Abstract
Methicillin-resistant Staphylococcus aureus (MRSA) is a serious pathogen in clinical settings and early detection is critical. Here, we investigated the MRSA discrimination potential of matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) using 320 clinical S. aureus isolates obtained in 2005–2014 and 181 isolates obtained in 2018. We conducted polymerase chain reactions (PCR) for staphylococcal cassette chromosome mec (SCCmec) typing and MALDI-TOF MS to find specific markers for methicillin resistance. We identified 21 peaks with significant differences between MRSA and methicillin-susceptible S. aureus (MSSA), as determined by mecA and SCCmec types. Each specific peak was sufficient to discriminate MRSA. We developed two methods for simple discrimination according to these peaks. First, a decision tree for MRSA based on six MRSA-specific peaks, three MSSA-specific peaks, and two SCCmec type IV peaks showed a sensitivity of 96.5%. Second, simple discrimination based on four MRSA-specific peaks and one MSSA peak had a maximum sensitivity of 88.3%. The decision tree applied to 181 S. aureus isolates from 2018 had a sensitivity of 87.6%. In conclusion, we used specific peaks to develop sensitive MRSA identification methods. This rapid and easy MALDI-TOF MS approach can improve patient management.
Keywords: Staphylococcus aureus, MRSA discrimination, MALDI-TOF MS
1. Introduction
Staphylococcus aureus is a major human pathogen that causes various diseases, including food poisoning, toxic shock syndrome, abscess, pneumonia, and sepsis [1]. Resistance to methicillin is caused by the mecA gene, which encodes an alternative penicillin-binding protein (PBP2a). Methicillin-resistance of S. aureus is attributed to the insertion of the staphylococcal cassette chromosome mec (SCCmec), a mobile genetic element, carrying the mecA into the chromosome of susceptible strains [2], with the SCCmec type reflecting the clonality of S. aureus [3]. Since the initial spread of methicillin-resistant S. aureus (MRSA) in hospital settings in the late 1980s, the infection rate has increased sharply, account for a huge number of deaths worldwide [4,5]. For proper antibiotic treatment, rapid discrimination between MRSA and methicillin-susceptible S. aureus (MSSA) is required.
Culture-based antimicrobial susceptibility testing, a standard method for the detection of methicillin resistance in clinical laboratories, is time-consuming [6]. Several alternative MRSA detection methods have been developed, such as selective chromogenic media, PCR assays, and, more recently, the matrix-assisted laser desorption ionization time-of-flight mass spectrometry (MALDI-TOF MS) [6,7,8,9]. Detection of MRSA is divided into two general type of techniques such as the performance/efficacy criteria or convenience/efficiency criteria. The polymerase chain reaction (PCR)-based methods have the advantages of high performance and efficacy, and qPCR or RT-qPCR, ddPCR, and modified 16S sequencing require only a few hours [10,11]. In contrast, the MALDI-TOF MS, has the potential to become a convenient and efficient type of detection method for MRSA identification, as it is already routinely used in many clinical microbiology laboratories; indeed, The detection of antibiotic resistance by MALDI-TOF MS is receiving substantial attention [12]. This technique can be performed in a few minutes with a single colony, costs only a few US dollars, and there is no additional cost if a laboratory already has a MALDI-TOF MS setup for bacterial identification. MALDI-TOF MS could be used to identify bacterial genera and species as well as intraspecific properties, such as clonal complexes and/or spa types of S. aureus [13,14,15,16]. Since susceptibility testing by MADI-TOF MS was first reported in 2000 [6], MRSA spectra have been evaluated extensively, including analyses of single peaks, clusters, or whole spectra [17,18,19]. The PBP2a has a direct effect on methicillin resistance, but cannot be detected by MALDI-TOF MS owing to its molecular weight of 76 kDa [20]. Some researchers have used MRSA-related special peaks, such as PSM-mec, which is related to methicillin resistance [19,21,22,23,24]. Several peaks can powerfully discriminate some MRSA, but may not be sufficient to detect all MRSA. Thus, this study aimed to identify specific markers for discrimination between MRSA and MSSA by MALDI-TOF MS, and to develop discrimination methods that can be applied to clinical S. aureus isolates.
2. Results
2.1. Specific Peaks for MRSA and MSSA
Methicillin resistance was evaluated by multiplex PCR (M-PCR) for mecA gene and SCCmec type.
We used two sets of S. aureus as the database set and test set. The database set comprised of 320 S. aureus isolated during 10 years, while the test set contained 181 recently isolated S. aureus. Among the 320 isolates of the database set, 213 were MRSA and 107 were MSSA. Among the 213 MRSA isolates, 72.8% were SCCmec type II (155 cases), 11.7% were type III (25 cases), and 15.5% were type IV (33 cases). All 320 S. aureus isolates were identified by MALDI-TOF MS at the species level with a score value of >2.0. After a spectral analysis, we obtained 21 peaks by MALDI-TOF MS that differed significantly with respect to mecA or SCCmec type (P < 0.01). Thirteen peaks were identified as MRSA-specific markers. Among them, peaks at m/z 2204, 2410, 2592, 4607, and 9216 were predominantly detected in MRSA strains and in some MSSA isolates (Table 1). Eight peaks were MSSA-specific markers and peaks at m/z 2339, 3034, and 5509 had specificities of greater than 91% (Table 1). Distinguishing MSSA and MRSA strains can serve as an effective transition to further characterize MRSA strains and test whether the MALDI-TOF method can distinguish between those strains.
Table 1.
Specific peaks for the discrimination of S. aureus SCCmec type.
| Type | Peak (m/z) | SCCmec | MSSA | SE | SP | PPV | NPV | P | Decision Tree | Reference | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| II | III | IV | ||||||||||
| MRSA SCCmec type II | 1975 | 81 | 12 | 6 | 26 | 80.6 | 80.0 | 79.1 | 81.5 | <0.001 | node 3 | |
| 2134 | 65 | 52 | 27 | 30 | 65.2 | 67.3 | 65.2 | 67.3 | <0.001 | |||
| 2592 | 24 | 12 | 3 | 4 | 23.9 | 95.2 | 82.2 | 57.1 | <0.001 | node 3 | ||
| 3890 | 55 | 0 | 0 | 3 | 54.8 | 98.2 | 96.6 | 69.8 | <0.001 | node 3 | [9] *, [10] †, [12] †, [20] † | |
| MRSA SCCmec type III | 2204 | 3 | 68 | 0 | 0 | 68.0 | 98.3 | 77.3 | 97.3 | 0.001 | ||
| 2410 | 43 | 100 | 18 | 15 | 100.0 | 69.8 | 21.9 | 100.0 | <0.001 | node 2, 3 | [15] †, [17,18,19,20] † | |
| 2874 | 6 | 64 | 9 | 5 | 64.0 | 94.2 | 48.5 | 96.9 | <0.001 | |||
| 4607 | 1 | 100 | 0 | 0 | 100.0 | 99.7 | 96.2 | 100.0 | <0.001 | node 2, 3 | ||
| 6594 | 3 | 92 | 9 | 12 | 92.0 | 93.2 | 53.5 | 99.3 | <0.001 | [12] †, [20] †, [25] ※, [26] † | ||
| 9216 | 1 | 100 | 0 | 0 | 100.0 | 99.7 | 96.2 | 100.0 | <0.001 | |||
| MRSA SCCmec type IV | 5053 | 98 | 80 | 21 | 91 | N/A | <0.001 | node 1 | ||||
| 5541 | 1 | 0 | 76 | 24 | 75.8 | 90.2 | 47.2 | 97.0 | <0.001 | node 1 | [10] † | |
| 5579 | 0 | 0 | 70 | 11 | 69.7 | 95.8 | 65.7 | 96.5 | <0.001 | |||
| MSSA | 2194 | 2 | 24 | 76 | 55 | 55.1 | 84.0 | 63.4 | 78.9 | <0.001 | node 3 | |
| 2232 | 1 | 8 | 55 | 37 | 37.4 | 90.1 | 65.6 | 74.1 | <0.001 | node 3 | ||
| 2301 | 6 | 56 | 94 | 74 | 73.8 | 74.6 | 59.4 | 85.0 | <0.001 | |||
| 2339 | 1 | 0 | 52 | 34 | 33.6 | 91.5 | 66.7 | 73.3 | <0.001 | |||
| 2631 | 5 | 44 | 85 | 66 | 66.4 | 77.9 | 60.2 | 82.2 | <0.001 | node 3 | ||
| 2668 | 17 | 44 | 67 | 42 | 42.1 | 72.3 | 43.3 | 71.3 | <0.001 | |||
| 3034 | 1 | 24 | 6 | 16 | 15.9 | 95.8 | 65.4 | 69.4 | <0.001 | [10] † | ||
| 5509 | 5 | 0 | 0 | 20 | 19.6 | 96.2 | 72.4 | 70.4 | <0.001 | [26] † | ||
Percentages of bacterial isolates showing each peak are shown (type II isolates, 155; type III isolates, 25; type IV isolates, 33; MSSA isolates, 107). SE, SP, PPV, and NPV were calculated by dividing each SCCmec type by the total sample number. SE: sensitivity, SP: specificity, PPV: positive predictive value, NPV: negative predictive value, P-value for cross-tabulation with other SCCmec type. *Reference paper did not include MRSA strains. ※Reference paper used MRSA and MSSA isolates. †Reference paper used only MRSA isolates.
2.2. Specific Peaks According to MRSA SCCmec Type
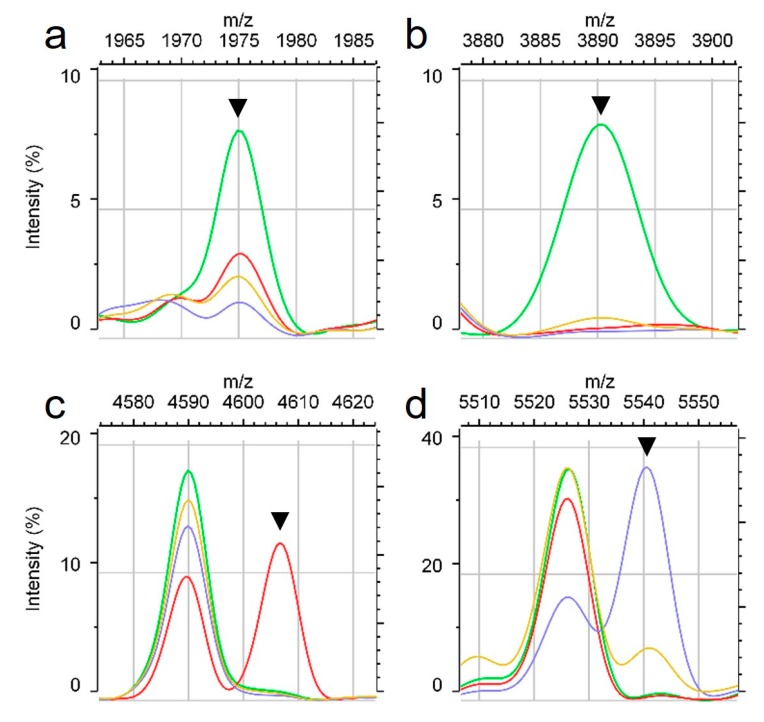
We detected peaks with different characteristics depending on SCCmec type. Peaks at m/z 1975, 2592, and 3890 were specific to SCCmec type II and had specificities and positive-predictive values of greater than 80% (Table 1, Figure 1a,b). The peaks at m/z 2204, 2410, 4607, 6594, and 9216 were specific to SCCmec type III. In particular, 100% of SCCmec type III carriers exhibited peaks at m/z 2410, 4607 (Figure 1c), and 9216. The PSM-mec peak was detected between m/z 2407 and 2412 (median, m/z 2410). For SCCmec type IV prediction, we selected three peaks. However, MALDI spectra for SCCmec type IV strains were similar to those for MSSA strains, with overlapping prediction peaks. Similarly, most MSSA prediction peaks were found in some SCCmec type IV isolates, but peaks at m/z 5541 and 5579 were more frequently observed in SCCmec type IV than in MSSA isolates (Table 1 and Figure 1d). Peaks at m/z 5541 and 5579 were detected in greater than 90% of SCCmec type IV isolates.
Figure 1.
Average spectra for S. aureus according to SCCmec type. Light green line indicates MRSA SCCmec type II, red line is type III, blue line is type IV, and yellow line is MSSA. Peaks are indicated by arrows specific to an SCCmec type, including a type II-specific peak at m/z 1975 (a) and m/z 3890 (b); type III-specific peak at m/z 4607 (c); type IV-specific peak at m/z 5541 (d).
2.3. Decision Tree for MRSA and MSSA
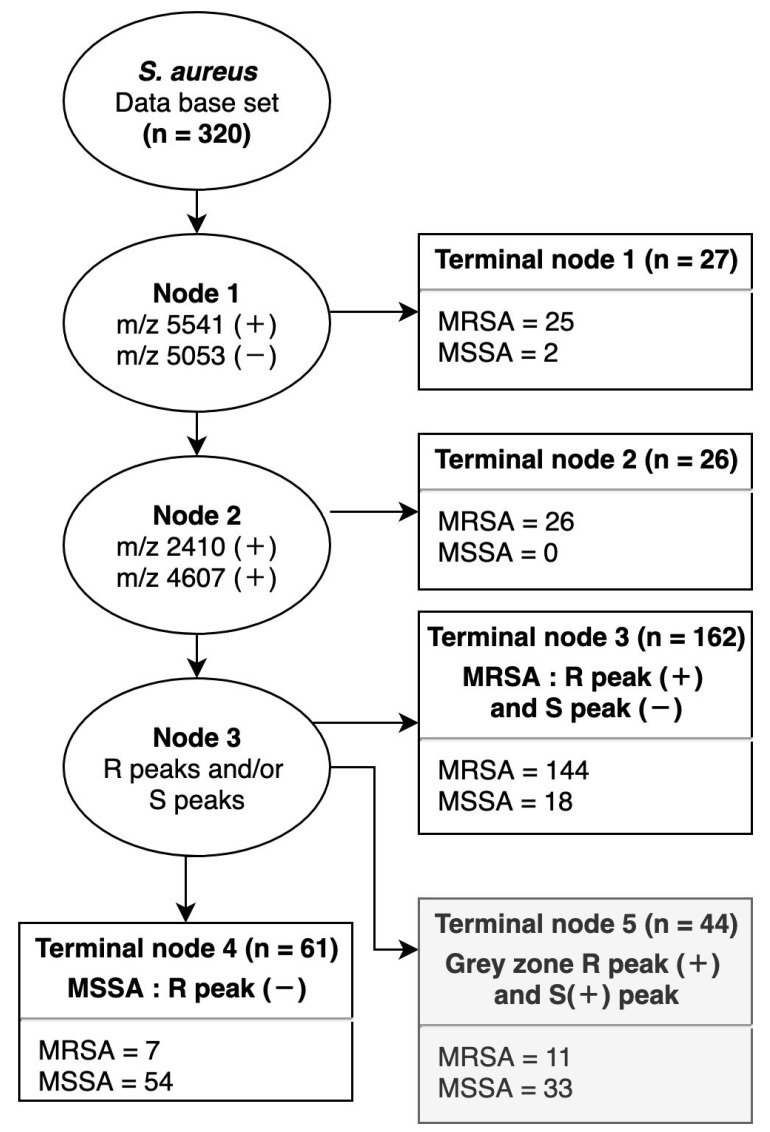
To identify MRSA by specific peaks, we made a decision tree with three nodes containing 11 peaks with high sensitivity and specificity (Figure 2). First, node 1 was used for the classification of SCCmec type IV based on the absence of the peak at m/z 5053 and the presence of the peak at m/z 5541. A total of 320 spectra were classified using node 1 into 25 MRSA and 2 MSSA. All 25 MRSA were SCCmec type IV strains. The remaining 293 spectra were classified using node 2 based on m/z 2410 and 4607, specific for SCCmec type III. Node 2 classified 26 MRSA, including one strain of type II and 25 strains of type III. Then, 267 spectra were classified as MRSA or MSSA (node 3). If at least one out of six peaks for MRSA prediction was present (m/z 1975, 2410, 3890, 4607, and 6594) and no peak for MSSA prediction was present (m/z 2194, 2339, and 2631), the isolate was classified as MRSA (terminal node 3). Spectra with no peak for MRSA prediction were classified as MSSA (terminal node 4). If peaks for both MRSA and MSSA were detected, the isolate was unclassified (i.e., assigned to a grey zone; terminal node 5). By node 3, 144 MRSA and 54 MSSA were classified. According to this decision tree, 96.5% of MRSA and 73.0% of MSSA were finally identified. Including a decision tree for a benchmarked method (e.g., the multiplexed PCR at the beginning of Section 2.1) will add to the validation of this assay by demonstrating that the MALDI-TOF method performs as well as a conventional test.
Figure 2.
Decision tree for MRSA and MSSA. Decision tree for MRSA and MSSA applied to the database set. At node 1, peaks at m/z 5541 and 5053 were used for SCCmec type IV classification. At node 2, peaks at m/z 2410 and 4607 were used for SCCmec type III classification. At node 3–5, R peaks (MRSA prediction peaks at m/z 1975, 2410, 2592, 3890, 4607, and 6594) and S peaks (MSSA prediction peaks at m/z 2194, 2339, and 2631) were used. Terminal node 3 identifies MRSA that expressed at least one or more R peaks and no S peaks. Terminal node 4 identifies MSSA with no expression of R peaks. Terminal node 5 determines unclassifiable isolates that express at least one or more R peaks and S peaks (referred to as the grey zone).
2.4. Simple Prediction Model Based on a Combination of Specific Peaks
To simplify prediction, we generated combinations of highly specific peaks for MRSA and MSSA. MRSA-specific peaks were selected at m/z 1975, 2410, 3890, and 5541, and MSSA-specific peaks were selected at m/z 2194, 2230, and 2339. Four-peak combinations (Table 2) showed improved MRSA prediction ability compared with that for single peaks (PSM-mec, 46%). Using only the combination of MRSA-specific peaks, the MSSA sensitivity was low, i.e., 45.8%. We added one MSSA-specific peak, but the difference in sensitivity was not substantial. The combination with the peak at m/z 2339 showed high predictive value for MRSA, with 88.3% sensitivity for MRSA and 67.3% for MSSA. Therefore, this simple method of discrimination is more sensitive than the one-peak method, but less sensitive than the decision tree.
Table 2.
Simple determination of MRSA-specific peaks and each MSSA-specific peak.
| Combined Peaks | Database Set | Test Set | |||
|---|---|---|---|---|---|
| MRSA (%) | MSSA (%) | MRSA (%) | MSSA (%) | ||
|
4-peak determination:
one or more peaks at m/z 1975, 2410, 3890, and 5541 (+) |
96.2 | 45.8 | 75.8 | 53.5 | |
| 4-peak determination with | 2194 (-) | 80.8 | 74.8 | 65.3 | 61.6 |
| 2230 (-) | 86.9 | 67.3 | 63.2 | 69.8 | |
| 2339 (-) | 88.3 | 67.3 | 67.4 | 58.1 | |
| 2630 (-) | 76.1 | 79.4 | 62.1 | 66.3 | |
Sensitivities of simple determination with 5-peak combinations are shown. For the prediction of MRSA, the inclusion of one or more peaks at m/z 1975, 2410, 3890, and 5541 and one MSSA-specific peak, such as m/z 2194, 2230, 2339, and 2630, was evaluated.
2.5. Evaluation of the Test Set
A total of 181 S. aureus collected in 2018 were used as the test set for evaluating the decision tree and simple discrimination method. The decision tree showed sensitivities of 87.6% for MRSA and 74.4% for MSSA (Table 3). Sensitivities for simple discrimination based on the 4 MRSA specific peaks and m/z 2339 peak were 67.4% for MRSA and 58.1% for MSSA, which were lower than those for the database set. Increasing MSSA sensitivity by simple determination could not be achieved.
Table 3.
Summary of decision tree results.
| Number of Isolates | SE (%) | SP (%) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|
| Database set | 320 | 96.5 | 73.0 | 90.7 | 88.5 |
| Test set | 181 | 87.6 | 71.4 | 78.0 | 83.3 |
Result of decision tree analyses (Figure 2) using the database and test sets. SE, SP, PPV, and NPV were calculated by dividing each SCCmec type by the total sample number. SE: sensitivity, SP: specificity, PPV: positive predictive value, NPV: negative predictive value.
3. Discussion
This study demonstrates the ability of the MALDI-TOF MS method to efficiently discriminate MRSA and MSSA strains through specific novel peaks that were identified. Previous studies have also identified specific peaks for MRSA, but a routine discrimination method for clinical settings based on these peaks is still lacking. Moreover, the reported peaks exhibit different patterns [14,25] or are applicable to only some MRSA strains [17,23,27]. In contrast, the discrimination methods described herein show high accuracy and good predictive value, and have the scope to be applied in routine clinical practice.
Previously, a PSM-mec peak at m/z 2415 ± 4 was reported as a prediction marker for MRSA [19,23]. In the current study, a PSM-mec peak was found at m/z 2410 ± 3 and appeared in the spectra for 46% of MRSA. The PSM-mec peak has the highest predictive value using the single peak prediction method. Other studies have reported a MRSA prediction peak at m/z 3890, which was also observed in our study [13,24]. In our MRSA isolates, this m/z 3890 peak was specific for SCCmec type II and did not appear in the spectra for type III or type IV isolates. These two peaks could identify MRSA, but showed a sensitivity of only 40–46%.
Peak shifts for S. aureus can be explained by different clonal groups or mutations [13,24,26]. The peak at m/z 6594 has been reported as a shift from m/z 6552, similar to peaks at m/z 6580 and 6568 [28]. In our study, most strains showed the original peak at m/z 6552 (m/z 6554 in this study), but 92% of SCCmec type III strains showed characteristic peak shifts to m/z 6594 (Table 1). This peak at m/z 6594 was specific to SCCmec type III. Additionally, peaks at m/z 4607 and 9216 were specific markers for MRSA, especially SCCmec type III that have 100% sensitivity (Table 1 and Figure 1c).
S. aureus SCCmec type IV is a community-acquired MRSA (CA-MRSA) and is genetically and phenotypically distinct from healthcare-associated MRSA (HA-MRSA). Infections with SCCmec type IV are increasing and causing diseases of the skin and soft tissue. CA-MRSA tends to be susceptible to most antibiotics other than methicillin and beta-lactamase [29]. Therefore, the accurate discrimination of not only MRSA but SCCmec types can enable the selection of a variety of effective antibiotics. In our experiment, it was not easy to discriminate between SCCmec type IV and MSSA owing to similar spectral patterns. The peak at m/z 5525 has three different shift forms at m/z 5507, 5551, and 5539 [30]. In this study, the original peak at m/z 5525 was found in 81.6% of S. aureus. The shifted peak at m/z 5507 (m/z 5509 in our study) was found in 20% of MSSA isolates and appeared to be MSSA-specific. The peak at m/z 5539 (m/z 5541 in this study) was found in most SCCmec type IV isolates. In particular, this peak was detected in 76% of SCCmec type IV isolates (Figure 1d) and 24% of MSSA isolates. The shifted peak at m/z 5541 was specific to SCCmec type IV, although it had a low positive predictive value (Table 1). We used this peak in node 1 of the decision tree for MRSA discrimination (Figure 2). Additionally, we used the peak at m/z 5053 in node 1 of the decision tree owing to its high positive predictive value of 98% for all isolates, except SCCmec type IV. As a result, we could discriminate MRAS SCCmec type IV more accurately by reducing similarity with spectral patterns for MSSA. These SCCmec type IV-specific peaks could be used as a basis for antibiotic therapy because type IV strains are susceptible to various antibiotics [31,32].
We found dense MRSA-specific peaks between m/z 2000–4000. Typically, MALDI-TOF MS spectra are analyzed in the range of m/z 2000–20000 after processing steps, such as baseline subtraction and peak selection. However, we found a specific peak at m/z 1975 before limiting the x-axis dimensions (Figure 1a). In our study, this peak was found in spectra for numerous MRSA isolates. The PSM-mec peak was found in 46% of MRSA isolates, but the peak at m/z 1975 was found in 61% of MRSA isolates. Though this peak was found in some MSSA, the peak at m/z 1975 can detect several strains that cannot be identified by the PSM-mec peak. Therefore, the peak at m/z 1975 can be broadly applied for the discrimination of MRSA. This result also demonstrated the potential for MRSA-specific peaks with low masses (i.e., under m/z 2000), emphasizing the importance of studying the low mass area.
Some of the specific peaks discovered in our analysis have been described previously, with the same or different interpretation. The use of MALDI-TOF MS to identify MRSA from unidentified strains has been controversial. Some studies have reported an inability to find a distinctive fingerprint for MRSA [11,17,33,34]. MALDI-TOF MS does not consistently yield the same intensities or peak patterns; therefore, various database of specific peaks for MRSA is needed.
We tested reference strains, including ATCC 43300, ATCC 29213, and RN4220 (Supplementary Figure S1 and Table S1). Unfortunately, MRSA ATCC 43300 and RN4220 strain could not be classified, as they did not express any of the peaks among the decision tree containing 11 peaks. The MSSA ATCC 29213 was classified to the grey zone, because this strain expressed specific peaks of both MRSA and MSSA. These reference strains were collected many years ago; therefore, the MALDI-TOF peaks may not reflect the protein expression profiles of recent clinical isolates. Nonetheless, additional analyses, such as spa typing or MLST analyses, are needed for the accurate detection of various types of S. aureus isolates from Korea as well as from other countries. Discovering specific peaks for each clonal type will increase accuracy.
We categorized peaks only by SCCmec type for easy and rapid classification. The specificity of some peaks reported in our study has already been described for each MLST type [14]. Previous studies have shown that the PSM-mec peak is specific to MRSA ST239 in Taiwan and the peak at m/z 3890 is specific to MRSA ST5 [24]; however, another study has reported that this latter peak has specificity for MSSA ATCC 29213 [13]. In our study, this peak was detected in 55% of SCCmec type II isolates. The peak at m/z 5509 was reported to be specific to CC30 of MRSA [30]. However, this peak was specific to MSSA in our study. The peak at m/z 6594 was previously reported to be specific to CC8, CC30, and ST239 of MRSA in Europe [24,28,30]; it was specific to MRSA SCCmec type III in our study.
Several studies have attempted to discriminate MRSA by single peaks or peak clustering. However, the sensitivity of PSM-mec peaks is only 39% [23], and that of two peaks in combination is only 63% [27]. Accordingly, discrimination based on only a few peaks is not highly accurate. Therefore, we constructed a decision tree using peaks with features associated with SCCmec type.
In conclusion, the 21 peaks and two discrimination methods described in our study provide exceptional predictive value, as high as 96.5%. These peaks can be used as basis for the accurate classification of MRSA. Using 181 clinical S. aureus isolates collected in 2018, the decision tree enabled the accurate identification of 87.6% of isolates. Thus, this approach is powerful for the discrimination of MRSA. MALDI-TOF, which can be used to identify strains and species, can also be used to distinguish between MRSA and MSSA. Therefore, the newly developed prediction methods will facilitate accurate and rapid diagnosis.
4. Materials and Methods
4.1. Bacterial Strains
Two sets of S. aureus clinical isolates, a database set and test set, were included. The database set included 320 S. aureus isolates that were randomly collected at Hallym University Kangdong Sacred Heart Hospital, Seoul, Korea from 2005 to 2014 (approximately 30 strains per year). The test set consisted of 181 isolates that were randomly selected in 2018 for the evaluation of the MRSA identification scheme. S. aureus was cultivated on blood agar plates for 18 h at 37 °C and 5% CO2.
4.2. MALDI-TOF Measurements of Bacterial Cells
Mass spectra were collected using a Microflex LT MALDI-TOF MS instrument (Bruker Daltonics, Bremen, Germany) operated by FlexControl v3.4. A bacterial colony was directly smeared on a steel MALDI plate without protein extraction. Dried samples were overlaid with 1 μl of formic acid and 1 μl of CHCA matrix solution (saturated solution of α-cyano-4-hydroxycinnamic acid in 50% acetonitrile with 2.5% trifluoroacetic acid), sequentially. The Bruker Bacterial Test Standard was used for mass calibration. The acquired raw spectra were exported from FlexAnalysis (Bruker Daltonics). Spectra were obtained in the range of 1960 to 20000 m/z.
4.3. Screening for the mecA Gene and SCCmec Type
Chromosomal DNA was extracted from S. aureus using the HiYield Genomic DNA Mini Kit (Real Biotech Corporation, Banqiao City, Taiwan) according to the manufacturer’s instructions. The mecA gene and SCCmec type were evaluated by two steps of M-PCR using the QIAGEN Multiplex PCR Master Mix (Qiagen, Hilden, Germany) and SCCmec element type primers [35]. M-PCR step 1 identified mecA and the ccr region. M-PCR step 2 identified the gene lineages of mecA-mecI, mecA-IS1272, and mecA-IS431.
4.4. Analysis of Mass Spectra
MALDI-TOF spectra were preprocessed using BioNumerics v7.6 (Applied Maths, Belgium) by recalibration, baseline subtraction (Top), and peak selection. Peak lists generated by BioNumerics software were analyzed using SPSS version 24. Correlations between peaks and mecA were evaluated by Pearson’s chi-square tests. A P-value of <0.05 indicated statistical significance.
4.5. Ethical Approval
The manuscript contains no data concerning animal studies, studies involving human subjects or inclusion of identifiable human data or clinical trials; thus, no ethical approval was required.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-0817/8/4/214/s1.
Author Contributions
Conceived and designed the experiment, J.-M.K., Y.C., M.H., J.-S.K.; performed the experiments, J.-M.K., S.H.C., I.K.; analyzed the data, J.-M.K., J.-S.K.; wrote the paper, J.-M.K., Y.C., M.H., J.-S.K.; edited by all co-authors.
Funding
This research was supported by a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number: HI17C2067). Furthermore, it was supported by the Hallym university research fund and Bio & Medical Technology Development Program of the National Research Foundation (NRF) funded by the Korean government, Ministry of Science and ICT (MSIT) 2017M3A9E4077232.
Conflicts of Interest
The authors declare that there are no conflicts of interest.
References
- 1.Tong S.Y., Davis J.S., Eichenberger E., Holland T.L., Fowler V.G., Jr. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015;28:603–661. doi: 10.1128/CMR.00134-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lakhundi S., Zhang K. Methicillin-Resistant Staphylococcus aureus: Molecular Characterization, Evolution, and Epidemiology. Clin. Microbiol. Rev. 2018;31 doi: 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asadollahi P., Farahani N.N., Mirzaii M., Khoramrooz S.S., van Belkum A., Asadollahi K., Dadashi M., Darban-Sarokhalil D. Distribution of the Most Prevalent Spa Types among Clinical Isolates of Methicillin-Resistant and -Susceptible Staphylococcus aureus around the World: A Review. Front. Microbiol. 2018;9:163. doi: 10.3389/fmicb.2018.00163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pastagia M., Kleinman L.C., Lacerda de la Cruz E.G., Jenkins S.G. Predicting risk for death from MRSA bacteremia. Emerg. Infect. Dis. 2012;18:1072–1080. doi: 10.3201/eid1807.101371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang C.I., Song J.H. Antimicrobial resistance in Asia: Current epidemiology and clinical implications. Infect. Chemother. 2013;45:22–31. doi: 10.3947/ic.2013.45.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards-Jones V., Claydon M.A., Evason D.J., Walker J., Fox A.J., Gordon D.B. Rapid discrimination between methicillin-sensitive and methicillin-resistant Staphylococcus aureus by intact cell mass spectrometry. J. Med. Microbiol. 2000;49:295–300. doi: 10.1099/0022-1317-49-3-295. [DOI] [PubMed] [Google Scholar]
- 7.Hiramatsu K., Katayama Y., Yuzawa H., Ito T. Molecular genetics of methicillin-resistant Staphylococcus aureus. Int. J. Med. Microbiol. 2002;292:67–74. doi: 10.1078/1438-4221-00192. [DOI] [PubMed] [Google Scholar]
- 8.Grmek-Kosnik I., Dermota U., Ribic H., Storman A., Petrovic Z., Zohar-Cretnik T. Evaluation of single vs pooled swab cultures for detecting MRSA colonization. J. Hosp. Infect. 2018;98:149–154. doi: 10.1016/j.jhin.2017.09.016. [DOI] [PubMed] [Google Scholar]
- 9.Walker J., Fox A.J., Edwards-Jones V., Gordon D.B. Intact cell mass spectrometry (ICMS) used to type methicillin-resistant Staphylococcus aureus: Media effects and inter-laboratory reproducibility. J. Microbiol. Methods. 2002;48:117–126. doi: 10.1016/S0167-7012(01)00316-5. [DOI] [PubMed] [Google Scholar]
- 10.Luo J., Li J., Yang H., Yu J., Wei H. Accurate Detection of Methicillin-Resistant Staphylococcus aureus in Mixtures by Use of Single-Bacterium Duplex Droplet Digital PCR. J. Clin. Microbiol. 2017;55:2946–2955. doi: 10.1128/JCM.00716-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Van Belkum A., Rochas O. Laboratory-Based and Point-of-Care Testing for MSSA/MRSA Detection in the Age of Whole Genome Sequencing. Front. Microbiol. 2018;9:1437. doi: 10.3389/fmicb.2018.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Patel R. MALDI-TOF MS for the diagnosis of infectious diseases. Clin. Chem. 2015;61:100–111. doi: 10.1373/clinchem.2014.221770. [DOI] [PubMed] [Google Scholar]
- 13.Bohme K., Morandi S., Cremonesi P., Fernandez No I.C., Barros-Velazquez J., Castiglioni B., Brasca M., Canas B., Calo-Mata P. Characterization of Staphylococcus aureus strains isolated from Italian dairy products by MALDI-TOF mass fingerprinting. Electrophoresis. 2012;33:2355–2364. doi: 10.1002/elps.201100480. [DOI] [PubMed] [Google Scholar]
- 14.Ostergaard C., Hansen S.G., Moller J.K. Rapid first-line discrimination of methicillin resistant Staphylococcus aureus strains using MALDI-TOF MS. Int. J. Med. Microbiol. 2015;305:838–847. doi: 10.1016/j.ijmm.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T., Ding J., Rao X., Yu J., Chu M., Ren W., Wang L., Xue W. Analysis of methicillin-resistant Staphylococcus aureus major clonal lineages by Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) J. Microbiol. Methods. 2015;117:122–127. doi: 10.1016/j.mimet.2015.08.002. [DOI] [PubMed] [Google Scholar]
- 16.Camoez M., Sierra J.M., Dominguez M.A., Ferrer-Navarro M., Vila J., Roca I. Automated categorization of methicillin-resistant Staphylococcus aureus clinical isolates into different clonal complexes by MALDI-TOF mass spectrometry. Clin. Microbiol. Infect. 2016;22 doi: 10.1016/j.cmi.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 17.Bernardo K., Pakulat N., Macht M., Krut O., Seifert H., Fleer S., Hunger F., Kronke M. Identification and discrimination of Staphylococcus aureus strains using matrix-assisted laser desorption/ionization-time of flight mass spectrometry. Proteomics. 2002;2:747–753. doi: 10.1002/1615-9861(200206)2:6<747::AID-PROT747>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- 18.Du Z., Yang R., Guo Z., Song Y., Wang J. Identification of Staphylococcus aureus and determination of its methicillin resistance by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Anal. Chem. 2002;74:5487–5491. doi: 10.1021/ac020109k. [DOI] [PubMed] [Google Scholar]
- 19.Josten M., Dischinger J., Szekat C., Reif M., Al-Sabti N., Sahl H.G., Parcina M., Bekeredjian-Ding I., Bierbaum G. Identification of agr-positive methicillin-resistant Staphylococcus aureus harbouring the class A mec complex by MALDI-TOF mass spectrometry. Int. J. Med. Microbiol. 2014;304:1018–1023. doi: 10.1016/j.ijmm.2014.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Van Belkum A., Welker M., Pincus D., Charrier J.P., Girard V. Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry in Clinical Microbiology: What Are the Current Issues? Ann. Lab. Med. 2017;37:475–483. doi: 10.3343/alm.2017.37.6.475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gagnaire J., Dauwalder O., Boisset S., Khau D., Freydiere A.M., Ader F., Bes M., Lina G., Tristan A., Reverdy M.E., et al. Detection of Staphylococcus aureus delta-toxin production by whole-cell MALDI-TOF mass spectrometry. PLoS ONE. 2012;7:e40660. doi: 10.1371/journal.pone.0040660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szabados F., Kaase M., Anders A., Gatermann S.G. Identical MALDI TOF MS-derived peak profiles in a pair of isogenic SCCmec-harboring and SCCmec-lacking strains of Staphylococcus aureus. J. Infect. 2012;65:400–405. doi: 10.1016/j.jinf.2012.06.010. [DOI] [PubMed] [Google Scholar]
- 23.Rhoads D.D., Wang H., Karichu J., Richter S.S. The presence of a single MALDI-TOF mass spectral peak predicts methicillin resistance in staphylococci. Diagn. Microbiol. Infect. Dis. 2016;86:257–261. doi: 10.1016/j.diagmicrobio.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Wang H.Y., Lien F., Liu T.P., Chen C.H., Chen C.J., Lu J.J. Application of a MALDI-TOF analysis platform (ClinProTools) for rapid and preliminary report of MRSA sequence types in Taiwan. PeerJ. 2018;6:e5784. doi: 10.7717/peerj.5784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lasch P., Fleige C., Stammler M., Layer F., Nubel U., Witte W., Werner G. Insufficient discriminatory power of MALDI-TOF mass spectrometry for typing of Enterococcus faecium and Staphylococcus aureus isolates. J. Microbiol. Methods. 2014;100:58–69. doi: 10.1016/j.mimet.2014.02.015. [DOI] [PubMed] [Google Scholar]
- 26.Sauget M., van der Mee-Marquet N., Bertrand X., Hocquet D. Matrix-assisted laser desorption ionization-time of flight Mass spectrometry can detect Staphylococcus aureus clonal complex 398. J. Microbiol. Methods. 2016;127:20–23. doi: 10.1016/j.mimet.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 27.Kwon S.S., Hong S.K., Kim M.S., Yong D., Lee K. Performance of Matrix-Assisted Laser Desorption Ionization Time-of-Fight Mass Spectrometry for Rapid Discrimination of Methicillin-Resistant Staphylococcus aureus (MRSA): First Report of a Relation Between Protein Peaks and MRSA spa Type. Ann. Lab. Med. 2017;37:553–555. doi: 10.3343/alm.2017.37.6.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Josten M., Reif M., Szekat C., Al-Sabti N., Roemer T., Sparbier K., Kostrzewa M., Rohde H., Sahl H.G., Bierbaum G. Analysis of the matrix-assisted laser desorption ionization-time of flight mass spectrum of Staphylococcus aureus identifies mutations that allow differentiation of the main clonal lineages. J. Clin. Microbiol. 2013;51:1809–1817. doi: 10.1128/JCM.00518-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zetola N., Francis J.S., Nuermberger E.L., Bishai W.R. Community-acquired meticillin-resistant Staphylococcus aureus: An emerging threat. Lancet Infect. Dis. 2005;5:275–286. doi: 10.1016/S1473-3099(05)70112-2. [DOI] [PubMed] [Google Scholar]
- 30.Wolters M., Rohde H., Maier T., Belmar-Campos C., Franke G., Scherpe S., Aepfelbacher M., Christner M. MALDI-TOF MS fingerprinting allows for discrimination of major methicillin-resistant Staphylococcus aureus lineages. Int. J. Med. Microbiol. 2011;301:64–68. doi: 10.1016/j.ijmm.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 31.Liu C., Bayer A., Cosgrove S.E., Daum R.S., Fridkin S.K., Gorwitz R.J., Kaplan S.L., Karchmer A.W., Levine D.P., Murray B.E., et al. Clinical practice guidelines by the infectious diseases society of america for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children: Executive summary. Clin. Infect. Dis. 2011;52:285–292. doi: 10.1093/cid/cir034. [DOI] [PubMed] [Google Scholar]
- 32.Udo E.E. Community-acquired methicillin-resistant Staphylococcus aureus: The new face of an old foe? Med. Princ. Pract. 2013;22(Suppl. 1):20–29. doi: 10.1159/000354201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lu J.J., Tsai F.J., Ho C.M., Liu Y.C., Chen C.J. Peptide biomarker discovery for identification of methicillin-resistant and vancomycin-intermediate Staphylococcus aureus strains by MALDI-TOF. Anal. Chem. 2012;84:5685–5692. doi: 10.1021/ac300855z. [DOI] [PubMed] [Google Scholar]
- 34.Kostrzewa M., Sparbier K., Maier T., Schubert S. MALDI-TOF MS: An upcoming tool for rapid detection of antibiotic resistance in microorganisms. Proteom. Clin. Appl. 2013;7:767–778. doi: 10.1002/prca.201300042. [DOI] [PubMed] [Google Scholar]
- 35.Ito T., Kuwahara-Arai K., Katayama Y., Uehara Y., Han X., Kondo Y., Hiramatsu K. Staphylococcal Cassette Chromosome mec (SCCmec) analysis of MRSA. Methods Mol. Biol. 2014;1085:131–148. doi: 10.1007/978-1-62703-664-1_8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.