Abstract
Plant cells form a multicellular symplast via cytoplasmic bridges called plasmodesmata (Pd) and the endoplasmic reticulum (ER) that crosses almost all plant tissues. The Pd proteome is mainly represented by secreted Pd-associated proteins (PdAPs), the repertoire of which quickly adapts to environmental conditions and responds to biotic and abiotic stresses. Although the important role of Pd in stress-induced reactions is universally recognized, the mechanisms of Pd control are still not fully understood. The negative role of callose in Pd permeability has been convincingly confirmed experimentally, yet the roles of cytoskeletal elements and many PdAPs remain unclear. Here, we discuss the contribution of each protein component to Pd control. Based on known data, we offer mechanistic models of mature leaf Pd regulation in response to stressful effects.
Keywords: plasmodesmata (Pd), Pd-associated proteins; (1,3)-β-glucanase; callose synthase; class 1 reversibly glycosylated polypeptides; formin-like protein 1 and 2; pectin methylesterase; Pd located proteins; synaptotagmin
1. Introduction
Due to their immobile lifestyle, plants are exposed to many abiotic and biotic stress factors [1]. A coordinated and generalized plant response to adverse environmental factors is accompanied by activation of both apoplastic and symplastic transport of macromolecules [2,3,4]. An understanding of the mechanisms of anti-stress reactions in plants has been facilitated by data obtained in recent years, indicating the role of intracellular traffic of secreted proteins with the participation of the endoplasmic reticulum (ER) and Golgi apparatus (GA) and the formation of an idea of alternative protein secretion pathways bypassing the GA [5,6,7,8]. Usually, with conventional secretion, secretory proteins with an N-terminal signal peptide are transported in secretory vesicles or secretory granules to the apoplast region via the ER, GA, and the subsequent endomembrane system. Passing through distinct GA compartments they undergo carbohydrate modifications and proteolytic processing [5,7,9,10,11]. However, recent plant secretome analysis has revealed considerable amount (up to 50%) of secreted proteins lacking a signal sequence [12]. These leaderless secretory proteins (LSPs) are not translocated into the ER and therefore cannot use the conventional protein secretion pathway with the participation of the GA. An alternative secretion route was termed as unconventional protein secretion (UPS) [6,7,13,14]. This definition also includes cases when plant proteins harbouring a signal peptide can traffic from this compartment to the extracellular space bypassing the GA [8,15]. Recent UPS models suggest that the ER is the central hub guiding proteins to an “external space” by circumventing the GA [8]. The extracellular space as well as the secretory route compartments’ lumen and the mitochondrial and plastid inner spaces are all considered as “external spaces” [7,8]. In contrast, proteomic studies have also revealed that the majority of LSPs are related to stress or pathogen infection [16], implicating the essence of the UPS pathway in dealing with various stress impacts [11]. Moreover, the increasing number of studies indicating a link between abiotic and biotic stress responses and secretion pathways offers research avenues towards understanding mechanisms involving the participation of plasmodesmata (Pd) in the plant stress tolerance [9,10,11]. With Pd, there is a direct connection between the symplast and the phloem that serves as a long-distance conductor in the vasculature, thus promoting rapid movement of materials between tissues both over short distances and over long distances throughout the plant. Pd provide a controlled symplastic exchange of both low molecular weight compounds and larger molecules, such as signaling molecules, transcription factors, non-cell-autonomous plant proteins [17,18], small regulatory RNAs and messenger RNAs [19,20], between plant cells, allowing general reactions in their growth processes and in response to stressful environmental factors [21,22,23]. Through Pd and phloem, photosynthesis products from the mesophyll of source leaves are exported to sink leaves and the plant growth point [24,25,26]. Sucrose, as a primary carbohydrate transported long distances in many plant species, is loaded into the phloem and unloaded into distal sink tissues. The productivity of agricultural plants largely depends on the efficiency of sucrose and other carbohydrates export from the mesophyll to the phloem through Pd, and further to the ripening fruits and grains [17,27,28,29,30,31,32,33,34].
The important role of Pd in stress response is universally recognized [21,22,23]. At the same time, the mechanisms for regulating Pd functions and permeability after stress impacts are still not fully understood. If the negative role of callose in the control of intercellular molecular flow via Pd is convincingly confirmed experimentally [35,36,37], the role of cytoskeletal elements and protein components, called Pd-associated proteins (PdAPs), is mostly unclear [32,38]. The analysis of the Pd proteome revealed that most PdAPs are secreted proteins [39] that might be involved in controlling Pd permeability under abiotic and biotic stress.
In this review, we analysed the characteristics of intracellular PdAP trafficking and assessed the contribution of each protein component to controlling Pd. A mechanistic view on Pd aperture regulation via callose and secreted proteins in plants is also presented.
2. Substructural Architecture of Plasmodesmata
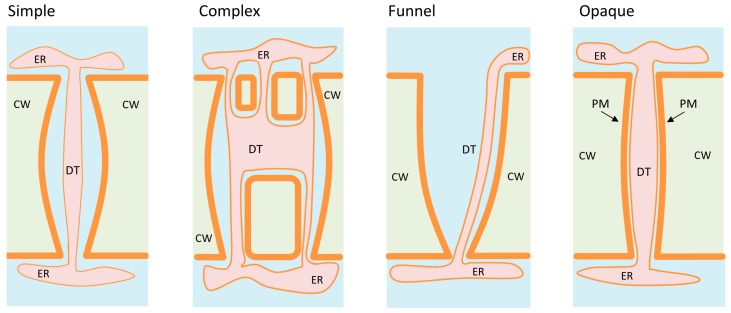
The structure of a typical simple Pd (Figure 1), whose length is approximately 100 nm with a diameter of 30 nm, is only clearly distinguishable by transmission electron microscopy [40]. In addition to the plasma membrane (PM) lining the inner surface of holes in the cell wall, Pd include a centrally located desmotubule (DT) which is derived from the compaction of ER membranes [41]. The cytoplasmic sleeve between the DT and PM defines the space available for intercellular trafficking and is filled with the combined cytoplasm of neighbouring cells (Figure 1).
Figure 1.
Different types of Pd. The Pd vary from simple with one channel connecting the adjacent cells to complex Pd with several channels that could merge. Also, there are some specific types of Pd such as funnel-shaped ones which are characteristic of the root tissues and opaque Pd found in the mitotically active tissue in the cell walls after cytokinesis. PM, plasma membrane (bold orange line); CW, cell wall (pale green); DT, desmotubule; ER, endoplasmic reticulum.
Primary Pd in plants are formed at the end of cell division when the ER penetrates into the developing cell plate, creating a cytoplasmic nanopore [42]. However, depending on the nature of the tissue and its age, the structure and configuration of Pd vary. Beyond Pd with a single nanochannel, referred to as simple Pd, there are also more complex Pd with double or even multiple branched nanochannels [40] (Figure 1).
Another form of Pd called a “funnel Pd” is found in Arabidopsis roots between protophloem sieve element (PSE) cells and phloem-pole pericycle (PPP) cells [43]. The location of the wide aperture of the “funnel Pd” inside the PSE is thought to be more effective than a simple Pd configuration for unloading dissolved substances in PPP (Figure 1).
Finally, in mitotically active tissue in the cell walls after cytokinesis, in addition to simple Pd, it is possible to discern Pd called “opaque”, in which there is such a tight contact between the ER and PM that there is no visible intermembrane space (Figure 1). However, in the absence of a visible sleeve in “opaque” Pd, intercellular movement of macromolecules such as GFP is still allowed [44].
Pd permeability is determined by the size of the largest molecule that can pass through it, creating a criterion termed the size exclusion limit (SEL) which is based on the dimensions of the folded molecule (or Stokes radius). Pd permeability testing methods have been evaluated in detail recently [32]. Branched secondary Pd have a decreased SEL compared with the primary Pd, restricting the movement of large molecules [45]. It is believed that the transition from immature to mature tissues correlates with increased Pd branching and a restriction in transport ability. The Pd SEL can be modulated in response to biotic/abiotic stresses, but Pd symplastic connectivity is strongly regulated in the space and over time [17,22,32,46,47].
3. Plasmodesmata-Associated Proteins (PdAPs)
The analysis of the current data enabled us to select PdAPs involved in the regulation of Pd permeability (Table 1). PdAPs can be divided into the following two groups: non-secreted cytoplasmic proteins including ER-resident proteins and secreted proteins.
Table 1.
Pd-associated proteins (PdAPs) involved in regulation of Pd permeability.
| Protein | Function/Description | Signal Sequence | Predicted N-Glycosylation Sites | Pd Localization | Protein Relocation After Stress Impact | References |
|---|---|---|---|---|---|---|
| Pd non-secretory proteins | ||||||
| Actin, myosin and tubulin | Actin-myosin filaments and tubulin are localized within the Pd cytoplasmic sleeve and negatively control Pd permeability | No | No | Cytoplasmic sleeves in the Pd cavity | No | [48,49,50] |
| A. thaliana calreticulin-1 (AtCRT1) (UniProt O04151) | Ca2+-sequestering protein chaperone and ER protein that negatively control Pd permeability | Yes | Asn59, Asn154, Asn399 | Associates with the desmotubule | Yes | [51,52,53] |
| Tobacco non-cell autonomous pathway protein (NCAPP) (UniProt Q947H5) | NCAPP has homology to aldose 1-epimerase; positively controls Pd permeability and PME/methanol production | Yes | Asn76 and Asn100 | Localizes in the ER | N/A | [54,55] |
| Remorin | Associates with PM raft-like structures and probably serves as a negative regulator of Pd permeability. Antagonist of Potato virus X triple gene block protein 1 | No | No | In the cytosolic surface of the Pd plasma membrane | N/A | [56] |
| A. thaliana reticulons, AtRTNLB3 (UniProt Q9SH59) and AtRTNLB6 (UniProt Q6DBN4) | ER-localized proteins with three TM domains that negatively control Pd permeability | No | No | Accumulates in the desmotubule | N/A | [57,58] |
| Arabidopsis synaptotagmin SYTA (AtSYTA) (UniProt - Q9SKR2) | ER-PM tethering and endocytic recycling | No | No | Interacts with TMV movement protein (MP) Pd localization signal (PLS) for cell-to-cell transport and participates in the formation of virus replication sites | Suggested that SYTA relocates to the Pd cavity after TMV infection | [59,60,61,62] |
| Pd secretory proteins | ||||||
| A. thaliana (1,3)-β-glucanase 1 (AtBG1) (UniProt - Q9M2M0) | Callose-degrading enzymes that positively control Pd permeability | Yes | Asn291 | Colocalizes with callose near Pd orifices | No | [36,63,64] |
| A. thaliana (1,3)-β-glucanase 2 (AtBG2) (UniProt P33157) | Yes | No | Colocalizes with callose near Pd orifices in the extracellular space | AtBG2 is not delivered to the extracellular space in TMV-infected cells, but associates with viral MP in Pd cytoplasmic sleeve | [36,64] | |
|
A. thaliana (1,3)-β-glucanase 3 (AtBG3) (UniProt F4j270) |
Yes | No | Constitutive Pd-associated enzyme but not stress-regulated | No | [36,64] | |
| *A. thaliana β-1,3-glucanase_putative Pd-associated protein (AtBG_ppap) (UniProt Q9FHX5) | Yes | No | Localizes to the Pd neck region | No | [35,36,65] | |
| A. thaliana Class 1 reversibly glycosylated polypeptide (AtC1RGP) (UniProt Q9SRT9) | AtC1RGP acts as a negative Pd regulator. Despite having no signal sequence, it is found in the GA and ultimately in the Pd | No | No | N/A | [66,67,68] | |
| A. thaliana Callose synthase (CalS) (UniProt Q9AUE0) | Callose-synthesizing enzyme encoded by glucan synthase-like (GSL) gene that negatively controls Pd permeability | No (needs exocyst-positive organelle (EXPO)-mediated secretion for Pd localization) |
No | CalS localizes at callose depositions | No | [36,37,69] |
|
A. thaliana formin-like protein 1 (UniProt Q9SE97) and 2 (UniProt O22824) (AtFH1 and AtFH2) |
Negatively regulates Pd permeability by interacting with actin filaments | Yes | Multiple Asn sites | Localizes in the Pd cavity and interacts with actin | N/A | [70,71] |
| A. thaliana β-1,6-N-acetylglucosaminyl transferase-like enzyme (GnTL) (UniProt Q9SUZ8) | Interacts with calreticulin and probably serves as a negative regulator of Pd permeability | Yes | Asn287 and Asn316 | Colocalizes with callose-binding protein near Pd orifices | N/A | [72] |
| *A. thaliana Pd callose-binding proteins 1,2 and 3 (PDCB1-3) (At5g61130) |
Pd callose-binding protein that negatively controls Pd permeability | Yes | Asn154 and Asn179 | Localizes to the Pd neck | No | [73,74] |
|
A. thaliana Pd-located protein 1 (PDLP1) (UniProt Q8GXV7) |
Membrane receptor-like protein with two extracellular DUF26 domains. PDLP1 overexpression causes restricted cell-to-cell trafficking. Acts as a negative Pd regulator by promoting callose deposition. Stimulates the transport of viruses that use tubule-guided movement by redundantly interacting with tubule-forming MPs within Pds | Yes | No | PDLP1 is targeted to Pd via the Brefeldin A–sensitive secretory pathway and resides at Pd with its C-terminus in the cytoplasmic space and its N-terminus in the apoplast. | No | [75,76,77,78] |
|
A. thaliana Pd located protein 5 (PDLP5) (UniProt Q8GUJ2) |
A member of the PDLP family that has 30% amino acid sequence identity to PDLP1. Contains sphingolipid binding motif in the TMD. Acts as a negative Pd regulator by promoting callose deposition. Delays systemic movement of TMV | Yes | Asn69 and Asn132 | PDLP5 localizes inside the central Pd region similar to TMV MP. However, PDLP5/MP overlap is not complete | Transmembrane secretory protein with ectopic localization | [79,80,81,82] |
| Tobacco pectin methylesterase (PME) (UniProt Q9LEBO) |
Non-direct regulator of Pd permeability that participates in the de-methylesterification of cell wall HG through the formation of methanol | Yes | Asn43, Asn101 and Asn220 in proPME | Immunogold localization of PME is preferentially around Pd | No | [83,84] |
* glycosylphosphatidylinositol-anchored protein (GPI-AP) [85].
3.1. Non-Secretory Pd Proteins
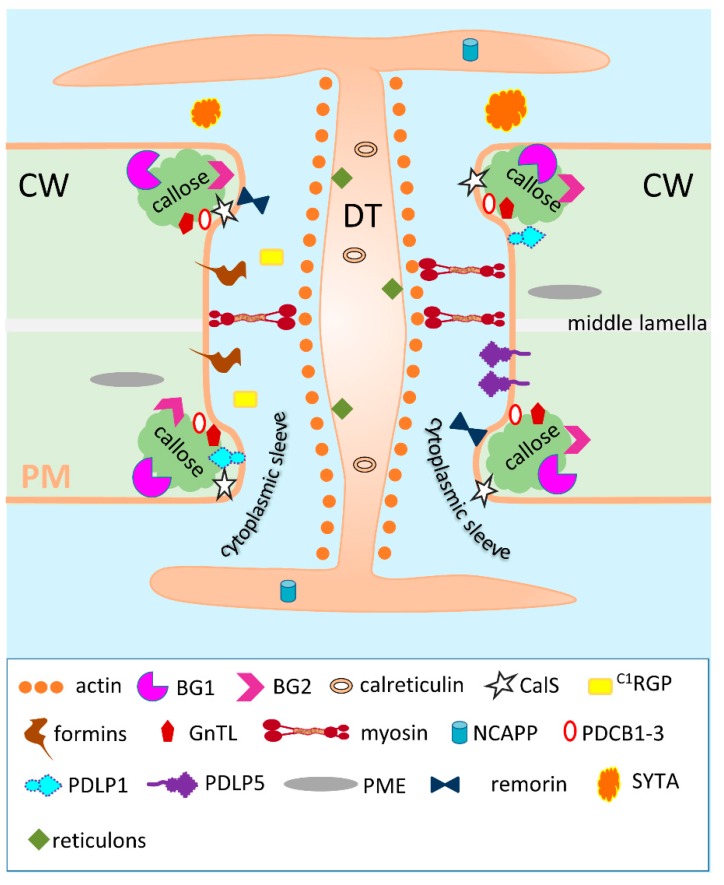
This group of proteins include non-secreted cytoplasmic proteins found in the Pd space as well as proteins associated with the DT and Pd plasma membrane (Figure 2).
Figure 2.
The structure of a typical simple Pd. Plasmodesmata is a pore in the cell wall (CW) lined with a plasma membrane (PM). It contains a centrally located desmotubule (DT) which originates from the endoplasmic reticulum of the adjacent cells. The space between the PM and the DT, a cytoplasmic sleeve, is available for the intercellular trafficking. Molecules of actin and myosin are resident Pd proteins as they are components of the cell cytoskeleton that extends from cell to cell through Pd and is involved in regulating Pd permeability. Callose (designated with a green cloud-like shape) is a linear β-1,3-glucan molecule and it is considered as a key plasmodesmal marker that controls Pd permeability. BG1 and 2, (1,3)-β-glucanase 1 and 2, callose-degrading enzymes; calreticulin, Ca2+-sequestering protein chaperone; CalS, callose synthase; C1RGP, Class 1 reversibly glycosylated polypeptide; formins, proteins involved in actin stabilization and anchoring cytoskeletal structures to membranes; GnTL, β-1,6-N-acetylglucosaminyl transferase-like enzyme; NCAPP, non-cell autonomous pathway protein; PDCB1-3, Pd callose binding proteins 1-3; PDLP1 and 5, Pd-located protein 1 and 5; PME, pectin methylesterase, an enzyme that performs de-methyl esterification of pectin; remorin, a protein associated with PM raft-like structures; SYTA, A. thaliana synaptotagmin which is a tethering protein that maintains ER morphology and stabilizes the formation of ER–PM contacts; reticulons, ER-tubulating proteins participating in the formation of the Pd DT.
3.1.1. Actin, Myosin and Tubulin
The cytoskeletal proteins actin and myosin have been implicated with the structure and function of Pd (Figure 2). The presence of these proteins in cytoplasmic sleeves in the Pd cavity was discovered long ago using immunodetection method [48,49,50]. Compared with actin and myosin, the role of tubulin in the functioning of Pd is not well understood. There are only speculations about tubulin’s participation in long distance transport [48,86].
3.1.2. Synaptotagmins
Analysis of the Arabidopsis Pd proteome revealed several synaptotagmins (SYTA) [39]. In addition to endosome recycling, AtSYTA participates in plant viral movement protein (MP)-mediated trafficking of begomovirus Cabbage leaf curl virus (CaLCuV) and Tobacco mosaic virus (TMV) genomes through Pd. In AtSYTA knockdown Arabidopsis lines, CaLCuV systemic infection was delayed, and cell-to-cell spread of TMV and CaLCuV movement proteins was inhibited [60].
The mechanism of AtSYTA participation in tobamovirus cell-to-cell movement was clarified by a study by Levy et al. [59], which showed that tobamovirus Turnip vein-clearing virus (TVCV) MP was able to (i) interact with SYTA, (ii) remodel ER-PM contact sites and (iii) form virus replication sites for further cell-to-cell movement. It has been suggested that TVCV MP-mediated recruitment of SYTA leads to its accumulation in the Pd cavity [59]. Most likely, such recruitment begins with the binding of the 50-aa TMV MP Pd-localization sequence [87,88] with SYTA, as was recently shown [62]. This event can be considered as the beginning of the process, leading to the dilatation of Pd. According to the hypothetical mechanism proposed by Pitzalis & Heinlein, MPs interact with one SYTA form, SYT1, the length of which, as the tethering molecule, is controlled through interaction with myosin VIII. The model assumes that the chain of events triggered by the interaction of MP and SYT1 leads to compression of the DT lumen, an increase in the size of the cytoplasmic channel and, accordingly, Pd SEL [89].
3.1.3. Remorin
Remorin is another non-secreted cytoplasmic protein lacking a signal sequence. Remorin is extremely hydrophilic, has a proline-rich N-terminus and is synthesized in the cytosol and targeted to the cytoplasmic surface of the Pd PM [90]. The C-terminal part of the protein was predicted to be a coiled-coil, suggesting that the protein interacts with other macromolecules. The localization of remorin was studied using immunogold labelling coupled with electron microscopy, which allowed its visualization not only in plant PM domains but also at the Pd cavity [56]. Remorin participation in virus cell-to-cell traffic was confirmed by data indicating the following: (1) its ability to interact physically with the Potato virus X (PVX) triple gene block protein 1 and (2) its expression levels negatively correlate with the cell-to-cell movement of PVX [56].
3.1.4. Calreticulin
Calreticulin, a Ca2+-sequestering protein chaperone responsible for the ER unfolded protein response and highly conserved between different species, also accumulates within the Pd in plants [51,91,92,93,94]. In general, calreticulin binds to misfolded ER luminal proteins and prevents them from being exported from the ER to the GA, functioning as a quality-control chaperone. Arabidopsis calreticulin-1 (AtCRT1) is 425 aa in length with 22-aa signal sequence mandatory for Pd localization [95], three N-glycosylation sites and the C-terminal retention sequence HDEL that prevents its exit from the ER lumen. AtCRT1 binds TMV MP in vitro and in vivo and colocalizes with this protein in Pd [52]. If, in response to stressful effects, CRT accumulates in the ER lumen in the area of the DT in the Pd central cavity, it can not only block the movement of molecules along Pd sleeves but also prevent the appearance of TMV MP in Pd. This assumption is supported by experiments by Chen et al., which showed that overexpression of AtCRT1 hinders TMV cell-to-cell movement and blocks MP accumulation in the Pd [52]. As an alternative explanation, it is possible that the presence of large quantities of AtCRT1 within plant cells redirects TMV MP from Pd to the microtubular network [52].
3.1.5. Non-Cell-Autonomous Pathway Proteins (NCAPPs)
NtNCAPP, which is found in tobacco [54] in two highly homologous forms, NCAPP1 and NCAPP2, has high homology to aldose 1-epimerase-like protein and all the features of a protein secreted by the GA, including a 24-aa signal sequence and predicted N-glycosylation sites. Immunogold labelling analysis showed the presence of NCAPP1 in cells near the Pd orifice but not in the central cavity. This PdAP was identified as an interaction partner of the 16-kD Cucurbita maxima (pumpkin) phloem non-cell autonomous protein CmPP16 which moves from cell to cell and participates in the RNAs intercellular transport [96]. In experiments using a dominant-negative mutant NCAPP1Δ1–22, its important role in the intercellular transport of CmPP16 and TMV MP was shown. A model [97] has been proposed where NCAPP1 promotes the translocation of some NCAPs through Pd after their reciprocal phosphorylation and glycosylation [98].
3.1.6. Reticulons
Reticulons (RTNLB), as family of membrane ER-tubulating proteins play a role in the formation of the Pd DT, which regulates the movement of small molecules via cytoplasmic sleeves [57,99,100]. Of the seven Arabidopsis RTNLBs, Pd essentially enriched only with RTNLB3 and RTNLB6. Although all AtRTNLBs lack an ER signal peptide, their translocation into the ER is likely directed by internal transmembrane domains [101]. It is unclear whether the GA is involved in the secretion of Arabidopsis reticulons, but the related human RTN3 uses it [102]. The results from studies of AtRTNLB3 and AtRTNLB6 showed high degree of colocalization of these proteins with TMV MP in the Pd central cavity [57]. It has been suggested that RTNLBs from the Pd central cavity could promote a mechanism for reducing Pd permeability through the expansion of the DT while viral MPs provide Pd gating by disrupting the DT scaffold and perturbing protein-lipid or protein-protein interactions.
3.2. Secreted PdAPs
This category includes PdAPs that obey the principles of traditional or conventional secretion, regardless of their final localization in Pd (Figure 2).
3.2.1. Callose-Degrading β-1,3-glucanases (BG)
The callose contents at Pd depends on the activity of (1,3)-β-glucanases (BGs). Based upon the protein domain structure and sequence, Arabidopsis BGs can be classified into five groups with at least 50 members [63]. BGs contain an N-terminal secretion signal. Some members contain a hydrophobic C-terminal sequence that includes a predicted glycosylphosphatidylinositol (GPI)-anchor attachment motif for targeting the protein to the cell membrane [73] and to Pd [85]. Callose degradation at the A. thaliana Pd depends on the activity of three GPI-anchored BG proteins, including β-1,3-glucanase_putative Pd-associated protein (AtBG_ppap), plasmodesmal-localized β-1,3-glucanase 1 (PdBG1), and plasmodesmal-localized β-1,3-glucanase 2 (PdBG2) [35,73]. PdBG3 is also localized at Pd, but its role in callose degradation is not proven [73].
Arabidopsis BGs are conventionally secreted proteins associated with PM and Pd. However, in detailed analysis using the plasmolysis method, AtBG_ppap, for example, could be seen to recede from the cell wall along with PM [35]. Thomas et al. suggest that unlike PDLP1a, β-1,3-glucanase moves to Pd via the PM and maintains only a superficial physical association with Pd [78].
Biotic and abiotic stress have significant impacts that affect the expression and localization of the BGs. Most BGs, including AtBG2 and AtBG3, are regulated at the transcriptional level during stress [65]. For example, while TVCV did not affect AtBG_ppap transcription, the transcription of an AtBG2-encoding gene was enhanced [64]. Analysis also revealed that AtBG2 is present in the extracellular space and colocalizes with callose near Pd orifices in healthy cells, while its secretion ceases and it associates with viral MP in Pd in the TMV-infected cells [64].
In general, the link between BGs activity and viral infection has been noted for a long time [36]; even in 1972, Moore and Stone proposed hypothesis that “the fact that it can depolymerize callose suggests that it could have a role in facilitating the spread of virus through removal of callose deposits or by modification of cell wall structures” [103]. This hypothesis was confirmed in experiments with TMV-infected plants treated with methanol [104]. It has been shown that genes encoding BGs are sensitive to methanol released from wounded plants. In contrast, both methanol treatment and the introduction of a gene encoding BG into the cell resulted in the increased gating capacity of Pd, which created favourable conditions for viral cell-to-cell movement and ultimately led to increased reproduction of TMV in the plant.
3.2.2. Pd-Associated Callose Binding Proteins (PDCBs)
PDCBs are secreted proteins that appear in the extracellular space through participation of the GA. Arabidopsis has 11 PDCB-like proteins, including PDCB2 and PDCB3, which are the most studied [74]. All PDCBs comprise an N-terminal signal peptide, an X8 domain (CBM43), an unstructured region in the middle and a GPI-anchor motif at the C-terminus. The At5g61130 gene encodes a GPI-anchored PDCB1 protein with N- and C-terminal domains that are cleaved during processing from the preprotein. Mature PDCB1 has a length of 153 amino acids, two potential N-glycosylation sites and a GPI-anchor-amidated Ser172. It was determined that PDCB1 was located at the neck region of Pd where callose becomes deposited [74]. It was also established that PDCB1 binds to callose through its X8 domain. PDCB1 does not have callose-synthesis enzymatic activity; however, its overexpression, for unclear reasons, leads to an increase in callose accumulation, and decreased Pd permeability in a GFP-movement assay [74].
3.2.3. Plasmodesmata-Located Protein 1 (PDLP1)
PDLP1 was the first identified representative of the 8-protein family in Arabidopsis. These proteins are type I membrane proteins with a molecular mass ranging from 30.2 to 35.3 kDa and comprising an N-terminal signal peptide, a large region containing two similar domains annotated as domains of unknown function 26 (DUF26), a single transmembrane domain (TMD), and a short C-terminal tail. DUF26 domains have a conserved C-X8-CX2-C motif, which is distinct from the Cys-rich regions found in S-locus glycoproteins [78].
Studies of PDLP1, as the first representative of a new family of proteins, confirmed that it uses a traditional secretion mechanism involving the GA, which was sensitive to brefeldin A and showed sensitivity to disruption of COPII-mediated ER-export by the GTP-locked form of Sar1. Expression in transgenic Arabidopsis showed that PDLP1 fused with GFP was located as punctuated spots on the cell wall and that the fluorescence persisted on the cell wall after plasmolysis. Further evidence that these punctuate sites were Pd was obtained by colocalization methods with callose and TMV MP [78]. Functional tests confirmed that PDLP1 can regulate intercellular transport by affecting Pd. PDLP1 deletion analysis has shown that its single TMD is required for targeting to Pd. PDLP1 acts as a negative Pd regulator because its overexpression causes restricted cell-to-cell trafficking due to callose deposition [78]. PDLP1 also promotes the transport of viruses that use tubule-guided movement by redundantly interacting with tubule-forming Grapevine fanleaf virus [75] and Cowpea mosaic virus (CPMV) [105] MPs within Pd. As suggested, viruses can subvert the functions of PDLPs proteins to facilitate the cell-to-cell movement of entire virions [22].
3.2.4. Plasmodesmata-Located Protein 5 (PDLP5)
PDLP5 is another representative of the 8-member family of Arabidopsis PDLPs. Although it has only 30% amino acid sequence identity to PDLP1, it has all the features of secreted proteins of this family and is a type I transmembrane protein containing a cysteine-rich extracellular domain and a transmembrane domain with a short C-terminal tail [79,81]. For intracellular movement, PDLP5 uses the GA and it has potential N-glycosylation sites [80]. Similar to PDLP1, PDLP5 acts as a negative Pd regulator that suppresses Pd trafficking and confers enhanced innate immunity against bacterial pathogens in a salicylic acid–dependent manner [31]. For this topic, it is important to note the differences between PDLP1 and PDLP5 in Pd localization. If PDLP5 is localized to the Pd membranes, then PDLP1 is distributed both throughout the Pd membrane [106] and along the PM [107]. Recently, a possible cause for the differences in the behaviour and localization of PDLP1 and PDLP5 has been identified [82]. Pd-lining plasma membrane is known to contain specific regions in which sphingolipids interact with sterols to create specialized domains [106,108,109], where GPI-anchored proteins accumulate [39,56]. Moreover, sphingolipid biosynthesis is very important for modulating Pd function [110]. Thus, the loss of the function of the PHLOEM UNLOADING MODULATOR (PLM) gene, which encodes the protein involved in the synthesis of sphingolipids, leads to a change in the function and structure of the Pd – specifically, the disappearance of cytoplasmic sleeves in the Pd [110]. Ning-Jing et al [82] revealed that Pd membranes are enriched with specific t18:0-based sphingolipids, compared with these sphingolipid levels in the plasma membrane. In examining the Arabidopsis sld1 sld2 double mutant, which lacks sphingolipid long-chain base 8 desaturases 1 and 2, leading to the accumulation of t18:0-based sphingolipids in the Pd membranes, the authors found increased accumulation of PDLP5, which has specific binding affinity for phytosphinganine (t18:0), in leaf epidermal cells. The double mutants also showed increased callose accumulation, decreased Pd permeability and enhanced resistance to the fungal-wilt pathogen Verticillium dahlia and the bacterium Pseudomonas syringae pv tomato DC3000. Ning-Jing et al [82] attributed the specific binding of PDLP5 to t18:0-based sphingolipids by the existence of the sphingolipid-binding motif TXXILXXVF [111] in the PDLP5 TMD (LAIIIGIVTLIILLVVFLAFV). It is important to note that this motif is absent in the PDLP1 TMD (IALAVGGVFVLGFVIVCLLVL), which explains the difference in the localization of these proteins.
PDLP5 was found to colocalize with TMV MP at the central region of Pd channels. Quantitative analysis of immunogold staining for TMV MP and PDLP5 showed that the labelling was highly specific and convincingly confirm their association with Pd. In accordance with the function of a negative Pd regulator, ectopic expression of PDLP5 delayed the systemic movement of TMV-GFP in N. benthamiana. In contrast, downregulation of PDLP5 alone was sufficient to enhance Pd permeability [79]. By contrast, ectopic expression of PDLP5 in N. benthamiana did not affect the systemic movement of Cucumber mosaic virus (CMV), which may indicate a difference in the functioning mechanisms of TMV and CMV MPs [22]. In turn, Turnip crinkle virus (TCV) can also overcome PDLP5-mediated Pd constriction in A. thaliana plants [77]. Thus, plants with PDLP5 overexpression can be a differential test system to identify the differences in the mechanisms of virus cell-to-cell movement.
It is agreed [79] that the identical localization of PDLP5 and TMV MP at the central region of Pd channels raises questions and requires additional research. The data on the preferential localization of specific t18:0-based sphingolipids and the associated with them PDLP5 in the Pd membrane, as described above, support the idea of co-localization of PDLP5 and TMV MP. It is possible that not only callose deposition but also competition between PDLP5 and TMV MP for binding to common Pd factors are critical for Pd regulation. If PDLP5 and TMV MP indeed occupy the same Pd subdomain, this means that, contrary to the transmembrane nature of the secretory protein, PDLP5 should be at the same site as TMV MP, which is localized on the cytoplasmic surface of the ER [112]. In accordance with our hypothesis, this contradiction could be resolved through a mechanism involving specific t18:0-based sphingolipid traffic, that results in PDLP5 localization in the Pd cytoplasmic sleeve (Figure 3).
Figure 3.
Hypothetical model showing the targeting of the secreted PdAPs into the cytoplasmic sleeve. Schematic representation of PDLP5 vesicular transport involving specific t18:0-based sphingolipid traffic with the final destination in the Pd cavity. Formins are hypothesized to be delivered to the Pd in a similar way. C1RGP is secreted to the cell wall and reaches its location in the Pd cytoplasmic sleeve by an unknown mechanism. Sphingolipids are designated as purple rods. CW, cell wall; C1RGP, Class 1 reversibly glycosylated polypeptide; DT, desmotubule; GA, the Golgi apparatus; PDLP5, Pd-located protein 5.
3.2.5. β-1,6-N-acetylglucosaminyl Transferase-Like Enzyme (GnTL)
In the selection of proteins that bind to Pd-localized calreticulin, Zalepa-King et al. isolated β-1,6-N-acetylglucosaminyl transferase-like enzyme (GnTL) among other Pd-associated proteins in A. thaliana [72]. AtGnTL is 346-residues protein containing an amino-terminal signal peptide followed by a catalytic domain (GnT) characteristic of the glycosyltransferase protein family, which catalyses the transfer of a specific activated sugar moiety from a donor molecule to an acceptor via a glycosidic bond in the GA. Subcellular localization studies using confocal microscopy observed AtGnTL at Pd within living plant cells. AtGnTL was confirmed as a Pd-located protein in a plasmolysis test. AtGnTL-mCherry retained its punctate localization pattern within the cell wall. The authors suggest that AtGnTL and TMV MP are likely to have different sites in Pd that are available for binding to calreticulin [72]. Although the authors did not directly show that AtGnTL expression suppresses Pd permeability in this work, it can be assumed that AtGnTL is able to function as a negative regulator of Pd given its association with calreticulin, the expression of which blocks TMV infection [52].
3.2.6. Cell Wall Pectin and Pectin Methylesterase (PME) as Factors Controlling Pd Permeability
Primary Pd are grouped in clusters called pit fields [113]. The cell wall that surrounds Pd is also an important component of its structure [83]. It is known that the cell wall material directly around the Pd is rich in pectin and depleted in cellulose. It is believed that the cell wall enriched in pectin is more flexible than cellulose fibres, and this may allow dynamic changes in the Pd structure and functions that occur in response to a number of environmental signals [29,44,83]. This view has been confirmed by known data about the stress-induced enzyme, PME, involved in pectic homogalacturonan (HG) de-esterification in muro [114], showing its accumulation in the cell wall near Pd [84].
An important consequence of pectin demethylation that affects Pd permeability is the stimulation of multiple methanol-inducible genes (MIGs) expression [104]. Some MIGs are involved in defense reactions, while others affect cell-to-cell trafficking. Tobacco BG coding for the related to basic vacuolar BG isoform, a previously unidentified gene (MIG-21), and NCAPP increased the Pd gating capacity, which in turn stimulated TMV cell-to-cell movement [104]. It is known that PME interacts with TMV MP in vitro [115,116], suggesting that PME may be directly involved in the cell-to-cell movement of plant viruses [117].
3.2.7. Callose Synthase (CalS)
Callose synthase (CalS) is an enzyme involved in the metabolism of callose. A. thaliana AtCalSs are encoded by 12 glucan-synthase-like genes (AtGSL1 to AtGSL12) and are large proteins (1770–1950 amino acids) possessing a large central catalytic domain which includes a UDP-glucose catalytic site and a glycosyltransferase domain surrounded by multiple transmembrane domains [69,118]. CalSs are located in the PM and exhibit high substrate specificity for the nucleotide sugar uridine diphosphate glucose (UDP-glucose) [37,47,119]. Several isoforms of AtCalSs were found in the proteome of trans-Golgi-network-derived vesicles [120] indicating that these proteins are conventionally secreted. The final step of their secretion and fusion to the PM involves EXOCYST subunit EXO70 family proteins [121]. CalS performs its function in conjunction with UDP-glucose transferase 1 (UGT1), Rho-like GTPase (Rop), RabA4c, tubulin, phragmoplastin (Phr), sucrose synthase (SuSy), and annexin (ANN), thus forming a highly specialized protein complex for callose biosynthesis [122].
CalSs are capable of post-translational regulation. Thus, in response to various abiotic and biotic stresses, some Arabidopsis CalSs are phosphorylated, which affects their activity [122]. Since callose is involved in the control of Pd, it can be assumed that CalSs are also located in Pd [47]. Indeed, a study of mutant Arabidopsis gsl12/cals3 lines confirmed this assumption [123]. It has been shown that the GFP-CalS3 fusion protein can be localized in Pd in transfected leaves. Moreover, gain-of-function mutations in CalS3 resulted in increased accumulation of callose at the Pd, a decrease in the Pd aperture and reduced intercellular trafficking [123].
3.2.8. Formins
Plant formins are evolutionarily conserved eukaryotic proteins involved in actin nucleation and cytoskeletal organization. Typical plant Class I formins are integral membrane proteins that can anchor cytoskeletal structures to membranes. The Arabidopsis Class I formin AtFH1 accumulates in actin-rich regions of the cortical cytoplasm and associates with Pd [71]. The Arabidopsis Class II formin AtFH2 was also detected in Pd and is suggested to regulate Pd permeability by anchoring actin filaments to Pd [70]. Formins are secreted proteins that have a signal sequence and use the GA for intracellular movement. AtFH2 is 894 amino acids in length with 20-aa signal sequence and multiple N-glycosylation sites. Similar to PDLP1 and PDLP5, AtFH1 and AtFH2 also have 21 amino acid transmembrane domains. Moreover, AtFH2 TMD, in contrast to AtFH1, contains a 9 amino acid region very similar to the sphingolipid-binding motif. Despite having all the properties of a secreted protein, AtFH2 appears to interact with actin microfilaments in the Pd cavity. Moreover, AtFH2 caps and stabilizes actin filaments, thereby regulating cell-to-cell trafficking by tethering actin filaments to the membrane at Pd. Since the polymerization of actin filaments reduces the clearance of Pd, AtFH2-mediated stabilization of actin filaments can play a negative role in Pd permeability. Indeed, loss of function of AtFH2 increased Pd SEL. These mutant plants, which have more permeable Pd, became more susceptible to a Cucumber mosaic virus [70]. Thus, plant formins participate in the Pd closing mechanism.
3.2.9. Class 1 Reversibly Glycosylated Polypeptide (C1RGP)
C1RGP is a member of the reversibly glycosylated polypeptides family that also includes C2RGP [124], which self-glycosylates in the presence of certain nucleotide sugars [125,126,127]. Analysis of AtC1RGP showed that it is a secreted protein. Despite having no signal sequence, this protein is found in the GA and, ultimately, in the Pd [67]. In plasmolysed cells, the association of AtC1RGP fused with GFP (AtC1RGP-GFP) with the cell wall remains unchanged. The hypothesis that AtC1RGP is delivered to Pd via the GA was confirmed by experiments where brefeldin A which prevented the appearance of AtC1RGP-GFP in Pd [67]. To explain the mechanism of association of C1RGP with Pd, Sagi et al. suggested that “upon fusion of the GA to the PM of a Pd or in the vicinity of a Pd, C1RGPs would be attached to the PM facing the cytoplasmic sleeve of the Pd. The integration of C1RGPs onto the cytosolic facing side of the plasmodesmal PM could be a factor in establishing the size exclusion limit for soluble proteins” [67]. Subsequently, this hypothesis was confirmed by experiments showing changes in the expression levels of the gene encoding AtC1RGP. It has been shown that AtC1RGP overexpression leads to a reduction in Pd permeability for photoassimilates and TMV cell-to-cell movement [68]; in contrast, reduced levels of AtC1RGP increase intercellular transport via Pd, and TMV exhibited accelerated systemic spread [66].
It is important to note here that when C1RGP was added to a class of secreted PdAPs (Table 1, Figure 2 and Figure 3), we proceeded from the data obtained by studying the ectopic expression of AtC1RGP in tobacco [67]. It is important to note that studies of the Rautengarten et al group [128] showed that AtC1RGP1 and AtC1RGP2 are cytosolic Golgi-associated enzymes belonging to the UDP-L-arabinopyranose (UDP-L-Ara) mutases, which catalyse the formation of UDP-Araf from UDP-Arap. In addition, the authors reported (results not shown) that they “did not observe any fluorescence originating from plasmodesmata, consistent with previous studies in Arabidopsis” [125,127,129]. This contradiction is apparently a consequence of the differences in the secretion mechanisms of tobacco and Arabidopsis; in our studies, the Nicotiana benthamiana RGP (NbC1RGP), which has 90% homology with AtC1RGP, was found in the Pd.
4. Mechanisms of PdAP Participation in Intercellular Cytoplasmic Connectivity
Plant intercellular cytoplasmic connectivity can be regulated by several mechanisms [17,27,28,31,34]. The first mode of regulating symplastic transport can be carried out by changing the density of primary Pd per unit area of the cell wall. As cells grow and the surface of their cell walls expand, the density of primary Pd is decreased.
The second way to change the intercellular conductivity is the modification of Pd structure when a transition from primary to secondary Pd occurs. The study of the sink-source transition in tobacco leaves has shown how this mechanism works [28] when a large number of simple primary Pd are lost and the contents of branched secondary Pd increase [26,40].
The third mechanism involves a change in Pd permeability based on the function of its structural elements and PdAPs. This mode is especially important for secondary Pd from mature leaves during rapid reactions to stress [21,22,23]. This mechanism includes mechano-sensing Pd closure in response to tissue damage or osmotic shock, and has recently been reviewed [130]. It is believed that intracellular pressure forces acting on the dumbbell-shaped ER DT cause the displacement of this complex, leading to Pd closure.
Although a relatively large number of potential Pd components have been identified [31,32,33,38], the current understanding of the mechanisms for regulating the Pd conductivity is based on the participation of callose, which was revealed by electron microscopy studies near and within the Pd channel [131]. Our analysis of PdAPs (Table 1) shows that features of secreted proteins also play an important role in the functioning of Pd. We assume that the impact of ER-GA secretion in the function of Pd-associated proteins increases under stress when the plant needs to provide sink leaves and a growth point with carbohydrates synthesized in mesophyll from source leaves. In general, all the PdAPs presented in Table 1 can be conditionally divided into two unequal groups. The first group includes PdAPs, which can be considered as negative Pd regulators and includes actin, remorin, PDLP1 and 5, GnTL, C1RGP, formins, reticulons, calreticulin. The small second group includes positive Pd regulators such as myosin, SYTA, BGs, PME and NCAPP. Many viral MPs can be also included in this group [132].
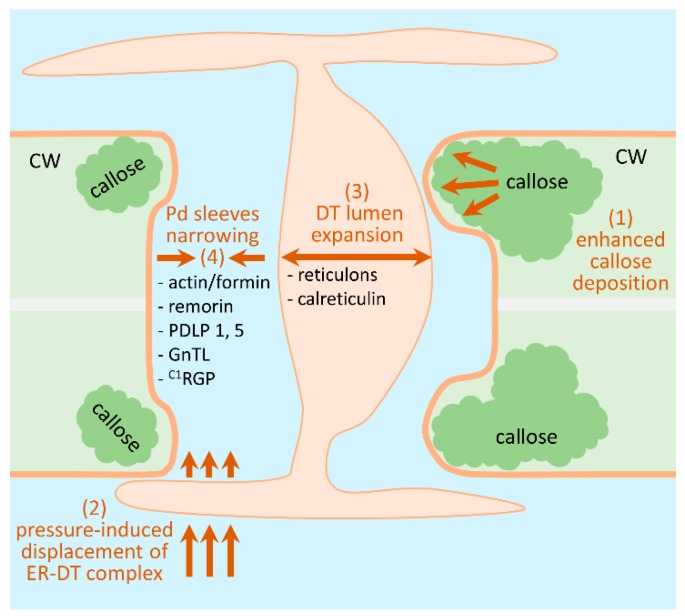
If we proceed from the Pd structure (Figure 1 and Figure 2) along channels in the cell wall that contain the dumbbell-shaped ER DT inside them and ducts, or Pd sleeves, surrounding DT, then there are several ways to block the movement of molecules through these structures (Figure 4).
Figure 4.
Schematic representation of plasmodesmata permeability negative regulation. (1) callose depositions around Pd neck; (2) stress-induced increasing of the intracellular pressure leading to the compression of the ER DT and blocking Pd entrance; (3) ER DT lumen expansion as a result of calreticulin and reticulons deposition; (4) narrowing of the cytoplasmic sleeves.
First, is stimulating callose deposition around Pd [36,37]. Second, by increasing the intracellular pressure, thereby compressing the ER DT and blocking the entrance to Pd in accordance with the model proposed by Park et al. [130]. Third, the deposition of calreticulin and reticulons in the DT lumen suggested by Knox et al. leads to the expansion of the ER DT lumen [57]. Fourth, narrowing the Pd sleeves, as probably occurs through the participation of the actin-formin complex [70], as well as the accumulation of remorin [56], PDLP1 and 5 [75,76,77,78,79], GnTL [72], and C1RGP [66,67,68]. Pd opening, as a process opposite to closing, can include (1) the removal of callose and (2) the displacement of negative regulators from the Pd structure. PME and NCAPP can also have an indirect positive effect on Pd dilation. Since most PdAPs are secreted proteins, modification of the intracellular protein trafficking pathways is also involved in Pd control.
5. Conclusions and Outlook
Although the important role of Pd in the development of tissues and plant organs is universally recognized, the mechanisms for regulating Pd functions after stress impact are still not fully understood. The reactions of mature leaves Pd to stress are controversial due to the nature of the stressful agents, and, most importantly, to the different time points after stress impact. It is generally accepted that stress responses include Pd closure as a result of reversible callose deposition.
Our analysis indicates a variety of protein factors involved in generalized Pd-mediated stress responses. It is still unclear which genes and protein factors encoded by stress-induced genes are activated in source leaf cells to provide a fast and generalized stress response, leading to an increase in Pd conductivity and the outflow of photoassimilates to sink leaves. We assumed that the search for these genes should be carried out among genes with different levels of mRNA accumulation in leaves and roots as well as after stress challenge. The analysis of the PdAPs presented in Table 1 allows to evaluate the role of secreted proteins in the functioning of Pd. The proteomic study of a Pd-enriched Arabidopsis cell wall fraction showed that most PdAPs are secreted proteins. The Pd structure, including its extracellular and endocellular parts, involves both cytoplasmic non-secreted proteins and secreted proteins control, such as in the case of callose metabolism. It was previously believed that the list of cytoplasmic PdAPs located directly in the intracellular cytoplasmic sleeve was limited by actin and myosin. However, PdAPs that have all the properties of proteins secreted by the GA have recently been revealed, though their final localization is in the Pd intracellular space. With the recent discovery of the specific role of t18:0-based sphingolipids in PDLP5 localization in the Pd cytoplasmatic sleeve this mechanism may also be suggested for formins (Figure 3).
The putative mechanisms for the participation of PdAPs in the regulation of Pd presented in Figure 4 are diverse, but ultimately come down to the means of closing/opening the molecule passage through Pd sleeves surrounding the DT.
Acknowledgments
We appreciate the Managing Editors and three anonymous reviewers for their corrections, suggestions, and in-depth comments, which greatly improved the manuscript.
Author Contributions
Y.L.D. created the concept of the manuscript. Y.L.D., N.M.E., E.V.S., T.V.K. wrote, revised and finalized the manuscript. N.M.E., E.V.S., and T.V.K. illustrated the manuscript. All the authors read and approved the manuscript.
Funding
The analysis of protein localization and glycosylation was funded by the Russian Science Foundation (project No. 19-74-20031). The analysis of intra- and intercellular transport of macromolecules was supported by the Russian Foundation for Basic Research (project No. 17-29-08012).
Conflicts of Interest
The authors confirm that there are no known conflict of interest associated with this publication.
References
- 1.Jeandroz S., Lamotte O. Editorial: Plant Responses to Biotic and Abiotic Stresses: Lessons from Cell Signaling. Front. Plant Sci. 2017;8:1772. doi: 10.3389/fpls.2017.01772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aubry E., Dinant S., Vilaine F., Bellini C., Le Hir R. Lateral Transport of Organic and Inorganic Solutes. Plants. 2019;8:20. doi: 10.3390/plants8010020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee J.-Y. Plasmodesmata: A signaling hub at the cellular boundary. Curr. Opin. Plant Biol. 2015;27:133–140. doi: 10.1016/j.pbi.2015.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sevilem I., Yadav S.R., Helariutta Y. Plasmodesmata: Channels for intercellular signaling during plant growth and development. In: Heinlein M., editor. Plasmodesmata: Methods and Protocols. Volume 1217. Springer; New York, NY, USA: 2015. pp. 3–24. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 5.Chung K.P., Zeng Y. An Overview of Protein Secretion in Plant Cells. In: Jiang L., editor. Plant Protein Secretion: Methods and Protocols. Volume 1662. Springer; New York, NY, USA: 2017. pp. 19–32. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 6.Davis D.J., Kang B.-H., Heringer A.S., Wilkop T.E., Drakakaki G. Unconventional Protein Secretion in Plants. In: Pompa A., De Marchis F., editors. Unconventional Protein Secretion: Methods and Protocols. Volume 1459. Springer; New York, NY, USA: 2016. pp. 47–63. Methods in Molecular Biology. [DOI] [PubMed] [Google Scholar]
- 7.Goring D.R., Di Sansebastiano G.P. Protein and membrane trafficking routes in plants: Conventional or unconventional? J. Exp. Bot. 2017;69:1–5. doi: 10.1093/jxb/erx435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bellucci M., De Marchis F., Pompa A. The endoplasmic reticulum is a hub to sort proteins toward unconventional traffic pathways and endosymbiotic organelles. J. Exp. Bot. 2018;69:7–20. doi: 10.1093/jxb/erx286. [DOI] [PubMed] [Google Scholar]
- 9.Fan L., Li R., Pan J., Ding Z., Lin J. Endocytosis and its regulation in plants. Trends Plant Sci. 2015;20:388–397. doi: 10.1016/j.tplants.2015.03.014. [DOI] [PubMed] [Google Scholar]
- 10.Rosquete M.R., Drakakaki G. Plant TGN in the stress response: A compartmentalized overview. Curr. Opin. Plant Biol. 2018;46:122–129. doi: 10.1016/j.pbi.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Zhang L., Xing J., Lin J. At the intersection of exocytosis and endocytosis in plants. New Phytol. 2019 doi: 10.1111/nph.16018. [DOI] [PubMed] [Google Scholar]
- 12.Agrawal G.K., Jwa N.-S., Lebrun M.-H., Job D., Rakwal R. Plant secretome: Unlocking secrets of the secreted proteins. Proteomics. 2010;10:799–827. doi: 10.1002/pmic.200900514. [DOI] [PubMed] [Google Scholar]
- 13.Ding Y., Robinson D.G., Jiang L. Unconventional protein secretion (UPS) pathways in plants. Curr. Opin. Cell Biol. 2014;29:107–115. doi: 10.1016/j.ceb.2014.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Ding Y., Wang J., Wang J., Stierhof Y.-D., Robinson D.G., Jiang L. Unconventional protein secretion. Trends Plant Sci. 2012;17:606–615. doi: 10.1016/j.tplants.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 15.Marchis F.D., Bellucci M., Pompa A. Unconventional pathways of secretory plant proteins from the endoplasmic reticulum to the vacuole bypassing the Golgi complex. Plant Signal. Behav. 2013;8:e25129. doi: 10.4161/psb.25129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickel W., Seedorf M. Unconventional mechanisms of protein transport to the cell surface of eukaryotic cells. Annu. Rev. Cell Dev. Biol. 2008;24:287–308. doi: 10.1146/annurev.cellbio.24.110707.175320. [DOI] [PubMed] [Google Scholar]
- 17.Brunkard J.O., Zambryski P.C. Plasmodesmata enable multicellularity: New insights into their evolution, biogenesis, and functions in development and immunity. Curr. Opin. Plant Biol. 2017;35:76–83. doi: 10.1016/j.pbi.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 18.Ruiz-Medrano R., Xoconostle-Cazares B., Kragler F. The plasmodesmatal transport pathway for homeotic proteins, silencing signals and viruses. Curr. Opin. Plant Biol. 2004;7:641–650. doi: 10.1016/j.pbi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 19.Liu L., Chen X. Intercellular and systemic trafficking of RNAs in plants. Nat. Plants. 2018;4:869–878. doi: 10.1038/s41477-018-0288-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reagan B.C., Ganusova E.E., Fernandez J.C., McCray T.N., Burch-Smith T.M. RNA on the move: The plasmodesmata perspective. Plant Sci. Int. J. Exp. Plant Biol. 2018;275:1–10. doi: 10.1016/j.plantsci.2018.07.001. [DOI] [PubMed] [Google Scholar]
- 21.Cheval C., Faulkner C. Plasmodesmal regulation during plant-pathogen interactions. New Phytol. 2018;217:62–67. doi: 10.1111/nph.14857. [DOI] [PubMed] [Google Scholar]
- 22.Ganusova E.E., Burch-Smith T.M. Review: Plant-pathogen interactions through the plasmodesma prism. Plant Sci. Int. J. Exp. Plant Biol. 2019;279:70–80. doi: 10.1016/j.plantsci.2018.05.017. [DOI] [PubMed] [Google Scholar]
- 23.Otero S., Helariutta Y., Benitez-Alfonso Y. Symplastic communication in organ formation and tissue patterning. Curr. Opin. Plant Biol. 2016;29:21–28. doi: 10.1016/j.pbi.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 24.Julius B.T., Leach K.A., Tran T.M., Mertz R.A., Braun D.M. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017;58:1442–1460. doi: 10.1093/pcp/pcx090. [DOI] [PubMed] [Google Scholar]
- 25.Liesche J. Sucrose transporters and plasmodesmal regulation in passive phloem loading. J. Integr. Plant Biol. 2017;59:311–321. doi: 10.1111/jipb.12548. [DOI] [PubMed] [Google Scholar]
- 26.Liesche J., Gao C., Binczycki P., Andersen S.R., Rademaker H., Schulz A., Martens H.J. Direct Comparison of Leaf Plasmodesma Structure and Function in Relation to Phloem-Loading Type. Plant Physiol. 2019;179:1768–1778. doi: 10.1104/pp.18.01353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Burch-Smith T.M., Stonebloom S., Xu M., Zambryski P.C. Plasmodesmata during development: Re-examination of the importance of primary, secondary, and branched plasmodesmata structure versus function. Protoplasma. 2011;248:61–74. doi: 10.1007/s00709-010-0252-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burch-Smith T.M., Zambryski P.C. Plasmodesmata paradigm shift: Regulation from without versus within. Annu. Rev. Plant Biol. 2012;63:239–260. doi: 10.1146/annurev-arplant-042811-105453. [DOI] [PubMed] [Google Scholar]
- 29.Faulkner C. Plasmodesmata and the symplast. Curr. Biol. 2018;28:R1374–R1378. doi: 10.1016/j.cub.2018.11.004. [DOI] [PubMed] [Google Scholar]
- 30.Patrick J.W., Botha F.C., Birch R.G. Metabolic engineering of sugars and simple sugar derivatives in plants. Plant Biotechnol. J. 2013;11:142–156. doi: 10.1111/pbi.12002. [DOI] [PubMed] [Google Scholar]
- 31.Sager R., Lee J.-Y. Plasmodesmata in integrated cell signalling: Insights from development and environmental signals and stresses. J. Exp. Bot. 2014;65:6337–6358. doi: 10.1093/jxb/eru365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sager R.E., Lee J.-Y. Plasmodesmata at a glance. J. Cell Sci. 2018;131 doi: 10.1242/jcs.209346. [DOI] [PubMed] [Google Scholar]
- 33.Sun Y., Huang D., Chen X. Dynamic regulation of plasmodesmatal permeability and its application to horticultural research. Hortic. Res. 2019;6:47. doi: 10.1038/s41438-019-0129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilsner J., Nicolas W., Rosado A., Bayer E.M. Staying Tight: Plasmodesmal Membrane Contact Sites and the Control of Cell-to-Cell Connectivity in Plants. Annu. Rev. Plant Biol. 2016;67:337–364. doi: 10.1146/annurev-arplant-043015-111840. [DOI] [PubMed] [Google Scholar]
- 35.Levy A., Erlanger M., Rosenthal M., Epel B.L. A plasmodesmata-associated beta-1,3-glucanase in Arabidopsis. Plant J. Cell Mol. Biol. 2007;49:669–682. doi: 10.1111/j.1365-313X.2006.02986.x. [DOI] [PubMed] [Google Scholar]
- 36.Zavaliev R., Ueki S., Epel B.L., Citovsky V. Biology of callose (β-1,3-glucan) turnover at plasmodesmata. Protoplasma. 2011;248:117–130. doi: 10.1007/s00709-010-0247-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu S.-W., Kumar R., Iswanto A.B.B., Kim J.-Y. Callose balancing at plasmodesmata. J. Exp. Bot. 2018;69:5325–5339. doi: 10.1093/jxb/ery317. [DOI] [PubMed] [Google Scholar]
- 38.Ueki S., Citovsky V. Plasmodesmata-associated proteins: Can we see the whole elephant? Plant Signal. Behav. 2014;9:e27899. doi: 10.4161/psb.27899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez-Calvino L., Faulkner C., Walshaw J., Saalbach G., Bayer E., Benitez-Alfonso Y., Maule A. Arabidopsis plasmodesmal proteome. PLoS ONE. 2011;6:e18880. doi: 10.1371/journal.pone.0018880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roberts I.M., Boevink P., Roberts A.G., Sauer N., Reichel C., Oparka K.J. Dynamic changes in the frequency and architecture of plasmodesmata during the sink-source transition in tobacco leaves. Protoplasma. 2001;218:31–44. doi: 10.1007/BF01288358. [DOI] [PubMed] [Google Scholar]
- 41.Tilsner J., Amari K., Torrance L. Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma. 2011;248:39–60. doi: 10.1007/s00709-010-0217-6. [DOI] [PubMed] [Google Scholar]
- 42.Nicolas W.J., Grison M.S., Bayer E.M. Shaping intercellular channels of plasmodesmata: The structure-to-function missing link. J. Exp. Bot. 2017;69:91–103. doi: 10.1093/jxb/erx225. [DOI] [PubMed] [Google Scholar]
- 43.Ross-Elliott T.J., Jensen K.H., Haaning K.S., Wager B.M., Knoblauch J., Howell A.H., Mullendore D.L., Monteith A.G., Paultre D., Yan D., et al. Phloem unloading in Arabidopsis roots is convective and regulated by the phloem-pole pericycle. eLIFE. 2017;6:e24125. doi: 10.7554/eLife.24125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nicolas W.J., Grison M.S., Trépout S., Gaston A., Fouché M., Cordelières F.P., Oparka K., Tilsner J., Brocard L., Bayer E.M. Architecture and permeability of post-cytokinesis plasmodesmata lacking cytoplasmic sleeves. Nat. Plants. 2017;3:17082. doi: 10.1038/nplants.2017.82. [DOI] [PubMed] [Google Scholar]
- 45.Oparka K.J., Roberts A.G., Boevink P., Santa Cruz S., Roberts I., Pradel K.S., Imlau A., Kotlizky G., Sauer N., Epel B. Simple, but not branched, plasmodesmata allow the nonspecific trafficking of proteins in developing tobacco leaves. Cell. 1999;97:743–754. doi: 10.1016/S0092-8674(00)80786-2. [DOI] [PubMed] [Google Scholar]
- 46.Amsbury S., Benitez-Alfonso Y. Tightening the pores to unload the phloem. Nat. Plants. 2019;5:561–562. doi: 10.1038/s41477-019-0450-8. [DOI] [PubMed] [Google Scholar]
- 47.Han X., Huang L.-J., Feng D., Jiang W., Miu W., Li N. Plasmodesmata-Related Structural and Functional Proteins: The Long Sought-After Secrets of a Cytoplasmic Channel in Plant Cell Walls. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20122946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blackman L.M., Overall R.L. Immunolocalisation of the cytoskeleton to plasmodesmata of Chara corallina. Plant J. 1998;14:733–741. doi: 10.1046/j.1365-313x.1998.00161.x. [DOI] [Google Scholar]
- 49.Radford J.E., White R.G. Localization of a myosin-like protein to plasmodesmata. Plant J. Cell Mol. Biol. 1998;14:743–750. doi: 10.1046/j.1365-313x.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- 50.White R.G., Badelt K., Overall R.L., Vesk M. Actin associated with plasmodesmata. Protoplasma. 1994;180:169–184. doi: 10.1007/BF01507853. [DOI] [Google Scholar]
- 51.Baluska F., Samaj J., Napier R., Volkmann D. Maize calreticulin localizes preferentially to plasmodesmata in root apex. Plant J. Cell Mol. Biol. 1999;19:481–488. doi: 10.1046/j.1365-313X.1999.00530.x. [DOI] [PubMed] [Google Scholar]
- 52.Chen M.-H., Tian G.-W., Gafni Y., Citovsky V. Effects of calreticulin on viral cell-to-cell movement. Plant Physiol. 2005;138:1866–1876. doi: 10.1104/pp.105.064386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye C.-M., Chen S., Payton M., Dickman M.B., Verchot J. TGBp3 triggers the unfolded protein response and SKP1-dependent programmed cell death. Mol. Plant Pathol. 2013;14:241–255. doi: 10.1111/mpp.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee J.-Y., Yoo B.-C., Rojas M.R., Gomez-Ospina N., Staehelin L.A., Lucas W.J. Selective trafficking of non-cell-autonomous proteins mediated by NtNCAPP1. Science. 2003;299:392–396. doi: 10.1126/science.1077813. [DOI] [PubMed] [Google Scholar]
- 55.Sheshukova E.V., Komarova T.V., Pozdyshev D.V., Ershova N.M., Shindyapina A.V., Tashlitsky V.N., Sheval E.V., Dorokhov Y.L. The intergenic interplay between aldose 1-epimerase-like protein and pectin methylesterase in abiotic and biotic stress control. Front. Plant Sci. 2017;8:1646. doi: 10.3389/fpls.2017.01646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raffaele S., Bayer E., Lafarge D., Cluzet S., German Retana S., Boubekeur T., Leborgne-Castel N., Carde J.-P., Lherminier J., Noirot E., et al. Remorin, a solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell. 2009;21:1541–1555. doi: 10.1105/tpc.108.064279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Knox K., Wang P., Kriechbaumer V., Tilsner J., Frigerio L., Sparkes I., Hawes C., Oparka K. Putting the Squeeze on Plasmodesmata: A Role for Reticulons in Primary Plasmodesmata Formation. Plant Physiol. 2015;168:1563–1572. doi: 10.1104/pp.15.00668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kriechbaumer V., Botchway S.W., Slade S.E., Knox K., Frigerio L., Oparka K., Hawes C. Reticulomics: Protein-Protein Interaction Studies with Two Plasmodesmata-Localized Reticulon Family Proteins Identify Binding Partners Enriched at Plasmodesmata, Endoplasmic Reticulum, and the Plasma Membrane1. Plant Physiol. 2015;169:1933–1945. doi: 10.1104/pp.15.01153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levy A., Zheng J.Y., Lazarowitz S.G. Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr. Biol. 2015;25:2018–2025. doi: 10.1016/j.cub.2015.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lewis J.D., Lazarowitz S.G. Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc. Natl. Acad. Sci. USA. 2010;107:2491–2496. doi: 10.1073/pnas.0909080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Uchiyama A., Shimada-Beltran H., Levy A., Zheng J.Y., Javia P.A., Lazarowitz S.G. The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front. Plant Sci. 2014;5:584. doi: 10.3389/fpls.2014.00584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yuan C., Lazarowitz S.G., Citovsky V. The Plasmodesmal Localization Signal of TMV MP Is Recognized by Plant Synaptotagmin SYTA. mBio. 2018;9 doi: 10.1128/mBio.01314-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Doxey A.C., Yaish M.W.F., Moffatt B.A., Griffith M., McConkey B.J. Functional divergence in the Arabidopsis beta-1,3-glucanase gene family inferred by phylogenetic reconstruction of expression states. Mol. Biol. Evol. 2007;24:1045–1055. doi: 10.1093/molbev/msm024. [DOI] [PubMed] [Google Scholar]
- 64.Zavaliev R., Levy A., Gera A., Epel B.L. Subcellular dynamics and role of Arabidopsis β-1,3-glucanases in cell-to-cell movement of tobamoviruses. Mol. Plant-Microbe Interact. MPMI. 2013;26:1016–1030. doi: 10.1094/MPMI-03-13-0062-R. [DOI] [PubMed] [Google Scholar]
- 65.Levy A., Guenoune-Gelbart D., Epel B.L. β-1,3-Glucanases. Plant Signal. Behav. 2007;2:404–407. doi: 10.4161/psb.2.5.4334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Burch-Smith T.M., Cui Y., Zambryski P.C. Reduced levels of class 1 reversibly glycosylated polypeptide increase intercellular transport via plasmodesmata. Plant Signal. Behav. 2012;7:62–67. doi: 10.4161/psb.7.1.18636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sagi G., Katz A., Guenoune-Gelbart D., Epel B.L. Class 1 reversibly glycosylated polypeptides are plasmodesmal-associated proteins delivered to plasmodesmata via the golgi apparatus. Plant Cell. 2005;17:1788–1800. doi: 10.1105/tpc.105.031823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zavaliev R., Sagi G., Gera A., Epel B.L. The constitutive expression of Arabidopsis plasmodesmal-associated class 1 reversibly glycosylated polypeptide impairs plant development and virus spread. J. Exp. Bot. 2010;61:131–142. doi: 10.1093/jxb/erp301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cui W., Lee J.-Y. Arabidopsis callose synthases CalS1/8 regulate plasmodesmal permeability during stress. Nat. Plants. 2016;2:16034. doi: 10.1038/nplants.2016.34. [DOI] [PubMed] [Google Scholar]
- 70.Diao M., Ren S., Wang Q., Qian L., Shen J., Liu Y., Huang S. Arabidopsis formin 2 regulates cell-to-cell trafficking by capping and stabilizing actin filaments at plasmodesmata. eLIFE. 2018;7 doi: 10.7554/eLife.36316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Oulehlovï D., Kollï Rovï E., Cifrovï P., Pejchar P., Žï Rskï V., Cvrčkovï F. Arabidopsis Class I Formin FH1 Relocates between Membrane Compartments during Root Cell Ontogeny and Associates with Plasmodesmata. Plant Cell Physiol. 2019;60:1855–1870. doi: 10.1093/pcp/pcz102. [DOI] [PubMed] [Google Scholar]
- 72.Zalepa-King L., Citovsky V. A plasmodesmal glycosyltransferase-like protein. PLoS ONE. 2013;8:e58025. doi: 10.1371/journal.pone.0058025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Benitez-Alfonso Y., Faulkner C., Pendle A., Miyashima S., Helariutta Y., Maule A. Symplastic intercellular connectivity regulates lateral root patterning. Dev. Cell. 2013;26:136–147. doi: 10.1016/j.devcel.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 74.Simpson C., Thomas C., Findlay K., Bayer E., Maule A.J. An Arabidopsis GPI-Anchor Plasmodesmal Neck Protein with Callose Binding Activity and Potential to Regulate Cell-to-Cell Trafficking. Plant Cell. 2009;21:581–594. doi: 10.1105/tpc.108.060145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amari K., Boutant E., Hofmann C., Schmitt-Keichinger C., Fernandez-Calvino L., Didier P., Lerich A., Mutterer J., Thomas C.L., Heinlein M., et al. A Family of Plasmodesmal Proteins with Receptor-Like Properties for Plant Viral Movement Proteins. PLoS Pathog. 2010;6:e1001119. doi: 10.1371/journal.ppat.1001119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bayer E., Thomas C., Maule A. Symplastic domains in the Arabidopsis shoot apical meristem correlate with PDLP1 expression patterns. Plant Signal. Behav. 2008;3:853–855. doi: 10.4161/psb.3.10.6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lim G.-H., Shine M.B., de Lorenzo L., Yu K., Cui W., Navarre D., Hunt A.G., Lee J.-Y., Kachroo A., Kachroo P. Plasmodesmata Localizing Proteins Regulate Transport and Signaling during Systemic Acquired Immunity in Plants. Cell Host Microbe. 2016;19:541–549. doi: 10.1016/j.chom.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 78.Thomas C.L., Bayer E.M., Ritzenthaler C., Fernandez-Calvino L., Maule A.J. Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 2008;6:e7. doi: 10.1371/journal.pbio.0060007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lee J.-Y., Wang X., Cui W., Sager R., Modla S., Czymmek K., Zybaliov B., van Wijk K., Zhang C., Lu H., et al. A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell. 2011;23:3353–3373. doi: 10.1105/tpc.111.087742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vaattovaara A., Brandt B., Rajaraman S., Safronov O., Veidenberg A., Luklová M., Kangasjärvi J., Löytynoja A., Hothorn M., Salojärvi J., et al. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2019;2:56. doi: 10.1038/s42003-019-0306-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wang X., Zhu P., Qu S., Zhao J., Singh P.K., Wang W. Ectodomain of plasmodesmata-localized protein 5 in Arabidopsis: Expression, purification, crystallization and crystallographic analysis. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2017;73:532–535. doi: 10.1107/S2053230X1701250X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ning-Jing L., Tao Z., Zhao-Hui L., Xin C., Hui-Shan G., Bai-Hang J., Yuan-Yuan Z., Guo-Zhu L., Qiang-Hui Z., Yong-Mei Q., et al. Phytosphinganine affects plasmodesmata permeability via facilitating pdlp5-stimulated callose accumulation in arabidopsis. Mol. Plant. 2019 doi: 10.1016/j.molp.2019.10.013. [DOI] [PubMed] [Google Scholar]
- 83.Knox J.P., Benitez-Alfonso Y. Roles and regulation of plant cell walls surrounding plasmodesmata. Curr. Opin. Plant Biol. 2014;22:93–100. doi: 10.1016/j.pbi.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 84.Morvan O., Quentin M., Jauneau A., Mareck A., Morvan C. Immunogold localization of pectin methylesterases in the cortical tissues of flax hypocotyl. Protoplasma. 1998;202:175–184. doi: 10.1007/BF01282545. [DOI] [Google Scholar]
- 85.Zavaliev R., Dong X., Epel B.L. Glycosylphosphatidylinositol (GPI) Modification Serves as a Primary Plasmodesmal Sorting Signal. Plant Physiol. 2016;172:1061–1073. doi: 10.1104/pp.16.01026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mirabet V., Krupinski P., Hamant O., Meyerowitz E.M., Jönsson H., Boudaoud A. The self-organization of plant microtubules inside the cell volume yields their cortical localization, stable alignment, and sensitivity to external cues. PLoS Comput. Biol. 2018;14:e1006011. doi: 10.1371/journal.pcbi.1006011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yuan C., Lazarowitz S.G., Citovsky V. Identification of a Functional Plasmodesmal Localization Signal in a Plant Viral Cell-To-Cell-Movement Protein. mBio. 2016;7:e02052-15. doi: 10.1128/mBio.02052-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yuan C., Lazarowitz S.G., Citovsky V. Identification of Plasmodesmal Localization Sequences in Proteins in Planta. J. Vis. Exp. JoVE. 2017 doi: 10.3791/55301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pitzalis N., Heinlein M. The roles of membranes and associated cytoskeleton in plant virus replication and cell-to-cell movement. J. Exp. Bot. 2017;69:117–132. doi: 10.1093/jxb/erx334. [DOI] [PubMed] [Google Scholar]
- 90.Reymond P., Kunz B., Paul-Pletzer K., Grimm R., Eckerskorn C., Farmer E.E. Cloning of a cDNA encoding a plasma membrane-associated, uronide binding phosphoprotein with physical properties similar to viral movement proteins. Plant Cell. 1996;8:2265–2276. doi: 10.1105/tpc.8.12.2265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Verchot J. The ER quality control and ER associated degradation machineries are vital for viral pathogenesis. Front. Plant Sci. 2014;5 doi: 10.3389/fpls.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baluska F., Cvrcková F., Kendrick-Jones J., Volkmann D. Sink plasmodesmata as gateways for phloem unloading. Myosin VIII and calreticulin as molecular determinants of sink strength? Plant Physiol. 2001;126:39–46. doi: 10.1104/pp.126.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Laporte C., Vetter G., Loudes A.-M., Robinson D.G., Hillmer S., Stussi-Garaud C., Ritzenthaler C. Involvement of the secretory pathway and the cytoskeleton in intracellular targeting and tubule assembly of Grapevine fanleaf virus movement protein in tobacco BY-2 cells. Plant Cell. 2003;15:2058–2075. doi: 10.1105/tpc.013896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Christensen A., Svensson K., Thelin L., Zhang W., Tintor N., Prins D., Funke N., Michalak M., Schulze-Lefert P., Saijo Y., et al. Higher plant calreticulins have acquired specialized functions in Arabidopsis. PLoS ONE. 2010;5:e11342. doi: 10.1371/journal.pone.0011342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wyatt S.E., Tsou P.-L., Robertson D. Expression of the high capacity calcium-binding domain of calreticulin increases bioavailable calcium stores in plants. Transgenic Res. 2002;11:1–10. doi: 10.1023/A:1013917701701. [DOI] [PubMed] [Google Scholar]
- 96.Xoconostle-Cázares B., Xiang Y., Ruiz-Medrano R., Wang H.L., Monzer J., Yoo B.C., McFarland K.C., Franceschi V.R., Lucas W.J. Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science. 1999;283:94–98. doi: 10.1126/science.283.5398.94. [DOI] [PubMed] [Google Scholar]
- 97.Lucas W.J., Ham B.-K., Kim J.-Y. Plasmodesmata-bridging the gap between neighboring plant cells. Trends Cell Biol. 2009;19:495–503. doi: 10.1016/j.tcb.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 98.Taoka K.-I., Ham B.-K., Xoconostle-Cázares B., Rojas M.R., Lucas W.J. Reciprocal phosphorylation and glycosylation recognition motifs control NCAPP1 interaction with pumpkin phloem proteins and their cell-to-cell movement. Plant Cell. 2007;19:1866–1884. doi: 10.1105/tpc.107.052522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., Mueller C., Frigerio L., Hawes C. Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell. 2010;22:1333–1343. doi: 10.1105/tpc.110.074385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kriechbaumer V., Maneta-Peyret L., Fouillen L., Botchway S.W., Upson J., Hughes L., Richardson J., Kittelmann M., Moreau P., Hawes C. The odd one out: Arabidopsis reticulon 20 does not bend ER membranes but has a role in lipid regulation. Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-20840-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nziengui H., Bouhidel K., Pillon D., Der C., Marty F., Schoefs B. Reticulon-like proteins in Arabidopsis thaliana: Structural organization and ER localization. FEBS Lett. 2007;581:3356–3362. doi: 10.1016/j.febslet.2007.06.032. [DOI] [PubMed] [Google Scholar]
- 102.Wakana Y., Koyama S., Nakajima K., Hatsuzawa K., Nagahama M., Tani K., Hauri H.-P., Melançon P., Tagaya M. Reticulon 3 is involved in membrane trafficking between the endoplasmic reticulum and Golgi. Biochem. Biophys. Res. Commun. 2005;334:1198–1205. doi: 10.1016/j.bbrc.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 103.Moore A.E., Stone B.A. Effect of infection with TMV and other viruses on the level of a β-1,3-glucan hydrolase in leaves of Nicotiana glutinosa. Virology. 1972;50:791–798. doi: 10.1016/0042-6822(72)90433-3. [DOI] [PubMed] [Google Scholar]
- 104.Dorokhov Y.L., Komarova T.V., Petrunia I.V., Frolova O.Y., Pozdyshev D.V., Gleba Y.Y. Airborne signals from a wounded leaf facilitate viral spreading and induce antibacterial resistance in neighboring plants. PLoS Pathog. 2012;8:e1002640. doi: 10.1371/journal.ppat.1002640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.den Hollander P.W., Kieper S.N., Borst J.W., van Lent J.W.M. The role of plasmodesma-located proteins in tubule-guided virus transport is limited to the plasmodesmata. Arch. Virol. 2016;161:2431–2440. doi: 10.1007/s00705-016-2936-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Grison M.S., Brocard L., Fouillen L., Nicolas W., Wewer V., Dörmann P., Nacir H., Benitez-Alfonso Y., Claverol S., Germain V., et al. Specific Membrane Lipid Composition Is Important for Plasmodesmata Function in Arabidopsis. Plant Cell. 2015;27:1228–1250. doi: 10.1105/tpc.114.135731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Caillaud M.-C., Wirthmueller L., Sklenar J., Findlay K., Piquerez S.J.M., Jones A.M.E., Robatzek S., Jones J.D.G., Faulkner C. The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 2014;10:e1004496. doi: 10.1371/journal.ppat.1004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ali U., Li H., Wang X., Guo L. Emerging Roles of Sphingolipid Signaling in Plant Response to Biotic and Abiotic Stresses. Mol. Plant. 2018;11:1328–1343. doi: 10.1016/j.molp.2018.10.001. [DOI] [PubMed] [Google Scholar]
- 109.Iswanto A.B.B., Kim J.-Y. Lipid Raft, Regulator of Plasmodesmal Callose Homeostasis. Plants. 2017;6:15. doi: 10.3390/plants6020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Yan D., Yadav S.R., Paterlini A., Nicolas W.J., Petit J.D., Brocard L., Belevich I., Grison M.S., Vaten A., Karami L., et al. Sphingolipid biosynthesis modulates plasmodesmal ultrastructure and phloem unloading. Nat. Plants. 2019;5:604–615. doi: 10.1038/s41477-019-0429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Contreras F.-X., Ernst A.M., Haberkant P., Björkholm P., Lindahl E., Gönen B., Tischer C., Elofsson A., von Heijne G., Thiele C., et al. Molecular recognition of a single sphingolipid species by a protein’s transmembrane domain. Nature. 2012;481:525–529. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- 112.Peiró A., Martínez-Gil L., Tamborero S., Pallás V., Sánchez-Navarro J.A., Mingarro I. The Tobacco mosaic virus movement protein associates with but does not integrate into biological membranes. J. Virol. 2014;88:3016–3026. doi: 10.1128/JVI.03648-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Faulkner C., Akman O.E., Bell K., Jeffree C., Oparka K. Peeking into Pit Fields: A Multiple Twinning Model of Secondary Plasmodesmata Formation in Tobacco. Plant Cell. 2008;20:1504–1518. doi: 10.1105/tpc.107.056903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dorokhov Y.L., Sheshukova E.V., Komarova T.V. Methanol in Plant Life. Front. Plant Sci. 2018;9:1623. doi: 10.3389/fpls.2018.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Chen M.H., Sheng J., Hind G., Handa A.K., Citovsky V. Interaction between the tobacco mosaic virus movement protein and host cell pectin methylesterases is required for viral cell-to-cell movement. EMBO J. 2000;19:913–920. doi: 10.1093/emboj/19.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Dorokhov Y.L., Mäkinen K., Frolova O.Y., Merits A., Saarinen J., Kalkkinen N., Atabekov J.G., Saarma M. A novel function for a ubiquitous plant enzyme pectin methylesterase: The host-cell receptor for the tobacco mosaic virus movement protein. FEBS Lett. 1999;461:223–228. doi: 10.1016/S0014-5793(99)01447-7. [DOI] [PubMed] [Google Scholar]
- 117.Chen M.-H., Citovsky V. Systemic movement of a tobamovirus requires host cell pectin methylesterase. Plant J. Cell Mol. Biol. 2003;35:386–392. doi: 10.1046/j.1365-313X.2003.01818.x. [DOI] [PubMed] [Google Scholar]
- 118.Amsbury S., Kirk P., Benitez-Alfonso Y. Emerging models on the regulation of intercellular transport by plasmodesmata-associated callose. J. Exp. Bot. 2017;69:105–115. doi: 10.1093/jxb/erx337. [DOI] [PubMed] [Google Scholar]
- 119.Shi X., Han X., Lu T. Callose synthesis during reproductive development in monocotyledonous and dicotyledonous plants. Plant Signal. Behav. 2016;11:e1062196. doi: 10.1080/15592324.2015.1062196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Drakakaki G., van de Ven W., Pan S., Miao Y., Wang J., Keinath N.F., Weatherly B., Jiang L., Schumacher K., Hicks G., et al. Isolation and proteomic analysis of the SYP61 compartment reveal its role in exocytic trafficking in Arabidopsis. Cell Res. 2012;22:413–424. doi: 10.1038/cr.2011.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kulich I., Vojtíková Z., Sabol P., Ortmannová J., Neděla V., Tihlaříková E., Žárský V. Exocyst Subunit EXO70H4 Has a Specific Role in Callose Synthase Secretion and Silica Accumulation. Plant Physiol. 2018;176:2040–2051. doi: 10.1104/pp.17.01693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Schneider R., Hanak T., Persson S., Voigt C.A. Cellulose and callose synthesis and organization in focus, what’s new? Curr. Opin. Plant Biol. 2016;34:9–16. doi: 10.1016/j.pbi.2016.07.007. [DOI] [PubMed] [Google Scholar]
- 123.Vatén A., Dettmer J., Wu S., Stierhof Y.-D., Miyashima S., Yadav S.R., Roberts C.J., Campilho A., Bulone V., Lichtenberger R., et al. Callose biosynthesis regulates symplastic trafficking during root development. Dev. Cell. 2011;21:1144–1155. doi: 10.1016/j.devcel.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 124.Langeveld S.M.J., Vennik M., Kottenhagen M., van Wijk R., Buijk A., Kijne J.W., de Pater S. Glucosylation Activity and Complex Formation of Two Classes of Reversibly Glycosylated Polypeptides. Plant Physiol. 2002;129:278–289. doi: 10.1104/pp.010720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Delgado I.J., Wang Z., de Rocher A., Keegstra K., Raikhel N.V. Cloning and characterization of AtRGP1. A reversibly autoglycosylated arabidopsis protein implicated in cell wall biosynthesis. Plant Physiol. 1998;116:1339–1350. doi: 10.1104/pp.116.4.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Dhugga K.S., Ulvskov P., Gallagher S.R., Ray P.M. Plant polypeptides reversibly glycosylated by UDP-glucose. Possible components of Golgi beta-glucan synthase in pea cells. J. Biol. Chem. 1991;266:21977–21984. [PubMed] [Google Scholar]
- 127.Dhugga K.S., Tiwari S.C., Ray P.M. A reversibly glycosylated polypeptide (RGP1) possibly involved in plant cell wall synthesis: Purification, gene cloning, and trans-Golgi localization. Proc. Natl. Acad. Sci. USA. 1997;94:7679–7684. doi: 10.1073/pnas.94.14.7679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Rautengarten C., Ebert B., Herter T., Petzold C.J., Ishii T., Mukhopadhyay A., Usadel B., Scheller H.V. The Interconversion of UDP-Arabinopyranose and UDP-Arabinofuranose Is Indispensable for Plant Development in Arabidopsis. Plant Cell. 2011;23:1373–1390. doi: 10.1105/tpc.111.083931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Drakakaki G., Zabotina O., Delgado I., Robert S., Keegstra K., Raikhel N. Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 2006;142:1480–1492. doi: 10.1104/pp.106.086363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Park K., Knoblauch J., Oparka K., Jensen K.H. Controlling intercellular flow through mechanosensitive plasmodesmata nanopores. Nat. Commun. 2019;10:3564. doi: 10.1038/s41467-019-11201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Maule A.J., Benitez-Alfonso Y., Faulkner C. Plasmodesmata-membrane tunnels with attitude. Curr. Opin. Plant Biol. 2011;14:683–690. doi: 10.1016/j.pbi.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 132.Reagan B.C., Burch-Smith T.M. Viruses Reveal the Secrets of Plasmodesmal Cell Biology. Mol. Plant-Microbe Interact. MPMI. 2019 doi: 10.1094/MPMI-07-19-0212-FI. [DOI] [PubMed] [Google Scholar]