fif (black bars) plants. Error bars indicate the standard deviation of the mean. The statistical significance (nNo-0 = 33, nfif = 25) was testet by two-sited t-test (***: p = 2 × 10−23). (d) Relative distribution of stem length in wild type No-0, wild type Col-0 and fif mutant plants 31 days after sowing (nCol-0 = 7, nNo-0 = 16, nfif = 89). (e) Number of rosette leaves at the onset of senescence for wild type No-0, wild type Col-0 and fif mutant plants. The data are presented in Box-and-Whisker plots including the median (thick line), the upper and lower quartile (+/− 25%, white boxes) and the maximum and minimum (dottet line). The statistical significance (nNo-0 = 4, nCol-0 = 9, nfif = 38) was tested with ANOVA followed by a Tukey honest significant difference post-hoc test (*: p < 0.05; **: p < 0.01; ***: p < 0.001).
2.2. The Transposon Insertion is Not Responsible for the fif Phenotype
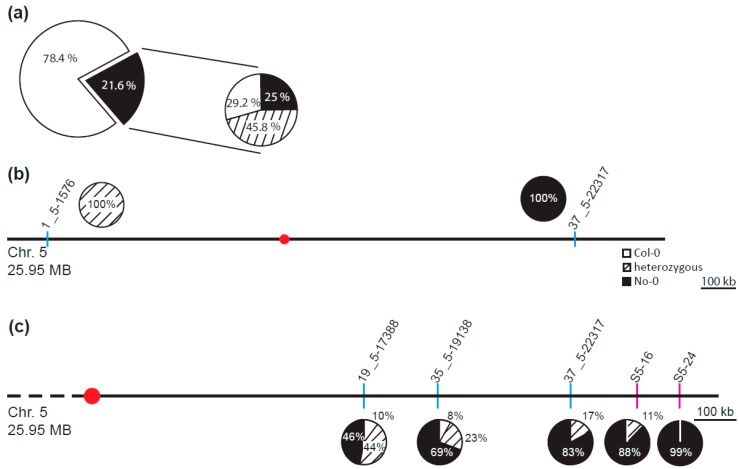
According to the RIKEN RAPID and our own genotyping results, the Ds transposon was located in the second exon of the gene At1g20990 that codes for a putative cysteine/histidine-rich C1 domain protein with a yet unknown function. To validate, whether the mutant allele of the At1g20990 locus is actually the cause for the fif phenotype, we performed a (co-) segregation analysis by backcrossing the fif mutant with wild type No-0 in both directions (♀fif × ♂No-0, ♀No-0 × ♂fif). Irrespective of the direction, the crosses were successful as demonstrated by PCR on genomic DNA extracted from F1 plants using Ds transposon- and At1g20990-specific primers (Figure S1). All tested F1 plants were heterozygous for the Ds transposon and wild type At1g20990 and displayed wild type floral organs and growth (Figure S1). Therefore, the mutation that causes the fif phenotype is recessive. Next, six F1 plants were self-fertilized and 20 to 30 progenies each analysed for their pheno- and genotypes, respectively. As shown in Figure 3a, around one quarter of the F2 plants displayed the fif phenotype indicating that it is caused by a single mutant gene. Intriguingly, our genotyping results showed that the Ds transposon insertion did not co-segregate with the fif phenotype: 29% of the fif phenotype-displaying plants did not contain the transposon, an additional 45.8% contained the transposon insertion only heterozygously (Figure 3a). These results prove that the Ds insertion into the At1g20990 locus is not the cause for the fif phenotype.
Figure 3.
Segregation analysis and mapping of the mutant locus causal for the fif mutant phenotype. (a) Segregation of the floral phenotype and the Ds transposon insertion within the combined F2 population of (♀fif × ♂No-0) and (♀No-0 × ♂fif) backcrosses showing either the wild type (78.4%) or the fif floral phenotype (21.6%) (left) and distribution of the transposon insertions within the plants of the F2 population that displayed the fif floral phenotype (right); white circle outcut: no transposon insertion (29.2%), striped outcut: heterozygous for the Ds transposon insertion (45.8%), black outcut: homozygous for the Ds transposon insertion (25.0%). (b,c) Insertion and DELetion INDEL marker- and single nucleotide polymorphism (SNP)-based derived cleaved amplified polymorphic sequences (dCAP) marker-associated containment of the fif locus using a mapping population generated by a cross of the fif mutant (No-0) with wild type Col-0. Schematic representation of the Arabidopsis thaliana chromosome 5 (sizes in MB) and the localization of the chromosome-specific INDEL markers initially used for mapping (codes above blue lines) (b). Schematic representation of the q-arm of chromosome 5 and the localization of INDEL (codes above the blue lines) and SNP-based dCAP markers (codes above red lines) used for fine mapping (c). The pie charts show the distribution of the No-0 and Col-0 genotypes for chromosome 5 (b) and the q-arm of chromosome 5 (c). White circular outcut: homozygous for Col-0, striped outcut: heterozygous for Col-0/No-0, black outcut: homozygous for No-0; red dot: localization of the centromere.
2.3. The fif Phenotype is Caused by a Novel Allele of LEAFY (LFY)
To identify the mutant locus genetically responsible for the fif phenotype, we combined a classical mapping [25,26,27] with a mapping-by-sequencing approach [28,29]. To establish a mapping population, fif mutant plants (No-0) were crossed in both direction with plants of the Col-0 accession. Irrespective of the crossing direction, all the F1 plant displayed a wild type phenotype. Eight F1 plants were self-fertilized and 1582 F2 plants characterized phenotypically. In accordance with the self-crossing results described above, around 25% of the F2 plants (437 of the 1582) showed the fif phenotype. Leaf material was harvested from 425 of the 437 F2 plants in groups of 15 to 20 individuals; in addition, leaf material from 200 F2 plants was collected individually. Genomic DNA was extracted and used for classical mapping. Using chromosome-specific INsertion and DELetion (INDEL) markers [27] the mutant locus was mapped to the q-arm of chromosome 5 (Figure 3b). Two additional INDEL markers and two single nucleotide polymorphism (SNP)-based derived cleaved amplified polymorphic sequences (dCAP) markers [25,26] limited the quantitative trait locus (QTL) responsible for the fif phenotype to the terminal end of chromosome 5’s q-arm (Figure 3c, dCAP S5-24: 99% No-0).
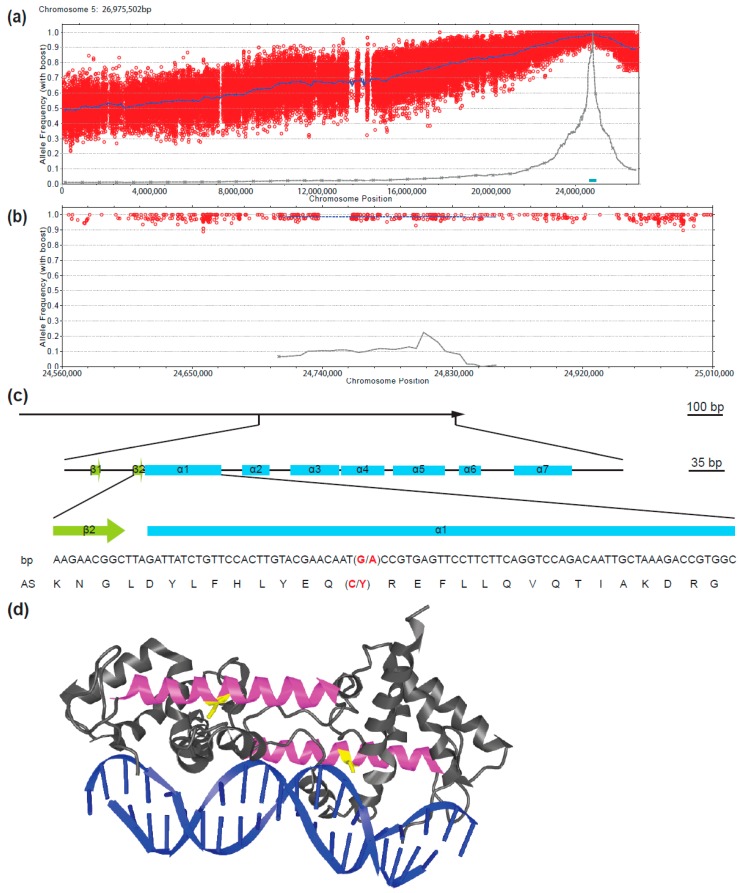
To establish the exact localization of the mutant locus, we deep-sequenced the total genome of 245 homozygous fif mutant plants derived from the fif (No-0) x WT (Col-0) crosses described above, and determined the frequencies of No-0 and Col-0 alleles along the chromosomes. Whereas the heterozygous distribution of No-0 and Col-0 sequences was found to be equal with respect to chromosomes 1 to 4 (Figure S2a–d), there was a very significant deviation towards No-0 sequences at the terminal end of chromosome 5 (Figure 4a). A detailed examination of this 300 kb stretch revealed 100% identity with the No-0 sequence (Figure 4b). This sequence stretch conformed with the QTL identified by the classical mapping.
Figure 4.
Identification of the fif-related SNP in the second exon of the LEAFY (LFY) locus on chromosome 5 by genome sequencing of a mapping population generated by a cross of the fif mutant (No-0) with wild type Col-0. (a) Allele frequency analysis of the No-0 genotype within chromosome 5 of the recombinant mutant pool. Each red circle refers to a SNP marker distinguishing the No-0 and Col-0 genotypes. The blue line refers to a 200 kb sliding window analysis of the allele frequencies. The brown line and blue box highlight the estimated mapping intervals (x-axis: genomic location; y-axis: Nos allele frequency). (b) Like (a), but only showing the 300 kb mapping interval. (c) Sequence of the LFY gene showing the fif SNP (G to A exchange, red) and the resulting amino acid exchange (C to Y, red) within the DNA-binding domain of the LFY protein. Green boxes: β-sheets; blue boxes: α-helices. (d) Structural representation of the wild type LFY dimer bound to DNA according to Hames and colleagues (2008). The α1 helix in the monomers is shown in magenta and the cysteine (C) in yellow that are mutated to tyrosine (Y) in the fif mutant.
A detailed comparison of the fif and wild type No-0 sequence in this 300 kb stretch revealed a single SNP, which did not result in a silent mutation but caused a change in a codon. This SNP was also found in all the 143 individually tested fif mutant plants and reflected a single guanine-to-adenine exchange in the second exon of the LEAFY (LFY) gene (At5g61850, Figure 4c). This mutation caused a cysteine-to-tyrosine amino acid exchange at position 263 in the DNA-binding domain of the LFY protein (Figure 4d). To prove that this point mutation causes the fif phenotype, we transformed the fif mutant (No-0) with constructs coding for wild type LFY-GFP under the control of the Arabidopsis ubiquitin 10 (UBI10) promoter. The functionality of C- (and N-terminal) GFP fusions of LFY was previously shown by the genetic complementation of the lfy-12 mutant phenotype [30]. Although expressed to a very low level, LFY-GFP was detectable inside the nucleus of cells of the transgenic fif mutant plants and complemented the fif phenotype (Figure 5).
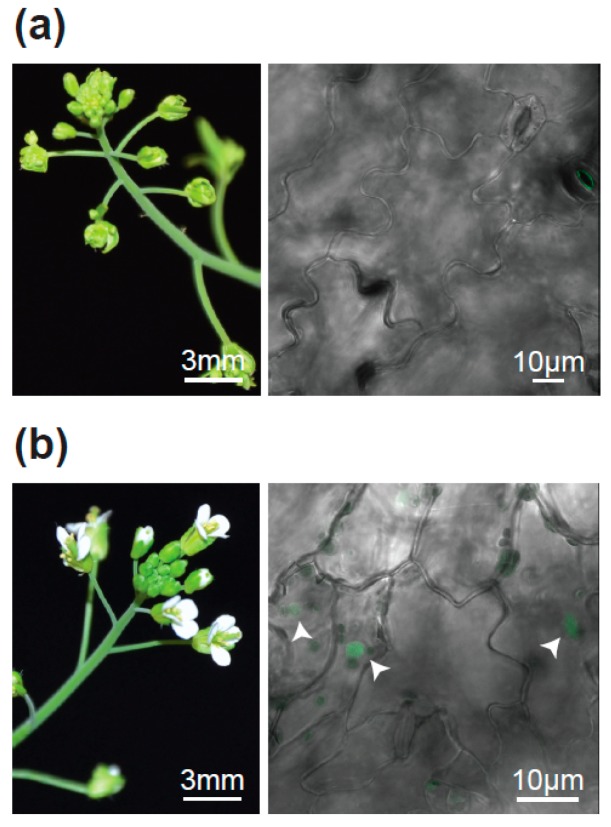
Figure 5.
Complementation of the fif mutant phenotype by the UBI10 promoter-driven expression of wild type LFY-GFP. (a) Inflorescence (left) and confocal image of epidermal cells (right) of a representative non-transformed fif mutant plant. (b) As in (a), but for a representative LFY-GFP transgenic plant in the fif mutant background. The fluorescent nuclei are highlighted by arrowheads. Three independent transgenic lines were obtained displaying nuclear localization of LFY-GFP and in parallel the complementation of the fif phenotype.
2.4. LFYFIF Impairs DNA-Binding Capability but Shows Wild Type Intracellular Localization and Homomerization
Having identified a new LFY allele to be responsible for the fif phenotype, we next analysed the putative consequences of the Cys263-to-Tyr exchange for LFY protein properties at molecular and cellular level.
To test a putative alteration in subcellular localization, a C-terminal GFP fusion of wild type LFY and a C-terminal RFP fusion of the mutant LFY version (LFYFIF) were co-expressed under the control of the UBQ10 promoter in transiently transformed Nicotiana benthamiana epidermal leaf cells. As shown in Figure 6a, LFY-GFP and LFYFIF-RFP localised to the cytoplasm and the nucleus in a similar manner. The observed fluorescence pattern of LFY-GFP and LFYFIF-RFP is in accordance with the pattern previously reported for the expression of fluorophore-tagged LFY fusion proteins in tobacco epidermal leaf cells [31].
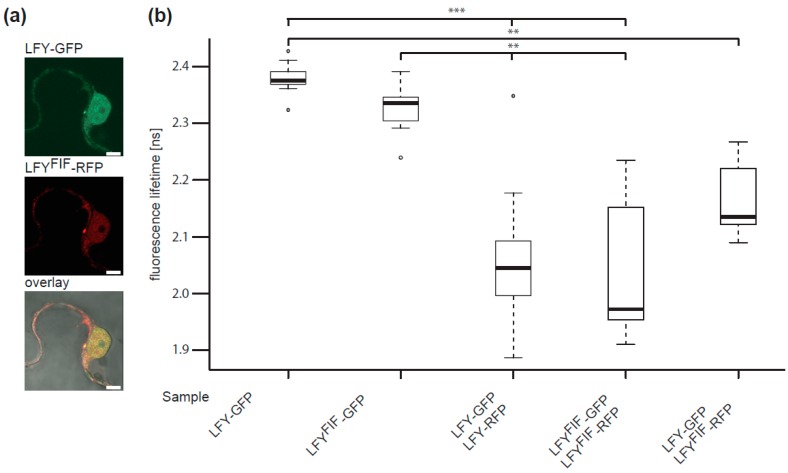
Figure 6.
Comparative analysis of the intracellular localization and homomerization capacity of LFY and LFYFIF. (a) Confocal fluorescence images of transiently transformed Nicotiana benthamina epidermal leaf cells expressing LFY-GFP and LFYFIF-RFP in the same cell. Size bar: 5 µm. (b) FRET-FLIM analysis of the homo- and heterotypic interaction of LFY and LFYFIF. LFY-GFP or LFYFIF-GFP were expressed either alone or together with the indicated RFP fusions and the fluorescence lifetime of the GFP fusions measured in the nuclei. A reduction of the GFP fluorescence lifetime indicates interaction. The data are presented in Box-and-Whisker plots including the median (thick line), the upper and lower quartile (+/− 25%, white boxes), the maximum and minimum (dottet line) and outlier points (n > 20, each). The variance was analyzed by a Levene test and statistical significance was determined with an all-pair, two-sided Kruskal–Wallis test followed by an all-pair Steel-Dwass test (**: p < 0.01; ***: p < 0.001).
Next, we tested by in vivo FRET-FLIM whether LFY protein–protein interaction, here especially LFY homomerization [31], was altered. To do so, C-terminal GFP fusions (FRET donors) and C-terminal RFP fusions (FRET acceptors) were transiently expressed, either individually (donor only) or in combination in N. benthamiana epidermal leaf cells and the fluorescence lifetime of the donor fusion was measured. As shown in Figure 6b, the fluorescence lifetimes of LFY-GFP and LFYFIF-GFP were similar in the absence of the acceptor fusions. However, the lifetimes of LFY-GFP and LFYFIF-GFP decreased significantly when they were co-expressed with either LFY-RFP or LFYFIF-RFP demonstrating homotypic (LFY-LFY, LFYFIF-LFYFIF) and heterotypic (LFY-LFYFIF) homomerization in planta (Figure 6b). There was no significant difference in the interaction properties of the homotypic and heterotypic homomers (Figure 6b).
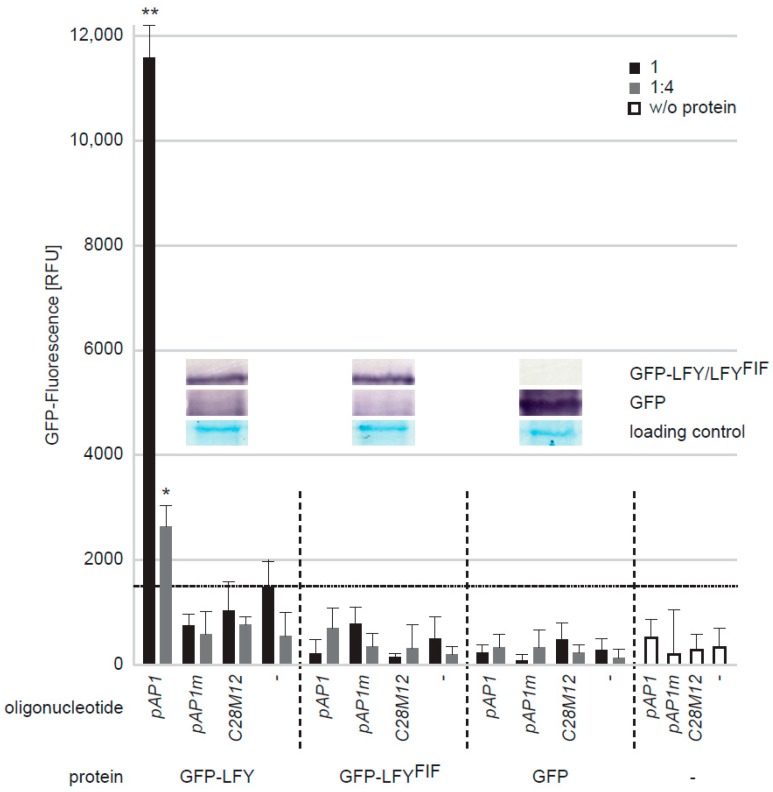
The Cys263-to-Tyr exchange is located in the first α-helix of the LFY DNA-binding domain (Figure 4d). We, therefore, used a quantitative DNA-protein interaction ELISA approach (qDPI-ELISA; [32]) to test whether the mutation interferes with the DNA-binding capability of LFY in vitro. We expressed N-terminally GFP-tagged full-length LFY, LFYFIF and GFP, in E. coli independently and applied the crude extracts containing the fusion proteins or GFP, in identical amounts, based on the GFP fluorescence and Western-blotting, to ELISA plates in two dilutions (Figure 7). The plates were covered with double-stranded (ds) DNA oligonucleotides representing either the LFY-binding sequence of the AP1 promoter (pAP1), a mutated pAP1 version (pAP1m) that is not recognized by LFY [17], a random sequence without any similarity to the LFY binding motif (C28M12), or were uncovered. The DNA-binding efficiency of the proteins was recorded by determining the GFP fluorescence of the bound proteins [32]. GFP-LFY exhibited a specific binding to pAP1 and no binding to any other oligonucleotide or to the oligonucleotide-free ELISA plate (Figure 7). In contrast, GFP-LFYFIF, like GFP or the E. coli crude extract without recombinant protein, was unable to recognize pAP1 or any other oligonucleotide (Figure 7). To exclude the possibility that the Cys263-to-Tyr exchange may alter the DNA-binding specificity we used a quantitative DPI-ELISA based in vitro approach to screen a dsDNA oligonucleotide library representing 4096 randomized DNA hexamers [33,34] with GFP-LFY- and GFP-LFYFIF-containing E. coli extracts. With this approach it is possible to narrow down the target dsDNA site of almost any DNA-binding GFP-tagged fusion protein of interest in vitro [32]. Whereas an in vitro DNA-binding sequence was obtained for GFP-LFY (5′-GGGC-3′/3′-CCCG-5′), there was no DNA-binding of GFP-LFYFIF to any oligonucleotide in the library.
Figure 7.
Comparative analysis of the in vitro DNA-binding capacity of LFY and LFYFIF using a GFP-fluorescence-based DPI-ELISA approach. GFP-LFY, GFP-LFYFIF and GFP were expressed in E. coli. After extraction, crude extracts containing either no recombinant protein (w/o protein) or, based on GFP fluorescence, equal amounts of GFP or the GFP fusion proteins were added to ELISA plates covered with either the double-stranded (ds) DNA oligonucleotide pAP1, which contains a LFY binding site, an altered version of pAP1 (pAP1m), in which the binding site was mutated, a dsDNA oligonucleotide unrelated to the pAP1 and pAP1m sequences (C28M12) or without any DNA-oligonucleotide. The amount of DNA-bound fusion protein was detected by reading out the GFP fluorescence. The crude extract was either used undiluted (black bars) or in a 1:4 dilution (grey bars). Error bars indicate the standard deviation of the mean (n = 3) and asterisk statistically significant differences to the background fluorescence (dotted horizontal line), determined by two-sided t-test (*: p < 0.05; **: p < 0.01). The inlet shows a Western-blot of the crude extracts using a GFP polyclonal antiserum for detection of GFP, GFP-LFY and GFP-LFYFIF as well as a Coomassie stain as loading control.
3. Discussion
In our search for novel floral genes in Arabidopsis thaliana we identified the fif Ds transposon insertion mutant in the No-0 accession in the RIKEN RAPID collection [23,24]. fif mutant plants display a striking floral phenotype and inflorescence architecture, as they develop aberrant and infertile primary flowers in combination with short stems that emerge from vegetative meristems in the axillars of the bracts and carry fertile secondary flowers.
The Ds transposon insertion in the genome of the fif mutant was annotated to gene At1g20990. However, as demonstrated here, the Ds transposon insertion into the At1g20990 locus is not the cause for the fif phenotype. Obviously, another mutant locus generated somewhere else in the genome, most likely during transposon movement, is responsible for the fif phenotype. Using combined classical and genome sequencing-based mapping approaches and complementation of the mutant phenotype by UBI10-driven expression of wild type LFY-GFP in the fif mutant, the causal mutation for the fif phenotype was found to be in the LFY gene. In contrast to LFY overexpression by the strong 35S promoter in Nossen [35], the UBI10-driven expression of wild type LFY-GFP in the fif mutant background did not cause primary shoot termination and conversion of axillary meristems into solitary flowers. We ascribe this to the very low accumulation of LFY-GFP that can hardly be detected in the nuclei of the stably transgenic Arabidopsis cells.
The fif phenotype-causing mutation is a single G-to-A exchange in the second exon of LFY, creating the novel, recessive lfy allele. The mutation causes a Cys-to-Tyr exchange at position 263 in the LFYFIF amino acid sequence. The cell biological analysis of LFY-GFP and LFYFIF-GFP revealed an intracellular localization in the cytoplasm and nucleus of tobacco epidermal leaf cells identical to that previously reported for LFY-GFP [31]. Thus, a mis-localisation cannot be the cause of the LFYFIF malfunction. In addition, as shown by quantitative FRET-FLIM interaction studies the mutation does not interfere with the oligomerization capacity of LFY that is essential for its floral function [36]. Especially the latter result was to be expected as the domain, required for oligomerization is located at the N-terminus of LFY [36].
However, our quantitative DPI-ELISA assay demonstrated that, in contrast to LFY-GFP, LFYFIF-GFP lost its capacity to bind to its DNA target in vitro, as it is present, for instance, in the AP1 promoter [17]. Furthermore, the in vitro DPI-ELISA based screen for the determination of putative alterations in binding specificity did not reveal any DNA-binding activity for LFYFIF-GFP, whilst a defined sequence was found for LFY-GFP. This sequence did not perfectly match the LFY consensus sequence identified by, for instance, in vivo chromatin immunoprecipitation [37]. However, this discrepancy is very likely due to the design of the dsDNA oligonucleotides (e.g., the region flanking the hexameric core) used for the library and the applied in vitro conditions [33,34] that could modify LFY-GFP’s binding characteristics.
According to the available crystal structure of the DNA-bound dimer, Cys263 is well conserved between the LFY homologs of many plant species but has never previously been reported to be crucial for DNA-binding [38]. Intriguingly, Cys263 does not contribute to the physical contact of LFY with DNA; however, the α1-helix, in which Cys263 is positioned, participates in the cooperative DNA-binding of LFY, as it facilitates the establishment and stabilization of the DNA-binding domains in the minor and major grove of DNA [38]. Therefore, the change of the relatively small Cys to the bulky, aromatic Tyr might prevent the folding of the α1-helix and thereby might restrict the cooperative binding of LFY to its target DNA.
The total failure of LFYFIF to bind to DNA explains the strong floral phenotype of especially the primary flowers. LFY is one of the master regulators in the floral initiation process of Arabidopsis (and other plant species) and controls, together with other factors and via a complex regulatory network, the spatiotemporal expression of downstream genes and also of the homeotic flower genes required for flower organ formation. Although only a single amino acid exchange is affected, fif mirrors in principle the flower phenotype of known strong lfy alleles. However, of the more than 15 described lfy alleles [12], the six alleles that show such a strong floral phenotype produce shortened LFY polypeptides caused by either premature stop codons (lfy-1, lfy-6, lfy-7, lfy-8, lfy-11) or a non-sense frame shift C-terminal of Gln196 (lfy-15). Hence, the strong phenotype of the fif allele needs a different explanation: LFYFIF may titrate out interaction partners by binding them and not binding to DNA. Thereby, LFYFIF increases its own phenotype.
The failure of LFYFIF to bind to DNA also explains the growth architecture of the fif mutant. It has been shown [10,12] that mutations in lfy can cause the emergence of axillary meristems instead of floral meristems resulting in an enhanced number of side branches. In addition, the expression of a nearly full-length LFY version with weaker in vitro DNA-binding capacity under the LFY promoter dramatically reduced in vivo transcriptional activity of [LFYHARA(∆40)] in the Col-0 accession. This causes the formation of shoot-like structures in positions where flowers should outgrow [39].
Taken together, our data demonstrate the general importance of Cys263 for LFY function not only in floral development but also in axillary meristem outgrowth in Arabidopsis.
Most intriguingly, the fif floral phenotype appears to be specific for the No-0 accession, as, to our knowledge, it has never been reported for the Col-0 or any other accession. However, the fif phenotype also becomes manifest in the Col-0 accession when the fif locus of No-0 is transferred to Col-0. This phenomenon might be explained by differences in the spatio-temporal transcriptional activity of the No-0 and Col-0 LFY loci during vegetative meristem and floral development. Therefore, the fif phenotype may only be visible in other accessions such as Col-0 when the No-0 locus is artificially introduced into them and drives LFYFIF accumulation there.
4. Materials and Methods
4.1. Plant Material and Arabidopsis thaliana Transformation
Seeds of the homozygous Ds transposon insertion line 15-3794-1 and the corresponding wild-type accession (No-0) were obtained from the RIKEN Arabidopsis Phenome Information database (RAPID; [22]. Agrobacterium tumefaciens-mediated transformation of the fif mutant with pUGT1-LFY leading to LHY-GFP expression and selection of transgenic lines were carried as described previously [40].
4.2. Plasmid Construction
Using gene-specific primers [sense (S): 5′-caccATGGATCCTGAAGGTTTCACG-3′, antisense (A): 5′-GAAACGCAAGTCGTCGCCG-3′) the cDNA of LFY was amplified from pSST14 (gift Jan Lohmann, University of Heidelberg, Germany) and cloned in pENTR™/D-TOPO®. Site-directed mutagenesis (SDM) was performed to produce the fif cDNA using the following primers (S: 5′-CTGTTCCACTTGTACGAACAATaCCGTGAGTTCCTTCTTCAG-3′, A: 5′-CTGAAGAAGGAACTCACGGtATTGTTCGTACAAGTGGAACAG-3′). With Gateway™ LR Clonase™ II Enzyme mix the LFY cDNA was inserted into pUGT1-Dest (A. Hahn, unpublished) and pB7RWG2-Dest [41] for plant expression and into pET-Dest42GFP [32] for E. coli expression.
4.3. Classical Mapping and Mapping by Genome Sequencing
Genetic mapping was accomplished using 100 phenotypic fif plants collected from a F2 population derived from a cross between fif (No-0) and Col-0. The mapping strategy and the molecular markers used to identify the causal locus were described by Pacurar, Pacurar, Street, Bussell, Pop, Gutierrez and Bellini [27]. After mapping of the chromosome arm and next-generation sequencing (NGS, see below) the point mutation was confirmed by derived cleaved-amplified polymorphic sequence primers designed by using the dCAPS Finder 2.0 software [25]. One or two mismatches were introduced in one of the used primers to incorporate an allele-specific restriction site into the PCR product. After amplification, the PCR products were digested (enzymes from Thermo Scientific, city, country, USA) following the manufacturer’s recommendations and separated on a 4% agarose gel. All used markers are listed in Table S1.
NGS mapping was performed using a pool of 425 phenotypic fif plants from the crossing described above. A pool of 40 wild-type No-0 plants was sequenced to generate a genome-wide marker list and to mine the fif genome for acquired mutations. Isolation of genomic DNA was performed in groups up to 20 plants using the DNeasy® Plant Mini Kit (QIAGEN, Venlo, Netherlands) following the manufacturer’s recommendations. DNA concentration was determined with the use of NanoDrop ND-1000 and the whole pool composed by using 100 µg DNA of each group. Sequencing was performed at the Max Planck-Genome-Centre Cologne by a HiSeq2500 (Illumina, San Diego, CA, USA) Sequencer producing ~35.000.000 read-pairs for each pool. Short reads of both pools were respectively aligned against the Col-0 reference sequence (TAIR10) and SNPs were called using shore pipeline (version v0.8) with GenomeMapper (version v0.4.4s) with default parameters [42,43]. Genome-wide SNP markers were defined with filtering for sequencing coverage and allele frequency using SHOREmap (version 3.0, [29,44,45]. Sliding window-based estimation of allele frequencies of the Nos allele in the pooled F2 samples and identification of a mapping interval were performed with SHOREmap (version 3.0) using default parameters. Comparison of the consensus calls of both pools in the 300 kb mapping interval revealed the mutation in LFY.
4.4. Localization and FRET-FLIM Studies
The indicated constructs and p19 as gene silencing suppressor were transformed into Agrobacterium tumefaciens strain GV3101 and infiltrated into Nicotiana benthamiana leaves. The localization of the fusion proteins was performed 3 days after infiltration using 488 nm or 561 nm lasers for GFP or RFP excitation, respectively, at the SP8 laser scanning microscope (Leica Microsystems GMBH) with LAS AF and SymPhoTime software using a 63x/1.20 water immersion objective [46]. FLIM data were derived from measurements of at least 20 probes for each fusion protein combination. To excite LFY-GFP and LFYFIF-GFP for FLIM experiments, a 470 nm pulsed laser (LDH-P-C-470) was used, and the corresponding emission was detected with a SMD Emission SPFLIM PMT from 495 to 545 nm by time-correlated single-photon counting using a Picoharp 300 module (PicoQuant, Berlin, Germany). Each time-correlated single-photon counting histogram was reconvoluted with the corresponding instrument response function and fitted against a monoexponential decay function for donor-only samples and a biexponential decay function for the other samples to unravel the GFP fluorescence lifetime of each probe. The average GFP fluorescence lifetimes as well as the standard error values were calculated using Microsoft Excel 2013. To test for homogenity of variance Levene’s test (df = 5/140, F = 26.298, p < 0.0001) was used and statistical significance was calculated by a two-tailed, all-pair Kruskal–Wallis test followed by a Steel-Dwass post hoc correction using JMP version 12.2.0 [47].
4.5. qDPI-ELISA, DPI-ELISA Based Screening and Western Blotting
qDPI-ELISA was performed using E. coli crude extracts containing GFP-tagged LFY or LFYFIF, GFP alone or no fluorescent protein according to Fischer, Böser, Hirsch and Wanke [32]. The sequences of the 5′-biotinylated dsDNA oligonucleotides AP1, mAP1 and C28M12 used for the immobilization on Streptavidin-coated 384 well microtiter plate are displayed in Table S2. Before addition to the microtiter plate, the equal content of GFP-tagged fusion protein in the crude extracts was adjusted according to the GFP fluorescence using a fluorescence reader (TECAN Safire, Männedorf, Switzerland). Western blotting with GFP antibodies was performed according to Brand, et al. [48].
The quantitative DPI-ELISA based specificity screening, using a dsDNA oligo array on a 384 well microtiter plate covering all possible 4096 hexanucleotide DNA motifs was performed as described previously [32,33,34].
Acknowledgments
The authors would like to thank J. Lohmann (Universität Heidelberg, Germany) for the LFY cDNA, M. Fischer for technical support, F. de Courcy for proofreading the manuscript and the members of the multidisciplinary graduate school “Morphological Variability of Organisms under Environmental Stress” for discussion.
Supplementary Materials
The following are available online at https://www.mdpi.com/2223-7747/8/12/599/s1, Figure S1: Proof of the successful backcross of the fif mutant with wild type No-0, Figure S2: Allele frequency analysis of the No-0 allele within the recombinant mutant pool, Table S1: Names and sequences of the INDEL and dCAP primers. Table S2: Sequences of the dsDNA oligonucleotides used in the DPI-ELISA.
Author Contributions
Conceptualization, A.M., Ü.K. and K.H.; methodology, A.M., H.S., N.G. and S.H.; software, N.G., H.S. and K.S.; validation, Ü.K., K.S. and K.H.; formal analysis, Ü.K., K.S. and K.H.; investigation, A.M., H.S. and S.H.; resources, Ü.K., K.S. and K.H.; data curation, Ü.K. and K.S.; writing—original draft preparation, K.H.; writing—review and editing, Ü.K., K.S. and K.H.; visualization, A.M. and N.G.; supervision, K.H.; project administration, K.H.; funding acquisition, A.M. and K.H.
Funding
This research was funded by the Ministerium für Wissenschaft, Forschung und Kunst Baden-Württemberg (Germany), partially by the Deutsche Forschungsgemeinschaft (DFG), HA 2146/22–1 and by the Open Access Publishing Fund of the University of Tübingen.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Pineiro M., Coupland G. The control of flowering time and floral identity in Arabidopsis. Plant Physiol. 1998;117:1–8. doi: 10.1104/pp.117.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ma H. To be, or not to be, a flower–control of floral meristem identity. Trends Genet. 1998;14:26–32. doi: 10.1016/S0168-9525(97)01309-7. [DOI] [PubMed] [Google Scholar]
- 3.Pidkowich M.S., Klenz J.E., Haughn G.W. The making of a flower: Control of floral meristem identity in Arabidopsis. Trends Plant Sci. 1999;4:64–70. doi: 10.1016/S1360-1385(98)01369-7. [DOI] [PubMed] [Google Scholar]
- 4.Denay G., Chahtane H., Tichtinsky G., Parcy F. A flower is born: An update on Arabidopsis floral meristem formation. Curr. Opin. Plant Biol. 2017;35:15–22. doi: 10.1016/j.pbi.2016.09.003. [DOI] [PubMed] [Google Scholar]
- 5.O’Maoileidigh D.S., Graciet E., Wellmer F. Gene networks controlling Arabidopsis thaliana flower development. New Phytol. 2014;201:16–30. doi: 10.1111/nph.12444. [DOI] [PubMed] [Google Scholar]
- 6.Prunet N. My favourite flowering image: An Arabidopsis inflorescence expressing fluorescent reporters for the APETALA3 and SUPERMAN genes. J. Exp. Bot. 2018 doi: 10.1093/jxb/ery098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wagner D. Key developmental transitions during flower morphogenesis and their regulation. Curr. Opin. Genet. Dev. 2017;45:44–50. doi: 10.1016/j.gde.2017.01.018. [DOI] [PubMed] [Google Scholar]
- 8.Wellmer F., Graciet E., Riechmann J.L. Specification of floral organs in Arabidopsis. J. Exp. Bot. 2014;65:1–9. doi: 10.1093/jxb/ert385. [DOI] [PubMed] [Google Scholar]
- 9.Irish V.F., Sussex I.M. Function of the Apetala-1 Gene during Arabidopsis Floral Development. Plant Cell. 1990;2:741–753. doi: 10.1105/tpc.2.8.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huala E., Sussex I.M. Leafy Interacts with Floral Homeotic Genes to Regulate Arabidopsis Floral Development. Plant Cell. 1992;4:901–913. doi: 10.2307/3869458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowman J.L., Alvarez J., Weigel D., Meyerowitz E.M., Smyth D.R. Control of flower development in Arabidopsis thaliana by APETALA1 and interacting genes. Development. 1993;119:721–743. [Google Scholar]
- 12.Weigel D., Alvarez J., Smyth D.R., Yanofsky M.F., Meyerowitz E.M. LEAFY controls floral meristem identity in Arabidopsis. Cell. 1992;69:843–859. doi: 10.1016/0092-8674(92)90295-N. [DOI] [PubMed] [Google Scholar]
- 13.Mandel M.A., Gustafsonbrown C., Savidge B., Yanofsky M.F. Molecular Characterization of the Arabidopsis Floral Homeotic Gene Apetala1. Nature. 1992;360:273–277. doi: 10.1038/360273a0. [DOI] [PubMed] [Google Scholar]
- 14.Kempin S.A., Savidge B., Yanofsky M.F. Molecular-Basis of the Cauliflower Phenotype in Arabidopsis. Science. 1995;267:522–525. doi: 10.1126/science.7824951. [DOI] [PubMed] [Google Scholar]
- 15.Blazquez M.A. Illuminating flowers: CONSTANS induces LEAFY expression. Bioessays. 1997;19:277–279. doi: 10.1002/bies.950190403. [DOI] [PubMed] [Google Scholar]
- 16.Moyroud E., Minguet E.G., Ott F., Yant L., Pose D., Monniaux M., Blanchet S., Bastien O., Thevenon E., Weigel D., et al. Prediction of regulatory interactions from genome sequences using a biophysical model for the Arabidopsis LEAFY transcription factor. Plant Cell. 2011;23:1293–1306. doi: 10.1105/tpc.111.083329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winter C.M., Austin R.S., Blanvillain-Baufume S., Reback M.A., Monniaux M., Wu M.F., Sang Y., Yamaguchi A., Yamaguchi N., Parker J.E., et al. LEAFY target genes reveal floral regulatory logic, cis motifs, and a link to biotic stimulus response. Dev. Cell. 2011;20:430–443. doi: 10.1016/j.devcel.2011.03.019. [DOI] [PubMed] [Google Scholar]
- 18.Coen E.S., Meyerowitz E.M. The war of the whorls: Genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- 19.Lohmann J.U., Weigel D. Building beauty: The genetic control of floral patterning. Dev. Cell. 2002;2:135–142. doi: 10.1016/S1534-5807(02)00122-3. [DOI] [PubMed] [Google Scholar]
- 20.Shinya T., Galis I., Narisawa T., Sasaki M., Fukuda H., Matsuoka H., Saito M., Matsuoka K. Comprehensive analysis of glucan elicitor-regulated gene expression in tobacco BY-2 cells reveals a novel MYB transcription factor involved in the regulation of phenylpropanoid metabolism. Plant Cell Physiol. 2007;48:1404–1413. doi: 10.1093/pcp/pcm115. [DOI] [PubMed] [Google Scholar]
- 21.Miwa H., Betsuyaku S., Iwamoto K., Kinoshita A., Fukuda H., Sawa S. The Receptor-Like Kinase SOL2 Mediates CLE Signaling in Arabidopsis. Plant Cell Physiol. 2008;49:1752–1757. doi: 10.1093/pcp/pcn148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuromori T., Wada T., Kamiya A., Yuguchi M., Yokouchi T., Imura Y., Takabe H., Sakurai T., Akiyama K., Hirayama T., et al. A trial of phenome analysis using 4000 Ds-insertional mutants in gene-coding regions of Arabidopsis. Plant J. 2006;47:640–651. doi: 10.1111/j.1365-313X.2006.02808.x. [DOI] [PubMed] [Google Scholar]
- 23.Ito T., Motohashi R., Kuromori T., Mizukado S., Sakurai T., Kanahara H., Seki M., Shinozaki K. A new resource of locally transposed Dissociation elements for screening gene-knockout lines in silico on the Arabidopsis genome. Plant Physiol. 2002;129:1695–1699. doi: 10.1104/pp.002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuromori T., Hirayama T., Kiyosue Y., Takabe H., Mizukado S., Sakurai T., Akiyama K., Kamiya A., Ito T., Shinozaki K. A collection of 11 800 single-copy Ds transposon insertion lines in Arabidopsis. Plant J. 2004;37:897–905. doi: 10.1111/j.1365.313X.2004.02009.x. [DOI] [PubMed] [Google Scholar]
- 25.Neff M.M., Turk E., Kalishman M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002;18:613–615. doi: 10.1016/S0168-9525(02)02820-2. [DOI] [PubMed] [Google Scholar]
- 26.Kover P.X., Valdar W., Trakalo J., Scarcelli N., Ehrenreich I.M., Purugganan M.D., Durrant C., Mott R. A Multiparent Advanced Generation Inter-Cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pacurar D.I., Pacurar M.L., Street N., Bussell J.D., Pop T.I., Gutierrez L., Bellini C. A collection of INDEL markers for map-based cloning in seven Arabidopsis accessions. J. Exp. Bot. 2012;63:2491–2501. doi: 10.1093/jxb/err422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.James G.V., Patel V., Nordstrom K.J.V., Klasen J.R., Salome P.A., Weigel D., Schneeberger K. User guide for mapping-by-sequencing in Arabidopsis. Genome Biol. 2013;14:R61. doi: 10.1186/gb-2013-14-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schneeberger K. Using next-generation sequencing to isolate mutant genes from forward genetic screens. Nat. Rev. Genet. 2014;15:662–676. doi: 10.1038/nrg3745. [DOI] [PubMed] [Google Scholar]
- 30.Wu X., Dinneny J.R., Crawford K.M., Rhee Y., Citovsky V., Zambryski P.C., Weigel D. Modes of intercellular transcription factor movement in the Arabidopsis apex. Development. 2003;130:3735–3745. doi: 10.1242/dev.00577. [DOI] [PubMed] [Google Scholar]
- 31.Siriwardana N.S., Lamb R.S. A conserved domain in the N-terminus is important for LEAFY dimerization and function in Arabidopsis thaliana. Plant J. 2012;71:736–749. doi: 10.1111/j.1365-313X.2012.05026.x. [DOI] [PubMed] [Google Scholar]
- 32.Fischer S.M., Böser A., Hirsch J.P., Wanke D. Quantitative Analysis of Protein-DNA Interaction by qDPI-ELISA. Methods Mol. Biol. 2016;1482:49–66. doi: 10.1007/978-1-4939-6396-6_4. [DOI] [PubMed] [Google Scholar]
- 33.Brand L.H., Fischer N.M., Harter K., Kohlbacher O., Wanke D. Elucidating the evolutionary conserved DNA-binding specificities of WRKY transcription factors by molecular dynamics and in vitro binding assays. Nucleic Acids Res. 2013;41:9764–9778. doi: 10.1093/nar/gkt732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brand L.H., Henneges C., Schussler A., Kolukisaoglu H.U., Koch G., Wallmeroth N., Hecker A., Thurow K., Zell A., Harter K., et al. Screening for protein-DNA interactions by automatable DNA-protein interaction ELISA. PLoS ONE. 2013;8:e75177. doi: 10.1371/journal.pone.0075177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weigel D., Nilsson O. A developmental switch sufficient for flower initiation in diverse plants. Nature. 1995;377:495–500. doi: 10.1038/377495a0. [DOI] [PubMed] [Google Scholar]
- 36.Sayou C., Nanao M.H., Jamin M., Pose D., Thevenon E., Gregoire L., Tichtinsky G., Denay G., Ott F., Peirats Llobet M., et al. A SAM oligomerization domain shapes the genomic binding landscape of the LEAFY transcription factor. Nat. Commun. 2016;7:11222. doi: 10.1038/ncomms11222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.William D.A., Su Y., Smith M.R., Lu M., Baldwin D.A., Wagner D. Genomic identification of direct target genes of LEAFY. Proc. Natl. Acad. Sci. USA. 2004;101:1775–1780. doi: 10.1073/pnas.0307842100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hames C., Ptchelkine D., Grimm C., Thevenon E., Moyroud E., Gerard F., Martiel J.L., Benlloch R., Parcy F., Muller C.W. Structural basis for LEAFY floral switch function and similarity with helix-turn-helix proteins. EMBO J. 2008;27:2628–2637. doi: 10.1038/emboj.2008.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chahtane H., Vachon G., Le Masson M., Thevenon E., Perigon S., Mihajlovic N., Kalinina A., Michard R., Moyroud E., Monniaux M., et al. A variant of LEAFY reveals its capacity to stimulate meristem development by inducing RAX1. Plant J. 2013;74:678–689. doi: 10.1111/tpj.12156. [DOI] [PubMed] [Google Scholar]
- 40.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 41.Karimi M., Inze D., Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 42.Ossowski S., Schneeberger K., Clark R.M., Lanz C., Warthmann N., Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneeberger K., Hagmann J., Ossowski S., Warthmann N., Gesing S., Kohlbacher O., Weigel D. Simultaneous alignment of short reads against multiple genomes. Genome Biol. 2009;10 doi: 10.1186/gb-2009-10-9-r98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun H.Q., Schneeberger K. Plant Functional Genomics: Methods and Protocols. 2nd ed. Volume 1284. Springer; Berlin/Heidelberg, Germany: 2015. SHOREmap v3.0: Fast and Accurate Identification of Causal Mutations from Forward Genetic Screens; pp. 381–395. [DOI] [PubMed] [Google Scholar]
- 45.Schneeberger K., Ossowski S., Lanz C., Juul T., Petersen A.H., Nielsen K.L., Jorgensen J.E., Weigel D., Andersen S.U. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat. Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 46.Ladwig F., Dahlke R.I., Stuhrwohldt N., Hartmann J., Harter K., Sauter M. Phytosulfokine Regulates Growth in Arabidopsis through a Response Module at the Plasma Membrane That Includes CYCLIC NUCLEOTIDE-GATED CHANNEL17, H+-ATPase, and BAK1. Plant Cell. 2015;27:1718–1729. doi: 10.1105/tpc.15.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ohmi Y., Ise W., Harazono A., Takakura D., Fukuyama H., Baba Y., Narazaki M., Shoda H., Takahashi N., Ohkawa Y., et al. Sialylation converts arthritogenic IgG into inhibitors of collagen-induced arthritis. Nat. Commun. 2016;7:11205. doi: 10.1038/ncomms11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brand L.H., Kirchler T., Hummel S., Chaban C., Wanke D. DPI-ELISA: A fast and versatile method to specify the binding of plant transcription factors to DNA in vitro. Plant Methods. 2010;6:25. doi: 10.1186/1746-4811-6-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.