Abstract
Epithelial cells express keratins, which are essential for the structural integrity and mechanical strength of the cells. In the junctional epithelium (JE) of the tooth, keratins such as K16, K18, and K19, are expressed, which is typical for non-differentiated and rapidly dividing cells. The expression of K17, K4, and K13 keratins can be induced by injury, bacterial irritation, smoking, and inflammation. In addition, these keratins can be found in the sulcular epithelium and in the JE. Our aim was to estimate the changes in K4, K13, K17, and K19 expression in gingival epithelial cells exposed to Aggregatibacter actinomycetemcomitans. An organotypic gingival mucosa and biofilm co-culture was used as a model system. The effect of the biofilm after 24 h was assessed using immunohistochemistry. The structure of the epithelium was also studied with transmission electron microscopy (TEM). The expression of K17 and K19, as well as total keratin expression, decreased in the suprabasal layers of epithelium, which were in close contact with the A. actinomycetemcomitans biofilm. The effect on keratin expression was biofilm specific. The expression of K4 and K13 was low in all of the tested conditions. When stimulated with the A. actinomycetemcomitans biofilm, the epithelial contact site displayed a thick necrotic layer on the top of the epithelium. The A. actinomycetemcomitans biofilm released vesicles, which were found in close contact with the epithelium. After A. actinomycetemcomitans irritation, gingival epithelial cells may lose their resistance and become more vulnerable to bacterial infection.
Keywords: Aggregatibacter actinomycetemcomitans, keratins, organotypic gingival mucosa
1. Introduction
Periodontal disease is an inflammatory disease caused by growing biofilm, which gradually develops to a bacterial community rich in inflammophilic Gram-negative species [1]. The formation of such biofilm and the subsequent inflammation affects the supporting tissues of the tooth, including the epithelium and the connective tissue. Cytoskeletal intermediate filaments are essential for the structural integrity and function of the epithelial cells. Intermediate filaments constitute the acidic (Type I) and basic (Type II) keratins. The molecular weight of Type I keratin ranges from 40 to 57 kDa and encompasses keratins K9–K20. The molecular weight of Type II keratin ranges from 50 to 70 kDa and encompasses keratins K1–K8.
Keratins play an important functional role in the integrity and mechanical stability of both individual epithelial cells and cell–cell contacts in tissues. Thus, they contribute not only to the stability of the epithelium itself but also to the basement membrane attachment to the connective tissue underlying the epithelium. In the epithelia of internal organs, which are under little mechanical stress, there are only a few loosely-distributed keratin filaments in the cytoplasm. Conversely, keratins are abundant and are densely bound in the lining of outer surfaces. The composition of keratins varies depending on the epithelial cell type and the differentiation status of the epithelial cells. The keratin composition may also be affected by external stimuli, inflammation, or other types of disease development (e.g., cancer). The oral epithelium is a keratinizing form of epithelium and provides an effective physical barrier to microbial invasion. In the oral epithelium, basal dividing cells express simple epithelial cell keratins (K5, K14, and K19), whereas suprabasal cells express keratins typical of differentiated cells (K1, K10, K6, and K16). The keratin profile of the junctional epithelium (JE) differs from the oral epithelium, as only keratins typical for non-differentiated and rapidly dividing cells are expressed both basally (K5, K14, and K19) and suprabasally (K19). In addition, keratins such as K4, K13, and K17 have been found in the sulcular area of the dentogingival junction and may be induced by acute injury, bacterial irritation, smoking, and inflammation [2,3].
In contrast, in patients with severe and rapidly progressive periodontitis (previous term aggressive periodontitis), K17 gene expression was found to be repressed in disease site gingival samples compared to healthy site samples [4]. An open issue of significant interest is whether the expression of those keratins is altered by irritation from biofilm bacteria, such as Aggregatibacter actinomycetemcomitans, in periodontal diseases. The Gram-negative bacterium A. actinomycetemcomitans is an aggressive pathogen that is frequently associated with subgingival biofilms and has been strongly implicated in the development of rapidly progressive periodontal disease involving the invasion of A. actinomycetemcomitans into epithelial layers. Although A. actinomycetemcomitans represents only one species in multispecies periodontal biofilm, it most likely is able to suppress the host defense with its virulence factors. To gain insight into this question, we examined keratin K4, K13, K17, and K19 expression and distribution in an organotypic gingival tissue culture model co-cultured with a periodontopathogenic A. actinomycetemcomitans biofilm. Keratin K19 was chosen since it is typical and most dominant keratin in the JE [5,6] and we wanted to investigate how well the tissue culture model mimics the JE. We found that the expression of K17 and K19, as well as total keratin expression, decreased in the suprabasal layers of epithelium, which were in close contact with the A. actinomycetemcomitans biofilm. The decreased keratin expression may lead to decreased resistance of gingival epithelial cells to bacterial infection.
2. Results
2.1. Control Cultures Showed Strong Expression of K17 and K19 and Only Weak or No Expression of K4 and K13
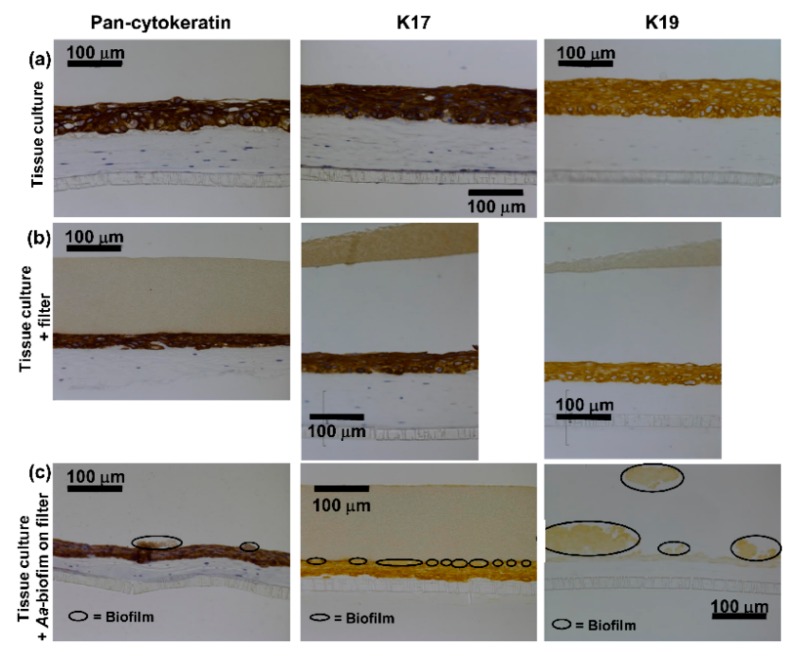
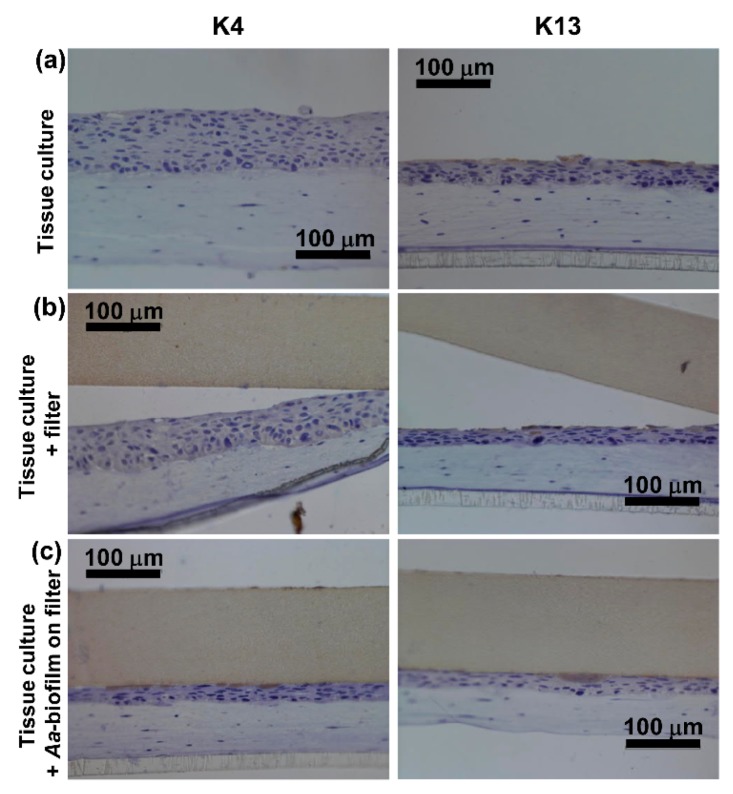
Both control cultures grown without anything on the top of the epithelium or with an empty sterile filter disc showed similar immunohistochemical staining. Pancytokeratin staining was strong and was evenly distributed throughout the epithelium (Figure 1a,b). Similarly, the specific cytokeratins K17 and K19 were found to be expressed from the basal layer throughout the epithelium to the surface (Figure 1a,b). Keratin 4 was not found in the control cultures (Figure 2a,b), and K13 expression was only observed occasionally in single cells (Figure 2a,b).
Figure 1.
A. actinomycetemcomitans biofilm decreased the total keratin as well as K17 and K19 expression in the suprabasal layers of the epithelium. (a,b) Immunohistochemical staining (peroxidase-3,3′-diaminobenzidine (DAB)) with anti-pan-cytokeratin/anti-cytokeratin 17/anti-cytokeratin 19 shows strong staining in the control cultures. (c) Pan-cytokeratin/K17/K19 expression is decreased in the co-cultures with the A. actinomycetemcomitans biofilm.
Figure 2.
The expression of K4 and K13 was low in all of the tested conditions. (a,b) Immunohistochemical staining (DAB) with anti-cytokeratin 4 in the control cultures or (c) in the co-cultures with the A. actinomycetemcomitans biofilm shows no expression of K4. (a,b) Immunohistochemical staining (DAB) with anti-cytokeratin 13 shows only a few stained cells in the control cultures. (c) In the co-cultures with A. actinomycetemcomitans biofilm, no expression of K13 is observed.
2.2. A. actinomycetemcomitans Biofilm Decreased the Expression of Keratin 17 and 19, Which Was in Accordance with the Decreased Expression of Total Keratin
When exposed to pre-grown A. actinomycetemcomitans biofilm for 24 h, the human gingival keratinocytes showed no or very weak expression of K17 and K19 throughout the epithelium and especially in areas in close contact to the biofilm (Figure 1c). Some expression of K17 was evident further away from the biofilm-epithelium contact site (Figure 1c) as well as in the basal layer adjacent to the connective tissue. K19 expression was almost totally absent (Figure 1c). No expression of K4 or K13 was observed in the co-cultures with the biofilm (Figure 2c). The decrease in the expression levels of total keratins (pan-cytokeratin staining, Figure 1c) in the co-cultures with the A. actinomycetemcomitans biofilm was in accordance with the decrease in specific keratin expression.
2.3. A. actinomycetemcomitans Biofilm Caused Necrosis of the Epithelial Surface
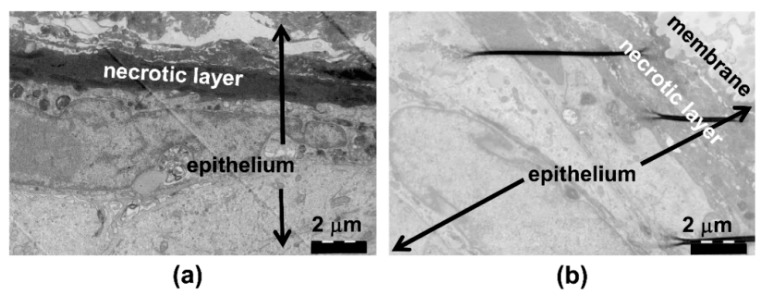
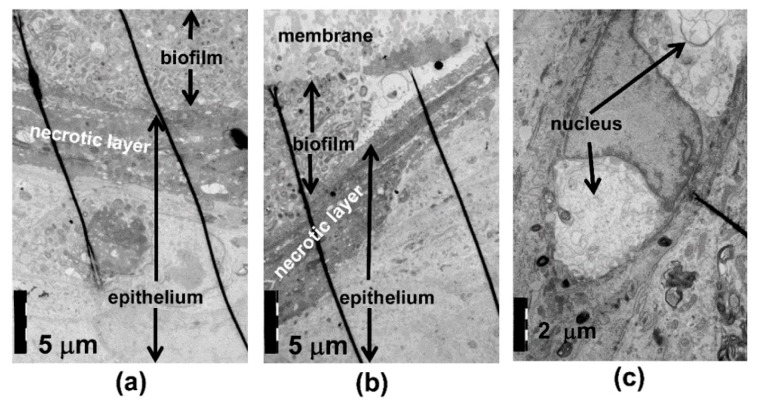
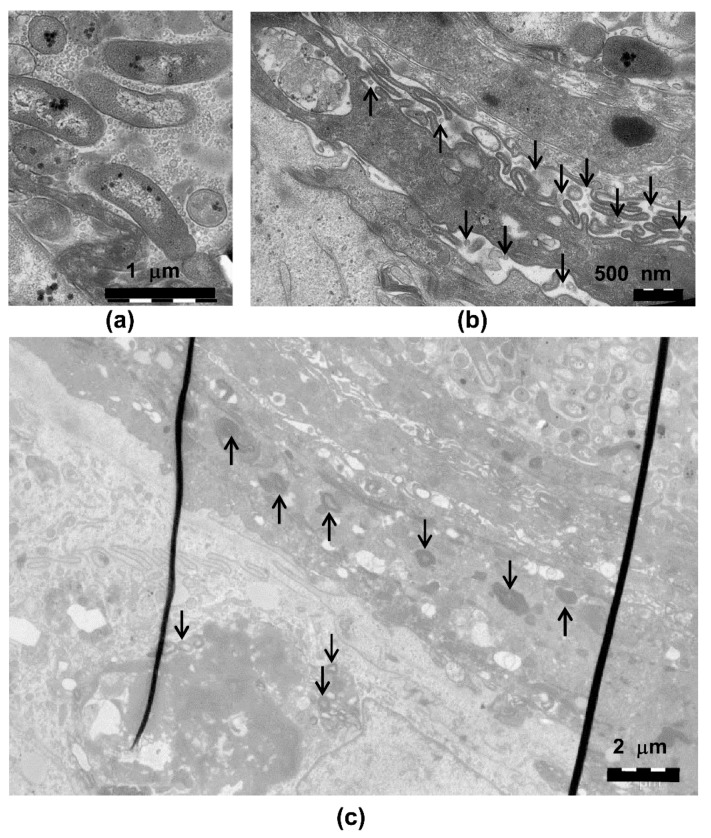
Using TEM analysis, the control culture surface showed a thin layer of exfoliating, necrotic cells (Figure 3a,b). The cell structures and cell–cell contacts appeared normal. When exposed to the A. actinomycetemcomitans biofilm, the necrotic area of the epithelial surface was thick (Figure 4a,b), and the thickness of the necrotic area increased in areas in close contact to the biofilm and with an increasing amount of biofilm (Figure 4b). Necrosis of the epithelium in contact with the biofilm was consistently seen in EM, as visualized in Figure 4b, where right beneath the increasing mass of A actinonmycetemcomitans (on the left side of the picture) the necrotic epithelial layer increases clearly in thickness. This is a descriptive result, but was consistenly seen in EM. In the co-cultures, nuclear breakdown was observed in areas in close contact to the biofilm (Figure 4a,c). When in close contact with the epithelium, the biofilm bacteria released vesicles, and similar structures could be observed intraepithelially (Figure 5a,b). The necrotic area appeared to act as a barrier to the biofilm bacteria; however, a few structures which resembled bacteria were observed inside the epithelium after the 24 h co-culture (Figure 5c).
Figure 3.
Normal structure of the epithelial cells and the epithelial cell nuclei. (a) Transmission electron microscopy (TEM) of the control cultures grown with nothing on the surface and (b) with the sterile membrane on the epithelium shows the normal structure of the cells and nuclei. On the top of the epithelium, a thin area of exfoliating and necrotic cells is observed (dark area).
Figure 4.
A. actinomycetemcomitans biofilm caused necrosis of the epithelial cells. (a,b) TEM of the co-cultures with A. actinomycetemcomitans shows the thickening of the necrotic layer in areas in close contact to the biofilm and (a,c) disruption of the nuclei beneath the biofilm.
Figure 5.
Vesicle-like structures are released by A. actinomycetemcomitans biofilm. (a) A. actinomycetemcomitans cells in the biofilm release vesicles, and (b) similar structures can be observed inside the epithelium. (c) (arrows) A few particles resembling single A. actinomycetemcomitans cells are observed inside the necrotic epithelium and inside the epithelial cells beneath the necrotic layer.
3. Discussion
Our major findings were that the A. actinomycetemcomitans biofilm caused decreased expression of cytokeratins, which is typical for the dentogingival junction and necrosis of the epithelial surface layers in close contact to the biofilm. Our tissue culture model appeared to mimic JE well, as K19, the typical keratin for JE, was highly expressed in the control cultures [6]. Our finding of decreased K19 expression adjacent to the A. actinomycetemcomitans biofilm is in disagreement with earlier reports showing that the inflammation in the periodontal pocket increases K19 expression [3,7]. However, our result may reflect the specific nature of the A actinomycetemcomitans biofilm–host tissue interaction. Previous work has shown that A. actinomycetemcomitans can invade buccal epithelial cells [8] and gingival tissue [9]. By decreasing the expression of one major structural protein, K19, in JE cells, A. actinomycetemcomitans could disturb the epithelial integrity, ease the invasion of the bacteria into deeper tissues, and lead to subsequent periodontal connective tissue destruction. In fact, structures resembling single A. actinomycetemcomitans cells were observed in the TEM analysis of epitheliums of co-cultures. Tissue destruction may even be further accelerated by decreased epithelial cell proliferation, which we showed in a previous study of tissue cultures exposed to an A. actinomycetemcomitans biofilm [10].
K17 was also strongly expressed in our control cultures. In vivo, K17 has been found in the sulcular epithelium of the dentogingival junction [2]. In addition, JE seems to express the genes encoding K17 [11]. Furthermore, it has been previously suggested that short chain fatty acids produced by some Gram-negative periodontal pathogens increase the expression of K17 [2] and that the inflammatory mediators would play a role in the regulation of the K17 expression [12]. Our finding that an A. actinomycetemcomitans biofilm decreased the expression of K17 seems to conflict with the earlier studies showing increased K17 expression after treatment with short chain fatty acids [2]. However, A. actinomycetemcomitans, although resistant to the antimicrobial activity of short chain fatty acids [13], produces only long chain fatty acids [14], which may at least partly explain these contradictory findings. Furthermore, K17 gene expression has been found depressed in clinical specimens from patients with severe and rapidly progressive periodontitis, which is in agreement with our results [4].
The model showed no evidence of K4 or K13 expression, which have been shown to be expressed in healthy oral sulcular epithelium [15]. Although K4 is typically absent in JE, its expression can be observed in JE of smoking periodontitis patients [15]. In our model, the A. actinomycetemcomitans biofilm did not increase or decrease K4 or K13 expression, of which the latter has been shown increasingly expressed in inflamed tissues [3]. However, the increase in K13 expression in inflammation may require the presence of host immune cells, such as macrophages, which were not included to our tissue co-culture model.
A closer investigation of the epithelial surface structure by TEM revealed that A. actinomycetemcomitans biofilm exposure caused necrosis of the epithelial surface in the co-culture models. The thick necrotic layer most likely inhibited the invasion of A. actinomycetemcomitans cells into the deeper layers of epithelium, as we could only detect a few structures resembling A. actinomycetemcomitans cells in the surface layers of the epithelium. However, the A. actinomycetemcomitans biofilm secreted high amounts of vesicles, which could have greater invasive potential than whole bacterial cells. For instance, A. actinomycetemcomitans vesicles have been shown to invade human HeLa and gingival fibroblast cells and release cytolethal distending toxin to the host cell nucleus [16,17]. A. actinomycetemcomitans vesicle-like structures could be observed in the epithelium. However, these vesicles may also originate from the host epithelial cells, and thus their origin needs to be confirmed in further studies.
In conclusion, A. actinomycetemcomitans biofilm decreases the expression of K17 and K19 and the total expression levels of keratins in an organotypic gingival mucosa model. However, the epithelium appears to utilize additional measures to withstand attack by the A. actinomycetemcomitans biofilm, which is suggested by the thicker necrotic layer between the epithelial cells and biofilm. Whether host immune cells and macrophages, in particular, change the keratin expression pattern during biofilm attack requires further investigation in a more complex environment.
4. Materials and Methods
4.1. A. actinomycetemcomitans Biofilm Culture
The A. actinomycetemcomitans biofilm cultures were generated as described previously [10]. Briefly, A. actinomycetemcomitans strain D7S was first cultured from trypticase soy agar blood plates. From the plates, an even bacterial suspension was made [18], and 5 × 107 cells/well were added to a 48-well plate containing porous filter discs. The biofilms were first grown in a rich trypticase soy broth medium for 24 h on a filter disc, and after which they were washed with a 0.85% NaCl solution. The cultivation was continued for an additional 24 h in glutamine supplemented RPMI-1640 medium. The 48-well plate also contained wells with sterile filter discs in the appropriate media for use as controls.
4.2. Gingival Mucosa Co-Culture Models
The A. actinomycetemcomitans biofilm/gingival mucosa co-culture models were constructed as described previously [10]. Briefly, human gingival fibroblasts [19] were grown in a collagen suspension with a cell culture insert (ThinCert™, Greiner Bio-One GmbH, Germany) for one day before 4 × 105 spontaneously immortalized human gingival keratinocyte cells [20] were seeded on top of the fibroblast-collagen matrix. The epithelial cells were cultured for one day submerged in the growth medium before the tissue model was lifted to the air–liquid interface. The model was air exposed for five days before a separately cultured A. actinomycetemcomitans D7S [21] biofilm (see above) was added on top of the tissue culture. The co-cultures were incubated without antibiotics in the cell culture medium. The gingival mucosa was co-cultured with the biofilm/control disc/no added components for 24 h, and the co-cultures were the fixed with a 10% formalin solution overnight. After the fixation step, the samples were embedded in paraffin and were sectioned.
4.3. Immunohistochemical Staining of Keratins K4, K13, K17, and K19
Before staining, the specimens were deparaffinized and heat-mediated antigen retrieval in 10 mM citrate buffer (pH 6.0) with microwaving was performed, which was followed by proteinase K treatment [22]. The staining was performed with a Dako TechMate™ 500 Plus Autostainer (Dako, Glostrup; Denmark) using the primary antibodies listed in Table 1 and the Dako REAL™ Detection System, Peroxidase/DAB+, Rabbit/Mouse (Code K5001; Dako) according to manufacturer’s instructions.
Table 1.
Primary anti-keratin antibodies used in the study.
| Keratin | Dilution | Host | Type | Code | Manufacturer |
|---|---|---|---|---|---|
| Keratin 4 | 1/10 | mouse | monoclonal | MON3015-1 | Sanbio, Holland |
| Keratin 13 | 1/50 | mouse | monoclonal | MUB0340S | Nordic MUbio, Holland |
| Keratin 17 | 1/20 | mouse | monoclonal | M704601-2 | Dako, Denmark |
| Keratin 19 | 1/100 | mouse | monoclonal | M088801-2 | Dako, Denmark |
| Pan-cytokeratin 5,6,8,17,(19) | 1/50 | mouse | monoclonal | M0821 | Dako, Denmark |
4.4. Transmission Electron Microscopic (TEM) Studies of the Biofilm-Epithelium Contact Site
The A. actinomycetemcomitans biofilm gingival mucosa co-culture models were assembled as described above. The samples were prefixed for TEM with a freshly prepared 5% glutaraldehyde solution (5% glutaraldehyde, 0.16 M s-collidin-HCl buffer, pH 7.4) for at least 3 h at room temperature. After prefixation, the samples were washed with s-collidin-HCl buffer three times for 3 min each. Then, the samples were postfixed (1% OsO4, 1.5% K-ferrocyanide) for 2 h [23], washed with s-collidin buffer three times for 5 min each, and were dehydrated with ethanol (70% ethanol, 1 min at 4 °C; 96% ethanol, 1 min, at 4 °C; 100% ethanol, 30 min, at 4 °C; 100% ethanol, three times, 30 min each, at 20 °C). The dehydrated samples were embedded in epoxy using the following series: propylene oxide, two times, 15 min each; propylene oxide + epoxy resin + DMP (10:10:0.15), 2 h; epoxy resin + DMP (10:0.15), 12 h; epoxy resin + DMP (10:0.15), at 60 °C, 36 h. The sectioning of the samples was accomplished with an ultramicrotome to a thickness of approximately 70 nm. After sectioning, the samples were stained with uranyl acetate (1% uranyl acetate in pure water for 30 min) and were rinsed three times with pure water for 30 s. Finally, the samples were stained with lead citrate (0.3% lead citrate in pure water for 3 min) and were rinsed with water as in the previous step. The samples were examined with a JEM-1400 Plus Transmission Electron Microscope (JEOL USA, Inc., Peabody, MA, USA).
Acknowledgments
We thank Katja Sampalahti, Marja-Riitta Uola, and Essi Hautamäki for their skillful technical assistance in the organotypic tissue cultures and histological staining. The transmission electron microscopic studies were performed in the EM core facility, and the light microscopic imaging was performed at the Cell Imaging Core (Turku Centre for Biotechnology, University of Turku and Åbo Akademi University).
Author Contributions
Conceptualization, R.I. and M.P.; formal analysis, A.B., R.I., and M.P.; investigation, A.B. and A.T.; methodology, A.T. and M.P.; visualization, R.I. and M.P., project administration, R.I.; resources, R.I. and M.P.; supervision, R.I. and M.P.; writing―original draft, A.B., A.T., R.I., and M.P.; and writing―review and editing, R.I. and M.P., funding acquisition, R.I.
Funding
This work was supported by the Academy of Finland grant numbers 126557, 265609, 272960, and 322817 to R.I. A.B. was funded by The Scientific and Technological Research Council of Turkey (TUBITAK).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Lamont R.J., Koo H., Hajishengallis G. The oral microbiota: Dynamic communities and host interactions. Nat. Rev. Microbiol. 2018;16:745–759. doi: 10.1038/s41579-018-0089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pöllänen M.T., Salonen J.I. Effect of short chain fatty acids on human gingival epithelial cell keratins in vitro. Eur. J. Oral Sci. 2000;108:523–529. doi: 10.1034/j.1600-0722.2000.00881.x. [DOI] [PubMed] [Google Scholar]
- 3.Mackenzie I.C., Gao Z. Patterns of cytokeratin expression in the epithelia of inflamed human gingiva and periodontal pockets. J. Periodontal Res. 1993;28:49–59. doi: 10.1111/j.1600-0765.1993.tb01050.x. [DOI] [PubMed] [Google Scholar]
- 4.Kebschull M., Guarnieri P., Demmer R.T., Boulesteix A.L., Pavlidis P., Papapanou P.N. Molecular differences between chronic and aggressive periodontitis. J. Dent. Res. 2013;92:1081–1088. doi: 10.1177/0022034513506011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pöllänen M.T., Salonen J.I., Uitto V.J. Structure and function of the tooth-epithelial interface in health and disease. Periodontol 2000. 2003;31:12–31. doi: 10.1034/j.1600-0757.2003.03102.x. [DOI] [PubMed] [Google Scholar]
- 6.Jiang Q., Yu Y., Ruan H., Luo Y., Guo X. Morphological and functional characteristics of human gingival junctional epithelium. BMC Oral Health. 2014;14:30. doi: 10.1186/1472-6831-14-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nagarakanti S., Ramya S., Babu P., Arun K.V., Sudarsan S. Differential expression of E-cadherin and cytokeratin 19 and net proliferative rate of gingival keratinocytes in oral epithelium in periodontal health and disease. J. Periodontol. 2007;78:2197–2202. doi: 10.1902/jop.2007.070070. [DOI] [PubMed] [Google Scholar]
- 8.Johnson J.D., Chen R., Lenton P.A., Zhang G., Hinrichs J.E., Rudney J.D. Persistence of extracrevicular bacterial reservoirs after treatment of aggressive periodontitis. J. Periodontol. 2008;79:2305–2312. doi: 10.1902/jop.2008.080254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christersson L.A., Albini B., Zambon J.J., Wikesjo U.M., Genco R.J. Tissue localization of Actinobacillus actinomycetemcomitans in human periodontitis. I. Light, immunofluorescence and electron microscopic studies. J. Periodontol. 1987;58:529–539. doi: 10.1902/jop.1987.58.8.529. [DOI] [PubMed] [Google Scholar]
- 10.Paino A., Lohermaa E., Sormunen R., Tuominen H., Korhonen J., Pöllänen M.T., Ihalin R. Interleukin-1beta is internalised by viable Aggregatibacter actinomycetemcomitans biofilm and locates to the outer edges of nucleoids. Cytokine. 2012;60:565–574. doi: 10.1016/j.cyto.2012.07.024. [DOI] [PubMed] [Google Scholar]
- 11.Hayashi Y., Matsunaga T., Yamamoto G., Nishii K., Usui M., Yamamoto M., Tachikawa T. Comprehensive analysis of gene expression in the junctional epithelium by laser microdissection and microarray analysis. J. Periodontal Res. 2010;45:618–625. doi: 10.1111/j.1600-0765.2010.01276.x. [DOI] [PubMed] [Google Scholar]
- 12.Zhang W., Dang E., Shi X., Jin L., Feng Z., Hu L., Wu Y., Wang G. The pro-inflammatory cytokine IL-22 up-regulates keratin 17 expression in keratinocytes via STAT3 and ERK1/2. PLoS ONE. 2012;7:e40797. doi: 10.1371/journal.pone.0040797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang C.B., Alimova Y., Myers T.M., Ebersole J.L. Short- and medium-chain fatty acids exhibit antimicrobial activity for oral microorganisms. Arch. Oral Biol. 2011;56:650–654. doi: 10.1016/j.archoralbio.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braunthal S.D., Holt S.C., Tanner A.C., Socransky S.S. Cellular fatty acid composition of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. J. Clin. Microbiol. 1980;11:625–630. doi: 10.1128/jcm.11.6.625-630.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritlove-Carson S., Charlesworth S., Morgan P.R., Palmer R.M. Cytokeratin phenotypes at the dento-gingival junction in relative health and inflammation, in smokers and nonsmokers. Oral Dis. 1997;3:19–24. doi: 10.1111/j.1601-0825.1997.tb00004.x. [DOI] [PubMed] [Google Scholar]
- 16.Rompikuntal P.K., Thay B., Khan M.K., Alanko J., Penttinen A.M., Asikainen S., Wai S.N., Oscarsson J. Perinuclear localization of internalized outer membrane vesicles carrying active cytolethal distending toxin from Aggregatibacter actinomycetemcomitans. Infect. Immun. 2012;80:31–42. doi: 10.1128/IAI.06069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thay B., Damm A., Kufer T.A., Wai S.N., Oscarsson J. Aggregatibacter actinomycetemcomitans outer membrane vesicles are internalized in human host cells and trigger NOD1- and NOD2-dependent NF-kappaB activation. Infect. Immun. 2014;82:4034–4046. doi: 10.1128/IAI.01980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karched M., Paul-Satyaseela M., Asikainen S. A simple viability-maintaining method produces homogenic cell suspensions of autoaggregating wild-type Actinobacillus actinomycetemcomitans. J. Microbiol. Methods. 2007;68:46–51. doi: 10.1016/j.mimet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Oksanen J., Hormia M. An organotypic in vitro model that mimics the dento-epithelial junction. J. Periodontol. 2002;73:86–93. doi: 10.1902/jop.2002.73.1.86. [DOI] [PubMed] [Google Scholar]
- 20.Mäkelä M., Salo T., Larjava H. MMP-9 from TNF alpha-stimulated keratinocytes binds to cell membranes and type I collagen: A cause for extended matrix degradation in inflammation? Biochem. Biophys. Res. Commun. 1998;253:325–335. doi: 10.1006/bbrc.1998.9641. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y., Shi W., Chen W., Chen C. Type IV pilus gene homologs pilABCD are required for natural transformation in Actinobacillus actinomycetemcomitans. Gene. 2003;312:249–255. doi: 10.1016/S0378-1119(03)00620-6. [DOI] [PubMed] [Google Scholar]
- 22.Paino A., Tuominen H., Jaaskelainen M., Alanko J., Nuutila J., Asikainen S.E., Pelliniemi L.J., Pöllönen M.T., Chen C., Ihalin R. Trimeric form of intracellular ATP synthase subunit beta of Aggregatibacter actinomycetemcomitans binds human interleukin-1beta. PLoS ONE. 2011;6:e18929. doi: 10.1371/journal.pone.0018929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karnovsky M.J. Use of ferrocyanide-reduced osmium tetroxide in electron microscopy; Proceedings of the 11th Annual Meeting of American Society for Cell Biology; New Orleans, LA, USA. 17–20 November 1971; p. 146. [Google Scholar]