Abstract
The carbapenemase OXA-244 is a derivate of OXA-48, and its detection is very difficult in laboratories. Here, we report the identification and genomic analysis of an Escherichia coli isolate (28Eco12) harboring the blaOXA-244 gene identified in Colombia, South America. The 28Eco12 isolate was identified during a retrospective study, and it was recovered from a patient treated in Colombia. The complete nucleotide sequence was established using the PacBio platform. A comparative genomics analysis with other blaOXA-244–harboring Escherichia coli strains was performed. The 28Eco12 isolate belonged to sequence type (ST) 38, and its genome was composed of two molecules, a chromosome of 5,343,367 bp and a plasmid of 92,027 bp, which belonged to the incompatibility group IncY and did not harbor resistance genes. The blaOXA-244 gene was chromosomally encoded and mobilized by an ISR1-related Tn6237 composite transposon. Notably, this transposon was inserted and located within a new genomic island. To our knowledge, this is the first report of a blaOXA-244–harboring Escherichia coli isolate in America. Our results suggest that the introduction of the OXA-244-producing E. coli isolate was through clonal expansion of the ST38 pandemic clone. Other isolates producing OXA-244 could be circulating silently in America.
Keywords: blaOXA-244, Escherichia coli, carbapenems, resistance, Colombia
1. Introduction
The World Health Organization WHO has recognized carbapenem-resistant Enterobacteriaceae as pathogens with critical priority for the development of new antibiotics [1]. OXA-244, a carbapenemase belonging to the Class D family, is a derivate of OXA-48 and encoded by the blaOXA-244 gene. Although there are multiple reports of OXA-48-producing isolates, reports of isolates harboring OXA-244 are less frequent, perhaps because their detection is difficult due to their reduced carbapenem activity. The blaOXA-244 gene was initially described in 2011, within a Klebsiella pneumoniae isolate, which was identified in Spain [2]. It has already been identified in Escherichia coli isolates recovered from Germany [3], France [4,5], the United Kingdom [6], Southeast Asia [7], and Egypt [5]. The molecular characterization of some of these E. coli isolates have shown that the majority of them belong to sequence type (ST) 38, although recently other STs have been found (ST361, ST1722, and ST3541) [5]; and they contain other β-lactamases, such as TEM, CTX-M, and CMY. The blaOXA-244 gene is located in the chromosome within a truncated Tn1999.2 transposon, which is immersed into an ISR1-based Tn6237 transposon [4,8]. Here, we provide a genomic analysis of an Escherichia coli isolate (28Eco12) containing the blaOXA-244 gene that was recovered from a patient in Colombia, South America. To our knowledge, this is the first report of a blaOXA-244–harboring Escherichia coli isolate in America.
2. Results
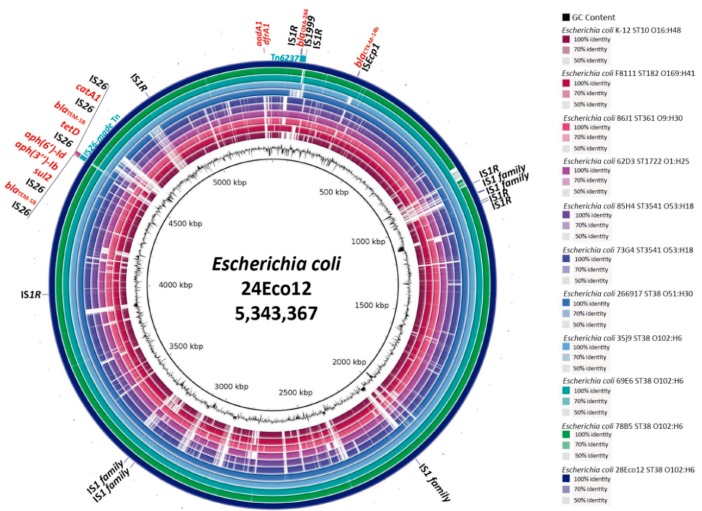
The 28Eco12 isolate was identified from a retrospective study in Bogotá, Colombia (see Materials and Methods), and we decided to establish its complete genome to determine its resistome and mobile genetic platform distribution (IS content). The genome was composed of two molecules, a chromosome of 5,343,367 bp and a plasmid of 92,027 bp (p28Eco12), which belonged to the incompatibility group IncY and did not harbor resistance genes. The resistance-genes arsenal of the isolate was composed of aph(3′’)-Ib, aph(6)-Id, aaaA1 (aminoglycosides), blaOXA-244, blaCTX-M-14b, blaTEM-1b (beta-lactams), catA1 (chloramphenicol), sul2 (sulphonamides), dfrA1 (trimethoprim), and tetD (tetracycline) genes, all chromosomally encoded (Figure 1). The 28Eco12 isolate belonged to ST38 [9]. The in silico serotyping of the isolate was O102:H6.
Figure 1.
BLASTn comparison of the blaOXA-244-containing Escherichia coli chromosomes. The K-12 (GenBank accession number NC_000913), F8111-1SC3 (GenBank accession number NZ_CP024269), and 266917_2 (GenBank accession number NZ_CP026723.1) strains were used as references. At the more external circle is shown the localization of the resistance genes and their putative genetic platforms of mobilization. The positions of the seven identical ISR1 and five IS1-family (89% of identity) sequences are also indicated. The strain positions on the figure are as follow (internal to external) (sequence type/serotype): K12 (ST10/O16:H48), F8111-1SC3 (ST182/O169:H41), 86J1 (ST361/O9:H30) MKGU01, 62D3 (ST1722/O1:H25) MKGY01, 85H4 (ST3541/O53:H18) MKGW01, 73G4 (ST3541/O53:H18) MKGV01, 266917_2 (ST38/O51:H30), 35J9 (ST38/O102:H6) MKGX01, 69E6 (ST38/O102:H6) MKGZ01, 78B5 (ST38/O102:H6) MKGT01, and 28Eco12 (ST38/O102:H6) NZ_CP038505.
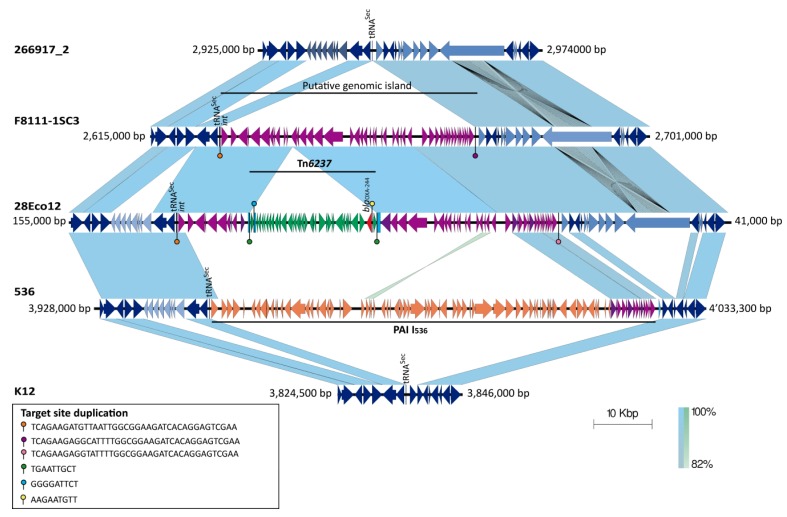
Using the complete genome sequence, the 28Eco12 isolate was found to have a close genetic relationship with the E. coli strain 266917_2 (ST38), described recently in the United Kingdom (90% coverage, 97% identity, GenBank accession number CP026723.1), which does not contain the blaOXA-244 gene. The genomic comparative analysis revealed that the blaOXA-244 gene was mobilized by the Tn6237 transposon, as it has previously been described in Escherichia coli strain VAL [4,8]. However, in the 28Eco12 isolate, the Tn6237 transposon was not inserted within the II536 pathogenicity island, as was previously reported to blaOXA-48 [8], but into a new putative genomic island, inserted within the tRNA-sec gene. Its insertion produced a 39 bp direct repeat sequence (TTCGACTCCTGTGATCTTCCGCCAATTAACATCTTCTGA). This event did not change the tRNA-sec gene sequence (Figure 2).
Figure 2.
Comparison of the region where the blaOXA-244 gene was inserted within Escherichia coli 28Eco12 isolate. The red arrow corresponds to the blaOXA-244 gene. The mobile genetics elements are shown in different colors. The putative genomic island is shown in purple and its insertion within the tRNA-sec gene is indicated respect to the E. coli strain 266917_2 (GenBank accession number CP026723.1), F8111-1SC3 (GenBank accession number NZ_CP024269), 536-EC15 (GenBank accession number HG977710.1), and K-12 (GenBank accession number NC_000913). The blue rectangles correspond to the gene where the Tn6237 transposon was inserted (green arrows). The pallets represent the target-site duplications. The int gene that encodes the phage integrase protein is shown. Blue shading between pairs of sequences indicates >90% of identity in a window of 400 bp. The scale bar indicates sequence length.
The putative island was also present in the blaOXA-244-negative enterotoxigenic E. coli F8111-1SC3 isolate (GenBank accession number NZ_CP024269). Interestingly, the tRNA-sec gene is a hot spot for DNA insertion, because it also serves as the insertion site of the I536 pathogenicity island in the uropathogenic strain E. coli 536 [10]. These results suggest that the Tn6237 transposon is active and moves to different sites in the E. coli chromosome. In addition, the isolate harbored 69 insertion sequences (IS) belonging to 17 different IS families (Table 1). Some of these present as single copy, partial form, or multiple copies. The most frequent IS families were IS1, IS200/IS605_ssgr_IS200, and IS3, with 13, 10, and 8 IS copies, respectively. Target site duplications (TSD) are signatures of transposition events, and among the 69 ISs, 25 presented TSDs and none were present within the E. coli F8111-1SC3 isolate, indicating that they were inserted by single-copy transposition. The TSD pattern analysis also revealed the presence of two composite transposons, the Tn6237 (mentioned previously) and a 15,730 bp IS26-made transposon, which was inserted within a gene that encodes a hypothetical protein and mobilizes the aph(3′’)-Ib, aph(6)-Id, blaTEM-1b (two copies), catA1, sul2, and tetD genes. Notably, this IS26 transposon was also inserted within another putative genomic island, which was inserted into the tRNA-leu gene. The comparative analysis suggested that this IS26 transposon was mobilized from a plasmid because it harbored the repA gene that corresponds to the incompatibility group IncQ-1 and possesses DNA fragments with a high percentage of identity to pD90-1 and pEC141 plasmids, which were identified in mcr-1-containing Salmonella enterica and E. coli strains, respectively [11]. With respect to the other resistance genes, the blaCTX-M gene was mobilized by ISEcp1 and an IS26 remnant, which were inserted within a gene that encodes a hypothetical protein.
Table 1.
Insertion sequences identified in 28Eco12 isolate. Target site duplications (TSD) are shown in bold and underlined.
| IS Family | IS | Position | Right and Left Flanking Sequences | Comments | |
|---|---|---|---|---|---|
| IS1 | IS1R | 102025..102792 | TGAATTGCT | AAGAATGTT | Composite transposon harboring the blaOXA-244 gene. |
| IS1R | 123120..123887 | GGGGATTCT | TGAATTGCT | ||
| IS1R | 936063..936830 | CAGACAACG | CAGACAACG | Single IS transposition. IS inserted within a putative prophage | |
| IS1-like | 975280..976060 | GTCGCAACC | TACAACGTT | IS inserted within a putative prophage | |
| IS1-like | 977300..978080 | GACAATGTC | CAATCTGCT | IS inserted within a putative prophage | |
| IS1R | 1007836..1008603 | TGCTTTTCT | TGCTTTTCT | Single IS transposition. IS inserted within an intergenic region | |
| IS1R | 1015519..1016286 | GCCAATTCG | GCCAATTCG | Single IS transposition. IS inserted within the cmtB gene | |
| IS1-like | 2087231.. 2087998 | CGGTTTTGG | GAAGAGTTC | IS inserted within the hchA gene | |
| IS1-like | 3237236..3237910 | - | GAAATCCCC | IS (truncated) inserted within a putative prophage | |
| IS1-like | 3266386..3267153 | CTGCAAATC | TACAACCGG | IS inserted within a putative prophage | |
| IS1R | 3972674..3973441 | CTGCTCCTG | CTGCTCCTG | Single IS transposition. IS inserted within a hypothetical gene | |
| IS1R | 4845817..4846584 | GACGGTATT | CGGATGCTG | IS inserted within the adiA gene | |
| IS1H | 5066636..5067399 | CCGGTAAAC | CTTCTGATG | IS inserted within an intergenic region | |
| IS200/ IS605_ssgr_IS200 |
IS200C | 1127230..1127936 | TTTT | TTTT | Single IS transposition. IS inserted within a T-rich region |
| IS200C | 1690413..1691121 | TTTT | TTTT | Single IS transposition. IS inserted within a T-rich region | |
| IS200C | 2442570..2443280 | TTAA | TTAA | Single IS transposition. IS inserted within a T-rich region | |
| IS200C | 2481694..2482403 | TTTT | TTAT | Single IS transposition. IS inserted within a T-rich region | |
| IS200C | 2990220..2990930 | AAAA | AAAA | Single IS transposition. IS inserted within a T-rich region | |
| IS200C | 3058643..3059351 | TAAA | AAAA | Single IS transposition. IS inserted within a T-rich region | |
| IS200C | 3060222..3060929 | AAAA | AAAA | Single IS transposition. IS inserted within a T-rich region | |
| IS200C | 3271558..3272271 | GCAA | AAAA | IS inserted within a putative prophage | |
| IS200C | 3939865..3940573 | CAAA | AAAA | Single IS transposition. IS inserted within a T-rich region | |
| IS200C | 3994005..3994713 | AAAA | AAAA | Single IS transposition. IS inserted within a T-rich region | |
| IS3 | IS600 | 3254256..3255501 | CAA | ACA | IS inserted within a genomic island |
| ISSd1 | 949559..950499 | CAGTT | - | IS (truncated) inserted within a putative prophage | |
| ISSd1 | 3267154..3267978 | - | GGT | IS (truncated) inserted within a genomic island | |
| ISSfl10 | 951719..952045 | - | GTT | IS (truncated) inserted within a putative prophage | |
| IS3 | 3259199..3260456 | TCAT | TTTA | IS inserted within a genomic island | |
| IS3 | 3236998..3237235 | - | CTTC | IS (truncated) inserted within a genomic island | |
| ISEc52 | 3249338..3250086 | - | - | IS (truncated) inserted within a genomic island | |
| ISEc52 | 3246586..3247067 | - | - | IS (truncated) inserted within a genomic island | |
| ISAs1 | ISEc1 | 369367..369900 | - | CCCT | IS (truncated, formerly Rhs-rearrangement hot-spots element) |
| ISEc1 | 2456311..2456957 | GATC | - | IS (truncated, formerly Rhs-rearrangement hot-spots element) | |
| ISEc1 | 3675287..3676199 | TGTTGTAG | TCCTTGGC | IS (formerly Rhs-rearrangement hot-spots element) | |
| ISEc1 | 3815490..3816780 | GATGTATA | CCTGCTCA | IS (formerly Rhs-rearrangement hot-spots element) | |
| ISEc1 | 4160599..4161889 | TTCCTTCC | CACTTCAC | IS (formerly Rhs-rearrangement hot-spots element) | |
| ISEc1 | 5069737..5071026 | AGACCAGT | GCATGTCA | IS (formerly Rhs-rearrangement hot-spots element) | |
| IS6 | IS26 | 4500893..4501712 | AAATCATG | ATATCAAG | Composite transposon harboring the blaTEM-1B (two copies), catA1, aph(6′)-id, aph(3′’)-ib, sul2, and tetD genes. |
| IS26 | 4503629..4504448 | ATATCGGC | GGTAAATC | ||
| IS26 | 4509192..4510011 | CCGGCAAT | GTAAGCTG | ||
| IS26 | 4513665..4514484 | ACCATTTG | CGCTGCGG | ||
| IS26 | 4515814..4516633 | CAACAGGG | AAATCATG | ||
| IS200/ IS605 |
IS609 | 3978710..3980457 | CTCA | ATAA | IS inserted within the yajI gene |
| IS609 | 4689442..4691189 | TGTG | ATAA | IS inserted within an intergenic region | |
| IS609 | 2110716..2111379 | - | - | IS (truncated) inserted within the yedK gene | |
| ISEc46 | 2191062..2192824 | TCAT | CTAA | IS inserted within an intergenic region | |
| IS3 ssgr IS150 | IS1397 | 1214273..1215704 | TCAA | TCAA | Single IS transposition within an intergenic region |
| IS1397 | 1368490..1369921 | TGGC | TGGC | Single IS transposition within an intergenic region | |
| IS150 | 259853..261295 | AAG | AAG | Single IS transposition within an intergenic region | |
| IS150 | 2414087..2415529 | GTT | GTT | Single IS transposition. IS inserted within a genomic island | |
| IS3_ssgr_IS2 | IS2 | 937126..938456 | GTGGT | TTGTC | IS inserted within a putative prophage |
| IS2 | 966497.. 967827 | CCGCC | ACGGT | IS inserted within a putative prophage | |
| IS2 | 2027528..2028858 | CCTTT | CCTTT | Single IS transposition. IS inserted within a genomic island | |
| IS2 | 4799912..4800262 | AAAAC | - | IS (truncated) inserted within a putative prophage | |
| IS21 | IS100Kyp | 2015511..2017464 | TTTGT | TTTGT | Single IS transposition. IS inserted within a genomic island |
| IS100Kyp | 3273162..3275115 | GTGATAAC | GATAACAT | IS inserted within a genomic island | |
| IS100Kyp | 4582722.. 4584675 | TTCAGATG | AGATGTAT | IS inserted within a putative prophage | |
| IS66 | IS682 | 924827..926816 | - | CATGTATC | IS (truncated) inserted within a putative prophage |
| ISEc22 | 923252..924827 | ACAGAAGG | - | IS (truncated) inserted within a putative prophage | |
| ISCro1 | 946022.. 948720 | TTTTATCT | TTTTATCT | Single IS transposition. IS inserted within a putative prophage | |
| IS3_ssgr_IS51 | IS629 | 570569..571878 | ATT | ATT | IS inserted within the acrF gene |
| IS1203 | 971759..973068 | GATTACTG | GTAATATC | IS inserted within a putative prophage | |
| ISL3 | ISKox3 | 970324..971101 | - | ATGTATCA | IS (truncated) inserted within a putative prophage |
| ISEc38 | 2022594..2024315 | AAAAGT | ACTTTT | Single IS transposition. IS inserted within a genomic island (inverted TSD) | |
| IS481 | ISErp1 | 891175.. 892368 | TATAATG | TATAATG | Single IS transposition. IS inserted within a putative prophage |
| IS30 | IS30D | 950498..951718 | GT | GT | Single IS transposition. IS inserted within a putative prophage |
| IS4 | IS10A | 105162..106490 | GGCCGAGC | GTGCTGAAC | IS inserted into IS1-composite transposon |
| IS1380 | ISEcp1 | 326913..330008 | TTTA | TTTA | Single IS transposition. IS inserted within a hypothetical gene |
| IS110 | IS5075 | 1568363..1569689 | TT | TT | Single IS transposition. IS inserted within a hypothetical gene |
3. Discussion
In this study, we perform the first report of an Escherichia coli isolate carrying the blaOXA-244 gene in Colombia, South America. These blaOXA-244-positive isolates are less frequent (or perhaps they circulate but are not detected) by their difficult detection and clonal dissemination. The multiresistant 28Eco12 isolate harbored only the phage-like IncY plasmid p28Eco12, which is genetically related to the plasmids p266917_2_02 (88% coverage, 99% identity, GenBank accession number CP026725.1), p1303_95 (91% coverage, 99% identity, GenBank accession number CP009168.1), p1 of Salmonella enterica strain ty3-243 (90% coverage, 93% identity, GenBank accession number LT905089.1), and the blaKPC-containing pCRKP-59-KPC (89% coverage, 94% identity, GenBank accession number KX928752.1). Although this plasmid does not transport resistance genes, it appears to be conserved in almost all blaOXA-244-containing E. coli strains included in our analysis, and its permanence is perhaps caused by the presence of the P1 phd-doc toxin-antitoxin system that participates in host post-segregational killing [12]. Currently, there is limited knowledge about this phage-like IncY-plasmid family (for instance, the 37% of their ORFs is encoding for hypothetical proteins), but it is also becoming a genetic platform to transport important resistance genes, such as blaCTX-M-15 and mcr-1; the latter confers resistance to colistin [13].
All resistant genes were chromosomally located and mobilized by active composite transposons, such as Tn6237, which has moved to different sites in the E. coli chromosome. In E. coli, the blaOXA-244 gene was disseminated mainly by ST38 clone in Europe and Asia [3,4,5,6,7]. However, non-ST38 E. coli isolates are starting to appear in other countries, showing some genetic differences (Figure 1). As it is known that ISs have an important impact on genetic variability, genome structure and function, and foreign DNA acquisition, we try to decipher the potential of the 28Eco12 isolate to capture and move more resistance genes through an analysis of the IS content and their TSD and flanking-sequences patterns. Notably, this isolate has incorporated at least 69 ISs, showing a IS massive expansion process [14]; the ISs-belonging family IS1 was the most active, with fifteen copies, in which four copies probably were recently mobilized as single transposition events (unique copies) and two mobilized as a composite transposon and responsible of the blaOXA-244-gene integration (Table 1). In spite of finding five IS26 copies, only two of these were mobilized as a composite transposon and transported seven resistance genes. A study conducted by He et al. reported the IS26 participation in the plasmid reorganization from clinical strains [15]. The high IS content found in this multiresistant E. coli isolate indicates a high likelihood to acquire more resistance genes.
Finally, our institution searched for the presence of the blaOXA-244 gene within other carbapenem-resistant E. coli isolates from 2013 to the present day, but none were positive. Considering the time of the identification of the isolate, we believe that the E. coli isolate could have been acquired in the remittent institution, suggesting an inter-institution dissemination. No additional information could be obtained from the other institution.
4. Materials and Methods
The 28Eco12 isolate was identified from a retrospective study, conducted to characterize the molecular mechanisms in carbapenem-resistant Enterobacteriaceae isolates, which were recovered between 2013 and 2017, from a health institution in Bogotá, Colombia. The 28Eco12 isolate was recovered from a male patient, in September 2013, who was transferred from another health institution in the same city. The patient had suffered multiple traumas caused by a fall from a height of 20 m, and he required treatment in the intensive-care unit for eleven days. The patient was transferred to our institution, however, on the next day; the patient had fever, dysuria, urethral pain, leukocytosis, and urethral purulent secretion, suggesting a possible catheter-associated urinary tract infection. From a urine sample, the carbapenem-resistant Escherichia coli isolate 28Eco12 was identified, which was also resistant to ampicillin/sulbactam, cefotaxime, ceftriaxone, cefepime, and aztreonam. The Hodge Test was positive, and synergy and double-disc tests with boronic acid and EDTA were negative. The patient was treated with meropenem (2 g every 8 h) and colistin (100 mg every 8 h), and thirteen days later, he responded well to the treatment. No history of travel by him or his relatives was reported.
The complete genome sequence of the blaOXA-244-positive 28Eco12 isolate was obtained using the PacBio RS II platform (Pacific Biosciences of California, Inc., Menlo Park, CA, USA) and assembled through the previously reported procedure [16]. Briefly, sequencing reads were de novo assembled, using the HGAP 3 protocol, and manually verified using BWA-MEM (Burrows–Wheeler Aligner with maximal exact matches) [17] and Tablet v1.15.09.01 [18]. Misassembled terminal repeat overlap sequences were identified with Gepard (Genome Pair Rapid Dotter) [19] and trimmed manually. The genome was annotated using Prokka v1.11 [20], and the relevant regions were manually confirmed using BLASTn and BLASTp and edited in Artemis [21]. The resistance-gene arsenal was identified using ARIBA (https://github.com/sanger-pathogens/ariba/wiki), ResFinder [22], CARD [23], and ARG-ANNOT databases [24]. The insertion sequences (IS) were found using ISsaga (http://issaga.biotoul.fr/), and their flanking sequences were manually determined.
The study was approved by the ethics committee of the Shaio Clinic. The 28Eco12 complete genome sequenced in this study is available in the DDBJ/EMBL/GenBank public databases, under the accession numbers CP038505.1 and CP038506.1.
5. Conclusions
The isolates producing OXA-244 could be circulating in America and may not yet be identified, perhaps due to their very low frequency, very difficult detection, or weakness in antimicrobial resistance surveillance programs in some countries (such as Colombia). It is necessary to strengthen the surveillance of last-line antibiotic resistance and move toward the implementation of molecular and genomic tools for the detection of resistance genes in clinical settings.
Acknowledgments
We gratefully acknowledge the clinical laboratory for your technical assistance and to the Vice Chancellery for Research of El Bosque University (especially to Miguel Otero for your invaluable support).
Author Contributions
J.E.-P., I.G.B.M., and N.V.G. designed research; I.G.M.B., D.F.J.M., and I.T.M. identified the isolate, performed microbiological analysis, and interpreted the clinical characteristics of the patient; D.A., R.A.M.O., and Z.L.C.R. performed the molecular analysis and genome sequencing; D.A., R.A.M.-O., J.E.-P., and Z.L.C.R. performed the bioinformatics analysis; D.A., I.G.B.M., R.A.M.-O., N.V.G., and J.E.-P. interpreted the data; D.A., I.G.B.M., and J.E.-P. wrote the paper.
Funding
This research was partially funded by the Departamento Administrativo de Ciencia, Tecnología e Innovación, Colciencias (grant number 1308-777-58007), Vice Chancellery for Research of El Bosque University (grant number PCI63-2014), and Fundacion-Clinica Shaio.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Tacconelli E., Carrara E., Savoldi A., Harbarth S., Mendelson M., Monnet D.L., Pulcini C., Kahlmeter G., Kluytmans J., Carmeli Y., et al. Discovery, research, and development of new antibiotics: The WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 2018;18:318–327. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 2.Oteo J., Hernandez J.M., Espasa M., Fleites A., Saez D., Bautista V., Perez-Vazquez M., Fernandez-Garcia M.D., Delgado-Iribarren A., Sanchez-Romero I., et al. Emergence of OXA-48-producing Klebsiella pneumoniae and the novel carbapenemases OXA-244 and OXA-245 in Spain. J. Antimicrob. Chemother. 2013;68:317–321. doi: 10.1093/jac/dks383. [DOI] [PubMed] [Google Scholar]
- 3.Valenza G., Nickel S., Pfeifer Y., Eller C., Krupa E., Lehner-Reindl V., Holler C. Extended-spectrum-beta-lactamase-producing Escherichia coli as intestinal colonizers in the German community. Antimicrob. Agents Chemother. 2014;58:1228–1230. doi: 10.1128/AAC.01993-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potron A., Poirel L., Dortet L., Nordmann P. Characterisation of OXA-244, a chromosomally-encoded OXA-48-like beta-lactamase from Escherichia coli. Int. J. Antimicrob. Agents. 2016;47:102–103. doi: 10.1016/j.ijantimicag.2015.10.015. [DOI] [PubMed] [Google Scholar]
- 5.Hoyos-Mallecot Y., Naas T., Bonnin R.A., Patino R., Glaser P., Fortineau N., Dortet L. OXA-244-Producing Escherichia coli Isolates, a Challenge for Clinical Microbiology Laboratories. Antimicrob. Agents Chemother. 2017;61:e00818-17. doi: 10.1128/AAC.00818-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Findlay J., Hopkins K.L., Loy R., Doumith M., Meunier D., Hill R., Pike R., Mustafa N., Livermore D.M., Woodford N. OXA-48-like carbapenemases in the UK: An analysis of isolates and cases from 2007 to 2014. J. Antimicrob. Chemother. 2017;72:1340–1349. doi: 10.1093/jac/dkx012. [DOI] [PubMed] [Google Scholar]
- 7.van Hattem J.M., Arcilla M.S., Bootsma M.C., van Genderen P.J., Goorhuis A., Grobusch M.P., Molhoek N., Oude Lashof A.M., Schultsz C., Stobberingh E.E., et al. Prolonged carriage and potential onward transmission of carbapenemase-producing Enterobacteriaceae in Dutch travelers. Future Microbiol. 2016;11:857–864. doi: 10.2217/fmb.16.18. [DOI] [PubMed] [Google Scholar]
- 8.Beyrouthy R., Robin F., Delmas J., Gibold L., Dalmasso G., Dabboussi F., Hamze M., Bonnet R. IS1R-mediated plasticity of IncL/M plasmids leads to the insertion of bla OXA-48 into the Escherichia coli Chromosome. Antimicrob. Agents Chemother. 2014;58:3785–3790. doi: 10.1128/AAC.02669-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wirth T., Falush D., Lan R., Colles F., Mensa P., Wieler L.H., Karch H., Reeves P.R., Maiden M.C., Ochman H., et al. Sex and virulence in Escherichia coli: An evolutionary perspective. Mol. Microbiol. 2006;60:1136–1151. doi: 10.1111/j.1365-2958.2006.05172.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brzuszkiewicz E., Bruggemann H., Liesegang H., Emmerth M., Olschlager T., Nagy G., Albermann K., Wagner C., Buchrieser C., Emody L., et al. How to become a uropathogen: Comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. USA. 2006;103:12879–12884. doi: 10.1073/pnas.0603038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang J., Li X., Li J., Hurley D., Bai X., Yu Z., Cao Y., Wall E., Fanning S., Bai L. Complete genetic analysis of a Salmonella enterica serovar Indiana isolate accompanying four plasmids carrying mcr-1, ESBL and other resistance genes in China. Vet. Microbiol. 2017;210:142–146. doi: 10.1016/j.vetmic.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 12.Yang Q.E., Walsh T.R. Toxin-antitoxin systems and their role in disseminating and maintaining antimicrobial resistance. FEMS Microbiol. Rev. 2017;41:343–353. doi: 10.1093/femsre/fux006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C., Feng Y., Liu F., Jiang H., Qu Z., Lei M., Wang J., Zhang B., Hu Y., Ding J., et al. A Phage-Like IncY Plasmid Carrying the mcr-1 Gene in Escherichia coli from a Pig Farm in China. Antimicrob. Agents Chemother. 2017;61 doi: 10.1128/AAC.02035-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Siguier P., Gourbeyre E., Chandler M. Bacterial insertion sequences: Their genomic impact and diversity. FEMS Microbiol. Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He S., Hickman A.B., Varani A.M., Siguier P., Chandler M., Dekker J.P., Dyda F. Insertion Sequence IS26 Reorganizes Plasmids in Clinically Isolated Multidrug-Resistant Bacteria by Replicative Transposition. MBio. 2015;6:e00762. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marquez-Ortiz R.A., Haggerty L., Olarte N., Duarte C., Garza-Ramos U., Silva-Sanchez J., Castro B.E., Sim E.M., Beltran M., Moncada M.V., et al. Genomic Epidemiology of NDM-1-Encoding Plasmids in Latin American Clinical Isolates Reveals Insights into the Evolution of Multidrug Resistance. Genome Biol. Evol. 2017;9:1725–1741. doi: 10.1093/gbe/evx115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milne I., Stephen G., Bayer M., Cock P.J., Pritchard L., Cardle L., Shaw P.D., Marshall D. Using Tablet for visual exploration of second-generation sequencing data. Brief. Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 19.Krumsiek J., Arnold R., Rattei T. Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics. 2007;23:1026–1028. doi: 10.1093/bioinformatics/btm039. [DOI] [PubMed] [Google Scholar]
- 20.Seemann T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 21.Rutherford K., Parkhill J., Crook J., Horsnell T., Rice P., Rajandream M.A., Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 22.Zankari E., Hasman H., Cosentino S., Vestergaard M., Rasmussen S., Lund O., Aarestrup F.M., Larsen M.V. Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McArthur A.G., Waglechner N., Nizam F., Yan A., Azad M.A., Baylay A.J., Bhullar K., Canova M.J., De Pascale G., Ejim L., et al. The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 2013;57:3348–3357. doi: 10.1128/AAC.00419-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gupta S.K., Padmanabhan B.R., Diene S.M., Lopez-Rojas R., Kempf M., Landraud L., Rolain J.M. ARG-ANNOT, a new bioinformatic tool to discover antibiotic resistance genes in bacterial genomes. Antimicrob. Agents Chemother. 2014;58:212–220. doi: 10.1128/AAC.01310-13. [DOI] [PMC free article] [PubMed] [Google Scholar]