Abstract
Background
The marine alga Ulva compressa is the dominant species in copper-polluted coastal areas in northern Chile. It has been shown that the alga tolerates micromolar concentrations of copper and accumulates copper at the intracellular level. Transcriptomic analyses were performed using total RNA of the alga cultivated with 10 μ M copper for 0, 1, 3 and 5 days using RNA-seq in order to identify processes involved in copper tolerance.
Results
The levels of transcripts encoding proteins belonging to Light Harvesting Complex II (LHCII), photosystem II (PSII), cytochrome b6f, PSI, LHCI, ATP synthase and proteins involved in repair of PSII and protection of PSI were increased in the alga cultivated with copper. In addition, the level of transcripts encoding proteins of mitochondrial electron transport chain, ATP synthase, and enzymes involved in C, N and S assimilation were also enhanced. The higher percentages of increase in the level of transcripts were mainly observed at days 3 and 5. In contrast, transcripts involved protein synthesis and degradation, signal transduction, and replication and DNA repair, were decreased. In addition, net photosynthesis and respiration increased in the alga cultivated with copper, mainly at days 1 to 3. Furthermore, the activities of enzymes involved in C, N and S assimilation, rubisco, glutamine synthase and cysteine synthase, respectively, were also increased, mainly at days 1 and 3.
Conclusions
The marine alga U. compressa tolerates copper excess through a concomitant increase in expression of proteins involved in photosynthesis, respiration, and C, N and S assimilation, which represents an exceptional mechanism of copper tolerance.
Keywords: Copper, Photosynthesis, Respiration, Transcriptomic analyses, Marine alga, Ulva compressa
Background
In photosynthetic organisms, copper is an essential heavy metal that it is required for the activity of several proteins and enzymes such as plastocyanin, cytochrome c oxidase, Cu/Zn superoxide dismutase (SOD), polyphenol oxidase, laccase and ascorbate oxidase, among others [1, 2]. Copper is required only in trace amount as in excess it produces an oxidative stress condition that becomes harmful for cellular macromolecules [2]. In photosynthetic organisms, copper excess may lead to the replacement of magnesium in chlorophylls inhibiting the release of energy from chlorophylls to PSII under low light, or it can also directly inhibit the reaction center in PSII in under high light [3, 4]. Copper excess can also inactivate enzymes by replacing zinc or other heavy metals [5].
The marine green macroalgae Ulva pertusa and U. armoricana cultivated with increasing concentrations of copper, from 0 to 250 μg L− 1 (3.9 μM) for 3 days displayed a differential behavior regarding photosynthesis and N assimilation [6]. U. pertusa showed no inhibition of photosynthetic parameters until 100 μg L− 1 (1.5 μM) and an increase in the activity of nitrate reductase activity, an enzyme involved in N assimilation [6]. In contrast, U. armoricana showed inhibition of photosynthetic parameters at 50–100 μg L− 1 of copper and an inhibition of nitrate reductase activity [6]. The green macroalga U. flexuosa cultivated with 0.8, 4 and 8 μM of copper for 5 days showed inhibition of photosynthesis when cultivated with 4 and 8 μM copper [7]. U. compressa L. Grev. (Cholorophyta) showed an increase in photosynthesis when cultivated with 10 μM of copper for 0 to 24 h [8]. Thus, green macroalgae from the same genus displayed differential responses to copper stress and it seems that, among Ulvaceae, U. compressa is the most tolerant species regarding copper stress. The red macroalga Gracilaria tenuistipitata cultivated with 16 nM of copper for 1 to 6 days showed an inhibition of photosynthesis from day 1 [9]. The red macroalga Porphyra haitiensis cultivated with 0.1 to 50 μM of copper for 3 days showed an increase in photosynthesis at concentrations of 0.1 to 1 μM and an increase in respiration with 0.1 to 50 μM suggesting that respiration is less sensitive to copper stress [10]. The brown macroalga Ectocarpus siliculosus showed an increase in photosynthesis when cultivated with 1.8 μM of copper for 8 h but a decrease in photosynthesis with 3.7 μM of copper [11]. Thus, marine macroalgae showed differential behaviors regarding photosynthesis and respiration in response to copper stress.
The marine alga U. compressa is highly tolerant to copper excess since the alga cultivated with 50 μM copper for 7 days displayed cellular viability [12]. The alga exposed to a sub-lethal concentration of copper (10 μM) showed the accumulation of superoxide anions beginning at day 3 and increasing until day 7 and the production of superoxide anions occurred mainly in chloroplasts and mitochondria [12]. In addition, the alga cultivated with copper showed the activation of antioxidant enzymes such as superoxide dismutase, ascorbate peroxidase and glutathione reductase, and the synthesis of antioxidant compounds such as ascorbate (ASC) and glutathione (GSH) [12–14]. In addition, U. compressa accumulate copper in its tissue, reaching 620 μg g− 1 when cultivated with 10 μM copper for 12 days [15]. Copper accumulation correlates with the synthesis of GSH, and phytochelatins (PCs), which are peptides produced through condensation of GSH units, and with the increase in expression of metallothioneins (MTs), which are small size cysteine-rich proteins that bind monovalent and divalent metal ions [16]. Initial transcriptomic analyses using RNAseq performed in U. compressa cultivated with 10 μM copper for 0 and 24 h allowed the identification of 7 potential MTs as well as transcripts that encode antioxidant enzymes, and enzymes involved in ASC and GSH synthesis [16]. The levels of transcripts encoding MTs, antioxidant enzymes, and enzymes involved in ASC and GSH synthesis, were increased in the alga exposed to copper excess [16]. Finally, transcripts encoding three MTs, MT1, MT2 and MT3, were cloned from U. compressa and their overexpression in bacteria allowed the accumulation of copper and zinc in vivo [17]. Thus, U. compressa may accumulate copper in its tissue through the binding of copper ions to GSH, PCs and MTs and this alga could represent a useful tool for phycoremediation of seawater contaminated with heavy metals.
Photosynthetic organisms displaying an increase in photosynthesis may produce an enhanced level of NADPH that may increase C, N and S assimilation, since the latter are reductive processes that require NADPH [18–20]. The marine macroalga U. compressa cultivated with 10 μM copper for 0 to 24 h showed an increase in photosynthesis and in the level of transcripts encoding enzymes of the Calvin-Benson cycle, suggesting that C assimilation may be increased [8]. In this work, we investigate whether the initial increase in photosynthesis is maintained along days, and whether there is a concomitant increase in respiration, and in C, N and S assimilation. To this end, the alga U. compressa was cultivated with 10 μM copper for 0, 1, 3, and 5 days and the levels of transcripts encoding proteins involved in photosynthesis and respiration, and those encoding enzymes involved C, N and S assimilation, were determined. In addition, the level of produced and consumed oxygen, reflecting photosynthesis and respiration, respectively, were determined. The activities of key enzymes involved in C, N and S assimilation such as rubisco, glutamine synthase and cysteine synthase, respectively, were also analyzed. Results indicate that the alga tolerates copper excess through a concomitant increase in expression of proteins involved in photosynthesis, respiration, and C, N and S assimilation, which represents an exceptional mechanism of copper tolerance.
Results
Assembly, annotation and classification of transcripts
The libraries obtained from samples of the alga cultivated in control condition (day 0) and treated with copper for 1, 3, and 5 days contained 785,500,000 M of reads, in total, and 98,187,500 reads, on average. The reads were subjected to quality control, they were trimmed and bacterial sequences were eliminated, resulting in 96,577,500 M of reads, on average, which correspond to 98.36% of the initial reads (Additional file 1: Table S1). Transcripts were assembled using Trinity software and resulted in 140,840 transcripts (contigs) of 300 to 5140 nucleotides in length, with an average length of 1575 nucleotides. The completeness of assembled transcriptomes was 93.4% (Additional file 1: Table S1). Transcripts were translated into amino acids, and annotated proteins having an e value of 1e− 3, or lower, were 65,494 (Additional file 5: Table S2). The latter proteins are involved in different biological processes (Additional file 2: Figure S1). Of the 65,494 proteins, 23,692 (36%) showed higher amino acid similarity with plant and green algae (Plantae) proteins; 15,073 (23%) displayed a higher similarity with animal proteins; and 26,729 (41%) proteins showed similarity to other organisms (Fungi, Protista and Prokaryotes) (Additional file 3: Figure S2). Proteins having similarity to animal proteins showed homology with human, mouse, rat, and other animal proteins (Additional file 3: Figure S2).
Transcripts differentially expressed in response to copper excess
Transcripts coding for 65,494 proteins previously mentioned that were differentially expressed were obtained considering times: 0 vs. 1, 0 vs. 3 and 0 vs. 5 (Additional file 4: Figure S3). At time point 0 vs. 1, the number of differentially expressed transcripts was 28,510, those up-regulated were 13,855 and those down-regulated were 14,655, which represent 48.6 and 51.4%, respectively. At time point 0 vs. 3, the number of differentially expressed transcripts was 8174, those up-regulated were 6047 and those down-regulated were 2127, which represent 74 and 26%, respectively. At time point 0 vs. 5, the number of differentially expressed transcripts was 30.589, those up-regulated were 17,218 and those down-regulated were 13,371, which represent 56.3 and 43.7%, respectively. Thus, the higher percentages of up-regulated transcripts were observed at days 3 and 5 in the alga exposed to copper excess.
Transcripts with increased levels of encoded proteins involved in photosynthesis and respiration
Regarding photosynthesis, the levels of transcripts encoding subunits of PSII corresponding to PsbA, PsbB, PsbC, PsbE, PsbH, PsbN, PsbO, PsbP, PsbR, PsbS, PsbW, PsbY and PsbZ were up-regulated in response to copper stress (Table 1). In addition, the levels of transcripts encoding subunits of Light Harvesting Complex II (LHCII), chlorophyll a/b-binding proteins LhcB1, LhcB4, LhcB5 and Cab1 as well as fucoxanthin-chlorophyll a/c-binding proteins, FcpA and FcpB, were increased. In addition, the subunits of the enzyme magnesium chelatase involved in chlorophyll synthesis, ChlD and ChlI, were up-regulated (Table 1). The levels of transcripts encoding subunits of cytochrome b6f corresponding to cytochrome b6 (petB), iron-sulfur Rieske subunit (PetC), an essential protein for assembly of cytb6f (PetG), and a carrier of electrons from cytb6f to PSI (PetJ), were also increased (Table 1). The levels of transcripts encoding subunits of PSI corresponding to PsaA, PsaB, PsaD, PsaF, PsaG and PsaL were also up-regulated (Table 1). Finally, the levels of transcript encoding a subunit of LHCI, LhcA, and the subunits of ATP synthase α, β, γ, δ, ε and γ subunits were also increased (Table 1).
Table 1.
Up-regulated genes related to photosynthesis and mitochondrial electron transport chain
| Process | ID Transcript | Proteins | Log2 Fold Change |
|---|---|---|---|
| PSII | Unigene17497_All | PsbA | 3.4 |
| CL2571.Contig1_All | PsbB | 2.0 | |
| CL11753.Contig1_All | PsbB | 2.4 | |
| CL11753.Contig2_All | PsbB | 4.5 | |
| Unigene33413_All | PsbC | 3.3 | |
| CL4160.Contig3_All | PsbE | 2.6 | |
| Unigene34274_All | PsbH | 3.3 | |
| Unigene29880_All | PsbN | 3.3 | |
| Unigene13168_All | PsbO | 1.6 | |
| Unigene29944_All | PsbP | 2.1 | |
| CL7636.Contig1_All | PsbP | 1.2 | |
| CL699.Contig2_All | PsbP | 2.3 | |
| CL4419.Contig1_All | PsbP | 1.8 | |
| CL4419.Contig5_All | PsbP | 1.2 | |
| CL3599.Contig1_All | PsbR | 2.0 | |
| CL10892.Contig2_All | PsbS | 3.6 | |
| Unigene33763_All | PsbW | 5.3 | |
| Unigene37851_All | PsbY | 1.6 | |
| Unigene1513_All | PsbZ | 2.5 | |
| LHCII | Unigene596_All | Chlorophyll a-b binding protein 1D (lhcB1) | 2.6 |
| CL4261.Contig2_All | Chlorophyll a-b binding protein CP26 (lhcB5) | 1.4 | |
| CL11211.Contig2_All | Chlorophyll a-b binding protein CP26 (lhcB5) | 1.9 | |
| CL1500.Contig3_All | Chlorophyll a-b binding protein L1818 (lhcb4) | 4.3 | |
| CL4399.Contig3_All | Chlorophyll a-b binding protein L1818 (lhcb4) | 6.8 | |
| CL9440.Contig1_All | Chlorophyll a-b binding protein L1818 (lhcb4) | 3.0 | |
| CL9440.Contig2_All | Chlorophyll a-b binding protein L1818 (lhcb4) | 4.7 | |
| Unigene29850_All | Chlorophyll a-b binding protein L1818 (lhcb4) | 1.5 | |
| CL1500.Contig2_All | Chlorophyll a-b binding protein L1818 (lhcb4) | 2.5 | |
| CL4399.Contig1_All | Chlorophyll a-b binding protein L1818 (lhcb4) | 1.4 | |
| CL12301.Contig27_All | Chlorophyll a-b binding protein Type I (CabII-1) | 2.9 | |
| CL12301.Contig28_All | Chlorophyll a-b binding protein Type I (CabII-1) | 2.5 | |
| Unigene22628_All | Fucoxanthin-chlorophyll a-c binding protein (FcpA) | 1.0 | |
| CL941.Contig8_All | Fucoxanthin-chlorophyll a-c binding protein (FcpE) | 2.4 | |
| CL2528.Contig4_All | Magnesium-chelatase subunit ChlD | 1.8 | |
| Cyt b6-f | Unigene41646_All | Iron-sulfur subunit (petB) | 1.8 |
| CL2893.Contig2_All | Iron-sulfur subunit (petC) | 1.5 | |
| CL2893.Contig4_All | Iron-sulfur subunit (petC) | 3.7 | |
| CL2893.Contig6_All | Iron-sulfur subunit (petC) | 5.7 | |
| CL2893.Contig9_All | Iron-sulfur subunit (petC) | 1.1 | |
| CL2893.Contig10_All | Iron-sulfur subunit (petC) | 4.9 | |
| Unigene174_All | Iron-sulfur subunit (petC) | 2.5 | |
| Unigene970_All | Iron-sulfur subunit (petG) | 2.5 | |
| Unigene33787_All | Iron-sulfur subunit (petJ) | 2.1 | |
| PSI | Unigene61010_All | PsaA | 7.7 |
| Unigene955_All | PsaD | 1.6 | |
| Unigene21099_All | PsaF | 1.3 | |
| Unigene38056_All | PsaG | 3.3 | |
| Unigene25315_All | PsaL | 1.3 | |
| LHC I ATP synthase | CL11079.Contig2_All | Chlorophyll a-b binding protein 5 (LhcA1 like) | 3.4 |
| Unigene33050_All | ATP synthase subunit alpha | 2.8 | |
| CL10342.Contig2_All | ATP synthase subunit alpha | 2.6 | |
| Unigene28940_All | ATP synthase subunit alpha | 1.4 | |
| Unigene21967_All | ATP synthase subunit alpha | 2.7 | |
| CL5620.Contig2_All | ATP synthase subunit alpha | 2.8 | |
| CL10342.Contig6_All | ATP synthase subunit alpha | 1.4 | |
| CL2062.Contig2_All | ATP synthase subunit beta | 8.1 | |
| Unigene284_All | ATP synthase subunit beta | 1.3 | |
| Unigene283_All | ATP synthase subunit beta | 1.9 | |
| Unigene286_All | ATP synthase subunit beta | 4.1 | |
| CL2062.Contig1_All | ATP synthase subunit beta | 1.2 | |
| Unigene33306_All | ATP synthase subunit beta | 2.1 | |
| CL4160.Contig1_All | ATP synthase subunit beta | 3.9 | |
| Unigene19914_All | ATP synthase subunit beta | 2.4 | |
| CL5278.Contig5_All | ATP synthase subunit gamma | 1.4 | |
| Unigene16523_All | ATP synthase subunit gamma | 2.2 | |
| CL5278.Contig5_All | ATP synthase subunit gamma | 1.4 | |
| CL5278.Contig10_All | ATP synthase subunit gamma | 1.4 | |
| Unigene16523_All | ATP synthase subunit gamma | 2.2 | |
| Unigene32932_All | ATP synthase subunit epsilon | 1.0 | |
| Unigene43520_All | ATP synthase subunit delta | 2.4 | |
| Assembly and Repair of PS II | Unigene29539_All | Met1 | 5.6 |
| CL1106.Contig1_All | Deg/HtrA Protease Do-like 1 | 4.2 | |
| CL1106.Contig2_All | Deg/HtrA Protease Do-like 1 | 2.6 | |
| CL1106.Contig5_All | Deg/HtrA Protease Do-like 1 | 4.5 | |
| CL1106.Contig7_All | Deg/HtrA Protease Do-like 1 | 2.1 | |
| CL1106.Contig8_All | Deg/HtrA Protease Do-like 1 | 2.5 | |
| CL1106.Contig9_All | Deg/HtrA Protease Do-like 1 | 2.4 | |
| CL1106.Contig13_All | Deg/HtrA Protease Do-like 1 | 2.6 | |
| CL3921.Contig1_All | Deg/HtrA Protease Do-like 1 | 3.6 | |
| CL3921.Contig3_All | Deg/HtrA Protease Do-like 1 | 2.0 | |
| CL6287.Contig3_All | Deg/HtrA Protease Do-like 2 | 1.4 | |
| CL6287.Contig4_All | Deg/HtrA Protease Do-like 2 | 1.0 | |
| Unigene12439_All | Deg/HtrA Protease Do-like 9 | 1.0 | |
| Unigene12443_All | Deg/HtrA Protease Do-like 9 | 1.1 | |
| Unigene7791_All | ATP-dependent zinc metalloprotease FtsH1 | 1.5 | |
| Unigene9549_All | ATP-dependent zinc metalloprotease FtsH1 | 1.1 | |
| Unigene71521_All | ATP-dependent zinc metalloprotease FtsH1 | 3.1 | |
| Unigene36205_All | ATP-dependent zinc metalloprotease FtsH1 | 9.1 | |
| CL8065.Contig1_All | ATP-dependent zinc metalloprotease FtsH2 | 2.5 | |
| Unigene16464_All | ATP-dependent zinc metalloprotease FtsH11 | 2.1 | |
| CL6915.Contig1_All | ABC1K1 | 3.2 | |
| CL6915.Contig7_All | ABC1K1 | 3.8 | |
| CL12141.Contig2_All | 2-carboxy-1,4-naphthoquinone phytyltransferase | 2.3 | |
| CL12141.Contig5_All | 2-carboxy-1,4-naphthoquinone phytyltransferase | 2.1 | |
| Assembly and Protection PSI | Unigene778_All | PGR5 1A | 6.4 |
| CL653.Contig10_All | Serine/threonine-protein kinase STN8 | 3.1 | |
| CL653.Contig13_All | Serine/threonine-protein kinase STN8 | 3.2 | |
| CL653.Contig38_All | Serine/threonine-protein kinase STN8 | 2.9 | |
| Unigene13129_All | YCF12 | 3.1 | |
| CL5916.Contig1_All | ATAB2 | 3.0 | |
| CL5916.Contig2_All | ATAB2 | 3.6 | |
| Mitochondrial | CL2273.Contig1_All | NADH dehydrogenase subunit 1 | 3.2 |
| Electron | Unigene75664_All | NADH dehydrogenase subunit 2 | 3.1 |
| Transport | CL8189.Contig1_All | cytochrome bc1 complex subunit V | 2.4 |
| Chain | CL5196.Contig1_All | Cytochrome bc1 complex subunit IV | 2.8 |
| Unigene965_All | Cytochrome bc1 complex subunit IV | 3.1 | |
| Unigene30266_All | ATP synthase subunit gamma | 3.1 |
On the other hand, the levels of transcripts encoding the chaperone MET1, involved in the insertion of PsbA (D1) in PSII; the ATP-independent serine proteases Deg 1, 2 and 9, and the ATP-dependent metalloproteases FtsH 1, 2 and 11, involved in degradation of damaged PsbA, were increased (Table 2). The levels of transcripts encoding bc1 complex kinase 1 (ABC1K1), involved in the synthesis of quinones and tocopherol (vitamin E) and the xanthophyll lutein which protect PSII to oxidative damage; the enzyme 2-carboxy-1,4-naphtoquinone phytyl transferase (ABC4), involved in the synthesis of phylloquinone (vitamine K) required in PSI, were up-regulated (Table 1). Moreover, transcripts encoding PGR5-1A, a protein involved in control of electron flow around PSI that protects PSI against photo-oxidation; YCF12, a protein involved in assembly and stabilization of PSI; the serine-threonine kinase STN8, involved in phosphorylation of LHCII allowing migration of subunits of PSII to PSI; and ATAB2, a light-regulated protein involved in the increased synthesis of photosystem proteins, were also increased (Table 1). Thus, the expression of a large number of proteins involved in photosynthesis and repair and protection of photosystems was increased in U. compressa exposed to copper excess (Additional file 6: Figure S4A).
Table 2.
Up-regulated genes related to Carbon, Nitrogen and Sulfur assimilation
| ID Transcript | Proteins | Log2 Fold Change | |
|---|---|---|---|
| Calvin-Benson Cycle | CL5634.Contig3_All | Ribulose bisphosphate carboxylase small chain 1 | 1.2 |
| CL5634.Contig2_All | Ribulose bisphosphate carboxylase small chain 1 | 8.9 | |
| CL12005.Contig2_All | Ribulose bisphosphate carboxylase small chain 1 | 4.7 | |
| CL12005.Contig1_All | Ribulose bisphosphate carboxylase small chain 1 | 1.4 | |
| Unigene16805_All | Ribulose bisphosphate carboxylase small chain 1 | 2.8 | |
| CL5634.Contig4_All | Ribulose bisphosphate carboxylase small chain 1 | 1.3 | |
| CL12005.Contig3_All | Ribulose bisphosphate carboxylase small chain 2 | 1.8 | |
| Unigene12933_All | Ribulose bisphosphate carboxylase large chain | 1.6 | |
| CL8954.Contig2_All | Phosphoglycerate kinase | 2.1 | |
| Unigene41371_All | Phosphoglycerate kinase | 3.4 | |
| CL1769.Contig1_All | Phosphoglycerate kinase | 1.0 | |
| CL1769.Contig2_All | Phosphoglycerate kinase | 1.5 | |
| Unigene32099_All | Phosphoglycerate kinase | 4.7 | |
| Unigene41512_All | Phosphoglycerate kinase | 5.1 | |
| CL1973.Contig6_All | Glyceraldehyde-3-phosphate dehydrogenase | 6.0 | |
| CL1973.Contig3_All | Glyceraldehyde-3-phosphate dehydrogenase | 1.3 | |
| CL8458.Contig1_All | Glyceraldehyde-3-phosphate dehydrogenase | 2.5 | |
| CL8458.Contig2_All | Glyceraldehyde-3-phosphate dehydrogenase | 3.0 | |
| CL1973.Contig7_All | Glyceraldehyde-3-phosphate dehydrogenase | 1.0 | |
| CL12146.Contig6_All | Glyceraldehyde-3-phosphate dehydrogenase | 1.6 | |
| Unigene26_All | Glyceraldehyde-3-phosphate dehydrogenase | 1.5 | |
| Unigene32957_All | Fructose-bisphosphate aldolase 1 | 7.0 | |
| Unigene32956_All | Fructose-bisphosphate aldolase 1 | 2.5 | |
| CL2940.Contig6_All | Fructose-bisphosphate aldolase 1 | 1.3 | |
| Unigene13983_All | Fructose-bisphosphate aldolase 5 | 3.8 | |
| CL2275.Contig2_All | Fructose-bisphosphate aldolase 8 | 1.0 | |
| Unigene29154_All | Fructose-1,6-bisphosphatase | 1.3 | |
| Unigene38319_All | Transketolase | 4.3 | |
| Unigene29707_All | Transketolase | 1.8 | |
| CL11843.Contig2_All | Phosphoribulokinase | 1.9 | |
| Unigene42641_All | Phosphoribulokinase | 2.9 | |
| Nitrogen Assimilation | Unigene26010_All | Nitrate reductase | 2.4 |
| CL10485.Contig1_All | Nitrate reductase | 3.0 | |
| Unigene32044_All | Glutamine synthetase | 5.3 | |
| Unigene79885_All | Glutamine synthetase | 1.2 | |
| Unigene10873_All | Glutamine synthetase | 2.3 | |
| CL1194.Contig12_All | Argininosuccinate lyase | 3.7 | |
| CL1194.Contig11_All | Argininosuccinate lyase | 9.2 | |
| CL1194.Contig13_All | Argininosuccinate lyase | 1.3 | |
| Unigene25622_All | Fumarate hydratase | 4.1 | |
| Unigene14757_All | Fumarate hydratase | 1.1 | |
| Sulfur assimilation | CL1413.Contig2_All | ATP sulfurylase | 1.8 |
| CL1413.Contig5_All | ATP sulfurylase | 1.2 | |
| CL1413.Contig8_All | ATP sulfurylase | 1.9 | |
| CL1413.Contig9_All | ATP sulfurylase | 2.7 | |
| Unigene66570_All | ATP sulfurylase | 3.1 | |
| Unigene1609_All | APS reductase | 1.8 | |
| CL7094.Contig2_All | APS reductase | 3.3 | |
| Unigene803_All | APS reductase | 1.9 | |
| Unigene15137_All | APS reductase | 5.4 | |
| Unigene17508_All | Sulfite reductase | 1.7 | |
| CL9121.Contig1_All | Sulfite reductase | 2.4 | |
| CL9650.Contig2_All | Sulfite reductase | 2.8 | |
| Unigene823_All | Cysteine synthase | 2.1 | |
| CL10220.Contig15_All | Cysteine synthase | 3.1 | |
| CL10220.Contig39_All | Cysteine synthase | 2.2 | |
| CL10220.Contig51_All | Cysteine synthase | 1.9 | |
| CL10220.Contig56_All | Cysteine synthase | 1.0 | |
| Unigene33426_All | Cysteine synthase | 3.9 | |
| Unigene1587_All | Cysteine synthase | 4.6 | |
| CL1730.Contig3_All | Glutamate-cysteine ligase | 2.1 | |
| CL2777.Contig3_All | Glutamate-cysteine ligase | 1.8 | |
| Unigene6218_All | Glutamate-cysteine ligase | 4.2 | |
| Unigene6958_All | Glutamate-cysteine ligase | 2.6 | |
| Unigene5542_All | Glutamate-cysteine ligase | 3.6 | |
| Unigene25762_All | Glutamate-cysteine ligase | 1.2 | |
| Unigene80578_All | Glutamate-cysteine ligase | 5.4 | |
| Unigene28997_All | Glutathione synthetase | 3.6 | |
| Unigene28998_All | Glutathione synthetase | 1.3 | |
| Unigene28999_All | Glutathione synthetase | 1.8 | |
| Unigene42348_All | Glutathione synthetase | 1.3 |
Regarding respiration, the levels of transcripts encoding subunit 1 and 2 of NADH dehydrogenase (complex I), subunit IV and V of cytochrome bc1 complex (complex III), and subunit γ of mitochondrial ATPase were increased (Table 1). Thus, the expression of a small number of proteins involved in respiration was increased in U. compressa compare to those involved in photosynthesis (Additional file 6: Figure S4B).
Transcripts with increased levels of encoded enzymes involved in C, N and S assimilation
The levels of transcripts encoding enzymes of the Calvin-Benson cycle involved in C assimilation, RbcS and RbcL corresponding to the small and large subunits of rubisco; phosphoglycerate kinase (PGK); glyceraldehyde 3-P dehydrogenase (G3PDH); fructose biphosphate aldolase (FBPA); fructose 1,6 biphosphatase (FBP), transketolase (TK); ribose 5-P isomerase (R5PI) and phosphoribulose kinase (PRK) were increased (Table 2). Thus, the expression of nine enzymes of the eleven enzymes of the Calvin-Benson cycle was increased in U. compressa exposed to copper excess.
The levels of transcripts encoding enzymes involved in N assimilation, nitrate reductase and glutamine synthase, were increased (Table 2). In addition, the levels of transcripts encoding enzymes of the urea cycle allowing detoxification of ammonium excess such as arginino-succinate lyase and fumarate hydratase were up-regulated (Table 2). Thus, the expression of two of the three enzymes involved in N assimilation was increased in U. compressa exposed to copper excess.
The levels of transcripts encoding enzymes involved in S assimilation, adenylyl-sulfate transferase (ATP sulfurylase); adenosine 5′-phosphosulfate reductase (APS reductase); sulfite reductase, and cysteine synthase (O-acetylserine thiol lyase) were increased (Table 2). In addition, enzymes involved in the synthesis of amino acids methionine, serine and alanine were also increased (data not shown). Furthermore, the levels of transcripts encoding enzymes involved in glutathione synthesis, glutamate cysteine ligase (γ-glutamyl cysteinyl synthase) and glutathione synthase, were also up-regulated (Table 2). Thus, the expression of the four enzymes involved in S assimilation was increased in U. compressa exposed to copper excess.
Kinetics of normalized reads of transcripts involved in photosynthesis and respiration
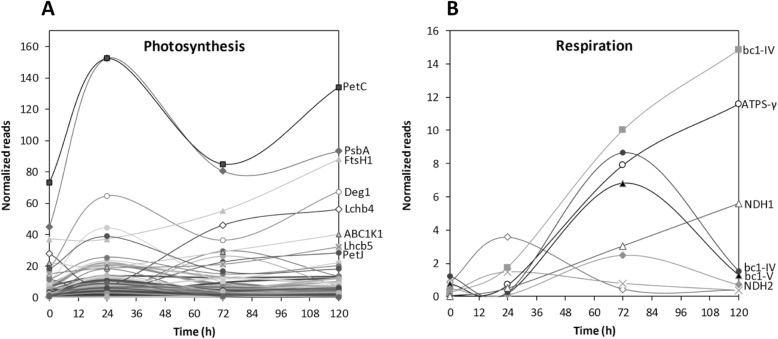
The levels of normalized reads of transcripts encoding proteins involved in photosynthesis that showed the highest increases were: subunit PetC of cytb6f that increased at day 1, decreased at day 3, and increased again at day 5; subunit PsbA of PSII that increased at day 1, decreased at day 3, and slightly increased at day 5; protease FtsH1 that increased from day 3 to day 5; proteases Deg1 that increased at day 1, decreased at day 3, and increased again at day 5; subunits LhB4 and LhB5 of LHCII that increased at day 3 and remained increased until day 5; subunit PetJ of cytb6f and the kinase ABC1K1 that increased at day 1, decreased at day 3 and remained decreased at day 5 (Fig. 1a).
Fig. 1.
Level of the highest increased transcripts encoding proteins involved in photosynthesis (a) and respiration (b) in U. compressa cultivated with 10 μM copper for 0, 1, 3 and 5 days. Transcripts encoding proteins involved in photosynthesis corresponding to subunit PetC of cytb6f complex (PetC), subunit PsbA of PSII (PsbA), the protease FstH1 (FstH1), the protease Deg1 (Deg1), subunit 4 of LHCII (Lhcb4), the kinase ABC1K1, subunit 5 of LHCII (Lhcb1) and subunit PetJ of cytb6f complex (PetJ) are indicated with an arrow (a). Transcripts encoding proteins of the mitochondrial electron transport chain (respiration) corresponding to subunit IV of bc1 complex (complex III), subunit γ of ATP synthase (ATP-γ), subunit 1 of NADH dehydrogenase (complex I), subunit V of bc1 complex (complex III) and subunit 2 of NADH dehydrogenase (complex I) are indicated with an arrow (b). The level of transcripts is expressed as the number of normalized reads and time in days
The levels of normalized transcripts encoding subunits of mitochondrial electron chain that showed the highest increases were: cytochrome c1 (subunit IV of complex III); subunit γ of ATP synthase and subunit 1 of NADH dehydrogenase (complex I) that increased at day 3 and remained increased until day 5; cytochrome c1 (subunit IV of complex III) and subunit γ of ATP synthase that increased at day 3 and decreased at day 5; subunit V of complex III and subunit 2 of NADH dehydrogenase that increased at day 1, decreased at day 3, and remained decreased at day 5 (Fig. 1b).
Kinetics of normalized reads of transcripts involved in C, N and S assimilation
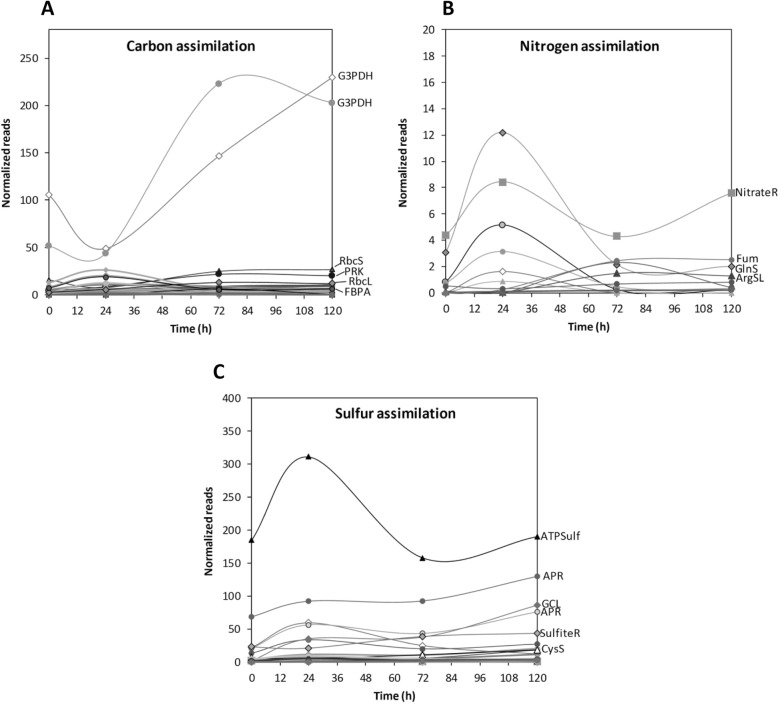
The levels of normalized reads of transcripts encoding enzymes involved in C assimilation that showed the highest increases were: G3PDH that increased mainly at day 3, and remained increased until day 5; rubisco small subunit RbcS, rubisco large subunit RbcL and enzymes PRK and FBPA that increased at day 3 and remained increased until day 5 (Fig. 2a).
Fig. 2.
Level of the highest increased transcripts encoding enzymes involved in C, N and S assimilation in U. compressa cultivated with 10 μM copper for 0, 1, 3 and 5 days. Transcripts encoding enzymes involved in C assimuilation corresponding to glyceraldehyde 3-P dehydrogenase (G3PDH), small subunit of rubisco (RbcL), phosphorribulo kinase (PRK), large subunit of rubisco (RbcL) and fructose biphosphate aldolase (FBPA) are indicated with an arrow (a). Transcripts encoding enzymes involved in N assimilation corresponding to nitrate reductase (NitrateR), fumarase (Fum), glutamine synthase (GlnS) and arginine-succinate lyase (ArgSL) are indicate with an arrow (b). Transcripts encoding enzymes involved in S assimilation corresponding to ATP sulfurylase (ATPS), APS reductase (APSR) glutamine cysteine ligase (GCL), sulfite reductase (SulfiteR) and cysteine synthase are indicated with an arrow (c). The level of transcripts is expressed as the number of normalized reads and time in days
The levels of normalized reads of transcripts encoding enzymes involved in N assimilation that showed the highest increases were: nitrate reductase that increased day 1, decrease at day 3, and increased again at day 5; glutamine synthase that increased at day 1, decreased at day 3, and remained decreased at day 5; fumarate hydratase and argino-succinate lyase that increased at day 3 and remained increased until day 5 (Fig. 2b).
The levels of normalized reads of transcripts encoding enzymes involved in S assimilation showing the highest increases were: ATP sulfurylase that increased at day 1, decreased at day 3, and slightly increased at day 5; APS reductase that increased at day 3 and continued to increase until day 5; glutamine cysteine ligase and APR reductase that increased at day 3 and remained increased until day 5; sulfite reductase that increased at day 1 and remained increased at days 3 and 5; and cysteine synthase that increased at day 3 and remained increased at day 5 (Fig. 2c). It is important to mention that the number of normalized reads of transcripts encoding enzymes of C and S assimilation showed higher levels (20 to 300 reads) compare to those of enzymes involved in N assimilation (1–12 reads).
Transcripts with decreased levels encode proteins involved in protein synthesis and degradation, signal transduction, and replication and DNA repair
The most down-regulated transcripts at time points 0 vs. 1, 0 vs. 3 and 0 vs. 5 were involved in protein synthesis and degradation, signal transduction and, replication and DNA repair. Transcripts encoding proteins involved in protein synthesis and degradation were: ribosomal proteins L2, L15, L16, S12 and S17, ribosomal protein S6 kinase, eukaryotic translation factor 5B, tRNA dehydrouridine synthase, elongation factor 2a kinase, cysteine tRNA ligase, glu-tRNA amidotransferase, peptidyl prolyl trans-isomerase CYP61, RING finger protein 32, protease ESD4, neurotrypsin, E3 ubiquitin protein ligase, signal peptide peptidase, prefoldin subunit 6, Hsp40 (DNAJ) and hsp70, among others (Additional file 5: Table S2). Transcripts encoding proteins involved in signal transduction were: calreticulin, the serine/threonine protein kinases PKWA, PRP4, SAPK8 and SAPK38, the MAPKKK11, myb-related protein 1, tyrosine protein kinase SRK3, the protein phosphatases 1 regulator subunit 7 and the bifunctional phophatase IMPL2, among others (Additional file 5: Table S2). Transcripts encoding proteins involved in replication and DNA repair were: histone deacetylase HD11, DNA polymerase ε subunit subunit B, DNA topoisomerase 6 subunit A, DNA helicase II, RNA helicase DExH10, RNA polymerase I subunit RPA12, DNA repair protein RAD45, and DNA mismatch repair protein MSH13, among others (Additional file 5: Table S2).
Copper-induced increases in net photosynthesis and respiration
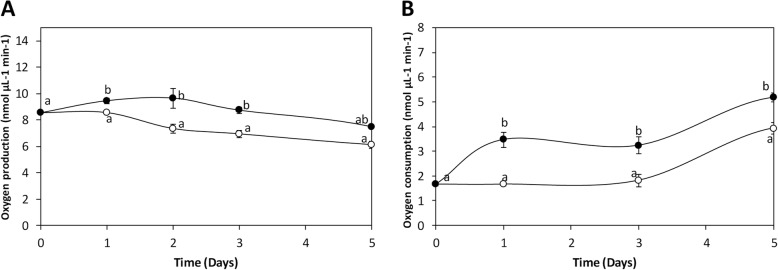
The level of produced oxygen under light (photosynthesis) decreased in control alga from 8.5 to 6.1 nmoles μL− 1 min− 1 whereas in treated alga it increased from 8.5 to 9.6 nmoles μL− 1 min− 1 at day 2, which represent a 32% of increase compare to control, and then decreased to 7.5 nmoles μL− 1 min− 1 at day 5, which represent a 23% of increase compare to control (Fig. 3a). The level of consumed oxygen in dark (respiration) in control algae was 1.7 nmoles μL− 1 min− 1 and did not change at day 1, but increased to 3.8 nmoles μL− 1 min− 1 at day 5 whereas in treated alga it increased from 1.7 to 3.5 nmoles μL− 1 min− 1 at day 1 (106% of increase), and to 4.5 nmoles μL− 1 min− 1 at day 5 (18% of increase) (Fig. 3b). Thus, net photosynthesis and respiration increased in U. compressa exposed to copper excess. It is important to mention that oxygen produced in high light was higher than oxygen consumed in dark, which is in accord with level of normalized reads observed in kinetic analyses.
Fig. 3.
Level of oxygen production under light (a) and oxygen consumption in the dark (b) in the marine alga U. compressa cultivated in control condition (open circles) and with 10 μM copper (black circles) for 5 days. The level of oxygen is expressed in nanomoles per microliter per minute, and time in days. Symbols represent the mean value of three independent experiments ± SD. Different letters indicate significant differences (P < 0.05)
Copper-induced increases in activities of enzymes involved in C, N and S assimilation
The activity of the enzyme rubisco in control algae was 2 μmol min− 1 mg− 1 of protein and it remained at this level until day 5 whereas in treated alga it increased to 4 μmol min− 1 mg− 1 of protein at day 3 and decreased to 3 μmol min− 1 mg− 1 of protein at day 5 (Fig. 4a). The activity of glutamine synthase increased in control alga from 0.15 to 0.22 nmol min− 1 mg− 1 of protein, and in treated alga it increased to 0.61 nmol min− 1 mg− 1 of protein at day 3 and then decreased to 0.1 nmol min− 1 mg− 1 of protein at day 5 (Fig. 4b). The activity of cysteine synthase in control algae increased from 1.8 to 2.3 μmol min− 1 mg− 1 of protein whereas in treated algae it increased to 3.3 μmol min− 1 mg− 1 of protein at days 3 and remained at this level at day 5 (Fig. 4c). Thus, the activity of key enzymes involved in C, N and S assimilation increased in U. compressa in response to copper excess. The activities of enzymes rubisco and cysteine synthase were higher than the activity of glutamine synthase, which is in accord with the level of normalized reads.
Fig. 4.
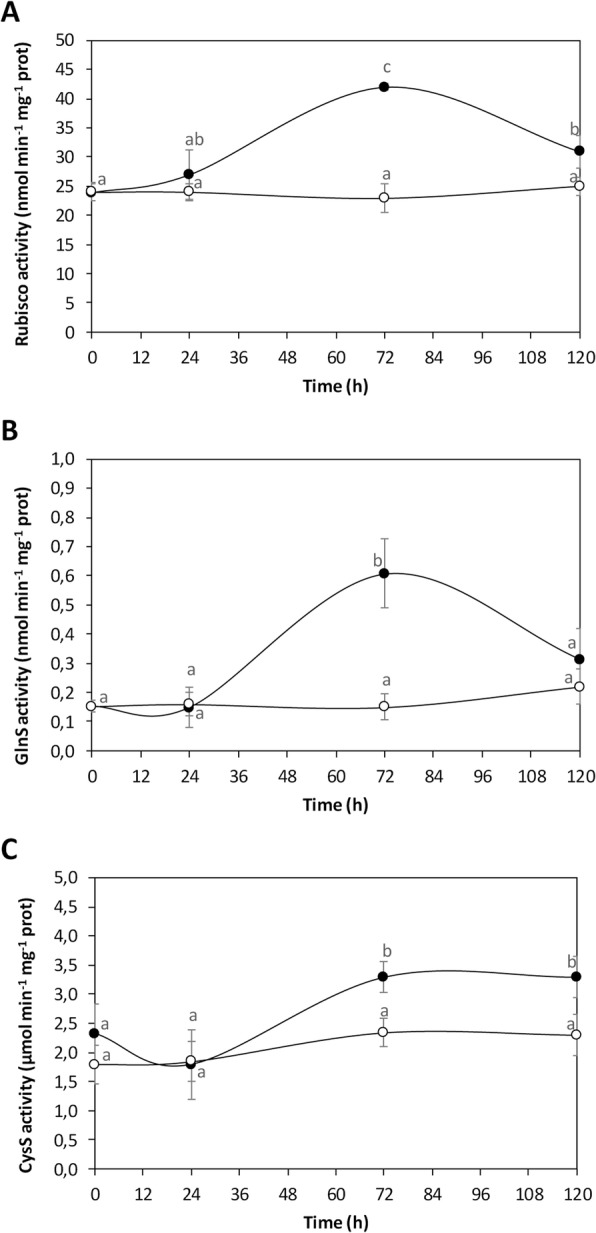
Activity of key regulatory enzymes involved in C assimilation, ribulose 1.5 biposphate carboxylase /oxygenase (rubisco, a), N assimilation, glutamine synthase (GlnS, b) and in S assimilation, cysteine synthase (CysS, c). The activity rubisco and GlnS is expressed in nanomoles per minute per milligram of proteins and the activity of CysS is expressed in micromoles per minute per milligram of proteins, and time in days. Symbols represent mean values of three independent experiments ± SD. Letters indicate significant differences (P < 0.05)
Discussion
Copper-induced increased expression of proteins involved in photosynthesis and in repair and protection of photosystems
In this work, we showed that the marine alga U. compressa cultivated with 10 μM copper for 5 days displayed an increased expression of transcripts encoding subunits of LHCII, PSII, cytb6f, LHCI, PSI and ATP synthase as well as proteins involved in repair of PSII and protection of PSI. It is important to mention that the level of transcripts encoding subunits of PSII, PSI, and proteins involved in PS repair and protection, showed higher increases compared with those observed in the alga cultivated with 10 μM copper for 0 to 24 h [8]. In this sense, the level of transcripts encoding subunits of PSII increased 2 Log2 FC in the alga cultivated with copper for 24 h whereas it increased 5.3 Log2 FC in the alga cultivated with copper for 5 days [8]. In particular, the level of transcripts encoding subunits PsbB and PsbW increased 2 and 3.3 Log2 FC, respectively, in the alga exposed to copper for 24 h, whereas the level of these proteins increased 4.5 and 5.3 times, respectively, in the alga cultivated with copper for 5 days [8]. The level of transcripts encoding the subunit of PSI, PsaA, increased 3.4 Log2 FC in the alga cultivated with copper for 24 h instead it increased 7.7 Log2 FC in the alga cultivated with copper for 5 days [8]. The level of transcripts encoding the chaperone MET1, involved in the replacement of oxidized PsbA, increased 3.4 Log2 FC in the alga cultivated with copper for 24 h whereas it increased 5.6 Log2 FC in the alga cultivated with copper for 5 days [8]. Thus, the expression of genes encoding proteins that constitute photosystems and those involved in repair and protection of photosystems was higher in the alga cultivated with copper for days compare with the alga cultivated with copper for hours.
Net photosynthesis increased in the alga cultivated with 10 μM copper for 1 to 5 days. This is in accord with the increase in the level of transcripts encoding proteins that constitute PS and those involved in repair and protection of PS. In this sense, photosynthesis also increased in the green macroalga U. pertusa cultivated with 4 μM of copper for 3 days and in U. flexuosa cultivated with 0.8 μM of copper for 8 days [6]. In addition, an increase in photosynthesis was observed in the red macroalga P. haitiensis cultivated with 1 μM of copper for 3 days and in the brown macroalga E. siliculosus cultivated with 0.8 μM of copper for 8 h [9–11]. Furthermore, it was determined that U. compressa cultivated 50 μM of copper for 7 days showed no sign of cell mortality [12]. Thus, U. compressa is more tolerant to copper excess than other green, red and brown marine macroalgae. In addition, it has been shown that U. compressa cultivated with 10 μM of copper for 0 to 7 days showed an increase in the level of superoxide anions beginning at day 3 and increasing until day 7 and that the production of superoxide anions occurred in chloroplats and mitochondria [12]. Thus, copper-induced oxidative stress may damage proteins present in photosystems and these proteins may be synthesized and replaced due to an increase in their expression. The kinetic analyses of the level of transcripts showed that the most increased transcripts encode the subunit PsbA of PSII which is highly susceptible to oxidation [21, 22], the protease MET1 involved in removing damaged subunit PsbA, the ATP-dependent protease FtsH1 and the ATP-independent protease Deg1 involved in degradation of damaged PsbA. In fact, the removing of oxidized PsbA and its degradation requires the concomitant action of FtsH and Deg proteases as well as other proteases such as Clp and SppA [23, 24]. Other higher expressed proteins were subunits LhcB4 and LhcB2 of LHCII, the iron-sulfur Rieske subunit PetC of cytb6f, and ABC1K1. Thus, subunit PsbA, proteins present in PSII, a subunit of cytb6f, and the kinase ABC1K1 are highly expressed proteins in response to copper stress and they may correspond to proteins that are more susceptible to oxidative damage as well as those required to remove and replace oxidized proteins.
Copper-induced increased expression of proteins involved in respiration
U. compressa cultivated with copper excess showed an increase in expression of transcripts encoding subunits of mitochondrial complexes I and III, and a subunit of ATP synthase, as well as an increase in respiration. This is in accord with findings in the red macroalga P. haitiensis cultivated with 0.1 to 50 μM of copper for 3 days that showed an increase in respiration with 0.1 to 50 μM, but displayed an inhibition in photosynthesis over 1 μM copper [10]. The number of proteins involved in respiration that were overexpressed in U. compressa is lower than those involved in photosynthesis suggesting that respiration is less sensitive to copper-induced oxidative stress than photosynthesis. In this sense, it has been reported that respiration is less sensitive to copper- and chromium-induced oxidative stress due to the accumulation of superoxide anions in organelles of Euglena gracilis [25]. The increase in respiration observed in U. compressa under copper stress may increase the level of ATP that is required for ATP-dependent synthesis and degradation of proteins, fatty acids and chlorophylls, as it has been observed in plants under abiotic stress [26]. Several enzymes involved in the synthesis and degradation of protein, fatty acids, chlorophylls appeared to be overexpressed in transcriptomes of U. compressa cultivated for 10 μM copper for 5 days (data not shown) indicating the high turnover of proteins, fatty acids and chlorophylls is occurring in response to copper stress.
Copper-induced increased expression of enzymes involved in C, N and S assimilation
The highest expressed transcripts encoding proteins involved in C, N and S assimilation were enzymes involved in the reduction of CO2, nitrate and sulfate as well as detoxification of ammonium and synthesis of glutathione. It is well known that enzymes involved in assimilation of C, N and S are regulated through thioredoxins and that the latter are regulated by cellular redox state [27]. Regarding C assimilation, it has been shown that the enzymes of the Calvin-Benson cycle G3PDH, PRK, FBP and sedoheptulose-1,7-biphosphatase are regulated by thioredoxins in plants [28] and the small and large subunit of rubisco, PGK and R5PE are regulated by thioredoxins in the green microalga Chamydomonas reinhardtii [29, 30]. Regarding N and S assimilation, it has been determined that glutamine synthase and adenosine-5′-phosphosulfate reductase can bind thioredoxins suggesting these enzymes may be regulated by these proteins [31]. On the other hand, it is well known that enzymes of the Calvin-Benson cycle enzymes and thioredoxins can be directly oxydized by Reactive Oxygen Species (ROS) such as hydrogen peroxide and superoxide anions [31]. As mentioned before, copper stress induced the accumulation of superoxide anions in U. compressa, beginning at day 3 and increasing until day 7 [12]. This wave of superoxide anions may directly oxidize and inhibit enzymes involved in C, N and S assimilation and/or thioredoxins. In order to restore the level of active enzymes, an increase in expression of the genes encoding enzymes involved in C, N and S assimilation, and thioredoxins (data not shown) was observed in the marine alga U. compressa in response to copper stress.
Copper-induced decrease in expression of genes involved in protein synthesis and degradation, cell signaling, and replication and DNA repair
The levels of transcripts that decreased the most were those encoding proteins involved in protein synthesis and degradation, signal transduction, and replication and DNA repair. Regarding protein synthesis, the level of transcripts encoding ribosomal proteins L2, L15, L16, S12 and S17 decreased. The latter contrast with the level of transcripts that increased in the alga cultivated with 10 μM for 3 days corresponding to ribosomal L9, 10, 23, 28, 31, 35 and S12, 15 and 20 [32]. In addition, the levels of chaperone proteins Hsp40 and Hsp70 were decreased whereas the level of Hsp14 was increased in the alga cultivated with copper for 3 days [32]. Regarding protein degradation, the level of transcripts encoding RING protein 32 appeared to be decreased whereas that the level of RING protein RBX1 increased in the alga cultivated with copper for 3 days [33]. Regarding signal transduction proteins, the level of transcripts encoding calreticulin, several serine/threonine kinases, a MAPKKK and a myb-related protein decreased whereas the level of a calmodulin, a histidine kinase and a phospholipase A increased in the alga cultivated with copper for 3 days [32]. Thus, transcripts that are decreased in response to copper excess encode proteins that differ from those that are increased in response to the copper stress. Regarding the MAPKKK, it has been shown that U. compressa cultivated with 10 μM copper, and with inhibitors of the MAPK corresponding to ERK, JNK and p-38, for 6 h to 6 days, displayed mostly an increase in the level of transcripts encoding antioxidant enzymes [34] which suggests that MAPK pathways mainly inhibit the expression of genes encoding antioxidant enzymes. In this sense, the level transcripts of MAPKKK11 that decreased in response to copper stress (this work) also suggest that MAPK pathways mostly inhibit the expression of genes encoding antioxidant enzymes.
The importance of S assimilation in U. compressa in response to copper stress
The activity of the enzyme cysteine synthase was higher than the activity of rubisco and much higher than the activity of glutamine synthase indicating the importance of S assimilation in U. compressa. In this sense, it has been shown that the alga exposed to increasing concentrations of copper for 0 to 12 days displayed an increase in the level of glutathione (GSH) and phytochelatins (PCs), which are sulfur containing peptides [15]. The nano-equivalents of thiol groups present in glutathione and phytochelatins correlated with the level of intracellular accumulated copper [15]. In addition, the level of transcripts of encoding MTs also increased in response to increasing concentration of copper [16]. Thus, cysteine-rich peptides and proteins are involved in copper accumulation and probably participate in the inhibition of copper-induced oxidative stress in U. compressa by sequestering copper ions [12]. Thus, increased S assimilation allowing an enhanced synthesis of thiol-rich peptides and the increased expression of thiol-rich proteins involved in sequestering copper ions may be essential for copper tolerance and accumulation in U. compressa.
Conclusions
The marine alga U. compressa showed an increase in net photosynthesis and respiration as well as in activities of enzymes involved C, N and S assimilation in response to copper excess and these responses were due, at least in part, to an enhanced expression of genes encoding proteins involved in these processes. The combination of these responses may represent an exceptional mechanism of copper tolerance among photosynthetic organisms.
Methods
Alga and seawater sampling
U. compressa was collected in Cachagua (32° 34S′), a costal site with no history of metal pollution, during spring 2018 and transported to the laboratory at 4 °C inside a cooler. Algae were identified visually based on their phenotype and have been previously identified based on the sequence of 18S cDNA. Algae were rinsed three times with filtered seawater collected in Quintay (33° 12′S), a pristine site. Algae were cleaned manually and sonicated three min in an ultrasound bath (Branson, Danbury, CT, USA) in order to remove epiphytic bacteria and organic debris.
In vitro cultures
U. compressa (100 mg of fresh tissue) was cultivated in seawater without copper addition (control, day 0) and with 10 μM CuCl2 for 1, 3 and 5 days under irradiance of 50 μmoles m− 2 s− 1 and a photoperiod of 12 h light:12 h darkness, and each sample in duplicate. All samples were washed with 2 ml of 100 mM Tris-10 mM EDTA pH = 7.0, twice for 10 min, in order to eliminate copper bound to algal cell walls. Samples were dried with paper, frozen in liquid nitrogen, and stores at − 80 °C.
RNA extraction and preparation of cDNA libraries
Total RNA was extracted from samples cultivated for 0, 1, 3 and 5 days using EZNA total RNA kit (Omega Biotek, GA, USA). To this end, the alga (100 mg) was frozen in liquid nitrogen and homogenized in 1 mL of TRK buffer with 20 μL of β-mercaptoetahnol. The homogenate was centrifuged at 12,000 rpm for 15 min and the supernatant was recovered. The supernatant was mixed with ethanol 70% and transferred to HiBind RNA mini column and washed with RNA washing solutions I and II. Total RNA, was eluted with 50 μL of distilled water treated with DEPC. Total RNA was cleaned using GenJet RNA cleanup and concentration micro kit (Thermo, MS, USA). Total RNA samples were send to BGI Genomic Center (Shenzen, China) where RNA quality was checked, stranded and pair-ended cDNA libraries were prepared and sequencing was performed using an Illumina 4000 sequencer.
De novo assembly and annotation
Reads obtained using Illumina sequencing were trimmed, cleaned and assembled using Trinity software at BGI Genome Center (Shenzen, China). Transcripts were translated into proteins using BlastX software and UniprotKB data base, annotated using an e-value of e− 3 or lower and classified according to Gene Ontology (GO) using OmicsBox software (Biobam, VA, Spain) and those having an e-value of e− 3 or lower were selected.
Identification of differentially expressed transcripts
Cleaned reads were mapped against transcriptomes using Bowtie2 software and raw reads were counted using eXpress (version 1.5.1). Reads were then normalized to CPM units using Trinity’s script abundance_estimates_to_matrix.pl under default settings. Differentially expressed transcripts were identified using EdgeR at an FDR < 0.01, and Log2 Fold of Change > 1 for up-regulated transcripts and Log2 Fold of Change < − 1 for down-regulated transcripts. Differentially expressed transcripts were identified at times: 0 vs.1, 0 vs. 3, 0 vs. 5 days. Differentially expressed transcripts were visualized as a heat-map using Spearman’s correlation coefficient on transcripts and samples, and hierarchical clustering (Additional file 4: Figure S3A). All biological replicates were more similar to each other than to other samples (Additional file 4: Figure S3B). To identify down-regulated transcripts, the one hundred more down-regulated transcripts at each sample time point were identified and they were classified in different processes such as protein synthesis and degradation, fatty acid synthesis and degradation, DNA synthesis and degradation, replication and DNA repair, transcription, splicing, secondary metabolism, cell growth, among others.
Kinetics of transcripts encoding proteins involved photosynthesis, respiration and C, N and S assimilation
Transcripts encoding proteins involved in photosynthesis and respiration and enzymes involved in C, N and S assimilation were selected based on the annotation and GO domain (biological processes). Reads of selected transcripts were normalized using TMM normalization method and they were analyzed using MEV software. Groups of transcripts showing similar temporal expression pattern were created and transcripts showing an increased expression were selected in these groups.
Quantification of photosynthesis and respiration
Photosynthesis was quantified cultivating 25 mg of fresh tissue (FT) of U. compressa in 2 mL of seawater in an oxygraph chamber (Hansatech, Norfolk, UK). O2 production was detected for 10 min, under a light intensity of 425 μmoles m− 2 s− 1 and respiration was determined cultivating 25 mg of the alga (FT) in 2 mL of seawater in the oxygraph chamber, and O2 consumption was detected for 10 min in darkness.
Preparation of protein extracts
One g of U. compressa (FT) was frozen in liquid nitrogen and homogenized in a mortar. Three mL of 100 mM phosphate buffer pH = 7.0 containing 5 mM β-mercaptoethanol were added and the homogenization was continued. The homogenate was centrifuged at 14,000 rpm for 15 min and the supernatant was recovered. Proteins in the supernatant were precipitated by addition of 0.6 g of ammonium sulfate per mL of extract and the mixture centrifuged at 15,000 rpm for 30 min. Protein pellets were solubilized in 200 μL of phosphate buffer pH = 7.0 containing 2 mM of β-mercaptoethanol and 20% glycerol. Final extracts contained around 3 mg mL− 1 of proteins and they were stored at − 80 °C.
Detection of enzymes activities involved in C, N and S assimilation
Rubisco activity was detected as described in [35]. One mL the reaction mixture containing 100 mM Tris-HCl pH 8.0, 1 mM ribulose 1,5-biphosphate, 10 mM KHCO3, 20 mM MgCl2, 5 mM creatine phosphate, 3 mM ATP, 10 U phosphoglycerate kinase, 10 U glyceraldehyde 3-phosphate dehydrogenase, 10 U creatine kinase, 0.15 mM NADH and 30 μg of protein extract was used to detect rubisco activity. The decrease in absorbance at 340 nm due to consumption of NADH was detected for 3 min and activity was calculated using the extinction coefficient of NADH (ε = 6.2 mM− 1 cm− 1).
Glutamine synthase (GlnS) activity was detected as described in [36]. One mL of the reaction mixture containing 200 mM HEPES buffer pH 7.0, 50 mM L- glutamate, 5 mM hydroxylamine, 5 mM MgCl2, 20 mM ATP and 150 μg of protein extract was used to detect GlnS activity. The reaction was incubated at 37 °C for 1 h and stopped by addition of 1 mL mixture containing 0.7 M ferric chloride, 20% (w/v) trichloroacetic acid and 0.3 M HCl. The mixture was centrifuged at 7400 g for 5 min and the supernatant was recovered. The absorbance of the supernatant was detected at 540 nm and activity was calculated using the extinction coefficient of γ-glutamyl-hydroxamate (ε = 0.85 mM− 1 cm− 1).
Cysteine synthase (CysS) activity was detected as described in [37]. One mL of the reaction mixture containing 50 mM phosphate buffer pH = 7.5, 10 mM O-acetylserine, 2 mM Na2S, 30 mM DTT, and 50 μg protein extract was used to detected CysS activity. The reaction was incubated at 37 °C for 1 h and the reaction was stopped by addition of 0.5 mL of 20% (w/v) trichloroacetic acid. Cysteine was detected by addition of 100 μL of acetic acid and 200 μL of ninhydrin reagent. The mixture was placed in boiling water for 10 min, rapidly cooled in ice and 550 μL of 95% (v/v) ethanol were added. The absorbance of the mixture was determined at 560 nm and activity was determined using the extinction coefficient the compound formed by ninhydrin and cysteine (ε = 25 mM− 1 cm− 1).
Statistical analyses
Significant differences in the detection production and consumption of oxygen and in activities of enzymes involved in C, N and S assimilation were determined using one-way ANOVA at 95% of confidence interval, followed by Tukey’s multiple comparison post-test using the statistical software Prism6 (Graphpad Sofware Inc., CA, USA).
Supplementary information
Additional file 1: Table S1. Sequencing, pre-processing of reads and assembly of Ulva compressa transcriptomes.
Additional file 2: Figure S1. Pie-Chart of the percentage of proteins involved in different biological processes in U. compressa.
Additional file 3: Figure S2. Pie-Chart of the percentage of proteins having similarity with plant and animal proteins.
Additional file 4: Figure S3. Heatmap of differentially expressed transcripts in U. compressa exposed to copper excess.
Additional file 5: Table S2. List of all transcripts in time points 0 vs. 1, 0 vs.3, 0 vs. 5, their differential expression and the encoded protein.
Additional file 6: Figure S4. Scheme of chloroplast Light Harvesting complex II (LHCII), photosystem II (PSII), cytochrome b6f, photosystem I (PSI), Light Harvesting Complex II and ATP synthase (A). Scheme of mitochondrial complex I, II, III and IV and ATP synthase (B). Subunits showing an increased expression are highlighted in black.
Acknowledgments
Not applicable.
Abbreviations
- APS reductase
Adenosine 5′-phosphosulphate reductase
- CysS
Cysteine synthase
- FBP
Fructose 1,6 biphosphatase
- FBPA
Fructose biphosphate aldolase
- FC
Fold of change
- FT
Fresh tissue
- G3PDH
Glyceraldehyde 3-P dehydrogenase
- GlnS
Glutamine synthase
- GSH
Glutathione
- MTs
Metallothioneins
- PCs
Phytochelatins
- PGK
Phosphoglycerate kinase
- PRK
Phosphoribulose kinase
- R5PI
Ribose 5-P isomerase
- rubisco
Ribulose 1,5- biposphate carboxylase /oxygenase
- TK
Transketolase
Authors’ contributions
DL did the bioinformatics work and determined activities of enzymes involved in C, N and S assimilation. AG did analyzed net photosynthesis and respiration and helped preparing figures and tables; FR and AZ prepared total RNA of samples in duplicate that were sequenced by RNA-seq. EC-N participate in bioinformatics analyses, CAS helped with discussion and revised the manuscript, and AM wrote the manuscript. All authors have read and approved the manuscript.
Funding
This work was funded by Fondecyt 1160013 to A.M., Fondecyt 3170511 to D. L, and Fondecyt 1160369 to C.A.S. The funding agencies did not play a role in the experimental design, results analysis or writing of the manuscript, but did provide financial support for the manuscript.
Availability of data and materials
Reads have been deposited in SRA database (NCBI), the accession number is PRJNA557176, and they will be released on 24th august 2020. Experimental data is available at the on line repository: 10.6084/m9.figshare.9108491
Ethics approval and consent to participate
Algae collection and experimental procedures were performed according to guidelines of Biosafety Committee of University of Santiago of Chile.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12870-019-2229-5.
References
- 1.Burkhead JI, Gogolin-Reynolds KA, Abdel-Ghany SF, Cohu CM, Pilon M. Copper homeostasis. New Phytol. 2009;182:799–816. doi: 10.1111/j.1469-8137.2009.02846.x. [DOI] [PubMed] [Google Scholar]
- 2.Bothe H. Plants in heavy metal soils. In: Sherameti J, Varna A, editors. Detoxification of Heavy Metals. Berlin: Springer; 2011. pp. 35–57. [Google Scholar]
- 3.Küpper H, Küpper F, Spiller M. Environmental relevance of heavy metal-substituted chlorophylls using the example of a water plant. J Exp Bot. 1996;47:259–266. doi: 10.1093/jxb/47.2.259. [DOI] [Google Scholar]
- 4.Küpper H, Setik I, Spiller M, Küpper F, Prasil O. Heavy-metal induced inhibition of photosynthesis targets in vivo heavy metal chlorophyll formation. J Phycol. 2002;38:429–441. [Google Scholar]
- 5.Valasata I, Rosato A, Fuhrman N, Thornton JM, Andreini C. To what extent do structural changes in catalytic metal sites affect enzyme function? J Inorg Chem. 2018;179:40–53. doi: 10.1016/j.jinorgbio.2017.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han T, Kang SH, Park JS, Lee HK, Brown MT. Physiological responses of Ulva pertusa and U. armoricana to copper exposure. Aquat Toxicol. 2008;86:176–184. doi: 10.1016/j.aquatox.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 7.Andrade LR, Farina M, Amado-Filho GM. Effect of copper on Enteromorpha flexuosa (Chlorophyta) in vitro. Ecotoxicol Environ Saf. 2004;58:117–125. doi: 10.1016/S0147-6513(03)00106-4. [DOI] [PubMed] [Google Scholar]
- 8.Rodríguez F, Laporte D, González A, Méndez K, Castro-Nallar E, Meneses C, et al. Copper-induced increased expression of genes involved photosynthesis, carotenoid synthesis and C assimilation in the marine alga Ulva compressa. BMC Genomics. 2018;19:829. doi: 10.1186/s12864-018-5226-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tonon-Pedroso A, Zaini PA, Dos Reis-Falcao V, Oliveira MC, Collén J, Boyen C, et al. Gracilaria tenustipitata (Rodophyta) tolerance to cadmium and copper exposure observed through gene expression and photosynthesis analyses. J Appl Phycol. 2018;30:1–13. doi: 10.1007/s10811-017-1198-z. [DOI] [Google Scholar]
- 10.Li YX, Zhou S, Zhao FJ, Liu Y, Fan PP, Wang GC. Physiological responses of Porphyra haitiensis to different copper and zinc concentrations. Braz J Oceanogr. 2010;58:261–267. doi: 10.1590/S1679-87592010000400001. [DOI] [Google Scholar]
- 11.Ritter A, Dittami SM, Goultiquer S, Correa JA, Boyen C, Potin P. Transcriptomic and metabolomics analysis of copper stress acclimation in Ectocarpus siliculosus highlights signaling and tolerance mechanisms in brown algae. BMC Plant Biol. 2014;14:116. doi: 10.1186/1471-2229-14-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.González A, Vera J, Castro J, Dennett G, Mellado M, Morales B, et al. A. co-occuring increases of calcium and organellar reactive oxygen species determine differential activation of antioxidant and defense enzymes in Ulva compressa (Chlorophyta) exposed to copper excess. Plant Cell Environ. 2010;33:1627–1640. doi: 10.1111/j.1365-3040.2010.02169.x. [DOI] [PubMed] [Google Scholar]
- 13.González A, Cabrera MA, Henríquez MJ, Contreras RA, Morales B, Moenne A. Cross talk among calcium, hydrogen peroxide and nitric oxide and activation of gene expression involving calmodulins and calcium-dependent protein kinases in Ulva compressa exposed to copper excess. Plant Physiol. 2012;158:1451–1462. doi: 10.1104/pp.111.191759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mellado M, Contreras RA, González A, Dennett G, Moenne A. Copper-induced synthesis of ascorbate, glutathione and phytochelatins in the marine alga Ulva compressa (Chlorophyta) Plant Physiol Biochem. 2012;51:102–108. doi: 10.1016/j.plaphy.2011.10.007. [DOI] [PubMed] [Google Scholar]
- 15.Navarrete A, González A, Gómez M, Contreras RA, Díaz P, et al. Copper excess detoxification is mediated by a coordinated and complementary induction of glutathione, phytochelatins and metallothioneins in the green seaweed Ulva compressa. Plant Physiol Biochem. 2019;135:423–431. doi: 10.1016/j.plaphy.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Laporte D, Valdés N, González A, Sáez CA, Zúñiga A, Navarrete A, et al. Copper-induced overexpression of genes encoding antioxidant system enzymes and metallothioneins in volve the activation of CaMs, CDPKs and MEK1/2 in the marine alga Ulva compressa. Aquat Toxicol. 2016;177:433–440. doi: 10.1016/j.aquatox.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Zúñiga A, Laporte D, González A, Gómez M, Sáez CA, Moenne A. Isolation and characterization of metallothioneins from the marine alga Ulva compressa (Chlorophyta). Int J Mol Sci. 2020;21:153. [DOI] [PMC free article] [PubMed]
- 18.Kopriva S, Mulheim R, Koprivova A, Trashsel N, Catalano C, Suter M, et al. Light regulation of assimilatory sulfate reduction in Arabidopsis thaliana. Plant Cell. 1999;20:37–41. doi: 10.1046/j.1365-313x.1999.00573.x. [DOI] [PubMed] [Google Scholar]
- 19.Kopriva S, Suter M, Von Baalmos P, Hesse H, Krähenbühl U, Rennenberg H. Interaction of sulfate assimilation with carbon and nitrogen metabolism in Lemna minor. Plant Physiol. 2002;130:1406–1413. doi: 10.1104/pp.007773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopriva S, Rennenberg H. Control of sulfate assimilation and glutathione synthesis: interaction with C and N metabolism. J Exp Bot. 2004;55:1831–1842. doi: 10.1093/jxb/erh203. [DOI] [PubMed] [Google Scholar]
- 21.Foyer C. Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ Exp Bot. 2018;154:134–142. doi: 10.1016/j.envexpbot.2018.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kato Y, Sun X, Zhang L, Sakamoto W. Cooperative D1 degradation in the photosystem II repair mediated by chloroplastic proteases in Arabidopsis. Plant Physiol. 2012;159:1428–1439. doi: 10.1104/pp.112.199042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dalal VK, Tripathi BC. Water-stress induced downsizing of light-harvesting antenna complex protects developing rice seedlings from photo-oxidative damage. Sci Rep. 2018;8:5955. doi: 10.1038/s41598-017-14419-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martinis J, Glauser G, Valimareanu S, Stettler M, Zeeman SC, Yamamoto H, et al. ABC1K1/PGR6 kinase: a regulatory link between photosynthetic activity and chloroplast metabolism. Plant J. 2014;77:269–283. doi: 10.1111/tpj.12385. [DOI] [PubMed] [Google Scholar]
- 25.Rochetta I, Küpper H. Chromium and copper-induced inhibition of photosynthesis in Euglena gracilis analyzed on the single-cell level by fluorescence kinetic microscopy. New Phytol. 2009;182:405–420. doi: 10.1111/j.1469-8137.2009.02768.x. [DOI] [PubMed] [Google Scholar]
- 26.Aráujo WL, Tohge T, Ishizaki K, Leaver CL, Fernie AR. Protein degradation, an alternative respiratory substrate for stressed plants. Trends Plant Sci. 2011;16:9. doi: 10.1016/j.tplants.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 27.Michelet L, Zaffagnini M, Morisse S, Sparla F, Pérez-Pérez F, Lemaire SD. Redox regulation of the Calvin-Benson cycle: something old, something new. Front Plant Sci. 2013;4:470. doi: 10.3389/fpls.2013.00470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemaire SD, Guillon B, Le Maréchal P, Keryere E, Migniac-Maslow M, Decottignies P. New thioredoxin targets in the unicellular photosynthetic eukaryote Chlamydomonas reinhardtii. Proc Natl Acad Sci U S A. 2004;101:7475–7480. doi: 10.1073/pnas.0402221101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morisse S, Michelet L, Bedhomme M, Marchand CH, Calvaresi M, Trost P, et al. Thioredoxin-dependent redox regulation of chloroplastic phosphoglycerate kinase from Chlamydomonas reinhardtii. J Biol Chem. 2014;289:30012–30024. doi: 10.1074/jbc.M114.597997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lemaire SD, Keryer E, Stein M, Shepens I, Issakidis-Bourget E, Gérard-Hirne C, et al. Heavy metal regulation of gene thioredoxin expression in Chlamydomonas reinhardtii. Plant Physiol. 1999;120:773–778. doi: 10.1104/pp.120.3.773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraiser WM. Reversible inhibition of the Calvin cycle and activation of oxidative pentose phosphate cycle in isolated intact chloroplast by hydrogen peroxide. Planta. 1979;145:377–382. doi: 10.1007/BF00388364. [DOI] [PubMed] [Google Scholar]
- 32.Contreras-Porcia L, Dennett G, González A, Vergara E, Medina C, Correa JA, et al. Identification of copper-induced genes in the marine alga Ulva compressa (Chlorophyta) Mar Biotechnol. 2011;13:544–556. doi: 10.1007/s10126-010-9325-8. [DOI] [PubMed] [Google Scholar]
- 33.Marri L, Zaffagnini M, Collin V, Issakidis-Bourget E, Lemaire SD, Pupillo P, et al. Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidospsis thaliana associated in the GSDPH/CP12/PRK supramolecular complex. Mol Plant. 2009;2:259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- 34.Rodríguez-Rojas F, Celis-Plá P, Méndez L, Moenne F, Muñoz P, et al. MAPK pathway under chronic copper excess in the green macroalga (Chlorophyta): involvement in the regulation of detoxification mechanisms. Int J Mol Sci. 2019;20:4546. doi: 10.3390/ijms20184546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lilley RM, Walker DA. An improved spectrophotometric assay for ribulosebiphosphate carboxylase. Biochim Biophys Acta. 1974;358:226–229. doi: 10.1016/0005-2744(74)90274-5. [DOI] [PubMed] [Google Scholar]
- 36.Barbosa JM, Singh NK, Cherry JH, Loci RD. Nitrate uptake and utilization is modulated by exogenous γ-aminobutiric acid in Arabidopsis thaliana seedlings. Plant Physiol Biochem. 2010;48:443–440. doi: 10.1016/j.plaphy.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 37.Lunn JE, Droux M, Martin J, Douce R. Localization of ATP sulfurylase and O-acetylserine (thiol) lyase in spinach leaves. Plant Physiol. 1990;94:1345–1352. doi: 10.1104/pp.94.3.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Sequencing, pre-processing of reads and assembly of Ulva compressa transcriptomes.
Additional file 2: Figure S1. Pie-Chart of the percentage of proteins involved in different biological processes in U. compressa.
Additional file 3: Figure S2. Pie-Chart of the percentage of proteins having similarity with plant and animal proteins.
Additional file 4: Figure S3. Heatmap of differentially expressed transcripts in U. compressa exposed to copper excess.
Additional file 5: Table S2. List of all transcripts in time points 0 vs. 1, 0 vs.3, 0 vs. 5, their differential expression and the encoded protein.
Additional file 6: Figure S4. Scheme of chloroplast Light Harvesting complex II (LHCII), photosystem II (PSII), cytochrome b6f, photosystem I (PSI), Light Harvesting Complex II and ATP synthase (A). Scheme of mitochondrial complex I, II, III and IV and ATP synthase (B). Subunits showing an increased expression are highlighted in black.
Data Availability Statement
Reads have been deposited in SRA database (NCBI), the accession number is PRJNA557176, and they will be released on 24th august 2020. Experimental data is available at the on line repository: 10.6084/m9.figshare.9108491