Abstract
Zanthoxylum schinifolium and Zanthoxylum piperitum are the sources of the well-known traditional Korean herbal medicines “sancho” (prickly ash) and “chopi” (Korean pepper), respectively. Sancho and chopi are often indiscriminately mixed due to the similar appearance of the herbal materials when used as spices and herbal medicines. Moreover, commercial sancho and chopi products often contain adulterants, which is insufficient to ensure food efficacy and safety. In this study, we developed hypervariable insertion/deletion (InDel) markers to distinguish between sancho and chopi products by comparing the complete chloroplast genome sequences of four Zanthoxylum species deposited in the National Center for Biotechnology Information (NCBI) GenBank. Comparative analyses of the nucleotide diversity (Pi) of these Zanthoxylum genomes revealed four hypervariable divergent sites (trnH-psbA, psbZ-trnG, trnfM-rps14, and trnF-ndhK) with Pi > 0.025 among 520 windows. Of these four regions, including two genic and two intergenic regions, only psbZ-trnG yielded accurate PCR amplification results between commercial sancho and chopi products from the Korean herbal medicine market. We therefore selected psbZ-trnG, an InDel-variable locus with high discriminatory powers, as a candidate DNA barcode locus. This InDel marker could be used as a valuable, simple, and efficient tool for identifying these medicinal herbs, thereby increasing the safety of these spices and herbal materials in the food market.
Keywords: chloroplast genome, chopi, InDel markers, nucleotide diversity, sancho, Zanthoxylum piperitum, Zanthoxylum schinifolium
1. Introduction
Zanthoxylum plants are thorny, deciduous shrubs and small trees with dense foliage, prickly trunks, and branches bearing edible fruits and leaves with a strong, pungent taste resembling lemon, anise, or mint. These plants, belonging to the Rutaceae family, comprise 250 species that are native to warm temperate and subtropical regions [1,2]. While members of Zanthoxylum are commonly found in the Himalayan region, they also occur in Central, South, Southeast, and East Asia, and some species are found in America and Africa [3,4,5]. Many of these species are used as traditional medicines to treat human and animal diseases in Africa, Asia, and South America [6,7]. Among these, Zanthoxylum schinifolium (“sancho” in Korean, “huajiao” in Chinese) and Zanthoxylum piperitum (“chopi” in Korean) are important traditional medicinal herbs for treating stomachache, helminths, and skin disorders in Korea and China [8,9,10] that are also used as spices. Korean Flora describes seven Zanthoxylum species from Korea, including Z. schinifolium var. inermis (Nakai) T.B. Lee, Z. piperitum f. pubescens (Nakai) W.T. Lee, Z. schinifolium SieboldandZucc., Z. piperitum (L.) DC., Z. schinifolium f. microphyllum (Nakai) W.T. Lee, Zanthoxylum planispinum SieblodandZucc., and Zanthoxylum coreanum Nakai [11,12,13]. Z. schinifolium and Z. piperitum are the earliest cultivated trees in southern regions of Korea. These trees are of major agricultural importance as sources of spices and traditional medicines [9,10]. Whereas Z. schinifolium thorns are arranged in an alternating pattern on branches, Z. piperitum thorns are arranged in an opposing pattern [11]. However, it is difficult to visually discriminate between dried and powdered herbal products made from Z. schinifolium and Z. piperitum. Furthermore, commercial herbal products commonly include a mixture of Z. schinifolium and Z. piperitum. Since these plants are used in herbal medicines and as health food supplements, accurate methods should be developed to identify and characterize the two species.
Several studies have focused on developing molecular markers that can discriminate between various Zanthoxylum species used as traditional medicines in different countries. These molecular markers include amplified fragment length polymorphism (AFLP) markers for distinguishing between Zanthoxylum acanthopodium and Zanthoxylum oxyphyllum [14], sequence-related amplified polymorphism (SRAP) markers [15], internal transcribed spacer (ITS) rDNA-specific markers [16], and ISSR markers [17]. However, the use of these markers is limited by population dynamics and their reproducibility as diagnostic markers in the food market. With the improvement of next-generation sequencing (NGS) technology, chloroplast (cp) genome assembly has become more accessible than Sanger sequencing. The development of molecular markers has also become cost effective through comparisons of cp genomes. The complete cp genomes of several Zanthoxylum species have been sequenced by de novo assembly using a small amount of whole-genome sequencing data [18,19,20].
In the current study, we compared the published cp genome sequences of four Zanthoxylum species and searched for species-specific regions with hypervariable nucleotide diversity amongmembers of Zanthoxylum. Our aim was to develop interspecies insertion/deletion (InDel) markers that could be used to discriminate between Z. schinifolium and Z. piperitum and prevent the indiscriminate mixing of these two materials in commercial products.
2. Materials and Methods
2.1. Comparison of Cp Genomes and Identification of Hypervariable Loci
All cp genome sequences in plants of the Zanthoxylum genus with complete genome sequence information were downloaded from GenBank (Table 1). The sequences were aligned using the Clustal W algorithm from MEGA7.0 [21] and CLC viewer 8.0 software (CLC bio, Aarhus, Denmark). The gene distribution patterns and similarities in the Zanthoxylum cp genomes were compared and visualized using mVISTA software (http://genome.lbl.gov/vista/mvista/submit.shtml) in Shuffle-LAGAN mode with the annotated Z. piperitum KT153018 cp genome as a reference. The variability of the aligned genomes was evaluated using the sliding window method with DnaSP ver. 5.0. software [22]. The window size was set to 600 base pairs (bp), the typical length of DNA markers. The step size was set to 300bp for relatively accurate positioning of hypervariable InDels. Only regions with a nucleotide diversity (Pi) value of >0.025 were considered. Hypervariable sites and genetic distance in the cp genome were calculated using MEGA 7.0. The InDel events were checked manually based on the aligned sequence matrix.
Table 1.
Chloroplast (cp) genomes of the four Zanthoxylum species used in this study.
| No. | Species | Research Group | GenBank Accession Number | Sequence Length (bp) |
|---|---|---|---|---|
| 1 | Z. schinifolium | Crop Science and Biotechnology, Department of Plant Sciences, Seoul National University, Korea | KT321318 | 158,963 |
| 2 | Z. piperitum | KT153018 | 158,154 | |
| 3 | Z. simulans | College of Forestry, Northwest A&F University, China | MF716524 | 158,461 |
| 4 | Z. sp. | MF716521 | 158,572 |
2.2. Sample Collection and Genomic DNA Isolation
Table 2 lists the Z. schinifolium and Z. piperitum collections from the National Institute of Biological Resources used in this study. Thirteen spice and powdered herbal materials (ten sancho and six chopi samples) were purchased from verified local market sources in Korea and China (Table 3). Species identification was performed by the National Institute of Biological Resources, Korea. Prior to total genomic DNA extraction, 50mg (dry weight) of each sample was added to a tube filled with stainless steel beads (2.38 mm in diameter) from a PowerPlantPro DNA Isolation Kit (Qiagen, Valencia, CA, USA), and the mixture was homogenized in a Precellys® Evolution homogenizer (Bertin Technologies, Montigny-le-Breonneux, France). Genomic DNA was extracted from the collected samples using the PowerPlantPro DNA Isolation Kit according to the manufacturer’s instructions.
Table 2.
Z. schinifolium and Z. piperitum samples used in this study.
| No. | Scientific Name | Common Name | Collection Site | Collection Number |
|---|---|---|---|---|
| 1 | Zanthoxylum schinifolium | Sanchonamu | Hyogok-ri, Ganjeon-myeon, Gurye-gun, Jeollanam-do, Korea | NIBRVP0000278863 |
| 2 | Galcheon-ri, Seo-myeon, Yangyang-gun, Gangwon-do, Korea | NIBRVP0000448009 | ||
| 3 | Daebubuk-dong, Danwon-gu, Ansan-si, Gyeonggi-do, Korea | NIBRVP0000538396 | ||
| 4 | Wondeok-ri, Jincheon-eup, Jincheon-gun, Chungcheongbuk-do, Korea | NIBRVP0000538545 | ||
| 5 | Sucheol-ri, Punggi-eup, Yeongju-si, Gyeongsangbuk-do, Korea | NIBRVP0000538696 | ||
| 6 | Cheongsu-ri, Hangyeong-myeon, Jeju-si, Jeju-do, Korea | NIBRVP0000538748 | ||
| 7 | Gujora-ri, Irun-myeon, Geoje-si, Gyeongsangnam-do, Korea | NIBRVP0000538812 | ||
| 8 | Zanthoxylum piperitum | Chopinamu | Hangyeong-myeon, Jeju-si, Jeju-do, Korea | NIBRVP0000354345 |
| 9 | Guseong-ri, Jugwang-myeon, Goseong-gun, Gangwon-do, Korea | NIBRVP0000470134 | ||
| 10 | Seongu-ri, Onjeong-myeon, Uljin-gun, Gyeongsangbuk-do, Korea | NIBRVP0000538333 | ||
| 11 | Jungjang-ri, Anmyeon-eup, Taean-gun, Chungcheongnam-do, Korea | NIBRVP0000538652 | ||
| 12 | Cheongsu-ri, Hangyeong-myeon, Jeju-si, Jeju-do, Korea | NIBRVP0000538652 | ||
| 13 | Husapo-ri, Bubuk-myeon, Miryang-si, Gyeongsangnam-do, Korea | NIBRVP0000538646 |
Table 3.
The 16 commercial sancho and chopi spices and herbal products used in this study.
| No. | Markets | Material Component | Origin |
|---|---|---|---|
| 1 | A | Dried or powdered seeds and pericarps: sancho products (50–300g) |
China |
| 2 | B | China | |
| 3 | C | China | |
| 4 | D | Korea | |
| 5 | E | China | |
| 6 | F | China | |
| 7 | G | China | |
| 8 | H | China | |
| 9 | I | Korea | |
| 10 | J | Korea | |
| 11 | K | Dried or powdered seeds and pericarps: chopi products (20–300g) |
Korea |
| 12 | L | Korea | |
| 13 | M | Korea | |
| 14 | N | Korea | |
| 15 | O | Korea | |
| 16 | P | Korea |
2.3. Development and Validation of the InDel Molecular Marker
To validate interspecies polymorphisms within the cp genomes and to develop DNA genetic markers for identifying these four Zanthoxylum species, primers were designed using Primer 3 Plus (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi), and National Center for Biotechnology Information (NCBI) Primer-BLAST online tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) was performed based on the mutational hotspot regions (hypervariable regions) found in these Zanthoxylum cp genomes. PCR amplifications were performed in a reaction volume of 50 μL containing 5 μL 10× Ex Taq buffer (with MgCl2), 4 μL dNTP mixture (each 2.5 mM), Ex Taq (5 U/μL) (TaKaRa Bio, Ostu, Japan), 10 ng genomic DNA templates, and 1 μL (10 pM) forward and reverse primers. The mixtures were denatured at 95 °C for 5 min and amplified for 40 cycles at 95 °C for 30 s, 55 °C for 20 s, and 72 °C for 30 s, with a final extension at 72 °C for 5 min. To detect PCR amplicons, the PCR products were separated by capillary electrophoresis (QIAxcel Advanced System, Qiagen, Hilden, Germany) using a QIAxcel DNA High Resolution Kit via the 0M500 method (Qiagen). The target DNA was extracted and purified using a MinElute PCR Purification Kit (Qiagen). Purified PCR products were sent to CosmoGenetech for sequencing (Seoul, Korea) with both forward and reverse primers. The sequencing results were analyzed by BLAST searches of the GenBank database. Sequence alignment and data visualization were carried out with CLC sequence viewer 8.0.
2.4. RFLP Analysis to Identify Suitable InDel Markers
PCR was performed using universal primers for DNA barcodes within trnH-psbA: forward primer 5′-GTTATGCATGAACGTAATGCTC-3′ and reverse primer 5′-CGCGCATGGTGGATTCACAATCC-3′ (approximate product size: 401 bp). The PCR was conducted in a 50 μL reaction mixture containing 5 μL 10× Ex Taq buffer (with MgCl2), 4 μL dNTP mixture (each 2.5 mM), Ex Taq® (5 U/μL) (TaKaRa Bio, Ostu, Japan), 10 ng genomic DNA template, and 1 μL (10 pM) forward and reverse primers. The mixtures were denatured at 94 °C for 5 min and amplified for 30 cycles of 94 °C for 60 s, 55 °C for 60 s, and 72 °C for 90 s, with a final extension at 72 °C for 7 min. The PCR-amplified products were separated by electrophoresis on 1.5% agarose gels for 30 min.
A 2 μL aliquot of each PCR-amplified trnH-psbA product (concentration 0.6 μg/μL to 1 μg/μL) was digested in 2 μL of 10× Cut Smart buffer, two units of PleI restriction enzyme (New England Biolabs, Ipswich, MA; NEB), and 15.8 μL distilled H2O in a final volume of 20 μL, followed by incubation at 37 °C for 20 min and inactivation at 65 °C for 20 min. The digested fragments were separated by electrophoresis on 1.5% agarose gels stained with ethidium bromide, and the fragment patterns were visualized under UV light.
3. Results
3.1. Comparative Analysis of the Cp Genomes of Various Zanthoxylum Species
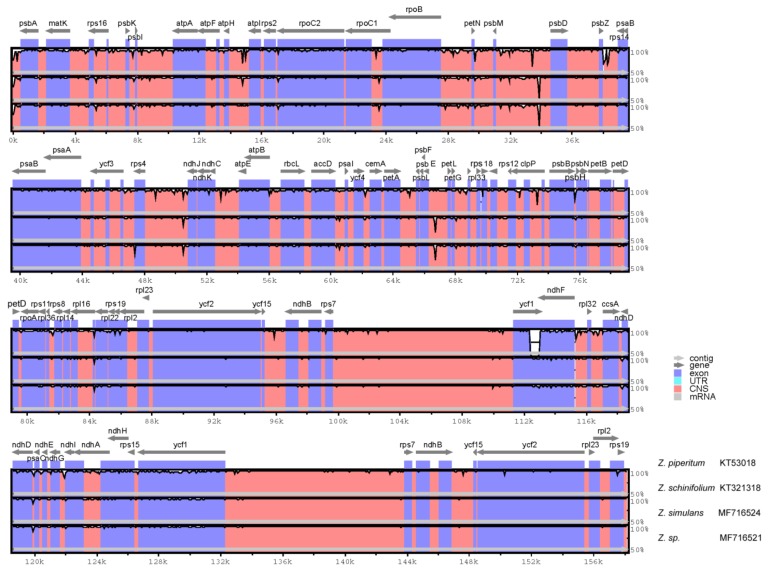
To investigate the level of sequence divergence among Zanthoxylum cp genomes, we performed a comparative analysis of three Zanthoxylum cp genomes with mVISTA using the annotated Z. piperitum sequence as a reference (Figure 1). The cp genomes of the Zanthoxylum species showed high sequence similarity, with identities of <90% in only a few regions, pointing to a high level of conservation among the cp genomes. However, sliding window analysis using DnaSP detected highly variable regions in the Zanthoxylum cp genomes (Figure 2). We calculated the nucleotide diversity (Pi) values in the four Zanthoxylum cp genomes as an indicator of divergence at the sequence level. The large single-copy (LSC) regions and small single-copy (SSC) regions were more divergent than the inverted repeat (IR) regions.
Figure 1.
Comparison of the four Zanthoxylum cp genomes using mVISTA. The complete cp genomes of four Zanthoxylum species obtained from GenBank were compared, with Z. piperitum used as a reference. Purple blocks: conserved genes; pink blocks: conserved non-coding sequences (CNS). White represents regions with high levels of sequence variation among the four Zanthoxylum species.
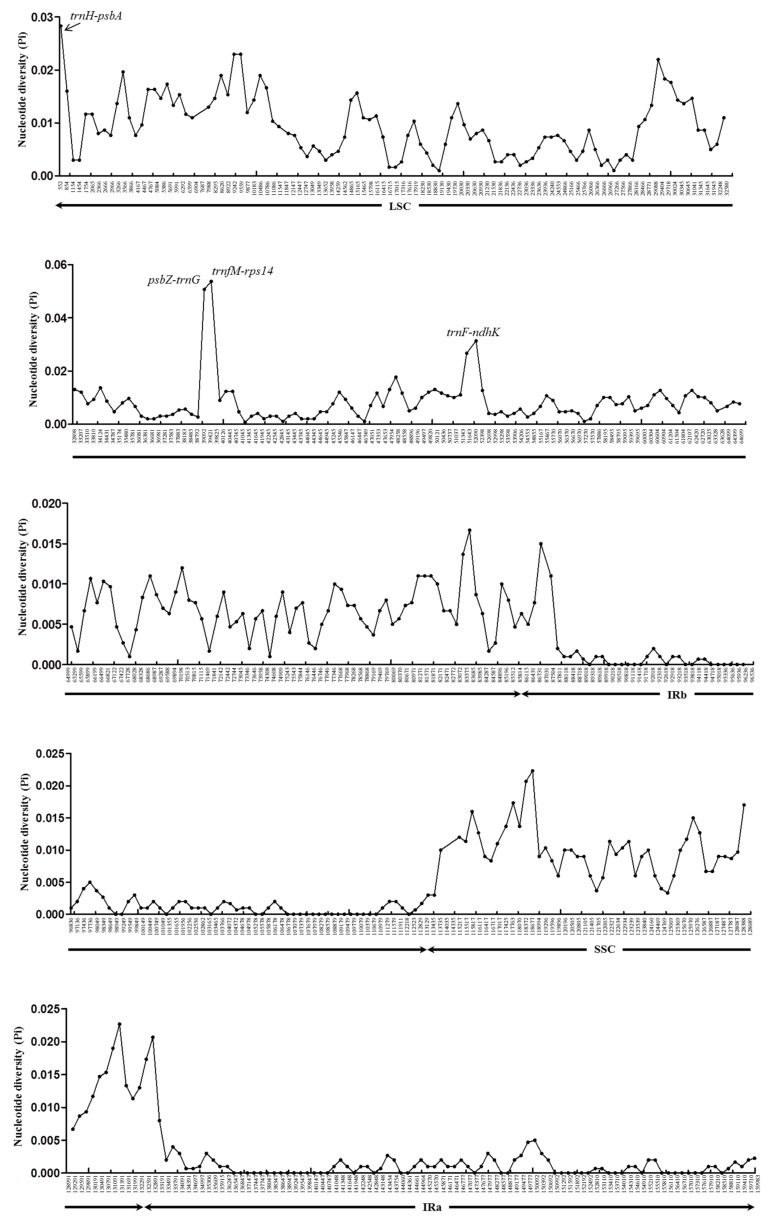
Figure 2.
Sliding window analysis of the whole cp genomes of four Zanthoxylum species (window size: 600 bp; step size: 300 bp). X-axis: position of the midpoint of a window, y-axis: nucleotide diversity (Pi) of each window.
Among the 520 windows examined, IR regions were more conserved than LSC and SSC regions, with average Pi values of 0.0015 and 0.0012 for IRa and IRb, respectively (for regions other than those with a Pi value = 0). The Pi values for the LSC regions averaged 0.0082, whereas the SSC regions had a Pi value of 0.0103, and the average Pi value for all regions was 0.00609. Four mutational hotspots in the cp genomes showed markedly higher Pi values (>0.025), including two intergenic regions (trnH-psbA, 0.02833; psbZ-trnG, 0.05067) and two genic regions (trnfM-rps14, 0.05367; trnF-ndhK, 0.02667 and 0.03133) (Figure 2). Although the SSC and IR regions were generally highly conserved, the four regions located in the LSC regions were particularly divergent.
3.2. Development of InDel Markers to Discriminate between Z. Schinifolium and Z. Piperitum
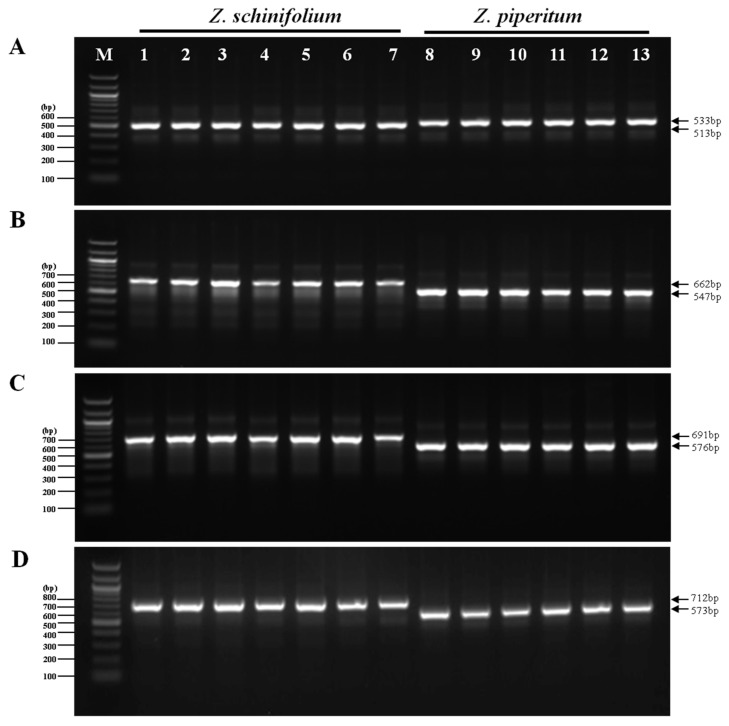
Based on multiple alignments of complete cp genome sequences, we selected the four most highly variable InDel loci as candidate DNA markers (Table 4). We confirmed these four InDel regions by PCR amplification and sequencing and investigated their utility for discriminating between Z. schinifolium and Z. piperitum (Figure 3). We produced four markers (ZanID1, ZanID2, ZanID3, and ZanID4) that were specific to Z. schinifolium and Z. piperitum and were derived from long InDels in the intergenic regions psbZ-trnG, trnfM-rps14, and trnF-ndhK, respectively. ZanID1 was derived from 11, 3, and 6bp InDels in the trnH-psbA locus and was specific to Z. schinifolium and Z. piperitum (Figure 3). ZanID2 was derived from 19, 28, and 5 bp InDels and 40 and 23 bp tandem repeats (TRs) in the psbZ-trnG locus. ZanID3 was derived from 19, 28, 39, and 6 bp InDels and a 23 bp TR in trnfM-rps14. ZanID4 was derived from 32, 6, 59, and 50 bp InDels and an 8 bp TR in trnF-ndhK.
Table 4.
Primers used to amplify and sequence four highly variable loci.
| Marker Name |
Locus | Location | Forward Primer (5′ to 3′) |
Reverse Primer (5′ to 3′) |
Product Range (bp) | Mean Pairwise Distance |
|---|---|---|---|---|---|---|
| ZanID1 | trnH-psbA | 35..578 | GATTCACAATCCACTGCCTTGAT | GCTCATAACTTCCCTCTAGACCT | 513–533 | 0.02833 |
| ZanID2 | psbZ-trnG | 38882..39545 | GTGGGTATCCTTAATTCTCTCATC | GATACTCTCTTCAGGGTAATTCCA | 547–662 | 0.05067 |
| ZanID3 | trnfM-rps14 | 39040..39730 | GGAGGGATCAAACTTCTGGAAC | AGAACCACTATACTATCACGGTCA | 576–691 | 0.05367 |
| ZanID4 | trnF-ndhK | 51439..52158 | GATTTGAGCAAGGAATCCCCATTTG | TCTTCATTTTGACCCGATATTCAA | 573–712 | 0.02667 0.03133 |
Figure 3.
Gel profiles of fragments amplified from seven Z. schinifolium and six Z. piperitum samples using four pairs of primers derived from the trnH-psbA (A), psbZ-trnG (B), trnfM-rps14 (C), and trnF-ndhK (D) loci in the four Zanthoxylum cp genomes. M: 100 bp ladder; 1–7: amplicons from Z. schinifolium DNA; 8–13: amplicons from Z. piperitum DNA.
3.3. Utilization of InDel Markers in the Korean Food Market
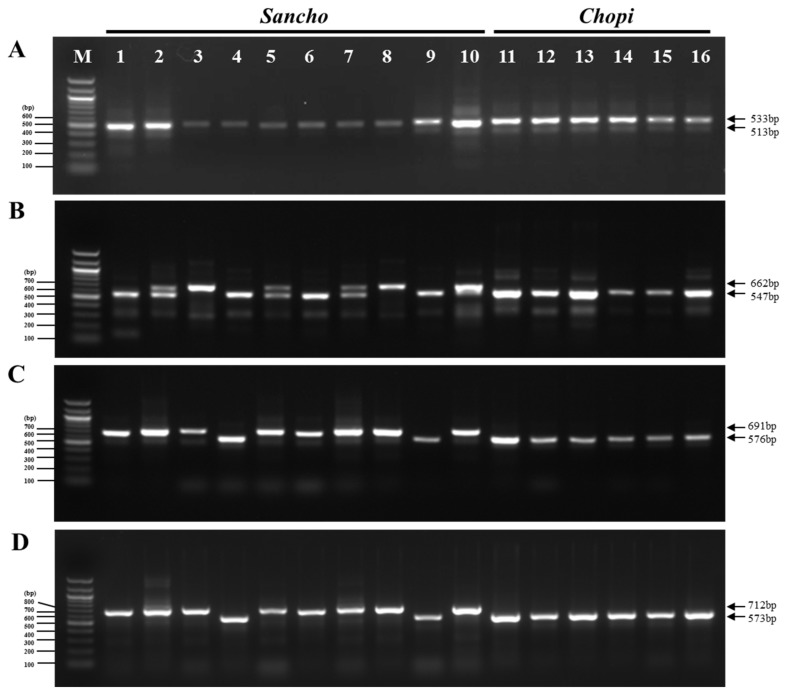
To validate the utility of our newly developed markers to identify commercial dried herbal materials, we extracted genomic DNA from powdered or dried samples of ten sancho and six chopi products and amplified them by PCR using the newly developed primers (Table 4). The banding patterns of the ZanID1 marker revealed that lanes 1, 2, 5, 6, 8, and 10 contained sancho samples, while lanes 3, 4, 9, and 11–16 were identified as chopi (Figure 4A). Different banding patterns were obtained using ZanID2, with lanes 2, 3, 5, 7, 8, and 10 containing sancho samples and the other samples identified as chopi (Figure 4B). Interestingly, samples 2, 5, and 7 clearly produced double bands in both species (Figure 4B). Furthermore, analysis of the banding patterns of the ZanID3 and ZanID4 markers revealed that lanes 2, 3, 5, 6, 7, 8, and 10 contained sancho samples, while the other samples were identified as chopi (Figure 4C,D). These three markers produced considerably different banding patterns, making it difficult to discriminate between sancho and chopi.
Figure 4.
PCR identification of 16 commercial sancho and chopi products comprising dried or powdered seeds and pericarps. Lanes on 1.5% agarose gel; M: 100bp ladder; 1–16: purchased commercial sancho and chopi products comprising dried or powdered seeds and pericarps (see Table 4 for details); 16 samples were amplified by PCR using four primer pairs; ZanID1_F and R (A), ZanID2_F and R (B), ZanID3_F and R (C), and ZanID4_F and R (D).
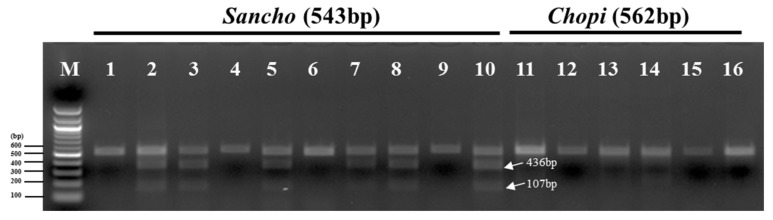
To select markers that could accurately discriminate between sancho and chopi, we performed PCR-RFLP analysis of the13 samples by developing a PCR-RFLP test to identify sancho samples. Many taxonomic studies of land plants have been based on the trnH-psbA region of cpDNA, as this DNA barcode exhibits high rates of sequence divergence among species. Based on the partial sequences of trnH-psbA in the cp genome that are shared between Z. schinifolium and Z. piperitum, we predicted that the PleI restriction enzyme would produce species-specific RFLP patterns and could therefore be used to identify Z. schinifolium based on the trnH-psbA locus. As shown in Figure 5, the fragment sizes for the two species were as follows: In Z. schinifolium, PleI produced two fragments (436 and 107 bp) from PCR products (lanes 2, 3, 5, 7, 8, and 10) of trnH-psbA (543 bp); in Z. piperitum, this enzyme did not digest the PCR products of trnH-psbA (562 bp). These results indicate that the psbZ-trnG marker is suitable for use as a reliable marker for differentiating between Z. schinifolium and Z. piperitum spices in the food market.
Figure 5.
PCR-RFLP profiles of partial regions of trnH-psbA from all samples digested with PleI. Numbers indicate sancho and chopi samples, as described in Table 4.
4. Discussion
Zanthoxylum formed a phylogenetic group in previous molecular phylogenetic studies [5,15,16]; however, these studies did not sufficiently resolve the relationships among some of its taxa. These studies were based on the ITS sequences of nuclear ribosomal DNA and the trnL-trnF, matK-trnK, atpB, atp-rbcL, and rbcL sequences of the cp genome [23,24,25]. Although these regions were considered to be universal DNA barcodes for higher plants, they did not allow the assessment of the usefulness of these loci in the barcoding of some taxa. Therefore, advances in NGS technologies have made it possible to sequence whole cp genomes and identify molecular markers. Highly variable markers derived from the cp genomes of different species at the genus level have uncovered many loci that are informative for systematic botany and DNA barcoding research [26,27].
Here, we retrieved the complete cp genome sequences of four Zanthoxylum species from the NCBI database and compared species-specific cp diversity in Zanthoxylum. We confirmed the variation among species, with an average nucleotide diversity value (Pi) of 0.00609 among the four Zanthoxylum species. Although the average Pi value for the SSC region was relatively high, high sequence divergence was detected at loci trnH-psbA, psbZ-trnG, trnfM-rps14, and trnF-ndhK in the LSC. Indeed, the trnH-psbA locus is highly variable in most plants and is known as a universal DNA barcoding region [28,29]. We used PCR amplification and sequencing to validate four hypervariable markers to distinguish between Z. schinifolium and Z. piperitum and to discriminate between sancho and chopi spice materials consumed in the online food market. However, PCR amplification of the four markers produced variable banding patterns, making it difficult to discriminate between sancho and chopi. Therefore, we designed a PCR-RFLP method using PleI digestion of the trnH-psbA DNA barcoding region, resulting in the production of two fragments (436 and 107 bp) only in Z. schinifolium. This marker, the ZanID2 marker from the psbZ-trnG region, is therefore suitable for discriminating between Z. schinifolium and Z. piperitum in sancho and chopi. The ZanID2 marker shows high sensitivity and specificity for detecting both sancho and chopi samples. Among the 10 sancho samples examined, three were successfully detected as sancho (30%), whereas three other samples (30%) produced a double band pattern that was clearly detected in both sancho and chopi samples. As expected, these results confirm the notion that products labeled as sancho that are sold in the spice and herbal medicine market often contain a mixture of sancho and chopi.
Although our results confirm that our newly developed InDel markers can be used to authenticate Z. schinifolium and Z. piperitum based on available cp genome data, more complete cp genome sequences are needed to comprehensively evaluate these InDel markers in the Zanthoxylum genus.
Acknowledgments
This research was supported by the Support Program for Creative Industry Institutes (Commercial Biotechnology Sophistication Platform Construction Program, R0003950), funded by the Ministry of Trade, Industry and Energy (MOTIE, Korea).
Author Contributions
Conceived and designed the experiments, Y.K. and C.C.; Collected the data, S.-S.C. and Y.-P.H.; Performed the experiments, Y.K. and J.S.; Analyzed the data and wrote the manuscript, Y.K. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest.
References
- 1.Sun X.W., Duan Z.X. Progress in the studies on medicinal plants of the genus Zanthoxylum Linn. Yao xuexuebao Acta Pharm. Sin. 1996;31:231–240. [PubMed] [Google Scholar]
- 2.Epifano F., Curini M., Carla Marcotullio M., Genovese S. Searching for novel cancer chemopreventive plants and their products: The genus Zanthoxylum. Curr. Drug Targets. 2011;12:1895–1902. doi: 10.2174/138945011798184128. [DOI] [PubMed] [Google Scholar]
- 3.Arun K., Paridhavi M. An ethno botanical phytochemical and pharmacological utilization of widely distributed species Zanthoxylum: A comprehensive overview. Int. J. Pharm. Invent. 2012;2:24–35. [Google Scholar]
- 4.Patiño L.O.J., Prieto R.J.A., Cuca S.L.E. Zanthoxylum genus as potential source of bioactive compounds. Bioact. Compd. Phytomed. InTech. 2012:185–218. doi: 10.5772/2374. [DOI] [Google Scholar]
- 5.Appelhans M.S., Reichelt N., Groppo M., Paetzold C., Wen J. Phylogeny and biogeography of the pantropical genus Zanthoxylum and its closest relatives in the proto-Rutaceae group (Rutaceae) Mol. Phylogenet. Evol. 2018;126:31–44. doi: 10.1016/j.ympev.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 6.Negi J.S., Bisht V.K., Bh A.K., Singh P., Sundriyal R.C. Chemical constituents and biological activities of the genus Zanthoxylum: A review. Afr. J. Pure Appl. Chem. 2011;5:412–416. [Google Scholar]
- 7.Supabphol R., Tangjitjareonkun J. Chemical constituents and biological activities of Zanthoxylum limonella (Rutaceae): A review. Trop. J. Pharm. Res. 2014;13:2119–2130. doi: 10.4314/tjpr.v13i12.25. [DOI] [Google Scholar]
- 8.Kim J., Jeong C.H., Bae Y.I., Shim K.H. Chemical components of Zanthoxylum schinifolium and Zanthoxylum piperitum leaves. Korean J. Food Preserv. 2000;7:189–194. [Google Scholar]
- 9.Ko Y.S., Han H.J. Chemical constituents of Korean chopi (Zanthoxylum piperitum) and sancho (Zanthoxylum schinifolium) Korean J. Food Sci. Technol. 1996;28:19–27. [Google Scholar]
- 10.Yang X. Aroma constituents and alkylamides of red and green huajiao (Zanthoxylum bungeanum and Zanthoxylum schinifolium) J. Agric. Food Chem. 2008;56:1689–1696. doi: 10.1021/jf0728101. [DOI] [PubMed] [Google Scholar]
- 11.Lee T.B. Coloured Flora of Korea. Volume 1 Hyangmunsa; Seoul, Korea: 2003. [Google Scholar]
- 12.Chung T.H. Korean Flora. Shinjisa; Seoul, Korea: 1957. p. 1025. (in Korean) [Google Scholar]
- 13.Nakai T. Notulæ and PlantasJaponiæ&Koreæ XXXIX. ShokubutsugakuZasshi. 1930;44:507–537. [Google Scholar]
- 14.Gupta D.D., Mandi S.S. Species specific AFLP markers for authentication of Zanthoxylum acanthopodium & Zanthoxylum oxyphyllum. J. Med. Plants. 2013;6:1–9. [Google Scholar]
- 15.Feng S., Yang T., Liu Z., Chen L., Hou N., Wang Y., Wei A. Genetic diversity and relationships of wild and cultivated Zanthoxylum germplasms based on sequence-related amplified polymorphism (SRAP) markers. Genet. Resour. Crop Evol. 2015;62:1193–1204. doi: 10.1007/s10722-015-0222-x. [DOI] [Google Scholar]
- 16.Kim W.J., Ji Y., Lee Y.M., Kang Y.M., Choi G., Moon B.C. Development of Molecular Markers for the authentication of Zanthoxyli Pericarpium by the analysis of rDNA-ITS DNA barcode regions. Korea J. Herbol. 2015;30:41–47. doi: 10.6116/kjh.2015.30.3.41. [DOI] [Google Scholar]
- 17.Feng S., Yang T., Li X., Chen L., Liu Z., Wei A. Genetic relationships of Chinese prickly ash as revealed by ISSR markers. Biologia. 2015;70:45–51. doi: 10.1515/biolog-2015-0005. [DOI] [Google Scholar]
- 18.Lee J., Lee H.J., Kim K., Lee S.C., Sung S.H., Yang T.J. The complete chloroplast genome sequence of Zanthoxylum piperitum. Mitochondrial DNA Part A. 2016;27:3525–3526. doi: 10.3109/19401736.2015.1074201. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y., Wei A. The complete chloroplast genome sequence of an economically important plant, Zanthoxylum bungeanum (Rutaceae) Conserv. Genet. Resour. 2017;9:25–27. doi: 10.1007/s12686-016-0609-9. [DOI] [Google Scholar]
- 20.Lee H.J., Koo H.J., Lee J., Lee S.C., Lee D.Y., Giang V.N.L., Kim M., Shim H., Park J.Y., Yoo K.O., et al. Authentication of Zanthoxylum species based on integrated analysis of complete chloroplast genome sequences and metabolite profiles. J. Agric. Food Chem. 2017;65:10350–10359. doi: 10.1021/acs.jafc.7b04167. [DOI] [PubMed] [Google Scholar]
- 21.Kumar S., Stecher G., Tamura K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Librado P., Rozas J. DnaSPv5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 23.Yan-Lin S., Wan-Geun P., Oh-Woung K., Soon-Kwan H. The internal transcribed spacer rDNA specific markers for identification of Zanthoxylum piperitum. Afr. J. Biotechnol. 2010;9:6027–6039. [Google Scholar]
- 24.Feng S., Liu Z., Chen L., Hou N., Yang T., Wei A. Phylogenetic relationships among cultivated Zanthoxylum species in China based on cpDNA markers. Tree Genet. Genomes. 2016;12:45. doi: 10.1007/s11295-016-1005-z. [DOI] [Google Scholar]
- 25.Zhao L.L., Feng S.J., Tian J.Y., Wei A.Z., Yang T.X. Internal transcribed spacer 2 (ITS 2) barcodes: A useful tool for identifying Chinese Zanthoxylum. Appl. Plant Sci. 2018;6:e01157. doi: 10.1002/aps3.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong W., Liu J., Yu J., Wang L., Zhou S. Highly variable chloroplast markers for evaluating plant phylogeny at low taxonomic levels and for DNA barcoding. PLoS ONE. 2012;7:e35071. doi: 10.1371/journal.pone.0035071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X., Yang Y., Henry R.J., Rossetto M., Wang Y., Chen S. Plant DNA barcoding: From gene to genome. Biol. Rev. 2015;90:157–166. doi: 10.1111/brv.12104. [DOI] [PubMed] [Google Scholar]
- 28.Kress W.J., Erickson D.L. A two-locus global DNA barcode for land plants: The coding rbcL gene complements the non-coding trnH-psbA spacer region. PLoS ONE. 2007;2:e508. doi: 10.1371/journal.pone.0000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Whitlock B.A., Hale A.M., Groff P.A. Intraspecific inversions pose a challenge for the trnH-psbA plant DNA barcode. PLoS ONE. 2010;5:e11533. doi: 10.1371/journal.pone.0011533. [DOI] [PMC free article] [PubMed] [Google Scholar]