Abstract
Honey bee venom has been established to have significant effect in immunotherapy. In the present study, (Z)-11-eicosenol-a major constituent of bee venom, along with its derivations methyl cis-11-eicosenoate and cis-11-eicosenoic acid, were synthesised to investigate their immune stimulatory effect and possible use as vaccine adjuvants. Stimuli that prime and activate the immune system have exerted profound effects on immune cells, particularly macrophages; however, the effectiveness of bee venom constituents as immune stimulants has not yet been established. Here, the abilities of these compounds to act as pro-inflammatory stimuli were assessed, either alone or in combination with lipopolysaccharide (LPS), by examining the secretion of tumour necrosis factor-α (TNF-α) and the cytokines interleukin-1β (IL-1β), IL-6 and IL-10 by THP-1 macrophages. The compounds clearly increased the levels of IL-1β and decreased IL-10, whereas a decrease in IL-6 levels suggested a complex mechanism of action. A more in-depth profile of macrophage behaviour was therefore obtained by comprehensive untargeted metabolic profiling of the cells using liquid chromatography mass spectrometry (LC-MS) to confirm the ability of the eicosanoids to trigger the immune system. The level of 358 polar and 315 non-polar metabolites were changed significantly (p < 0.05) by all treatments. The LPS-stimulated production of most of the inflammatory metabolite biomarkers in glycolysis, the tricarboxylic acid (TCA) cycle, the pentose phosphate pathway, purine, pyrimidine and fatty acids metabolism were significantly enhanced by all three compounds, and particularly by methyl cis-11-eicosenoate and cis-11-eicosenoic acid. These findings support the proposed actions of (Z)-11-eicosenol, methyl cis-11-eicosenoate and cis-11-eicosenoic acid as immune system stimulators.
Keywords: (Z)-11-eicosenol, methyl cis-11-eicosenoate, cis-11-eicosenoic acid, pro- and anti-inflammatory cytokines, LPS stimulation, THP-1 cells, macrophages, adjuvant vaccine
1. Introduction
Adjuvants, in the context of vaccines, are described as substances capable of enhancing and modulating antigen-specific immune responses to improve vaccine efficacy [1]. The immune stimulating effects of adjuvants was first established with the addition of aluminium potassium sulphate or aluminium salts to human vaccines [2,3]. Today, a better understanding of immune responses has increased the availability and variety of vaccine adjuvants to include virosomes, MF59 and AS04 [3]. Adjuvants are now in widespread use, but their development has been empirical, without a clear understanding of their molecular and cellular mechanisms of action. However, several studies have suggested that adjuvants act by enhancing T and B cell responses, by stimulating innate immunity and by increasing the magnitude of the adaptive responses to the vaccine [4,5,6]. Adjuvants also stimulate a strong and comprehensive immune response to antigen by mimicking natural defensive trigger molecules, such as endogenous immune-active substances (e.g., chemokines and cytokines) or other natural compounds (e.g., vitamin E and saponins) [3].
A crucial need exists for the development and investigation of new vaccine adjuvant compounds that have the ability to induce immune responses. However, the molecular and cellular mechanisms that regulate the activity of the majority of human-licensed adjuvants are still only partially characterised. The adjuvant derivatives of bacterial components, such as lipopolysaccharide (LPS), cytidine–phosphate–guanosine (CpG) oligonucleotides and monophosphoryl lipid A (MPL), are probably the most characterised at present [7]. MPL works as an agonist of Toll-like receptor (TLR4), which is expressed on antigen presenting cells (APCs), such as dendritic cells (DCs) and macrophages, and promotes cytokine expression, antigen presentation and migration of the APCs to the T-cell area [8,9]. These compounds act as microbial sensors called pathogen-associated molecular patterns (PAMPs), and they activate pattern recognition receptors (PRR) and subsequently TLR [10].
LPS is the most widely studied of the TLR4 ligands, and its adjuvant derivatives (which are less toxic) have been the basis of virtually all clinical trials of adjuvant TLR4 agonists [11]. The interactions of LPS derivatives with TLR4 are restricted mainly to their lipid A portion, which are composed of polyacylated diglucosamine lipids [12]. A combination of specific adjuvants with TLR agonists has been proposed to optimise vaccines [13]. Therefore, evaluation of the LPS enhancement of cytokine productions and a metabolic interpretation of LPS immune-modulatory effects would be a promising strategy for investigating proposed new adjuvants.
Honey bee (Apis mellifera) venom is a very complex mixture of substances, including organics, peptides and enzymes. These ingredients contribute a variety of biological activities towards the overall toxic shock [14], and have attracted attention as possible leads for drug discovery. Some components have been applied to the treatment of inflammation and cancer, for example, [15,16], but their use as vaccine adjuvants is still under investigation. Melittin, a major lytic peptide found in bee venom, has been proposed as a vaccine adjuvant due to its confirmed ability to enhance TNF-α, IL-1β and IL-6 cytokine production within the THP-1 monocyte-derived macrophage cell line [17,18].
An organic compound of interest in honey bee venom is (Z)-11-eicosen-1-ol. This is a major component of the alarm pheromone mixture co-secreted with the venom [19]. Insect pheromones are typically volatile organics that are released to warn of danger [20]. (Z)-11-eicosen-1-ol prolongs the effectiveness of isopentyl acetate, another key component of the alarm pheromone secretion, thereby increasing the speed of the aggressive response [14,19]. A similar compound, (Z)-9-eicosen-1-ol, was detected in A. mellifera venom, resembling the compound described by Pickett et al. and differing only with respect to the double bond position [18]. It was found to enhance the release of pro-inflammatory cytokines, other than IL-6 [18].
Immune metabolism is a rapidly growing area of research. Recently, several studies have shown the importance of metabolites, such as succinate and itaconate, in the modulation of the innate immune response of macrophages [21,22]. The exposure of macrophages to various immune stimuli, including pathogenic antigens and cytokines, initiates several signaling cascades by interactions with receptors (i.e., cytokine receptors or PRR) [23]. This intracellular and extracellular signaling is expected to elicit major changes in metabolites to promote the required alteration of the cell phenotype and changes in anabolic and catabolic pathways [23]. Immune cells use different metabolic pathways to provide adequate energy generation and cell survival during cell growth and proliferation. These metabolic pathways, which include fatty acid synthesis, rely on products from other pathways, such as glycolysis and the tricarboxylic acid (TCA) cycle, to provide key synthetic precursors [24]. For this reason, metabolomics analysis of immune cells can provide a more complete understanding of the physiological state of an organism and of the alterations occurring in the metabolome, as metabolites are often considered the end stage of biological processes.
A growing number of findings highlight the crucial role of metabolic reprogramming in macrophage activation. Immune cells utilise five metabolic pathways: glycolysis, the TCA cycle, the pentose phosphate pathway (PPP), fatty acid synthesis and amino acid metabolism [24]. The present study investigated the metabolic responses of phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 cells treated with three different forms of synthetic honey bee venom eicosenoid compounds: (Z)-11-eicosenol, methyl cis-11-eicosenoate and cis-11-eicosenoic acid. The overall goal was to determine how these compounds might trigger an immune response and further used as vaccine adjuvants. Comprehensive metabolic profiling and assessments of pro- and anti-inflammatory cytokines (TNF-α, IL-1β, IL-6 and IL-10) were also conducted following exposure of LPS-stimulated THP-1 macrophage cells with these compounds. The observation of a synergistic effect of the investigated compounds with LPS in enhancement of immune stimulatory activation and cytokine production suggested the potential use of honey bee venom components as immune-modulatory agents and as vaccine adjuvants, which support their use as pro- rather than anti-inflammatory agents.
2. Materials and Methods
2.1. Sample Preparation
Methyl cis-11-eicosenoate (CAS 2390-09-2) and cis-11-eicosenoic acid (CAS 5561-99-9) were purchased from Sigma-Aldrich and used directly.
Cis (Z)-11-eicosenol (CAS 62442-62-0) was obtained by selective reduction of methyl cis-11-eicosenoate with lithium aluminium hydride. Briefly, dry diethyl ether was added to the hydride and stirred in an ice bath for 30 min. The ester was added dropwise, stirred for 30 min, then refluxed gently for 60 min. The reaction mixture was worked up by quenching with wet ether and washing with potassium sodium tartrate solution and water. The extracted organic layer was dried over sodium sulphate, filtered and excess solvent evaporated with an air stream to obtain the product. Confirmation of the reduction was obtained via IR and the full structure of the intended product was confirmed by NMR, including by 1D 1H and 13C-{1H} NMR spectra and 2D [1H, 13C]-HSQC and 2D [1H, 1H] DQFCOSY and TOCSY NMR spectra Data and their assignments were confirmed directly with that of the natural product as previously assigned [18].
1H NMR (1D, 600 MHz, DMSO-d6): δH 0.84–0.86 (t, J = 13 Hz, 3H, C1), 1.24–1.30 (m, 26H, C2/C3/C4/C5/C6/C7/C12/C13/C14/C15/C16/C17/C18), 1.35–1.40 (m, 2H, C19) 1.96–1.99 (q, J = 19 Hz, 4H, C8/C11), 3.35–3.38 (m, 2H, C20), 4.30–4.31 (m, 1H, OH), 5.29–5.34 (sept, 2H, C9/10).
13C-{1H} NMR (1D, 150 MHz, DMSO- d6): δ 13.90 (C1), 22.07 (C2), 25.50 (C18), 26.53 (C8/C11), 28.56 (C5), 28.67 (C6), 28.81 (C7), 28.85 (C12), 28.95 (C14), 29.00 (C15), 29.07 (C16), 29.08 (C17), 31.28 (C3), 32.54 (C19), 60.70 (C20), 129.61 (C9/C10).
2.2. Cell Culture and Differentiation
THP-1 cells (American Type Culture Collection, ATCC®, Manassas, VA, USA) were cultured to a seeding density of 1 × 105 cells/mL in Roswell Park Memorial Institute (RPMI) medium 1640 (Thermo Fisher Scientific, Loughborough, UK) with addition of L-glutamine (2 mmol/L, Life Tech, Paisley, UK), foetal calf serum (FCS, 10% v/v, Life Tech) and penicillin/streptomycin (100 IU/100 µg/mL, Life Tech). Sub-cultures were prepared every 2 to 4 days in fresh media and incubated at 37 °C and 100% humidity in 5% CO2 prior to differentiation by addition of phorbol 12-myristate 13-acetate (PMA, 60 ng/mL final concentration, Sigma-Aldrich, Dorset, UK). The cells were then incubated for 48 h, after which the PMA-containing media were replaced with fresh media and the cells rested for an additional 24 h prior to examination by light microscopy.
2.3. Cell Viability Assay
THP-1 cells were cultured in 96-well plates at a seeding density of 1 × 105 cells/mL. The cells were incubated for 24 h at 37 °C and 100% humidity in 5% CO2 prior to treatment with eicosenoid compounds at various concentrations (1.2 to 150 µg/mL) followed by a further 24 h incubation. Controls included cells alone (no treatment), medium alone (background) and dimethyl sulphoxide (1.5% v/v DMSO, positive) were added. The plates were then incubated for an additional 24 h following addition of resazurin salt solution (0.1 mg/mL, 10% v/v final concentration) before examining the fluorescence at λEx = 560 nm and λEm = 590 nm (SpectraMax M5, Molecular Devices, Sunnyvale, CA, USA). A background correction was performed and the call viability for each treatment concentration was determined with respect to the mean viability of the negative control (n = 3). Mean inhibitory concentration (IC50) values and dose–response curves were obtained using GraphPad Prism for Windows v 5.00 (GraphPad Software, San Diego, CA, USA).
2.4. Cytokine Production
The differentiated cells were incubated for a further 24 h in the presence and absence of LPS (0.5 µg/mL, Sigma-Aldrich) with the final eicosenoid concentrations presented in Table 1. The prepared medium was obtained and frozen until needed for the enzyme-linked immunosorbent assay (ELISA) (n = 3).
Table 1.
Sample groups, IC50 concentrations and final chosen concentrations of synthetic bee venom compounds tested in phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 cells.
| Synthetic Compounds | IC50 (µg/mL) | Selected Final Concentration (µg/mL) | |
|---|---|---|---|
| Group ID | Chemical Name | ||
| 11E-OH | (Z)-11-eicosenol | 19.88 | 9.0 |
| 11E-ester | methyl cis-11-eicosenoate | >150 | 150 |
| 11E-acid | cis-11-eicosenoic acid | 90.98 | 40.0 |
IC: inhibitory concentration.
2.5. Enzyme-Linked Immunosorbent Assay (ELISA)
The production of inflammatory cytokines TNF-α, IL-1β, IL-6 and IL-10 were quantified using ELISA Ready-Set-Go kits (Thermo Fisher Scientific, Bremen, Germany) in accordance with the supplier’s protocol. Sulphuric acid solution (2N) was added to stop the reaction prior to reading the absorbance at 560 nm (SpectraMax M5, Molecular Devices) and subtracting the absorbance at 570 nm.
2.6. Metabolite Extraction
THP-1 cells differentiated with PMA were cultured in 6-well plates at a seeding density of 4.5 × 105 cells/mL (n = 6) for 48 h, after which the medium was removed by aspiration and replaced prior to incubation with LPS (0.5 µg/mL) either alone or together with (Z)-11-eicosenol (9 µg/mL), eicosenoate (150 µg/mL) and eicosenoic acid (40 µg/mL) for another 24 h. Extraction of the metabolites was carried out as described previously [17]. Seven different analytical standard solutions were prepared by adding each metabolite standard (10 µg/mL final concentration) containing glycine-13C2 [25]. Combined quality control (QC) samples were prepared by pipetting 20 µL from each individual sample, mixing and transferring to a high-performance liquid chromatography (HPLC) vial.
2.7. LC-MS Conditions
LC-MS was carried out using an Exactive Orbitrap (Thermo Fisher Scientific, Bremen, Germany); the conditions used were as described in our previous paper [17].
2.8. Data Extraction and Statistical Analysis
Data extraction was performed using the MZMatch software (SourceForge, La Jolla, CA, USA) and the metabolite peaks were filtered, compared and characterised using the Microsoft Excel IDEOM [26] as described in our previous paper [17]. The metabolite data were also subjected to multivariate analysis by fitting PCA-X and OPLS-DA using the SIMCA-P software v.14.0 (Umetrics, Umea, Sweden). Univariate comparisons were performed using Microsoft Excel and paired t-tests between treated and control cells and differences were considered significant at p < 0.05. Using ELISA, standard calibration curves were plotted by fitting the average optical density (OD) values of TNF-α, IL-1β, IL-6 and IL-10 to 4-parameter logistic (4-PL) regression curves. Each standard concentration assayed in duplicate (n = 2), as shown in Figures S1–S12.
3. Results
3.1. Cytotoxicity of Eicosenoid Compounds against PMA-Differentiated THP-1 Cells
The potential cytotoxicity of (Z)-11-eicosenol (11E-OH), eicosenoate (11E-ester) and eicosenoic acid (11E-acid) (Figure 1) on PMA-differentiated THP-1 cells was evaluated to select an appropriate final concentration for further tests. Clear dose-dependent toxicity to THP-1 cells was observed for 11E-OH and its 11E-acid form. The lowest IC50 value at 19.88 µg/mL was observed for 11E-OH, whereas this value was 90.98 µg/mL for 11E-acid (Figure 2A–C). By contrast, the 11E-ester form was nontoxic to THP-1 cells, with an IC50 value greater than 150 µg/mL. ELISAs were also conducted to assess the cytokine levels in THP-1 derived-macrophage cells upon treatment with these three compounds. As shown in Table 1, the final concentrations for cytokine assessments were chosen as those that were below the IC50 values and resulted in >90% of the cells remaining viable. There was no difference in effect between the solvent control DMSO alone (1.5% final concentration) and negative control media on cell viability.
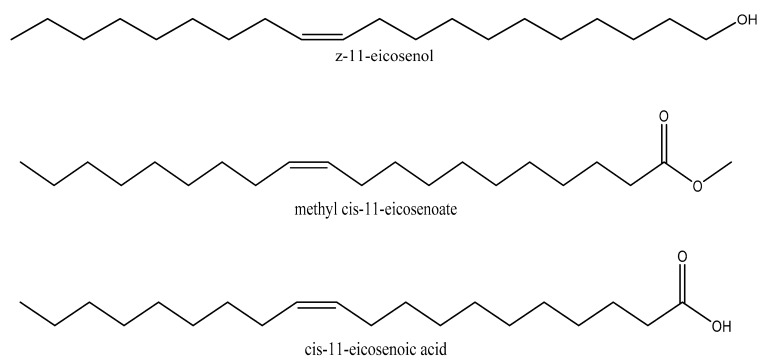
Figure 1.
Chemical structures of (Z)-11-eicosenol, methyl cis-11-eicosenoate and cis-11-eicosenoic acid.
Figure 2.
Cytotoxic effects of eicosenoid compounds at varying doses on phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 cells. (A) (Z)-11-eicosenol (11E-OH) compound was cytotoxic to PMA-treated cells. (B) Eicosenoate (11E-ester) compound was non-cytotoxic to PMA-treated cells. (C) Eicosenoic acid (11E-acid) compound was cytotoxic to PMA-treated cells. Each data point represents the mean ± SD (n = 3).
3.2. Effect of Eicosenoid Compounds on Pro-Inflammatory TNF-α Cytokine Production
Using ELISA, the effects of the eicosenoid compounds on the production of TNF-α cytokine are shown in Figure 3 and Table S1. The levels of secreted TNF-α by PMA-differentiated THP-1 cells were slightly or negligibly affected by all three forms of eicosanoid when combined with LPS. The increase was only statistically significant (p < 0.05) with 11E-acid when compared with LPS alone. (Z)-11-eicosenol compounds on their own significantly enhanced the production of TNF-α when compared with untreated cells.
Figure 3.
Effect of eicosenoid compounds on the production of TNF-α by PMA-differentiated THP-1 cells in the absence and presence of LPS (0.5 µg/mL). The TNF-α levels were significantly increased by 11E-acid when compared with LPS alone (n = 3). Ctr: Untreated control; LPS: Lipopolysaccharide; (11E-OH): (Z)-11-eicosenol; (11E-ester): Eicosenoate; (11E-acid): Eicosenoic acid; *: Significant (p < 0.05); **: Significant (p < 0.01).
3.3. Effect of Eicosenoid Compounds on Pro-Inflammatory IL-1β Cytokine Production
Compared to TNF-α, the enhancement in the production of IL-1β by (Z)-11-eicosenol, eicosenoate and eicosenoic acid in LPS co-stimulated THP-1 cells was much more pronounced. The release of IL-1β was greatly enhanced (ratio > 1.0) by approximately 84% and was statistically significant when compared with LPS alone. (Z)-11-eicosenol (~50%) enhanced the production of this cytokine upon stimulation with LPS, although the increase was not significant when compared with LPS alone (Figure 4 and Table S2). The level of IL-1β was enhanced by (Z)-11-eicosenol and eicosenoic acid alone, in the absence of LPS, when compared with the untreated control; however, the effects were not statistically significant.
Figure 4.
Effect of eicosenoid compounds on the production of IL-1β by phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 cells in the absence and presence of LPS (0.5 µg/mL). The IL-1β levels were significantly enhanced in 11E-ester and 11E-acid combination treatments when compared with LPS alone (n = 3). Ctr: Untreated control; LPS: Lipopolysaccharide; (11E-OH): (Z)-11-eicosenol; (11E-ester): Eicosenoate; (11E-acid): Eicosenoic acid; *: Significant (p < 0.05); **: Significant (p < 0.01).
3.4. Effect of Eicosenoid Compounds on Pro-Inflammatory IL-6 Cytokine Production
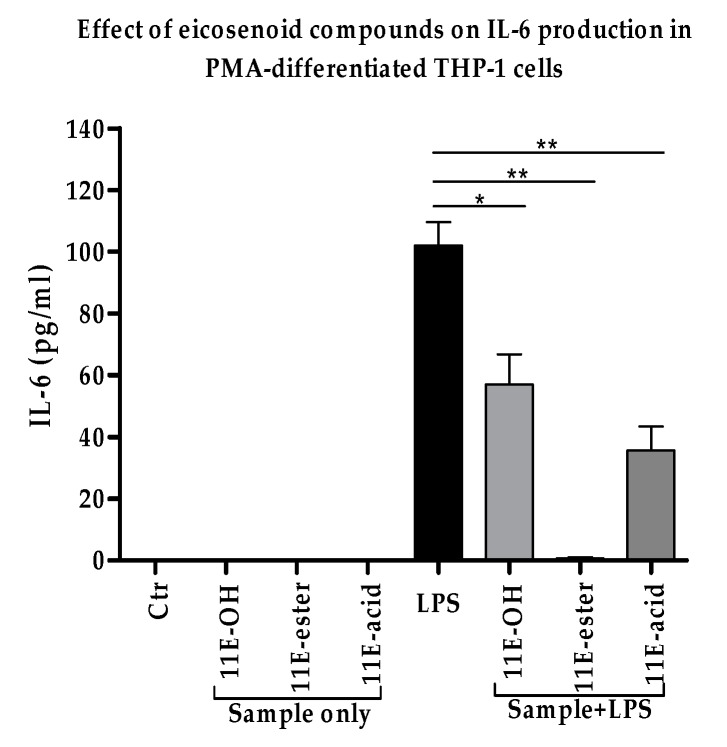
The levels of IL-6 cytokine were evaluated to confirm the previously reported decrease in response to eicosenoid compounds [18]. A decrease in IL-6 levels in THP-1 derived macrophage cells was also observed in the present study in response to all three forms of synthetically prepared eicosenoids. The levels of this cytokine were significantly decreased (p < 0.05) when compared with LPS alone. Surprisingly, no detectable amount of IL-6 was produced by the cells treated with eicosenoate (11E-ester) in the presence of LPS (Figure 5). Unstimulated macrophage-like THP-1 cells did not produce any IL-6. The same was true for cells stimulated with eicosenoids alone (Table S3).
Figure 5.
Effect of eicosenoid compounds on the production of IL-6 by phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 cells in the absence and presence of LPS (0.5 µg/mL). The IL-6 levels were significantly decreased in all three combination treatments when compared with positive control LPS (n = 3). Ctr: Untreated control; LPS: Lipopolysaccharide; (11E-OH): (Z)-11-eicosenol; (11E-ester): Eicosenoate; (11E-acid): Eicosenoic acid; *: Significant (p < 0.05); **: Significant (p < 0.01).
3.5. Effect of Eicosenoid Compounds on Anti-Inflammatory Il-10 Cytokine Production
A decrease in the levels of the anti-inflammatory IL-10 cytokine would support the use of the eicosenoid compounds as immune response stimulators. Combination treatments with LPS significantly decreased the level of this cytokine in PMA-differentiated THP-1 cells when compared with LPS alone. The extent of the reductions in the release of IL-10 were about 30%, 80% and 50% in response to (Z)-11-eicosenol, eicosenoate and eicosenoic acid treatments, respectively, when compared to treatment with LPS alone (Figure 6 and Table S4).
Figure 6.
Effect of eicosenoid compounds on the production of IL-10 by phorbol 12-myristate 13-acetate (PMA)-differentiated THP-1 cells in the absence and presence of LPS (0.5 µg/mL). The IL-10 levels were significantly decreased in all three combination treatments when compared with positive control LPS (n = 3). Ctr: Untreated control; LPS: Lipopolysaccharide; (11E-OH): (Z)-11-eicosenol; (11E-ester): Eicosenoate; (11E-acid): Eicosenoic acid; **: Significant (p < 0.01); ***: Significant (p < 0.001).
3.6. Effect of Eicosenoid Compounds on Polar THP-1 Cell Metabolites
Untargeted metabolic profiling of PMA-differentiated THP-1 cells was performed using LC-MS analysis. Samples were prepared by incubation of the macrophage cells with LPS and one of the three forms of the eicosenoids and compared to control untreated cells (C). Multivariate and univariate statistical analysis were used to visualise and examine the metabolite effects on the following treatment combinations: T1 (9 µg/mL (Z)-11-eicosenol + 0.5 µg/mL LPS), T2 (150 µg/mL eicosenoate + 0.5 µg/mL LPS) and T3 (40 µg/mL eicosenoic acid + 0.5 µg/mL LPS). The effect of LPS alone was also evaluated to confirm the previous findings [17] and to investigate new pathways that might be involved in immune stimulation by eicosenoids. LPS synergism was clearly evident from the cytokine assessment, particularly with IL-1β and IL-10. Further metabolic profiling of these combination treatments would aid in determining how they inhibit or stimulate the immune response.
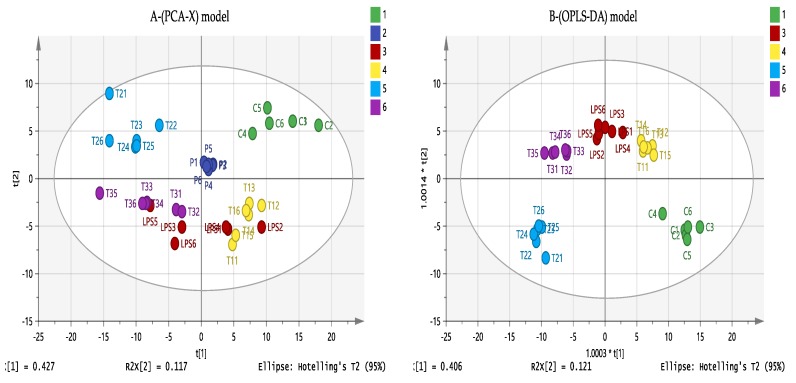
As shown in Figure 7A, principle component analysis (PCA) showed an absence of outliers. In addition, pooled quality control samples (QC, P1–6) produced a single tight cluster in the centre of the dataset, confirming the stability, precision and validity of the instrumental analytical method. Orthogonal partial least squares discriminant analysis (OPLS-DA), a supervised model for sample classification, showed a clear separation of each combination treatment, indicating unique metabolite profiling (Figure 7B). The OPLS-DA model parameters and validation of the plot suggest a strong model, with the p value associated with the cross-validation (CV)-ANOVA = 1.96 × 10−21, indicating that the model was valid.
Figure 7.
(A) Principle component analysis (PCA-X) vs. (B) Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) score plots of THP-1 cells. The figures show a clear separation between control, pooled and treatment groups based on 358 polar metabolites separated on a ZIC-pHILIC column (n = 6). PCA score plot (A) gives the goodness of fit (R2X) = 0.807, and the goodness of prediction (Q2) = 0.641. OPLS-DA score plot (B) gives R2X = 0.829, R2Y = 0.962, Q2 = 0.874. (C: Control; LPS: Lipopolysaccharides; T1(1–6): (11E-OH) (Z)-11-eicosenol + LPS; T2(1–6): (11E-ester) Eicosenoate + LPS; T3(1–6): (11E-acid) Eicosenoic acid + LPS; p = pooled samples).
Pooled quality control samples were injected at intervals (n = 6) during the course of the run and used for further filtration of the dataset based on relative standard deviations (RSD). Metabolites with RSD values >30% within the pooled samples were excluded. The univariate analysis shown in Table 2 reveals a large number of metabolic changes resulting from application of eicosenoid treatments. Greater effects were observed with the 11E-ester and 11E-acid forms when combined with LPS and compared to untreated control cells. Increases in abundance were detected for a large number of metabolites, including arginine and proline, Krebs cycle (TCA cycle) compounds and purine metabolites, which are all critically involved in inflammatory processes and immune responses of the cells.
Table 2.
Significantly changed polar metabolites in THP-1 cells treated with lipopolysaccharide (LPS), alone or in combination with one of three synthetic forms of honey bee eicosenoids (0.5 µg/mL LPS; 9 µg/mL 11E-OH; 150 µg/mL 11E-ester; 40 µg/mL 11E-acid). Data are compared with those from untreated control cells.
| Mass | Rt | Putative Metabolite | LPS/C | 11E-OH + LPS/C | 11E-Ester + LPS/C | 11E-Acid + LPS/C | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ratio | p Value | Ratio | p Value | Ratio | p Value | Ratio | p Value | |||
| Arginine and Proline Metabolism | ||||||||||
| 129.090 | 15.79 | 4-Guanidinobutanal | 1.523 | 0.030 | 1.580 | 0.008 | 0.880 | ns | 1.539 | 0.010 |
| 111.032 | 20.00 | Pyrrole-2-carboxylate | 1.200 | 0.012 | 1.384 | <0.001 | 1.068 | ns | 1.260 | 0.003 |
| 189.064 | 14.09 | N-Acetyl-l-glutamate * | 0.547 | 0.007 | 0.599 | 0.001 | 1.139 | ns | 0.757 | 0.031 |
| 132.054 | 14.39 | N-Carbamoylsarcosine | 1.100 | ns | 1.051 | ns | 1.395 | 0.003 | 1.367 | 0.011 |
| 240.122 | 16.51 | Homocarnosine | 1.382 | 0.049 | 0.900 | ns | 1.593 | <0.001 | 1.866 | <0.001 |
| 211.036 | 15.28 | Phosphocreatine * | 0.944 | ns | 0.970 | ns | 1.723 | 0.001 | 1.285 | 0.024 |
| 113.059 | 9.94 | Creatinine | 1.625 | ns | 0.939 | ns | 2.104 | <0.001 | 2.199 | <0.001 |
| 130.122 | 26.73 | Agmatine | 1.509 | 0.021 | 1.041 | ns | 2.149 | <0.001 | 1.884 | <0.001 |
| 129.043 | 10.08 | Oxoproline * | 1.732 | 0.013 | 0.850 | ns | 2.331 | <0.001 | 2.525 | <0.001 |
| 175.096 | 16.21 | L-Citrulline * | 1.554 | 0.011 | 0.923 | ns | 2.379 | <0.001 | 2.180 | <0.001 |
| 246.133 | 18.71 | N2-(d-1-Carboxyethyl)-l-arginine | 2.225 | 0.008 | 1.233 | ns | 3.317 | <0.001 | 3.245 | <0.001 |
| 290.122 | 16.97 | N-(l-Arginino)succinate | 0.794 | ns | 1.022 | ns | 1.153 | ns | 1.596 | 0.015 |
| 115.063 | 13.07 | l-Proline * | 1.102 | ns | 0.897 | ns | 0.995 | ns | 1.208 | 0.036 |
| 174.112 | 26.73 | l-Arginine * | 1.593 | 0.022 | 1.089 | ns | 2.172 | <0.001 | 2.160 | <0.001 |
| 132.090 | 26.73 | l-Ornithine * | 1.834 | 0.003 | 1.223 | ns | 2.665 | <0.001 | 2.501 | <0.001 |
| Glycolysis/TCA cycle | ||||||||||
| 260.030 | 16.93 | d-Glucose 1-phosphate * | 1.231 | ns | 1.450 | <0.001 | 0.833 | ns | 1.508 | 0.001 |
| 180.063 | 15.08 | d-Glucose * | 3.076 | <0.001 | 1.901 | <0.001 | 3.638 | <0.001 | 3.309 | <0.001 |
| 339.996 | 18.13 | d-Fructose 1,6-bisphosphate * | 1.789 | <0.001 | 1.930 | <0.001 | 2.217 | <0.001 | 1.734 | <0.001 |
| 260.030 | 16.10 | d-Fructose 6-phosphate * | 1.580 | <0.001 | 1.716 | <0.001 | 1.515 | <0.001 | 1.653 | <0.001 |
| 169.998 | 16.16 | d-Glyceraldehyde 3-phosphate * | 0.600 | <0.001 | 0.441 | <0.001 | 0.519 | <0.001 | 0.404 | <0.001 |
| 185.993 | 16.75 | 3-Phospho-d-glycerate | 0.775 | ns | 1.849 | ns | 0.886 | ns | 0.934 | ns |
| 169.998 | 15.33 | Glycerone phosphate | 1.244 | ns | 2.368 | 0.014 | 1.804 | <0.001 | 1.293 | 0.023 |
| 177.943 | 15.96 | Pyrophosphate | 0.832 | 0.002 | 0.946 | ns | 1.040 | ns | 1.015 | ns |
| 97.977 | 15.96 | Orthophosphate | 0.768 | 0.003 | 0.912 | ns | 1.031 | ns | 1.036 | ns |
| 192.027 | 18.13 | Citrate * | 1.594 | <0.001 | 1.187 | 0.021 | 1.939 | <0.001 | 1.904 | <0.001 |
| 134.022 | 15.88 | (S)-Malate * | 1.116 | ns | 0.943 | ns | 0.869 | 0.002 | 1.124 | ns |
| 174.016 | 18.13 | cis-Aconitate * | 1.265 | ns | 0.984 | ns | 1.662 | 0.001 | 1.633 | 0.001 |
| 192.027 | 19.36 | Isocitrate * | 1.388 | ns | 1.493 | ns | 2.417 | 0.002 | 1.788 | 0.014 |
| 116.011 | 15.02 | Fumarate * | 1.186 | ns | 0.831 | ns | 1.445 | 0.002 | 1.826 | <0.001 |
| 118.027 | 14.98 | Succinate * | 1.557 | 0.002 | 1.170 | ns | 1.346 | 0.005 | 1.819 | 0.001 |
| 131.070 | 14.97 | Creatine * | 1.437 | 0.001 | 1.061 | ns | 3.014 | <0.001 | 1.365 | 0.002 |
| 809.126 | 12.49 | Acetyl-CoA | 0.730 | 0.006 | 0.907 | ns | 1.001 | ns | 0.995 | ns |
| 665.125 | 13.44 | NADH * | 0.553 | <0.001 | 0.642 | <0.001 | 1.330 | ns | 0.752 | 0.008 |
| 663.109 | 14.39 | NAD+ * | 0.490 | <0.001 | 0.499 | <0.001 | 0.683 | <0.001 | 0.548 | <0.001 |
| 506.995 | 16.67 | ATP * | 0.689 | 0.002 | 0.769 | 0.001 | 0.584 | <0.001 | 0.724 | 0.001 |
| 427.030 | 15.30 | ADP * | 0.598 | <0.001 | 0.671 | <0.001 | 0.638 | <0.001 | 0.762 | <0.001 |
| 443.024 | 18.01 | GDP * | 0.772 | 0.011 | 0.978 | ns | 1.004 | ns | 0.931 | ns |
| 522.990 | 19.36 | GTP * | 0.975 | ns | 1.385 | 0.003 | 1.297 | 0.002 | 1.051 | ns |
| Oxidative Stress/Pentose Phosphate Pathway | ||||||||||
| 370.007 | 18.36 | d-Sedoheptulose 1,7-bisphosphate | 1.475 | 0.002 | 1.817 | <0.001 | 1.565 | <0.001 | 1.513 | <0.001 |
| 232.035 | 15.75 | d-Ribitol 5-phosphate * | 0.683 | 0.007 | 0.744 | ns | 0.563 | 0.002 | 0.862 | 0.027 |
| 276.025 | 17.73 | 6-Phospho-d-gluconate * | 1.606 | 0.006 | 1.480 | 0.004 | 0.568 | 0.001 | 0.921 | ns |
| 196.058 | 13.26 | d-Gluconic acid * | 0.886 | ns | 0.800 | <0.001 | 0.795 | 0.002 | 0.925 | ns |
| 150.053 | 13.64 | d-Ribose | 1.307 | 0.017 | 0.899 | ns | 1.286 | 0.018 | 1.289 | 0.011 |
| 290.040 | 16.33 | d-Sedoheptulose 7-phosphate | 1.481 | 0.001 | 2.016 | <0.001 | 1.654 | <0.001 | 1.884 | <0.001 |
| 230.019 | 15.35 | d-Ribulose 5-phosphate | 1.566 | ns | 4.634 | 0.030 | 1.410 | 0.010 | 1.438 | 0.007 |
| 230.019 | 15.75 | d-Ribose 5-phosphate * | 1.525 | <0.001 | 1.516 | <0.001 | 1.796 | <0.001 | 1.230 | <0.001 |
| 745.091 | 17.14 | NADPH * | 0.733 | 0.002 | 0.331 | <0.001 | 00.00 | n/a | 0.347 | 0.004 |
| 743.076 | 16.87 | NADP+ * | 0.722 | 0.024 | 1.983 | <0.001 | 1.879 | <0.001 | 2.419 | 0.001 |
| 152.068 | 13.11 | Xylitol * | 1.423 | 0.001 | 1.063 | ns | 0.907 | ns | 1.189 | 0.049 |
| 196.058 | 13.89 | d-Mannonate | 2.006 | ns | 0.860 | ns | 2.406 | <0.001 | 2.454 | <0.001 |
| 166.048 | 13.43 | d-Xylonate | 1.673 | 0.019 | 0.929 | ns | 3.156 | <0.001 | 2.964 | <0.001 |
| 150.053 | 13.64 | d-Ribose | 1.307 | 0.017 | 0.899 | ns | 1.286 | 0.018 | 1.289 | 0.011 |
| 307.084 | 14.37 | Glutathione | 0.691 | <0.001 | 0.774 | <0.001 | 0.683 | <0.001 | 0.738 | <0.001 |
| 612.152 | 17.52 | Glutathione disulfide * | 1.113 | ns | 1.679 | 0.003 | 3.605 | <0.001 | 3.221 | <0.001 |
| Purine Metabolism | ||||||||||
| 363.058 | 19.36 | Guanosine 3’-phosphate | 0.909 | ns | 1.551 | 0.005 | 1.406 | 0.001 | 1.043 | ns |
| 168.028 | 12.41 | Urate * | 1.487 | ns | 0.738 | ns | 1.778 | 0.003 | 2.285 | 0.001 |
| 268.081 | 11.11 | Inosine * | 1.804 | 0.004 | 5.853 | <0.001 | 12.553 | <0.001 | 9.126 | <0.001 |
| 283.092 | 12.83 | Guanosine * | 1.454 | ns | 4.991 | <0.001 | 16.908 | <0.001 | 4.976 | <0.001 |
| 136.038 | 10.39 | Hypoxanthine * | 33.258 | <0.001 | 31.216 | <0.001 | 56.447 | <0.001 | 67.586 | <0.001 |
| 284.075 | 12.01 | Xanthosine * | 1.564 | 0.004 | 1.306 | 0.020 | 1.651 | 0.003 | 1.825 | 0.001 |
| 152.033 | 11.31 | Xanthine * | 1.907 | 0.005 | 1.171 | ns | 2.376 | <0.001 | 2.492 | <0.001 |
| 363.058 | 16.78 | GMP * | 1.016 | ns | 1.100 | ns | 0.809 | ns | 1.201 | 0.006 |
| 363.058 | 19.36 | Guanosine 3’-phosphate | 0.909 | ns | 1.551 | 0.005 | 1.406 | 0.001 | 1.043 | ns |
| 283.092 | 12.83 | Guanosine * | 1.454 | ns | 4.991 | <0.001 | 16.908 | <0.001 | 4.976 | <0.001 |
| 347.063 | 13.89 | AMP * | 0.656 | <0.001 | 0.660 | 0.001 | 0.326 | <0.001 | 0.680 | <0.001 |
| Pyrimidine Metabolism | ||||||||||
| 483.968 | 17.92 | UTP * | 0.355 | <0.001 | 0.440 | <0.001 | 0.393 | <0.001 | 0.417 | <0.001 |
| 308.041 | 13.81 | dUMP | 0.357 | <0.001 | 0.394 | <0.001 | 0.548 | <0.001 | 0.394 | <0.001 |
| 482.984 | 18.48 | CTP * | 0.473 | <0.001 | 0.596 | <0.001 | 0.688 | <0.001 | 0.492 | <0.001 |
| 403.018 | 17.14 | CDP * | 0.363 | <0.001 | 0.440 | <0.001 | 0.822 | ns | 0.541 | <0.001 |
| 323.052 | 15.35 | CMP * | 0.763 | 0.001 | 0.882 | 0.040 | 1.148 | ns | 0.999 | ns |
| 128.058 | 10.57 | 5,6-Dihydrothymine | 1.664 | ns | 0.698 | 0.006 | 1.395 | 0.007 | 2.075 | 0.033 |
| 126.043 | 15.31 | Thymine | 1.439 | 0.031 | 1.125 | ns | 1.995 | <0.001 | 1.893 | 0.001 |
| 125.059 | 10.93 | 5-Methylcytosine | 1.826 | ns | 1.021 | ns | 2.013 | <0.001 | 2.139 | 0.002 |
| 244.069 | 12.15 | Pseudouridine | 1.780 | 0.007 | 1.058 | ns | 2.266 | <0.001 | 2.134 | <0.001 |
| 243.085 | 12.15 | Cytidine * | 3.051 | 0.009 | 1.812 | ns | 4.662 | <0.001 | 4.849 | <0.001 |
| 176.043 | 16.65 | N-Carbamoyl-l-aspartate | 0.576 | <0.001 | 0.446 | <0.001 | 0.295 | <0.001 | 0.351 | <0.001 |
| 324.036 | 15.18 | UMP * | 0.417 | <0.001 | 0.412 | <0.001 | 0.475 | <0.001 | 0.482 | <0.001 |
| 404.002 | 16.59 | UDP * | 0.218 | <0.001 | 0.347 | <0.001 | 0.413 | <0.001 | 0.350 | <0.001 |
| 536.044 | 16.19 | UDP-d-xylose | 0.593 | <0.001 | 0.699 | 0.001 | 1.017 | ns | 0.745 | 0.002 |
| 580.034 | 18.96 | UDP-glucuronate | 0.615 | <0.001 | 0.736 | <0.001 | 0.701 | <0.001 | 0.664 | <0.001 |
| 566.055 | 16.31 | UDP-glucose * | 0.539 | <0.001 | 0.662 | <0.001 | 0.628 | <0.001 | 0.620 | <0.001 |
| Tryptophan Metabolism | ||||||||||
| 175.063 | 10.37 | Indole-3-acetate | 1.537 | <0.001 | 1.545 | <0.001 | 0.417 | <0.001 | 1.527 | <0.001 |
| 236.079 | 10.37 | l-Formylkynurenine | 1.507 | 0.002 | 1.431 | <0.001 | 0.418 | <0.001 | 1.453 | <0.001 |
| 191.058 | 10.37 | 5-Hydroxyindoleacetate * | 1.512 | <0.001 | 1.506 | <0.001 | 0.523 | <0.001 | 1.547 | <0.001 |
| 208.085 | 11.15 | l-Kynurenine * | 1.483 | ns | 1.114 | ns | 1.565 | 0.003 | 2.087 | <0.001 |
| 220.085 | 9.99 | 5-Hydroxy-l-tryptophan isomer | 8.354 | <0.001 | 7.721 | <0.001 | 1.655 | 0.007 | 8.798 | <0.001 |
| 205.074 | 7.56 | Indolelactate | 2.315 | 0.009 | 1.399 | ns | 2.267 | <0.001 | 3.333 | <0.001 |
| 117.058 | 11.92 | Indole * | 1.413 | 0.011 | 1.216 | ns | 3.554 | <0.001 | 2.257 | <0.001 |
| 204.090 | 11.91 | l-Tryptophan * | 1.311 | ns | 1.050 | ns | 3.194 | <0.001 | 2.019 | 0.003 |
| Miscellaneous | ||||||||||
| 146.069 | 15.31 | l-Glutamine * | 1.445 | 0.001 | 1.105 | ns | 1.965 | <0.001 | 1.889 | <0.001 |
| 147.053 | 14.71 | d-Glutamate * | 1.035 | ns | 0.777 | 0.002 | 1.259 | 0.002 | 1.355 | 0.003 |
| 301.056 | 14.93 | N-Acetyl-d-glucosamine 6-phosphate * | 0.785 | 0.006 | 0.755 | <0.001 | 0.642 | <0.001 | 0.878 | ns |
| 103.100 | 20.65 | Choline * | 1.277 | ns | 1.336 | 0.002 | 1.268 | 0.005 | 1.944 | <0.001 |
| 141.019 | 15.91 | Ethanolamine phosphate * | 0.869 | ns | 0.987 | ns | 0.555 | <0.001 | 0.797 | 0.016 |
| 105.043 | 16.02 | l-Serine * | 1.646 | 0.004 | 1.228 | 0.030 | 1.835 | <0.001 | 1.927 | <0.001 |
| 119.058 | 14.68 | l-Threonine | 2.719 | <0.001 | 1.618 | 0.001 | 3.190 | <0.001 | 2.821 | <0.001 |
| 155.070 | 15.16 | l-Histidine * | 2.450 | <0.001 | 1.285 | 0.018 | 3.175 | <0.001 | 2.737 | <0.001 |
| 146.106 | 25.28 | l-Lysine * | 1.520 | 0.001 | 1.254 | 0.017 | 1.862 | <0.001 | 1.734 | <0.001 |
| 384.122 | 13.99 | S-Adenosyl-l-homocysteine * | 1.163 | ns | 1.047 | ns | 1.580 | 0.001 | 1.545 | 0.002 |
| 203.116 | 11.29 | O-Acetylcarnitine * | 0.702 | 0.001 | 0.688 | <0.001 | 0.644 | <0.001 | 0.689 | 0.001 |
| 175.048 | 14.53 | N-Acetyl-l-aspartate * | 0.758 | 0.003 | 0.628 | <0.001 | 0.797 | 0.003 | 0.970 | ns |
| 133.038 | 15.04 | l-Aspartate * | 1.136 | ns | 0.888 | ns | 1.306 | 0.003 | 1.618 | <0.001 |
| 89.048 | 14.97 | l-Alanine * | 0.671 | 0.001 | 0.731 | 0.004 | 1.041 | ns | 0.720 | 0.001 |
| 226.106 | 16.03 | Carnosine * | 1.088 | ns | 1.080 | ns | 1.203 | 0.016 | 1.201 | 0.012 |
| 132.054 | 15.48 | l-Asparagine * | 1.211 | ns | 0.798 | ns | 1.310 | 0.001 | 1.416 | 0.001 |
| 149.051 | 11.79 | l-Methionine * | 1.358 | ns | 0.910 | ns | 1.822 | <0.001 | 1.869 | <0.001 |
| 222.067 | 17.33 | Cystathionine * | 0.714 | ns | 0.833 | ns | 0.508 | <0.001 | 0.811 | ns |
| 181.074 | 13.24 | l-Tyrosine * | 1.360 | ns | 0.989 | ns | 1.930 | <0.001 | 1.840 | 0.001 |
| 131.095 | 11.11 | l-Leucine * | 1.383 | 0.007 | 0.962 | ns | 1.836 | <0.001 | 1.949 | <0.001 |
| 131.094 | 11.51 | l-Isoleucine * | 1.424 | ns | 0.915 | ns | 1.838 | <0.001 | 1.852 | <0.001 |
| 117.079 | 12.77 | l-Valine * | 1.739 | 0.006 | 1.045 | ns | 1.901 | <0.001 | 1.876 | <0.001 |
Rt: Retention time (min); LPS: Lipopolysaccharides; *: Matches the analytical standard retention time; ns: Non-significant; n/a: Not applicable.
3.7. Effect of Eicosenoid Compounds on Lipophilic Metabolites
In order to gain a comprehensive overview, further analysis was also carried out on the non-polar lipophilic metabolites in the cells using a reversed phase (RP) column. As shown in Figure 8A, PCA was employed for the 314 lipophilic compounds and shows the absence of the outliers. In addition, pooled quality control samples (QC, P1–6) clustered together which indicates the stability, precision and validity of the instrumental analytical method. OPLS-DA shows a clear separation of each combination treatment and represents a unique metabolite profile (Figure 8B). The OPLS-DA model parameters and validation of the plot suggest a strong model with the p CV-ANOVA = 0.0094, indicating that the model was valid (p < 0.5). From the data visualisation below (Figure 8B), strong effects can be predicted on the level of lipophilic metabolites when ester (T2) and acid (T3) treatments are applied to the THP-1 cells.
Figure 8.
(A) Principle component analysis (PCA-X) vs. (B) Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA) score plots of THP-1 cells. The figures show a clear separation between control, pooled and treatment groups based on 314 non-polar metabolites separated on an ACE C4 column (n = 4). PCA score plot (A) gives the goodness of fit (R2X) = 0.571, and the goodness of prediction (Q2) = 0.403. OPLS-DA score plot (B) gives R2X = 0.697, R2Y = 0.982, Q2 = 0.628. (C: Control; LPS: Lipopolysaccharide; T1(1–4): (11E-OH) eicosenol + LPS; T2(1–4): (11E-ester) eicosenoate + LPS; T3(1–4): (11E-acid) eicosenoic acid + LPS; p = pooled samples).
Table 3 summarises the list of metabolites separated on the ACE C4 column in cells treated with eicosenoids in combination with LPS. The metabolites were identified by matching the retention times to those of a standard mixture of known fatty acids.
Table 3.
Significantly changed non-polar metabolites in THP-1 cells treated with lipopolysaccharide (LPS), alone or in combination with one of three synthetic forms of honey bee eicosenoids (0.5 µg/mL LPS; 9 µg/mL 11E-OH; 150 µg/mL 11E-ester; 40 µg/mL 11E-acid). Data are compared with those from untreated control cells.
| Mass | Rt | Putative Metabolite | LPS/C | 11E-OH + LPS/C | 11E-Ester + LPS/C | 11E-Acid + LPS/C | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Ratio | p Value | Ratio | p Value | Ratio | p Value | Ratio | p Value | |||
| Fatty Acid and Related Metabolites | ||||||||||
| 306.256 | 19.23 | Icosatrienoic acid * | 1.963 | <0.001 | 2.976 | 0.001 | 7.296 | <0.001 | 2.941 | <0.001 |
| 328.240 | 18.11 | Docosahexaenoic acid * | 1.637 | 0.007 | 2.005 | 0.003 | 4.187 | <0.001 | 2.336 | <0.001 |
| 336.303 | 22.99 | Docosadienoic acid * | 1.058 | ns | 1.021 | ns | 2.301 | <0.001 | 1.027 | ns |
| 282.256 | 19.94 | Oleic acid * | 1.126 | ns | 1.189 | 0.034 | 2.938 | <0.001 | 1.391 | 0.003 |
| 216.136 | 4.28 | Undecanedioic acid | 1.181 | 0.041 | 1.463 | 0.001 | 9.890 | <0.001 | 1.487 | 0.003 |
| 214.193 | 14.92 | Tridecanoic acid * | 1.138 | ns | 1.102 | ns | 1.731 | 0.018 | 1.323 | 0.004 |
| 334.287 | 21.67 | Docosatrienoic acid * | 1.288 | 0.050 | 1.346 | 0.018 | 3.345 | <0.001 | 1.523 | 0.006 |
| 398.339 | 24.40 | Axillarenic acid * | 1.167 | ns | 1.118 | 0.023 | 1.331 | 0.039 | 1.190 | 0.007 |
| 366.350 | 26.47 | Tetracosenoic acid * | 1.322 | 0.001 | 1.632 | <0.001 | 6.416 | <0.001 | 4.085 | 0.018 |
| 230.152 | 7.29 | Dodecanedioic acid | 1.071 | ns | 1.313 | ns | 23.842 | <0.001 | 1.352 | 0.023 |
| 258.183 | 12.16 | Tetradecanedioic acid | 1.342 | ns | 1.231 | ns | 23.897 | 0.009 | 1.573 | 0.030 |
| 242.225 | 17.85 | Pentadecanoic acid * | 1.021 | ns | 1.084 | ns | 1.711 | 0.017 | 1.233 | 0.039 |
| 248.178 | 12.32 | Hexadecatetraenoic acid | 1.205 | ns | 1.837 | ns | 2.553 | 0.017 | 2.041 | 0.044 |
| 298.287 | 23.03 | Nonadecanoic acid * | 1.133 | 0.001 | 1.133 | ns | 1.310 | 0.001 | 1.219 | <0.001 |
| 332.272 | 20.18 | Docosatetraenoic acid * | 1.664 | 0.004 | 2.549 | <0.001 | 4.261 | <0.001 | 2.348 | <0.001 |
| 304.240 | 18.24 | Eicosatetraenoic acid * | 1.578 | 0.006 | 2.874 | <0.001 | 6.998 | <0.001 | 2.985 | <0.001 |
| 338.319 | 24.50 | Docosenoic acid * | 1.155 | 0.050 | 1.871 | <0.001 | 22.311 | <0.001 | 8.335 | <0.001 |
| 394.381 | 28.24 | Hexacosenoic acid * | 1.433 | <0.001 | 1.047 | ns | 1.903 | <0.001 | 1.685 | <0.001 |
| 364.334 | 25.02 | Tetracosadienoic acid * | 0.888 | ns | 1.409 | ns | 3.193 | <0.001 | 2.568 | <0.001 |
| 202.120 | 4.64 | Decanedioic acid | 1.134 | ns | 1.441 | 0.008 | 1.944 | 0.007 | 1.204 | ns |
| 226.193 | 15.21 | Tetradecenoic acid * | 0.820 | ns | 0.941 | ns | 1.660 | 0.009 | 0.963 | ns |
| 186.162 | 11.81 | Undecanoic acid * | 1.065 | ns | 0.898 | ns | 1.864 | 0.015 | 1.238 | ns |
| 158.131 | 8.54 | Nonanoic acid * | 1.084 | ns | 0.946 | ns | 1.714 | 0.019 | 1.370 | ns |
| 280.240 | 18.26 | Linoleate * | 1.169 | ns | 1.052 | ns | 1.617 | <0.001 | 1.202 | ns |
| 172.110 | 4.54 | 9-Oxononanoic acid | 1.213 | ns | 1.231 | ns | 1.497 | ns | 1.388 | 0.031 |
| 368.220 | 11.57 | Prostaglandin G2 | 1.358 | ns | 1.841 | 0.026 | 3.300 | 0.003 | 2.432 | 0.008 |
| 356.257 | 14.60 | Prostaglandin F1alpha | 1.203 | ns | 1.446 | 0.021 | 1.927 | <0.001 | 1.544 | 0.001 |
| Glycerophospholipids | ||||||||||
| 638.396 | 13.80 | PA (32:5) | 1.549 | ns | 1.510 | ns | 2.748 | 0.021 | 2.678 | ns |
| 847.645 | 29.88 | PC (42:6) | 0.963 | ns | 3.742 | <0.001 | 4.541 | <0.001 | 2.501 | 0.019 |
| 825.530 | 18.21 | PC (40:10) | 0.897 | 0.042 | 0.753 | 0.031 | 0.551 | 0.002 | 0.563 | <0.001 |
| 851.546 | 18.59 | PC (42:11) | 0.821 | 0.016 | 0.574 | <0.001 | 0.269 | <0.001 | 0.406 | <0.001 |
| 881.593 | 21.81 | PC (44:10) | 1.016 | ns | 1.743 | 0.002 | 2.051 | <0.001 | 1.090 | ns |
| 722.545 | 24.76 | PG (33:0) | 0.884 | ns | 0.933 | ns | 0.857 | ns | 0.515 | 0.001 |
| 484.280 | 22.13 | Lyso PG (16:0) | 1.056 | ns | 0.876 | ns | 0.817 | 0.002 | 0.942 | ns |
| 482.264 | 7.32 | Lyso PG (16:1) | 0.833 | ns | 1.097 | ns | 0.820 | 0.050 | 0.942 | ns |
| 572.296 | 7.29 | Lyso PI (16:0) | 1.028 | ns | 1.137 | 0.027 | 1.268 | 0.001 | 1.200 | 0.001 |
| 598.312 | 7.95 | Lyso PI (18:1) | 1.031 | ns | 1.241 | 0.007 | 1.563 | <0.001 | 1.303 | 0.004 |
| 622.312 | 7.71 | Lyso PI (20:3) | 1.119 | ns | 1.302 | 0.020 | 1.154 | ns | 1.255 | 0.031 |
| 620.296 | 7.04 | lyso PI (20:4) | 1.152 | ns | 1.187 | 0.045 | 1.891 | <0.001 | 1.770 | <0.001 |
| 495.260 | 6.38 | lyso PS (16:1) | 1.015 | ns | 0.885 | ns | 0.730 | 0.004 | 0.818 | 0.036 |
| 771.505 | 24.05 | PS (35:3) | 0.647 | 0.001 | 0.434 | <0.001 | 0.315 | <0.001 | 0.272 | <0.001 |
| 497.275 | 7.72 | Lyso PS (16:0) | 1.033 | ns | 1.139 | ns | 0.801 | 0.011 | 1.036 | ns |
| 517.244 | 12.63 | Lyso PS 18:4 | 1.689 | 0.040 | 1.922 | <0.001 | 2.208 | 0.020 | 1.390 | ns |
| 691.441 | 12.63 | PS (29:1) | 1.380 | ns | 2.276 | 0.038 | 2.380 | ns | 2.448 | ns |
Rt: Retention time (min); LPS: Lipopolysaccharides; *: Matches the analytical standard retention time; ns: Non-significant; n/a: Not applicable.
4. Discussion
A recognition of the crucial role of the regulation of the adaptive response and induction of the innate immune response has led to a reassessment of the role of adjuvants in vaccinology. Recent studies oninnate immunity activation of macrophages and DCs are now providing better glimpses into the mechanisms underlying adjuvant actions. These new insights now support the development of novel adjuvants and combinations of adjuvants to enhance the recognition of antigens by the immune system and to induce more potent cellular immune responses that exploit the advantages of each individual component. To the best of our knowledge, no study has yet investigated the eicosenoid effect on immune macrophage cells. Therefore, the aim of this study was to assess the ability of three eicosenoid derivatives to induce specific immunological functions of THP-1 macrophage cells by examining cytokine production and eicosenoid-induced alterations in the cell metabolome.
THP-1 cell viability in the presence of (Z)-11-eicosenol, methyl cis-11-eicosenoate and cis-11-eicosenoic acid revealed different IC50 values, indicating an effect of functional group substitution on cellular respiration. The lowest IC50 value was obtained with (Z)-11-eicosenol, which contains a hydroxyl group (Figure 1), in comparison with the ester and carboxylic acid forms. Several studies have assessed the effect of functional groups on different cell lines. For example, Sakagami et al. reported the structure–activity relationships of 11 piperic acid ester derivatives based on their cytotoxic effects against oral squamous cell carcinoma cell lines and found that addition of two hydroxyl groups had the highest cytotoxic effect [27]. Similarly, polyhydroxylated analogues of resveratrol, a natural polyphenol compound, showed higher cytotoxic effects [28].
Previously, (Z)-9-eicosenol was purified from honey bee venom and its stimulatory effect on TNF-α and IL-1β secretion was reported in the U937 cell line stimulated with LPS, where a surprising observation was significant inhibition of the level of IL-6 in response to this compound [18]. For this reason, in the present study, other eicosenoid derivatives were examined for their effects on the cellular immune response and in particular for their effects on the production of pro and anti- inflammatory cytokines by THP-1 macrophage cells. All three derivatives showed similar bioactivity with respect to the induction of cytokine levels, as described previously [18]. A small effect was observed for the secretion of TNF-α, whereas IL-1β production was enhanced significantly by the 11E-ester and 11E-acid forms, when combined with LPS. By contrast, the level of IL-6 decreased, suggesting the presence of a more subtle mechanism of action for eicosenoids. In addition, release of the anti-inflammatory IL-10 cytokine was largely inhibited, supporting their immune stimulatory effect.
The complexity of the IL-6 cytokine responses has been reported extensively [29]. This cytokine has dual properties pro- and anti-inflammatory, which are referred to as its classic and trans-signaling pathways [29]. IL-6 promotes a protective effect in some inflammatory diseases, such as inflammatory bone destruction and dextran sodium sulphate-induced colitis [30,31]. Several studies have also demonstrated an association between IL-6 and IL-10 and a requirement for IL-6 in IL-10 production by T cells to suppress inflammation [32,33]. Interestingly, in the current case, both IL-6 and IL-10 were decreased in the same manner (i.e., the 11E-ester form strongly decreased their secretion, with weaker decreases by the 11E-acid and still weaker effects by the 11E-OH form). Therefore, the important consequences on the therapeutic blockade of IL-6 as a treatment of chronic inflammatory diseases should be carefully considered.
The ability of synthetic eicosenoid derivatives to induce immune responses and to alter cytokine production was investigated further by a comprehensive untargeted metabolomics assessment of PMA-stimulated THP-1 cells. Induction of the characteristic morphology of activated macrophages as a result of Toll-like receptor (TLR4) activation by LPS has been previously described [34]. Several biomarkers, including nitric oxide (NO), are used to monitor the inflammatory status of these cells.
Inducible nitric oxide synthase (iNOS) is the main enzyme responsible for the release of large amounts of NO through the conversion of L-arginine to NO and citrulline [35]. Arginine metabolism is also involved in the regulation of inflammation through its breakdown into ornithine and urea by arginase [36]. Upregulation of arginine and proline metabolism was significantly identified by metabolomic analysis in response to treatment with the ester and acid forms, as indicated by the alterations in the levels of NO-related metabolites, including L-arginine, L-citrulline, N-(L-arginino) succinate, L-proline and L-ornithine (Table 2). L-proline and L-ornithine work as precursors for the formation of pro-proliferative polyamines and production of extracellular matrix, as a repair phase response [36]. The enhancement of cytokine production by the 11E-acid form is evident mainly by the alteration of several pathways, including arginine and proline metabolism, where a significantly higher number of metabolites was altered by this treatment (Figure S13).
Several studies have shown that metabolic reprogramming of the cell regulates macrophage activation. In the present study, the findings for LPS activation of THP-1 macrophages regarding specific pathway and biomarker metabolites are consistent with those of previous reports [17,37]. In general, pro-inflammatory stimuli cause the macrophages to undergo a metabolic switch from oxidative phosphorylation (OXPHOS) to glycolysis [38]. The lactate flux from pyruvate increases in response to a reduction in acetyl-coenzyme A (acetyl-CoA) levels [38,39] leading to dysfunctional activity of the TCA cycle, which is, in turn, compensated for by an increase in glycolytic flux to ensure a rapid regeneration of adenosine triphosphate (ATP) [40].
LPS enhanced activities clearly shown by the combination treatments with eicosenoid derivatives in this study. The levels of ATP were reduced in the treatments with LPS alone and in combination with eicosenoids, when compared with untreated control cells suggesting an increased requirement in the treated cells, leading to upregulation of most of the glycolysis metabolites, including fructose 1,6-bisphosphate and fructose 6-phosphate. In order to maintain a high cellular redox state, PPP activity was increased. LPS is reported to suppress the expression of carbohydrate kinase-like protein (CARKL), which is associated with a greater flux into glycolysis and away from the non-oxidative PPP [40]. This would lead to a significant decrease of sedoheptulose-7-phosphate (S7P) [41]; however, the opposite was observed for all three treatments with LPS when compared with the untreated control. The reason for this is not clear.
The TCA cycle intermediates, such as succinate and citrate, are critical in M1 macrophage activation and are positively associated with inflammation [38]. High levels of succinate were observed in the present study in response to LPS alone and with the 11E-acid and LPS combination. This increase has been attributed to glutamine metabolism following dysfunction of the TCA cycle, which normally would provide the source for succinate generation [42]. Succinate drives inflammation through the inhibition of prolyl hydroxylase (PHD) and further stabilisation of hypoxia-inducible factor-1α (HIF-1α) [38]. LPS triggers T-cell activation and the adaptive immune system through an increased expression of the citrate transport carrier, which could lead to cytosolic citrate accumulation. This citrate is then converted to acetyl-CoA and oxaloacetate for fatty acid synthesis and generation of NO and reactive oxygen species (ROS), respectively [43]. Interestingly, in the current study, the citrate levels were approximately 20% higher in the 11E-ester and 11E-acid combinations with LPS when compared with LPS alone. Clearly, citrate and succinate are important signaling molecules in innate and adaptive immunity [44]. Furthermore, the levels of itaconate, a metabolite synthesised via decarboxylation of cis-aconitate, were elevated in the LPS-activated macrophages. This metabolite has been suggested to work as an anti-microbial, to limit inflammation and to have a crucial role in macrophage-based immune responses [45,46,47]. Interestingly, cis-aconitate was significantly elevated following the ester and acid form treatments, while itaconate was not detected (Table 2).
An imbalance between cellular oxidants and antioxidants can potentially lead to oxidative stress and cell damage. Therefore, antioxidant defence is required for cellular adaptation to stress conditions [48]. Glutathione (GSH), an important protective antioxidant tripeptide, has been implicated in inflammatory responses and immune modulation [49]. GSH is oxidised to glutathione disulphide (GSSG) when it reacts with peroxide (H2O2) in the presence of glutathione peroxidase, an enzyme that facilitates the inactivation of peroxide [50]. GSSG can be reduced again using nicotinamide adenine dinucleotide phosphate (NADPH). Therefore, regulation of both the NADPH/NADP+ and GSH/GSSG ratios is tightly coupled to the control of oxidative stress (Figure 9) [51,52]. A decrease in the level of GSH and an increase in its product GSSG was observed in this study; this response has been reported as a hallmark of oxidative stress [53,54]. A large depletion in the level of NADPH and an increase in NADP+ were observed in the current study after the combination treatment of eicosenoids and LPS (Table 2). NADPH generates ROS through NADPH oxidase, and it also serves as a substrate for the conversion of arginine to citrulline and NO [55,56]; arginine and citrulline were also strongly elevated in response to the treatments with LPS in combination with 11E-ester or 11E-acid (Figure 9).
Figure 9.
Role of nicotinamide adenine dinucleotide phosphate (NADPH) in generating hydrogen peroxide (H2O2) and nitric oxide (NO) through glutathione metabolism and arginine biosynthesis in THP-1 macrophages and show (A) Enhanced activity of oxidative stress related metabolites after treatment with 11E-ester+LPS and 11E-acid+LPS when compared with LPS alone. (B) Overexpression (dark red) of most of the significant metabolites can be observed in a heat map visualising by 11E-ester+LPS and 11E-acid+LPS treatments. (C) GSH, GSSG and GSH/GSSG responses after each group treatments. (D) NADPH, NADP+ and NADPH/NADP+ responses after each group treatments. C: Untreated control; LPS: Lipopolysaccharide; (11E-OH): (Z)-11-eicosenol; (11E-ester): Eicosenoate; (11E-acid): Eicosenoic acid; (Red rows): Response of 11E-ester+LPS and 11E-acid+LPS combination treatments.
Enhanced activity of the PPP boosts the production of purine and pyrimidine nucleotides for further biosynthesis in activated cells [38]. An increase in inosine, guanosine, hypoxanthine, xanthine and urate levels was detected in the present study. In purine metabolism, xanthine oxidase catalyses the oxidation of hypoxanthine to xanthine and H2O2 and then to uric acid [23]. Xanthine oxidase inhibitors, such as allopurinol, can limit the inflammation in a mouse model of arthritis [57]. This suppression of inflammation is associated with an elevation of several metabolites, including inosine, guanosine and xanthosine, which are substrates of purine nucleoside phosphorylase (PNP). A loss of PNP activity or accumulation of one or more enzyme substrates has been linked extensively to immune deficiency [58]. However, the observation of upregulation in the level of substrates and products of xanthine oxidase and PNP enzymes in the present study (Figure 10) suggests an ability of eicosenoid derivatives (particularly 11E-ester and 11E-acid form) to modulate the activity of these enzymes and to perturb the purine nucleotide pathway in order to boost the immune system through the provision of ROS.
Figure 10.
Schematic representation of the purine metabolism. This reflects how 11E-ester + LPS and 11E-acid + LPS synergise the effect of LPS alone focusing on hypoxanthine, xanthine and uric acid, which accumulated with H2O2 and superoxide (O2−) during purine degradations. C: Untreated control; LPS: Lipopolysaccharide; (11E-OH): (Z)-11-eicosenol; (11E-ester): Eicosenoate; (11E-acid): Eicosenoic acid; (Red rows): Response of 11E-ester + LPS and 11E-acid + LPS combination treatments; ATP: Adenosine triphosphate; AMP: Adenosine monophosphate; **: Significant (p < 0.01); ***: Significant (p < 0.001).
Recent research has reflected a significantly increased interest in fatty acid metabolism and its effects on inflammation and immune cells. In activated macrophages, the importance of mitochondrial fatty acid metabolism or fatty acid oxidation (FAO) has been considered to reflect the regeneration of ATP and a compensation for the reduction in ATP levels [59]. Several studies have highlighted the possibility of attenuating inflammation by promoting macrophage FAO, which also reduces lipid-induced triglyceride accumulation [60]. Inhibition of FAO in THP-1 cells macrophages exacerbates palmitate-induced endoplasmic reticulum stress and inflammation responses [61]. In addition, LPS-induced inflammation results in elevated levels of kidney triglyceride, fatty acid and cholesteryl ester through a decrease in fatty acid beta oxidation [62]. Intriguingly, the levels of fatty acids were highly elevated in response to the treatments with LPS in combination with eicosenoids when compared with cells treated with LPS alone or untreated controls. In the present study, treatment with 11E-ester and 11E-acid in particular led to increases in a large number of fatty acids, such as arachidonic acid (i.e., eicosatetraenoic acid), docosenoic acid, tetracosenoic acid, eicosenoic acid and icosatrienoic acid (Table 3), which all are strongly correlated with inflammation and immune activation. The presence of elevated levels of several dioic acids (decandioic, dodecandioic and tetradecandoic acids) suggests that the eicosanoic acid and its ester are functioning as peroxisome proliferator ligands since dioic acids are products of peroxisome activity [63].
Lipid profiles (lipidomics) and full assessment of their levels, including eicosanoid, are effective ways to diagnose the severity and progression of several diseases [64,65]. Arachidonic acid (AA), a precursor of eicosanoids, is considered to represent a potent signal of cellular responses. AA metabolites are involved in the inflammatory and immune responses [66], and the activation of phospholipase A2 (PLA2) is critical for increasing the AA level and subsequent eicosanoid biosynthesis. TLR4-mediated priming has been observed to activate cytosolic calcium-dependent PLA2 (cPLA2) and to enhance cyclooxygenase (COX2) production via Nuclear factor-kappa B (NF-κB), which results in release of AA and pro-inflammatory eicosanoids [65]. In the present study, the level of AA was strongly elevated by all the combination treatments when compared to LPS alone; however, this elevation was more pronounced with 11E-ester form, which resulted in a seven-fold increase in the AA level.
Comprehensive assessment of lipid-correlated inflammatory signaling could provide a better understanding of cytokine integration with fatty acid metabolism and particular the production of eicosanoids. In general, the suppression of β-oxidation of fatty acids could result in the apparent boost in fatty acid synthesis and the reductions in ATP levels and citrate accumulation observed in the present study. A more targeted investigation is therefore required to confirm the proposed use of these eicosenoid derivatives as immune system stimulators and as vaccine adjuvants.
5. Conclusions
A deeper understanding of the modes of action that regulate the immune stimulatory properties of new or existing adjuvants is prerequisite for the rational design of more sophisticated vaccines. The present study identifies the main effects of synthetic compounds related to 11-eicosanol, which is found in bee venom, in modulating macrophage behaviour. In agreement with previous findings [17,37], a stimulatory effect of LPS was confirmed in PMA-differentiated THP-1 cells. Furthermore, the possible use of (Z)-11-eicosenol, methyl cis-11-eicosenoate and cis-11-eicosenoic acid to alter macrophage cytokine production was studied. Although these eicosenoid compounds enhanced the production of IL-1β and suppressed the production of IL-10, which mirrored their immune stimulatory effects, IL-6 release was inhibited, suggesting the presence of a more subtle mechanism of immune modulation. Further metabolic investigations were therefore performed to obtain a better understanding of changes in the macrophage metabolome. Alterations in metabolite levels in response to LPS treatment in the presence or absence of eicosenoids reflected the strength of the actions of each component and confirmed the potential of these compounds, particularly the ester and acid forms, to synergise LPS action. Overall, the findings supported their actions as pro- rather than anti-inflammatory agents.
Significant alterations were observed in the metabolite levels associated with different pathways, including redox cell signaling pathways, which have been extensively associated with transcriptional immune factors and immune functions [50,67]. These alterations occurred concomitantly with upregulation of the levels of several metabolites within the arginine and proline pathways, glycolysis, the TCA cycle and purine metabolism. Moreover, marked increases occurred in the levels of several fatty acids and inflammatory biomarker metabolites, including arachidonic acid. Cellular activation and enhanced immune response were confirmed by the effects of (Z)-11-eicosenol, methyl cis-11-eicosenoate and cis-11-eicosenoic acid, the findings suggested their possible use as immune-modulating agents and vaccine adjuvants. Taken together, these findings provide a better understanding of the mechanism of immune stimulation, and they support the potential use of new adjuvants in shaping a desired immune response. Comparison with existing adjuvants would need to be carried out in order to confirm any advantages of the eicosenoids.
Acknowledgments
We thank the Saudi Government for a scholarship for A.M.A.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-393X/7/4/142/s1, Figures S1–S12: Showing a representative 4-parameter logistic plot of TNF-α, IL-1β, IL-6, and IL-10 standard samples, Figure S13: Changes in metabolites by treatments and LPS when compared with untreated control, Tables S1–S4: Effect of eicosenoid compounds on the production of TNF-α, IL-1β, IL-6, and IL-10 cytokines in the presence and absence of LPS, Table S5: Additional changed non-polar metabolites in THP-1 cells, Table S6: List of abbreviation, Table S7: List of catalog/serial number of instruments and reagents.
Author Contributions
Conceptualization, A.M.A., D.G.W., J.A.P., D.J.N. and M.J.D.; Methodology, A.M.A., T.D., D.G.W. and V.A.F.; Software, A.M.A., T.D.; Validation, A.M.A. and D.G.W.; Formal Analysis, A.M.A.; Investigation, A.M.A.; Resources, D.G.W. and V.A.F.; Data Curation, A.M.A., M.J.D., J.A.P., V.A.F. and D.G.W.; Writing—Original Draft Preparation, A.M.A.; Writing—Review and Editing, D.G.W., J.A.P., D.J.N., V.A.F., and M.J.D.; Visualisation, A.M.A. and D.G.W.; Supervision, D.G.W. and V.A.F.; Project Administration, D.G.W.; Funding Acquisition, J.A.P., D.J.N., M.J.D. and D.G.W.
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Reed S.G., Orr M.T., Fox C.B. Key roles of adjuvants in modern vaccines. Nat. Med. 2013;1919:1597–1608. doi: 10.1038/nm.3409. [DOI] [PubMed] [Google Scholar]
- 2.McKee A.S., Marrack P. Old and new adjuvants. Curr. Opin. Immunol. 2017;47:44–51. doi: 10.1016/j.coi.2017.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garçon N., Leroux-Roels G., Cheng W.-F. Vaccine adjuvants. Perspect. Vaccinol. 2011;11:89–113. doi: 10.1016/j.pervac.2011.05.004. [DOI] [Google Scholar]
- 4.Coffman R.L., Sher A., Seder R.A. Vaccine adjuvants: Putting innate immunity to work. Immunity. 2010;33:492–503. doi: 10.1016/j.immuni.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKee A.S., Munks M.W., Marrack P. How do adjuvants work? Important considerations for new generation adjuvants. Immunity. 2007;27:687–690. doi: 10.1016/j.immuni.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 6.McCartney S., Vermi W., Gilfillan S., Cella M., Murphy T.L., Schreiber R.D., Murphy K.M., Colonna M. Distinct and complementary functions of MDA5 and TLR3 in poly (I:C)-mediated activation of mouse NK cells. J. Exp. Med. 2009;206:2967–2976. doi: 10.1084/jem.20091181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tritto E., Mosca F., de Gregorio E. Mechanism of action of licensed vaccine adjuvants. Vaccine. 2009;27:3331–3334. doi: 10.1016/j.vaccine.2009.01.084. [DOI] [PubMed] [Google Scholar]
- 8.De Gregorio E., D’Oro U., Wack A. Immunology of TLR-independent vaccine adjuvants. Curr. Opin. Immunol. 2009;21:339–345. doi: 10.1016/j.coi.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 9.Fraser C.K., Diener K.R., Brown M.P., Hayball J.D. Improving vaccines by incorporating immunological coadjuvants. Expert Rev. Vaccines. 2007;6:559–578. doi: 10.1586/14760584.6.4.559. [DOI] [PubMed] [Google Scholar]
- 10.Akira S., Uematsu S., Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 11.Steinhagen F., Kinjo T., Bode C., Klinman D.M. TLR-based immune adjuvants. Vaccine. 2011;29:3341–3355. doi: 10.1016/j.vaccine.2010.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.GALANOS C., Lüderitz O., Rietschel E.T., Westphal O., Brade H., Brade L., Freudenberg M., Schade U., Imoto M., Yoshimura H., et al. Synthetic and natural Escherichia coli free lipid A express identical endotoxic activities. Eur. J. Biochem. 1985;148:1–5. doi: 10.1111/j.1432-1033.1985.tb08798.x. [DOI] [PubMed] [Google Scholar]
- 13.Guy B. The perfect mix: Recent progress in adjuvant research. Nat. Rev. Microbiol. 2007;5:505–517. doi: 10.1038/nrmicro1681. [DOI] [PubMed] [Google Scholar]
- 14.El-Wahed A.A.A., Khalifa S.A., Sheikh B.Y., Farag M.A., Saeed A., Larik F.A., Koca-Caliskan U., AlAjmi M.F., Hassan M., Wahabi H.A., et al. Bee Venom Composition: From Chemistry to Biological Activity, in Studies in Natural Products Chemistry. Elsevier; Amsterdam, The Netherlands: 2018. pp. 459–484. [Google Scholar]
- 15.Hider R.C. Honeybee venom: A rich source of pharmacologically active peptides. Endeavour. 1988;12:60–65. doi: 10.1016/0160-9327(88)90082-8. [DOI] [PubMed] [Google Scholar]
- 16.Alonezi S., Tusiimire J., Wallace J., Dufton M.J., Parkinson J.A., Young L., Clements C.J., Park J.K., Jeon J.W., Ferro V., et al. Metabolomic profiling of the synergistic effects of melittin in combination with cisplatin on ovarian cancer cells. Metabolites. 2017;7:14. doi: 10.3390/metabo7020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alqarni A., Ferro V., Parkinson J., Dufton M., Watson D. Effect of Melittin on Metabolomic Profile and Cytokine Production in PMA-Differentiated THP-1 Cells. Vaccines. 2018;6:72. doi: 10.3390/vaccines6040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tusiimire J., Wallace J., Woods N., Dufton M.J., Parkinson J.A., Abbott G., Clements C.J., Young L., Park J.K., Jeon J.W., et al. Effect of Bee Venom and Its Fractions on the Release of Pro-Inflammatory Cytokines in PMA-Differentiated U937 Cells Co-Stimulated with LPS. Vaccines. 2016;4:11. doi: 10.3390/vaccines4020011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pickett J.A., Williams I.H., Martin A.P. (Z)-11-eicosen-1-ol, an important new pheromonal component from the sting of the honey bee, Apis mellifera L. (Hymenoptera, Apidae) J. Chem. Ecol. 1982;8:163–175. doi: 10.1007/BF00984013. [DOI] [PubMed] [Google Scholar]
- 20.Free J. The stimuli releasing the stinging response of honeybees. Anim. Behav. 1961;9:193–196. doi: 10.1016/0003-3472(61)90008-2. [DOI] [Google Scholar]
- 21.Tannahill G., Curtis A.M., Adamik J., Palsson-Mcdermott E.M., McGettrick A.F., Goel G., Frezza C., Bernard N.J., Kelly B., Foley N.H., et al. Succinate is an inflammatory signal that induces IL-1β through HIF-1α. Nature. 2013;496:238–242. doi: 10.1038/nature11986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lampropoulou V., Sergushichev A., Bambouskova M., Nair S., Vincent E.E., Loginicheva E., Cervantes-Barragan L., Ma X., Huang S.C.C., Griss T., et al. Itaconate links inhibition of succinate dehydrogenase with macrophage metabolic remodeling and regulation of inflammation. Cell Metab. 2016;24:158–166. doi: 10.1016/j.cmet.2016.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rattigan K.M., Pountain A.W., Regnault C., Achcar F., Vincent I.M., Goodyear C.S., Barrett M.P. Metabolomic profiling of macrophages determines the discrete metabolomic signature and metabolomic interactome triggered by polarising immune stimuli. PLoS ONE. 2018;13:e0194126. doi: 10.1371/journal.pone.0194126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O′Neill L.A., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang R., Watson D.G., Wang L., Westrop G.D., Coombs G.H., Zhang T. Evaluation of mobile phase characteristics on three zwitterionic columns in hydrophilic interaction liquid chromatography mode for liquid chromatography-high resolution mass spectrometry based untargeted metabolite profiling of Leishmania parasites. J. Chromatogr. A. 2014;1362:168–179. doi: 10.1016/j.chroma.2014.08.039. [DOI] [PubMed] [Google Scholar]
- 26.Scheltema R.A., Jankevics A., Jansen R.C., Swertz M.A., Breitling R. PeakML/mzMatch: A file format, Java library, R library, and tool-chain for mass spectrometry data analysis. Anal. Chem. 2011;83:2786–2793. doi: 10.1021/ac2000994. [DOI] [PubMed] [Google Scholar]
- 27.Sakagami H., Uesawa Y., Masuda Y., Tomomura M., Yokose S., Miyashiro T., Murai J., Takao K., Kanamoto T., Terakubo S., et al. Quantitative structure–cytotoxicity relationship of newly synthesized piperic acid esters. Anticancer Res. 2017;37:6161–6168. doi: 10.21873/anticanres.12065. [DOI] [PubMed] [Google Scholar]
- 28.Murias M., Jäger W., Handler N., Erker T., Horvath Z., Szekeres T., Nohl H., Gille L. Antioxidant, prooxidant and cytotoxic activity of hydroxylated resveratrol analogues: Structure–activity relationship. Biochem. Pharmacol. 2005;69:903–912. doi: 10.1016/j.bcp.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 29.Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. The pro-and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 30.Balto K., Sasaki H., Stashenko P. Interleukin-6 deficiency increases inflammatory bone destruction. Infect. Immun. 2001;69:744–750. doi: 10.1128/IAI.69.2.744-750.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grivennikov S., Karin E., Terzic J., Mucida D., Yu G.Y., Vallabhapurapu S., Scheller J., Rose-John S., Cheroutre H., Eckmann L., et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stumhofer J.S., Silver J.S., Laurence A., Porrett P.M., Harris T.H., Turka L.A., Ernst M., Saris C.J., O’Shea J.J., Hunter C.A. Interleukins 27 and 6 induce STAT3-mediated T cell production of interleukin 10. Nat. Immunol. 2007;8:1363–1371. doi: 10.1038/ni1537. [DOI] [PubMed] [Google Scholar]
- 33.Jin J.-O., Han X., Yu Q. Interleukin-6 induces the generation of IL-10-producing Tr1 cells and suppresses autoimmune tissue inflammation. J. Autoimmun. 2013;40:28–44. doi: 10.1016/j.jaut.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukhopadhyay S., Peiser L., Gordon S. Activation of murine macrophages by Neisseria meningitidis and IFN-γ in vitro: Distinct roles of class A scavenger and Toll-like pattern recognition receptors in selective modulation of surface phenotype. J. Leukoc. Biol. 2004;76:577–584. doi: 10.1189/jlb.0104014. [DOI] [PubMed] [Google Scholar]
- 35.Li H., Poulos T.L. Structure–function studies on nitric oxide synthases. J. Inorg. Biochem. 2005;99:293–305. doi: 10.1016/j.jinorgbio.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 36.Satriano J. Arginine pathways and the inflammatory response: Interregulation of nitric oxide and polyamines. Amino Acids. 2004;26:321–329. doi: 10.1007/s00726-004-0078-4. [DOI] [PubMed] [Google Scholar]
- 37.Alqarni A.M., Niwasabutra K., Sahlan M., Fearnley H., Fearnley J., Ferro V.A., Watson D.G. Propolis Exerts an Anti-Inflammatory Effect on PMA-Differentiated THP-1 Cells via Inhibition of Purine Nucleoside Phosphorylase. Metabolites. 2019;9:75. doi: 10.3390/metabo9040075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kelly B., Neill L.A.J. Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 2015;25:771–784. doi: 10.1038/cr.2015.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freemerman A.J., Johnson A.R., Sacks G.N., Milner J.J., Makowski L., Kirk E.L., Troester M.A., Macintyre A.N., Goraksha-Hicks P., Rathmell J.C. Metabolic reprogramming of macrophages: Glucose transporter 1 (GLUT1)-mediated glucose metabolism drives a proinflammatory phenotype. J. Biol. Chem. 2014;289:7884–7896. doi: 10.1074/jbc.M113.522037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haschemi A., Kosma P., Gille L., Evans C.R., Burant C.F., Starkl P., Knapp B., Haas R., Schmid J.A., Jandl C., et al. The Sedoheptulose Kinase CARKL Directs Macrophage Polarization through Control of Glucose Metabolism. Cell Metab. 2012;15:813–826. doi: 10.1016/j.cmet.2012.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blagih J., Jones R.G. Polarizing Macrophages through Reprogramming of Glucose Metabolism. Cell Metab. 2012;15:793–795. doi: 10.1016/j.cmet.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 42.Rodríguez-Prados J.C., Través P.G., Cuenca J., Rico D., Aragone J., Martín-Sanz P., Cascante M., Boscá L. Substrate fate in activated macrophages: A comparison between innate, classic, and alternative activation. J. Immunol. 2010;185:605–614. doi: 10.4049/jimmunol.0901698. [DOI] [PubMed] [Google Scholar]
- 43.Infantino V., Iacobazzi V., Palmieri F., Menga A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem. Biophys. Res. Commun. 2013;440:105–111. doi: 10.1016/j.bbrc.2013.09.037. [DOI] [PubMed] [Google Scholar]
- 44.Weinberg S.E., Sena L.A., Chandel N.S. Mitochondria in the regulation of innate and adaptive immunity. Immunity. 2015;42:406–417. doi: 10.1016/j.immuni.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Strelko C.L., Lu W., Dufort F.J., Seyfried T.N., Chiles T.C., Rabinowitz J.D., Roberts M.F. Itaconic acid is a mammalian metabolite induced during macrophage activation. J. Am. Chem. Soc. 2011;133:16386–16389. doi: 10.1021/ja2070889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mills E.L., Ryan D.G., Prag H.A., Dikovskaya D., Menon D., Zaslona Z., Jedrychowski M.P., Costa A.S., Higgins M., Hams E., et al. Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature. 2018;556:113–117. doi: 10.1038/nature25986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michelucci A., Cordes T., Ghelfi J., Pailot A., Reiling N., Goldmann O., Binz T., Wegner A., Tallam A., Rausell A., et al. Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc. Natl. Acad. Sci. USA. 2013;110:7820–7825. doi: 10.1073/pnas.1218599110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sies H. Oxidative stress: Oxidants and antioxidants. Exp. Physiol. 1997;82:291–295. doi: 10.1113/expphysiol.1997.sp004024. [DOI] [PubMed] [Google Scholar]
- 49.Rahman I., MacNee W. Oxidative stress and regulation of glutathione in lung inflammation. Eur. Respir. J. 2000;16:534–554. doi: 10.1034/j.1399-3003.2000.016003534.x. [DOI] [PubMed] [Google Scholar]
- 50.Ghezzi P. Role of glutathione in immunity and inflammation in the lung. Int. J. Gen. Med. 2011;4:105–113. doi: 10.2147/IJGM.S15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fan J., Kamphorst J.J., Shlomi T., Thompson C.B., Rabinowitz J.D. Quantitative flux analysis reveals folate-dependent NADPH production. Nature. 2014;510:298–302. doi: 10.1038/nature13236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreno-Sánchez R., Marín-Hernández Á., Gallardo-Pérez J.C., Vázquez C., Rodríguez-Enríquez S., Saavedra E. Control of the NADPH supply and GSH recycling for oxidative stress management in hepatoma and liver mitochondria. Biochim. Biophys. Acta Bioenerg. 2018;1859:1138–1150. doi: 10.1016/j.bbabio.2018.07.008. [DOI] [PubMed] [Google Scholar]
- 53.Kalkan I.H., Suher M. The relationship between the level of glutathione, impairment of glucose metabolism and complications of diabetes mellitus. Pak. J. Med. Sci. 2013;29:938–942. doi: 10.12669/pjms.294.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margonis K., Fatouros I.G., Jamurtas A.Z., Nikolaidis M.G., Douroudos I., Chatzinikolaou A., Mitrakou A., Mastorakos G., Papassotiriou I., Taxildaris K., et al. Oxidative stress biomarkers responses to physical overtraining: Implications for diagnosis. Free Radic. Biol. Med. 2007;43:901–910. doi: 10.1016/j.freeradbiomed.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 55.El Kasmi K.C., Stenmark K.R. Contribution of metabolic reprogramming to macrophage plasticity and function. Semin. Immunol. 2015;27:267–275. doi: 10.1016/j.smim.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neill L.A.J. A critical role for citrate metabolism in LPS signalling. Biochem. J. 2011;438:e5–e6. doi: 10.1042/BJ20111386. [DOI] [PubMed] [Google Scholar]
- 57.Alorainy M. Effect of allopurinol and vitamin E on rat model of rheumatoid arthritis. Int. J. Health Sci. 2008;2:59–67. [PMC free article] [PubMed] [Google Scholar]
- 58.Arpaia E., Benveniste P., Di Cristofano A., Gu Y., Dalal I., Kelly S., Hershfield M., Pandolfi P.P., Roifman C.M., Cohen A. Mitochondrial Basis for Immune Deficiency: Evidence from Purine Nucleoside Phosphorylase–deficient Mice. J. Exp. Med. 2000;191:2197–2208. doi: 10.1084/jem.191.12.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Newsholme P., Gordon S., Newsholme E.A. Rates of utilization and fates of glucose, glutamine, pyruvate, fatty acids and ketone bodies by mouse macrophages. Biochem. J. 1987;242:631–636. doi: 10.1042/bj2420631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malandrino M.I., Fucho R., Weber M., Calderon-Dominguez M., Mir J.F., Valcarcel L., Escoté X., Gómez-Serrano M., Peral B., Salvadó L., et al. Enhanced fatty acid oxidation in adipocytes and macrophages reduces lipid-induced triglyceride accumulation and inflammation. Am. J. Physiol. Endocrinol. Metab. 2015;308:E756–E769. doi: 10.1152/ajpendo.00362.2014. [DOI] [PubMed] [Google Scholar]
- 61.Namgaladze D., Lips S., Leiker T.J., Murphy R.C., Ekroos K., Ferreiros N., Geisslinger G., Brüne B. Inhibition of macrophage fatty acid β-oxidation exacerbates palmitate-induced inflammatory and endoplasmic reticulum stress responses. Diabetologia. 2014;57:1067–1077. doi: 10.1007/s00125-014-3173-4. [DOI] [PubMed] [Google Scholar]
- 62.Feingold K.R., Wang Y., Moser A., Shigenaga J.K., Grunfeld C. LPS decreases fatty acid oxidation and nuclear hormone receptors in the kidney. J. Llipid Res. 2008;49:2179–2187. doi: 10.1194/jlr.M800233-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wanders R.J., Waterham H.R. Biochemistry of mammalian peroxisomes revisited. Annu. Rev. Biochem. 2006;75:295–332. doi: 10.1146/annurev.biochem.74.082803.133329. [DOI] [PubMed] [Google Scholar]
- 64.Li P., Spann N.J., Kaikkonen M.U., Lu M., Fox J.N., Bandyopadhyay G., Talukdar S., Xu J., Lagakos W.S., Patsouris D., et al. NCoR Repression of LXRs Restricts Macrophage Biosynthesis of Insulin-Sensitizing Omega 3 Fatty Acids. Cell. 2013;155:200–214. doi: 10.1016/j.cell.2013.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dennis E.A., Norris P.C. Eicosanoid storm in infection and inflammation. Nat. Rev. Immunol. 2015;15:511–523. doi: 10.1038/nri3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis J.G., Adams D.O. Enhanced Release of Hydrogen Peroxide and Metabolites of Arachidonic Acid by Macrophages from SENCAR Mice following Stimulation with Phorbol Esters. Cancer Res. 1986;46:5696–5700. [PubMed] [Google Scholar]
- 67.Dröge W., Schulze-Osthoff K.L.A.U.S., Mihm S., Galter D., Schenk H.E., Eck H.P., Roth S., Gmünder H. Functions of glutathione and glutathione disulfide in immunology and immunopathology. FASEB J. 1994;8:1131–1138. doi: 10.1096/fasebj.8.14.7958618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.