Abstract
Background
With the advent of immunotherapy, substantial progress has been made in improving outcomes for patients with advanced cancer. However, not all patients benefit equally from treatment, and confounding immune‐related issues may have an impact. Several studies suggest that antibiotic use (which alters the gut microbiome) may result in poorer outcomes for patients treated with immune checkpoint inhibitors (ICI).
Materials and Methods
This is a large, single‐site retrospective review of n = 291 patients with advanced cancer treated with ICI (n = 179 melanoma, n = 64 non‐small cell lung cancer, and n = 48 renal cell carcinoma). Antibiotic use (both single and multiple courses/prolonged use) during the periods 2 weeks before and 6 weeks after ICI treatment was investigated.
Results
Within this cohort, 92 patients (32%) received antibiotics. Patients who did not require antibiotics had the longest median progression‐free survival (PFS), of 6.3 months, and longest median overall survival (OS), of 21.7 months. With other clinically relevant factors controlled, patients who received a single course of antibiotics had a shorter median OS (median OS, 17.7 months; p = .294), and patients who received multiple courses or prolonged antibiotic treatment had the worst outcomes overall (median OS, 6.3 months; p = .009). Progression‐free survival times were similarly affected.
Conclusion
This large, multivariate analysis demonstrated that antibiotic use is an independent negative predictor of PFS and OS in patients with advanced cancer treated with ICIs. This study highlighted worse treatment outcomes from patients with cumulative (multiple or prolonged courses) antibiotic use, which warrants further investigation and may subsequently inform clinical practice guidelines advocating careful use of antibiotics.
Implications for Practice
Antibiotic use is negatively associated with treatment outcomes of immune checkpoint inhibitors (ICI) in advanced cancer. Cumulative antibiotic use is associated with a marked negative survival outcome. Judicious antibiotic prescribing is warranted in patients receiving treatment with ICI for treatment of advanced malignancy.
Keywords: Immunotherapy, Cumulative antibiotics, Checkpoint inhibitors, Melanoma, Lung cancer, Renal cancer
Short abstract
Evidence suggests that antibiotic use may result in worse outcomes for patients treated with immune checkpoint inhibitors. This article assesses the effect of including cumulative antibiotic use, at the time of ICI treatment, on outcomes for patients with advanced or metastatic solid tumors.
Introduction
Immune checkpoint inhibitors (ICIs) act by modulating coinhibitory T cell signaling and are now commonly used for the treatment of certain cancers 1. The cancer immunogram was described in 2016 to provide a comprehensive framework based on tumor‐ and patient‐specific factors to understand ICI responses 2. However, responses to ICI therapy can be varied, and identification of predictive biomarkers for this class of agents has been challenging 3. Tumor mutational burden, mismatch repair status, and programmed cell death ligand 1 (PD‐L1) expression are currently used as predictive biomarkers for ICI 4, 5, 6. Other potential biomarkers have been proposed, such as tumor‐infiltrating lymphocytes (TILs), TIL‐derived interferon‐γ, neutrophil‐to‐lymphocyte ratio, and peripheral cytokines 2, 4. Preclinical studies have given some insights into mechanisms of resistance with ICIs. However, as clinical use increases, oncologists are developing an enhanced understanding of factors other than tissue biomarkers, such as the impact of concomitant medications, which may impact treatment outcomes, thereby highlighting primary and secondary treatment resistance mechanisms 7.
A retrospective multicenter study has demonstrated that baseline steroid use impacts negatively on outcomes for patients treated with ICIs 8, demonstrating that concomitant medications may negate or lessen efficacy of ICIs. The primary mechanism for this negative interaction is postulated to be immune modulation. Optimizing treatment response and limiting exposure to nonbeneficial agents is critical in the management of advanced cancer patients.
Gut microbiota (gastrointestinal microbiota or gut flora) is the complex community of microorganisms that live symbiotically in the digestive tract. In humans, the gut microbiota has the largest numbers of bacteria and the greatest number of species compared with other areas of the body 9. Treatment with antibiotics reduces the diversity of gut microbiota within days of exposure and recovery after broad‐spectrum antibiotic treatment can take >6 weeks. More protracted reductions in gut microbiota persisting up to 4 years have been noted following treatment for Helicobacter pylori eradication with 7 days of clarithromycin, metronidazole, and omeprazole 10.
Recent studies have demonstrated the impact of the gut microbiota on immunotherapy responses to ICIs in cancer. The gut microbiota has been shown to mediate inflammation‐driven tumors. In preclinical mouse models, multiple studies have demonstrated that the composition of the gut microbiota is an important factor in tumor immunity and responses to treatment with ICIs. Alteration of the composition of the gut microbiota (either by administrating antibiotics or using germ‐free mice) has been shown to have a negative impact on ICI treatment outcomes 11, 12, 13, 14. A link between specific gut microbiota, CD4+ regulatory T cells (Treg), levels of circulating gut‐tropic α4/β7 T cells, and responses to immune checkpoint inhibition with the cytotoxic T‐lymphocyte‐associated protein 4 (CTLA‐4) inhibitor ipilimumab has been established in melanoma 15. Interestingly, fecal analysis demonstrated a more diverse microbiome for patients with metastatic melanoma (a highly immunogenic tumor) who exhibited a treatment response to ICIs when compared with the nonresponder population 16, 17, 18. In addition, modulation of the gut microbiome with fecal microbiota transplantation has been shown to abrogate ICI‐associated colitis by increasing abundance and diversity of microbiota 19. Patients with melanoma treated with anti‐CTLA‐4 immune blockade who were rich in specific Bacteroidetes species did not develop colitis. This kind of toxicity may be associated with the known effects of these bacteria involving in the Treg differentiation 15.
Given the effect of antibiotic therapy on gut microbiota, this work was undertaken to assess the impact of antibiotic use at the time of ICI treatment on the outcomes for patients with advanced or metastatic solid tumors.
Materials and Methods
Permission was granted to collect retrospective patient data for this study by The Quality Improvement and Clinical Audit Committee of The Christie NHS Foundation Trust (The Christie), Manchester, U.K. on January 22, 2018 (reference SE18/2128).
Data Collection and Analysis
Inclusion criteria included patients with advanced melanoma, non‐small cell lung cancer, and renal cell cancer who received an ICI agent at the Christie NHS Foundation Trust, Manchester, U.K., between January 1, 2015, and April 1, 2017. Patients with progressive‐free survival (PFS) <14 days or overall survival (OS) <21 days were excluded. Data were collected from patients from 2 weeks before until 6 weeks after ICI treatment began. This time‐frame was chosen because of the potential duration of modification of gut microbiota following antibiotic therapy, which can be different for different classes of antibiotics and was alluded to above 10. Data were collected from The Christie electronic patient records (The Christie Web Portal) and the electronic prescribing system (ePrescribing, EMIS Health, Leeds, U.K.) within the Trust. Relevant patient demographics were captured, including age, gender, tumor type, number of prior systemic treatments, metastatic burden, Eastern Cooperative Oncology Group (ECOG) performance status (PS), comorbidities, date of commencement of ICI, ICI drug used, clinical trial involvement, antibiotic indication, type of antibiotic(s), and route of administration. The date of clinical or radiological progression was evaluated in trials patients using the Response Evaluation Criteria In Solid Tumors version 1.1 (RECIST 1.1) 20 or Immune‐related RECIST criteria (irRECIST) 21. Nontrial patients’ imaging results were recorded as per the radiologist's report and evaluated against clinical benefit. If the same patient was treated sequentially with two ICI drugs (e.g., first treated with ipilimumab, then pembrolizumab following disease progression), two separate data records were created for progression events whereas a single data record was created for death event using the later ICI treatment. Patient antibiotic use was gathered and was divided into no antibiotics, single course of antibiotics, or cumulative antibiotic use. Patients who did not use antibiotics during the specified time period were analyzed in the no antibiotic group. Patients who received up to 7 days of a single antibiotic (either intravenous or oral) were analyzed in the single course antibiotic group. Patients who received more than one intravenous or oral antibiotic (sequential antibiotic use for multiple sources of infection or concurrent use of more than 1 antibiotic for severe infection) or a single antibiotic for a prolonged duration of more than 7 days were analyzed in the cumulative antibiotic group.
All patients were followed up until death or the time of data lock on January 1, 2018. PFS was calculated as the time from the start of ICI treatment to the diagnosis of disease progression (clinical progression or radiological progression as per RECIST 1.1 or irRECIST) or death, whichever came first. OS was calculated as the time from commencement of ICI treatment to the date of death. A detailed description on how PFS and OS were calculated and where data censoring, competing risk, and other confounding factors were dealt with can be found in supplemental online Figure 1. Missing clinical information was imputed using a multiple imputation approach as implemented in the MICE package in R 22 where appropriate. Patients who received intravenous antibiotics and later converted to oral antibiotics had these data recorded as two separate antibiotic uses under the same patient. Similarly, where patients had more than one infection during the 8‐week period, this was recorded as two antibiotics uses. It was recognized that different intervals between two consecutive clinical visits may introduce bias in PFS evaluation 23. In this study, all patients were treated in a same hospital and followed a standardized patient pathway. A subset of patients involved in this study participated in clinical trials. The trial status was included as a covariate in modeling.
Statistical Analysis
The association between antibiotic use and other clinical factors was examined using chi‐squared tests. The factors that demonstrated significant association (number of metastatic sites, ECOG PS, comorbidities) were investigated for their interaction with antibiotic use in the subsequent proportional hazard regression analysis. A Cox proportional hazard model was used to assess the association between antibiotic use and patient survival (PFS and OS). The analysis started from a univariate analysis that included each clinical factor as a sole covariate in the model, with the significance of association evaluated using a Wald test. Assumption of proportionality was verified based on Schoenfeld residuals 24. A plot of the Martingale residuals from each marker specific analysis was examined for evidence of nonlinearity in the biomarker‐hazard relationship 25. Antibiotic use and clinical factors with univariate p values <.2 were selected for subsequent multivariate proportional hazard regression analysis. A backward stepwise method was applied to identify the optimum subset of clinical factors that were associated with survival. Interactions between antibiotic use and selected clinical factors were explored in the multivariate analysis.
The analysis present in this study follows the REMARK guideline 26. The p values <.05 were considered statistically significant. All analyses were implemented using R. Multiple comparison was not adjusted due to the exploratory nature of the study.
Results
Data were collected from 347 patients who were treated with ICI between Jan 1, 2015, and April 1, 2017, at The Christie. There were 291 evaluable patients included in the final analysis, as 56 patients were excluded (Fig. 1). Reasons for exclusion included 27 nonadministered prescriptions, 22 patients who received ICI as adjuvant treatment, 3 coding inconsistencies or alternative diagnoses, and 3 patients who had key data missing. The evaluable cohort comprised 179 patients with advanced or metastatic melanoma, 64 patients with non‐small cell lung cancer (NSCLC), and 48 with renal cell carcinoma (RCC). A total of 157 patients (54%) participated in clinical trials. The characteristics of these patients are summarized in Table 1. Among this cohort, 92 patients had antibiotic therapy during ICI treatment. The types of infections, choices of antibiotics, and ICI agent received can be found in supplemental online Table 1.
Figure 1.
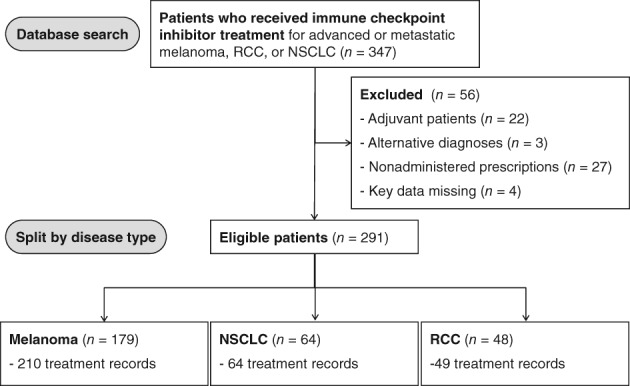
Patient enrolment CONSORT flow‐chart diagram. Patients were identified who had been treated at The Christie NHS Foundation Trust between January 1, 2015, and April 1, 2017.
Abbreviations: NSCLC, non‐small cell lung cancer; RCC, renal cell carcinoma.
Table 1.
Characteristics of patients evaluated in the study
| Characteristic | ABX+ (n = 92) | ABX− (n = 199) |
|---|---|---|
| Age, yr | ||
| Median | 66.0 | 66.0 |
| Mean | 63.9 | 65.3 |
| Gender n (%) | ||
| Female | 41 (37.3) | 69 (62.7) |
| Male | 51 (28.2) | 130 (71.8) |
| Tumor type n (%) | ||
| Melanoma | 54 (30.3) | 124 (69.7) |
| RCC | 13 (27.1) | 35 (72.9) |
| NSCLC | 25 (39.1) | 39 (60.9) |
| Number of metastatic sites (%) | ||
| None | 0 (0) | 10 (100) |
| 1 | 14 (28.6) | 35 (71.4) |
| 2 | 42 (31.8) | 90 (68.1) |
| > 2 | 36 (33.0) | 73 (67.0) |
|
ECOG performance status (%) |
||
| 0 | 33 (25.2) | 98 (74.8) |
| 1 | 28 (28.3) | 71 (71.7) |
| 2 | 15 (50.0) | 15 (50.0) |
| 3 | 2 (40.0) | 3 (60.0) |
| Not recorded | 14 (53.8) | 12 (46.2) |
| Participating in a clinical trial (%) | ||
| Yes | 33 (21.0) | 124 (79.0) |
| No | 59 (44.0) | 75 (56.0) |
| Comorbidity score (%) | ||
| 0 | 40 (31.3) | 88 (68.7) |
| 1 | 27 (27.0) | 73 (73.0) |
| 2 | 20 (38.5) | 32 (61.5) |
| Not recorded | 5 (45.5) | 6 (54.5) |
| Previous therapy n (%) | ||
| 0 | 39 (33.1) | 79 (66.9) |
| 1 | 32 (29.4) | 77 (70.6) |
| ≥ 2 | 21 (32.8) | 43 (67.2) |
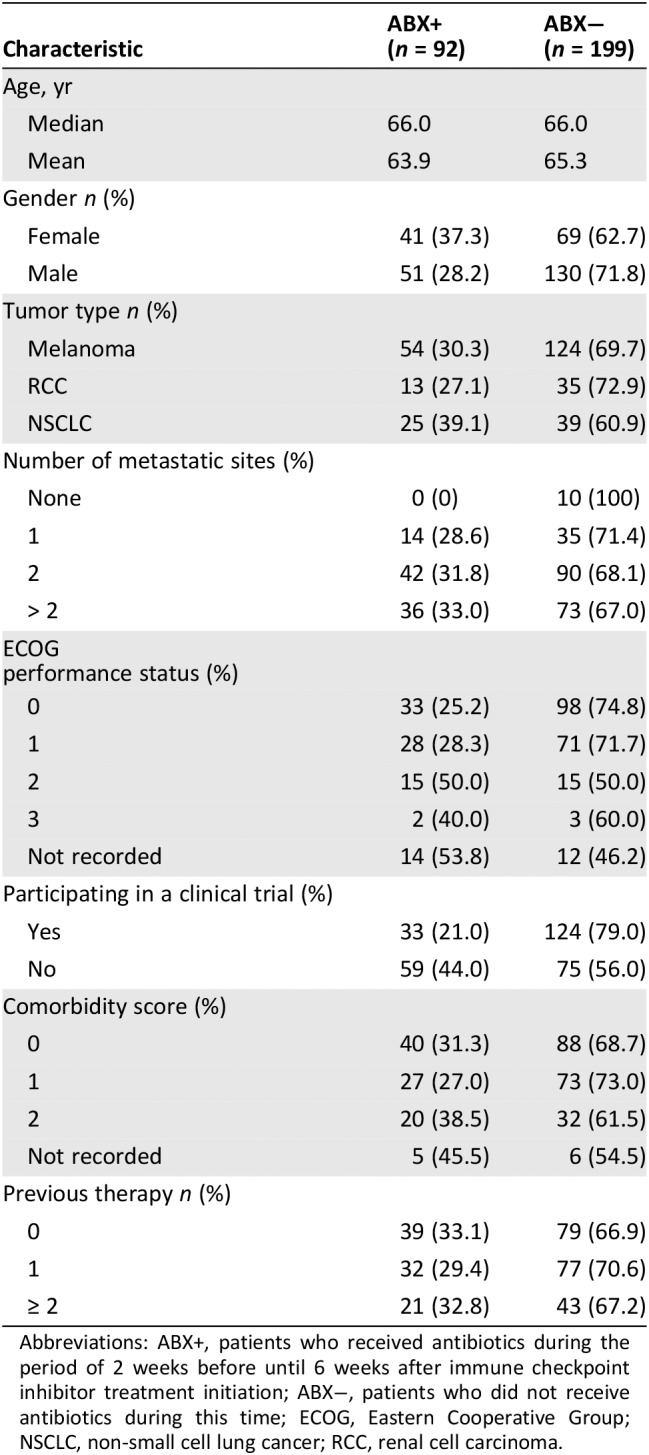
Abbreviations: ABX+, patients who received antibiotics during the period of 2 weeks before until 6 weeks after immune checkpoint inhibitor treatment initiation; ABX−, patients who did not receive antibiotics during this time; ECOG, Eastern Cooperative Group; NSCLC, non‐small cell lung cancer; RCC, renal cell carcinoma.
The association between antibiotic use and other clinical factors was examined using chi‐squared tests. ECOG performance status was the only factor significantly associated with antibiotic use, with greater levels of functional impairment noted in patients receiving antibiotics (p = .026, supplemental online Table 2). In the univariate analysis (Table 2), the use of antibiotics during ICI was significantly associated with shorter PFS (median PFS 3.1 vs. 6.3 months; hazard ratio [HR], 1.564; p = .003) and OS (median OS, 10.4 vs. 21.7 months; HR, 1.699; p = .002). Figure 2 demonstrates survival outcomes of patients with or without antibiotics. Other prognostic factors significant for both PFS and OS include ECOG PS (≥2 vs. 1 vs. 0; median PFS, 2.8 vs. 4.2 vs. 10.1 months; HR, 2.498, p = .001; median OS, 111 vs. 451 vs. 795 days; HR, 4.375; p = .001) and presence of comorbidities (median PFS, 3.7 vs. 8.7 months; HR, 1.420; p = .016; median OS, 14 vs. 26.8 months; HR, 1.526; p = .018). Patients with three or more metastatic sites (0–1 vs. 2 vs. 3+) had significantly shorter OS (median OS not evaluable because of censoring; HR, 2.537; p = .001) and a moderately shorter PFS (median PFS, 8.2 vs. 5.6 vs. 3.3 months; HR, 1.524; p = .048). Patients involved in clinical trials of ICI had significantly longer OS (median OS, 14 vs. 25.5 months; HR 0.627; p = .011), whereas a trend to improvement in PFS was not statistically significant (median OS, 5.1 vs. 8.1 months; HR, 0.823; p = .199).
Table 2.
Univariate analysis of the association between clinical factors and survival
| Variable | PFS | OS | ||
|---|---|---|---|---|
| HR | p value | HR | p value | |
| Gender, female vs. male | 1.300 | .071 | 1.126 | .495 |
| Age >57 yr, median | 1.001 | .940 | 1.012 | .076 |
| Cancer type | ||||
| Melanoma vs. RCC | 1.153 | .492 | 1.434 | .164 |
| NSCLC vs. RCC | 1.523 | .075 | 1.588 | .113 |
| Clinical trial, yes vs. no | 0.823 | .199 | 0.627 | .011 |
| Number of metastatic sites | ||||
| 2 vs. 0/1 | 1.276 | .233 | 1.373 | .263 |
| 3+ vs. 0/1 | 1.524 | .048 | 2.537 | .001 |
| Number of previous treatments | ||||
| 1 vs. 0 | 0.880 | .436 | 0.950 | .788 |
| 2+ vs. 0 | 0.922 | .661 | 0.840 | .439 |
| ECOG performance status | ||||
| 1 vs. 0 | 1.419 | .028 | 1.647 | .01 |
| 2+ vs. 0 | 2.498 | .001 | 4.375 | <.001 |
| Comorbidy, yes vs. no | 1.420 | .016 | 1.526 | .018 |
| Antibiotics, yes vs. no | 1.564 | .003 | 1.699 | .002 |
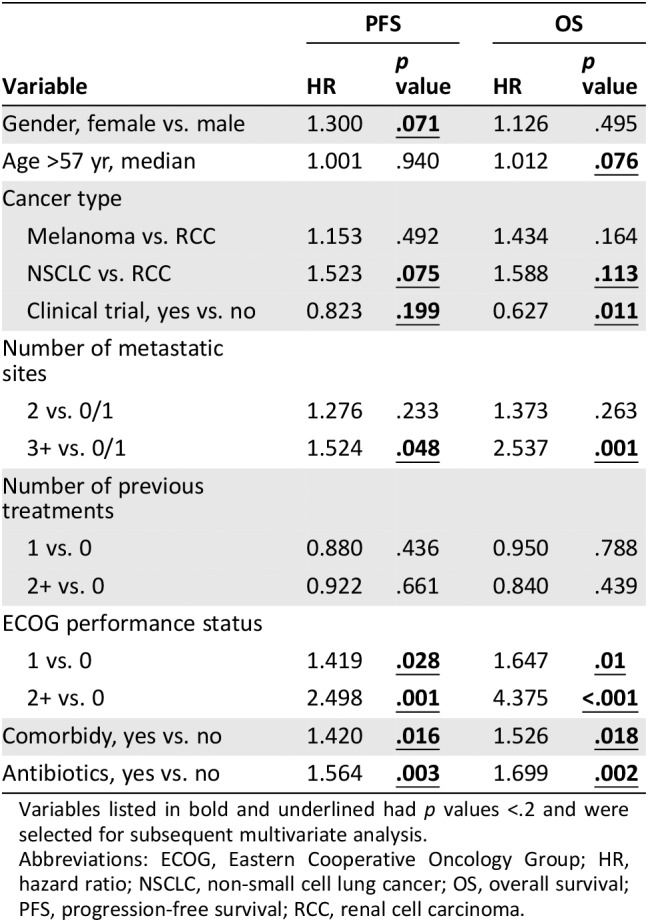
Variables listed in bold and underlined had p values <.2 and were selected for subsequent multivariate analysis.
Abbreviations: ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio; NSCLC, non‐small cell lung cancer; OS, overall survival; PFS, progression‐free survival; RCC, renal cell carcinoma.
Figure 2.
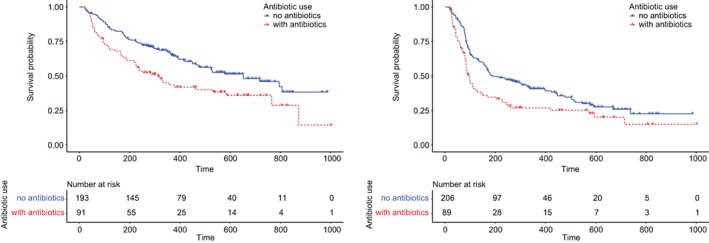
Survival outcomes for patients based on their exposure to antibiotics. Kaplan‐Meier survival estimators were plotted for patients who had been treated with immune checkpoint inhibitors with and without antibiotics. Patients treated with antibiotics during immune checkpoint inhibitor treatment demonstrated a median progression‐free survival (PFS) of 3.1 months and a median overall survival (OS) of 10.4 months, significantly less than those without antibiotics (p = .003 and .002, respectively). The latter demonstrated a median PFS of 6.3 months and a median OS of 21.7 months. The results have not been adjusted for other prognostic factors.
Clinical factors with univariate p values <.2 were included in multivariate analysis to further interrogate the impact of antibiotic use during ICI, and the results are summarized in Tables 3A and B. Type of malignancy (renal cancer vs. NSCLC vs. melanoma) did not impact significantly on survival, and the significance of other prognostic factors was not affected by including the additional covariate of cancer type in the model. PFS was significantly associated with a small number of clinical factors, namely antibiotics, comorbidity, and ECOG PS. OS was associated with patient trial status, number of metastatic sites, and all the PFS‐associated factors. Patients with higher ECOG PS had reduced PFS (HR, 1.929; p = .006) and OS (HR, 3.272; p < .001). A similar pattern was observed in patients with comorbidities (PFS HR, 1.391; p = .024 and OS HR, 1.542; p = .017). Patients involved in clinical trials had improved OS (HR, 0.549; p = .002), and OS was reduced in those with three or more metastatic sites (HR, 2.181; p = .006). Keeping these prognostic factors under control, patients treated with antibiotics during ICI demonstrated significantly reduced PFS (HR, 1.401; p = .033) and OS (HR, 1.473; p = .033) compared with those who were not. The interaction between antibiotic use and ECOG PS was investigated, based on their prognostic impact as described above, but the interaction between these variables was not prognostic.
Table 3A.
Multivariate analysis of the association between the use of antibiotics and progression‐free survival
| Variable | p value | HR (95% CI) |
|---|---|---|
| Antibiotics, yes vs. no | .033 | 1.401 (1.028–1.920) |
| Comorbidity, yes vs. no | .024 | 1.391 (1.045–1.853) |
| ECOG performance status | ||
| 1 vs. 0 | .069 | 1.324 (0.979–1.792) |
| 1 vs. 0 | .006 | 1.929 (1.203–3.092) |
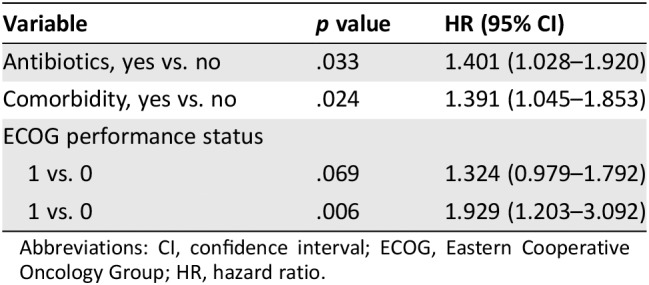
Abbreviations: CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio.
Table 3B.
Multivariate analysis of the association between the use of antibiotics and overall survival
| Variable | p value | HR (95% CI) |
|---|---|---|
| Antibiotics, yes vs. no | .033 | 1.473 (1.038–2.107) |
| Comorbidy, yes vs. no | .017 | 1.542 (1.082–2.198) |
| On trial, yes vs. no | .002 | 0.549 (0.378–0.797) |
| Number of metastatic sites | ||
| 2 vs. 0/1 | .771 | 1.088 (0.616–1.924) |
| 3+ vs. 0/1 | .006 | 2.181 (1.247–3.816) |
| ECOG performance status | ||
| 1 vs. 0 | .006 | 1.864 (1.277–2.722) |
| 2+ vs. 0 | <.001 | 3.272 (1.973–5.427) |

Abbreviations: CI, confidence interval; ECOG, Eastern Cooperative Oncology Group; HR, hazard ratio.
To better characterize the antibiotic treatment received, an exploratory analysis was carried out by categorizing patients as either having had “no antibiotics,” “single course” antibiotics, or “cumulative courses” antibiotics, where concurrent or successive antibiotics for >7 days were administered. Patients who did not receive antibiotics had the longest PFS and OS (median PFS, 6.3 months; median OS, 21.7 months). Administration of a single course of antibiotics was associated with a nonsignificant reduction in PFS (median PFS, 3.7 months; HR, 1.324; p = .279) and OS (median OS, 17.7 months; HR, 1.259; p = .294). However, patients who had received cumulative courses of antibiotics had significantly worse PFS (median PFS, 2.8 months; HR, 2.625; p = .026) and OS (median OS, 6.3 months; HR, 1.904, p = .009). As shown in Figure 3 and Tables 4A and B, cumulative use of antibiotics is an independent significant prognostic factor for clinical outcome of ICI.
Figure 3.
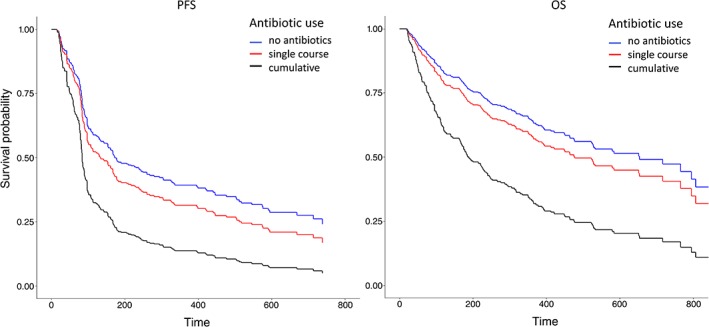
Survival of patients based upon antibiotic use. Patients were divided into three categories based on whether they had received antibiotics or not during the period of 2 weeks before until 6 weeks after immune checkpoint inhibitor treatment initiation. Kaplan‐Meier estimator graphs illustrate survival outcomes for patients who did not receive antibiotics (blue), patients who received a single course of antibiotics only (red), and patients who received prolonged or multiple courses of antibiotics (black). Other prognostic factors were controlled in this plot.
Abbreviations: OS, overall survival; PFS, progression‐free survival.
Table 4A.
Impact of cumulative use of antibiotics on progression‐free survival
| Covariates | p value | Hazard ratio (95% CI) |
|---|---|---|
| ECOG performance status | ||
| 1 vs. 0 | .198 | 1.270 (0.883–1.829) |
| 2+ vs. 0 | .0006 | 3.141 (1.638–6.025) |
| Comorbidies, yes. vs. no | .022 | 1.425 (1.052–1.930) |
| Cumulative use of antibiotics | ||
| Single course vs. no | .279 | 1.324 (0.796–2.200) |
| cumulative courses vs. no | .026 | 2.625 (1.245–6.127) |
| Interaction of ECOG PS and use of antibiotics | ||
| PS = 1 and single ABX | .856 | 0.929 (0.420–2.057) |
| PS ≥2 and single ABX | .385 | 0.599 (0.188–1.905) |
| PS = 1 and cumulative ABX | .937 | 0.959 (0.341–2.698) |
| PS ≥2 and cumulative ABX | .021 | 0.225 (0.063–0.798) |
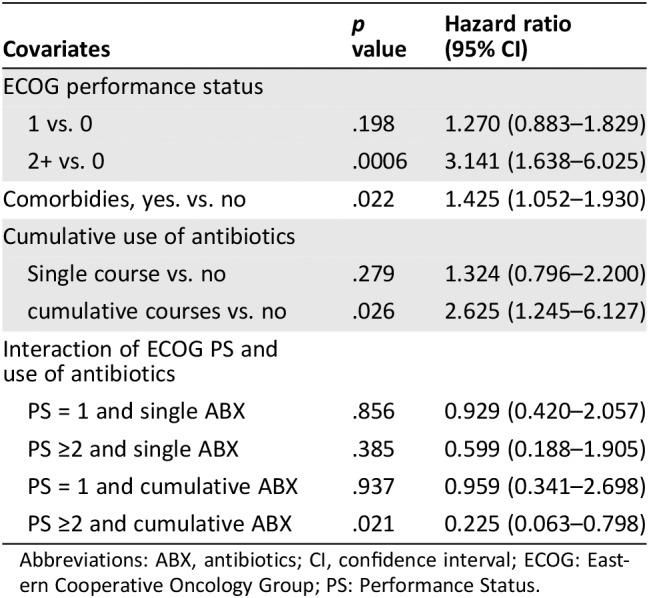
Abbreviations: ABX, antibiotics; CI, confidence interval; ECOG: Eastern Cooperative Oncology Group; PS: Performance Status.
Table 4B.
Impact of cumulative use of antibiotics on overall survival
| Name of covariates | p value | Hazard ratio (95% CI) |
|---|---|---|
| Participate in trials, yes vs. no | .002 | 0.559 (0.384–0.812) |
| Number of metastatic sites | ||
| 2 vs. 0/1 | .697 | 1.120 (0.633–1.983) |
| 3+ vs. 0/1 | .004 | 2.289 (1.304–4.019) |
| ECOG performance status | ||
| 1 vs. 0 | .002 | 1.812 (1.238–2.651) |
| 2+ vs. 0 | .00003 | 3.027 (1.801–5.090) |
| Comorbities, yes vs. no | .020 | 1.523 (1.069–2.169) |
| Cumulative use of antibiotics | ||
| Single course vs. no | .294 | 1.259 (0.819–1.934) |
| Cumulative courses vs. no | .009 | 1.904 (1.178–2.078) |
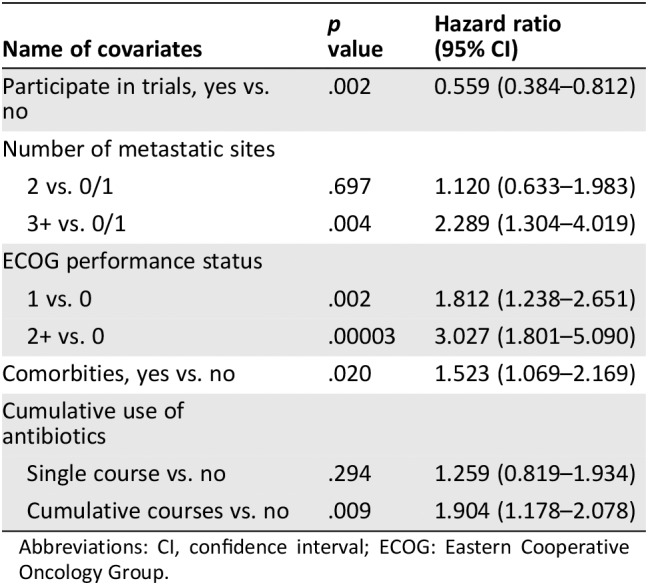
Abbreviations: CI, confidence interval; ECOG: Eastern Cooperative Oncology Group.
Discussion
This large multivariate analysis demonstrates that antibiotic use is an independently negative predictor of PFS and OS in patients with advanced cancer treated with ICIs, with cumulative antibiotic use resulting in further significant detriment in outcomes. To our knowledge, this is the first study to suggest an adverse effect of cumulative antibiotic use in patients receiving ICI treatment for advanced cancer.
Inclusion in this study was not limited to a particular subtype of malignancy and reflected the variation of patients presently receiving immunotherapy in routine clinical practice or clinical trials. A rigorous approach was adopted for calculating PFS and OS to ensure that no bias was introduced and that competing risks were resolved. During the analysis, the interaction between significantly associated ECOG PS and antibiotic use was investigated to ensure correct determination of the impact of antibiotics on patient survival. These efforts safeguard the robustness of the analysis. Nevertheless, on adjusting for these factors in multivariate analysis, antibiotic use emerged as an independent prognostic factor.
Treatment of, or prophylaxis from, infection is crucial to maintain patient well‐being in the context of immunosuppressive cancer therapy or disease. The risk of life‐threatening sepsis in patients with advanced cancer has led to a culture of frequent antibiotic administration 27. However, ICIs do not result in immunosuppression, and more judicious use of antibiotics may be warranted for patients treated with these newer agents.
Antibiotics substantially reduce the number and diversity of gut microbiota 10, which is known to be an important factor in regulating antitumor immunity 28. The gut microbiota can be manipulated and artificially improved through the use of fecal microbiota transplantation (FMT), probiotics (a single or defined combination of bacterial species), or prebiotics (designed to stimulate the growth and retention of specific beneficial species) to increase the number and diversity of organisms and to potentially improve ICI treatment outcomes in cancer. Direct manipulation of the microbiota offers a possibility for improving cancer therapy outcomes, minimizing toxicities and mitigating the impact of infectious diseases 27.
Since this work began, a series of studies have reported negative associations between administration of antibiotics and outcome of ICI treatment 11, 28, 29, 30, 31, 32. In the largest of these, the impact of antibiotic use in patients with RCC and NSCLC was examined across two cancer treatment centers, highlighting an increased risk of progressive disease, with shorter PFS (median 1.9 vs. 7.4 months) and OS (17.3 vs. 30.6 months) in patients who had received antibiotic therapy during their treatment 28. Other studies have assessed antibiotic therapy more specifically to the month prior of commencing ICI and showed a detrimental effect on response and survival 33. Preclinical work has demonstrated enhanced efficacy of ICIs in germ‐free or antibiotic‐treated mice receiving FMT from patients who responded to ICI compared with those receiving FMT from patients who did not. Analysis of these fecal samples confirmed significantly higher levels of Akkermansia muciniphila in the stools of responders, and introducing this species to mice resulted in improved responses to ICI 11. Similar outcomes have been noted following treatment with the CTLA‐4 inhibitor Ipilimumab following modulation of Bacteroides fragilis in mouse models 12. This suggests that gut microbiota is a modifiable factor which can be used as an aid to treatment. Melanoma patients stools were assessed for their microbiome diversity and composition. Significant differences were seen in the responders versus nonresponder microbiome in two distinct studies, highlighting the importance of a rich microbiome, with specific propensity for Bifidobacterium longum, Collinsella aerofaciens, and Enterococcus faecium 17, 18.
In contrast, two retrospective reviews reported negative findings, with antibiotic use not influencing responses to nivolumab in patients with lung cancer 34, 35.
Clinical trials investigating the impact of probiotic therapy on quality of life and outcomes for patients with cancer are ongoing. Using the search terms “probiotic” and “cancer” found over 50 studies listed to date (http://www.clincialtrials.gov). Randomized clinical trials examining the impact of probiotics are needed to fully evaluate their therapeutic potential when coadministered with ICIs.
This retrospective study did not include examination of the gut microbiota for the patient cohort explored. It will be important to consider how microbiota can most conveniently be evaluated in routine clinical practice, what interpatient variation is observed, how often sampling should be carried out, and what other factors can influence the gut microbiota (e.g., chemotherapy or radiotherapy) with potential impact upon the outcome of ICI therapy. Monitoring the evolution of microbiota throughout treatment and following intervention would also enhance our understanding of these interactions.
Microbiome sequencing is likely to become a more widely adopted and accessible technique. Techniques used to identify and compare bacteria within a sample are commonly used in clinical microbiology laboratories for the detection of bacteria in stool, sputum, or other microbiology samples. The composition of the gut microbiome may be analyzed by several modalities, for example, via 16S ribosomal RNA and shotgun sequencing‐based methods 36, 37. If correctly characterized, this detailed microbiome information could then be used as a predictive, prognostic, or toxicity biomarker.
This large retrospective analysis, demonstrating that antibiotic use is an independent negative predictor of PFS and OS in patients with advanced cancer treated with ICIs, is the first study to clearly demonstrate that multiple or prolonged antibiotic use results in poorer clinical outcomes than single course treatment. This work adds to an increasing body of evidence supporting a detrimental effect of antibiotic therapy in the treatment of patients receiving ICIs for advanced cancer.
It is challenging to make a causal link between the use of antibiotics and poorer outcomes for patients treated with ICIs. It may be that unwell patients are more likely to acquire infections that require treatment with antibiotics. These patients may be less likely to benefit from anticancer intervention, thereby shortening their progression‐free interval and survival. In addition, those patients who require multiple courses of antibiotics or escalation to intravenous antibiotics, thus resulting in cumulative use, may have more severe infections. This study did not explore the supporting evidence for commencement of antibiotic therapy in these patients, so questions remain regarding the potential detrimental impact of infection (and severity thereof) on outcomes for this cohort. It may also be possible that infections can have an unknown immunosuppressive effect, altering responses to ICI. For example, a decrease in lymphocyte counts has been associated with poor outcomes in CTLA‐4‐treated melanoma patients 2. However, the independent prognostic status of ECOG PS and antibiotic use in the multivariate analysis may give some indication that the therapy, rather than the clinical condition of the patient, was the major determinant of outcome.
Additional limitations of this study include its retrospective approach, relying on hospital records for patient information. Independent database verification was undertaken to minimize inconsistencies. We were unable to compare antibiotic use from before commencing ICI to post‐ICI because of low number of patients in this group. Given the possibility of pseudoprogression with this class of agents, patients who were treated through radiological progression but continued the same immunotherapy treatment were not regarded as having disease progression, which may introduce bias in the results. With the exception of those patients enrolled in a clinical trial, imaging was not RECIST or irRECIST evaluated 20, 38 and may not have been performed at standardized time points across disease sites, relying instead on radiologist interpretation. Other factors known to affect ICI responses such as PD‐L1 status, tumor mutational burden, mismatch repair, or baseline steroid use were not consistently available for our data set and therefore not analyzed.
A major challenge currently facing oncologists is to optimize the therapeutic benefits of ICIs while minimizing immune‐related side effects and understanding the complexity of factors that contribute to treatment responses. The processes involved in immunotherapy responses require further investigation to generate an enhanced understanding of accurate predictive and safety biomarkers (which may be inclusive of the gut microbiome).
Conclusion
This retrospective study demonstrates that antibiotic use is an independent predictor of shorter PFS and OS in patients with advanced cancers treated with immune checkpoint inhibitors. This is irrespective of other clinical factors such as cancer type, previous cancer treatment, performance status, comorbidities, or entry into clinical trial. This effect is significantly enhanced with cumulative antibiotic use and may be related to antibiotic‐related microbiota imbalance.
The findings of this study warrant further investigation and may subsequently inform clinical practice guidelines for ICI‐treated patients, advocating more judicious use of prophylactic antibiotics and clear indications for commencement of therapeutic antibiotics. It will be challenging in these already complex clinical situations to find the optimal balance between the need to reduce the risk of genuine infections in patients with cancer and the attempt to preserve the gut microbiota.
Author Contributions
Conception/design: Nadina Tinsley, Cong Zhou, Grace Tan, Samuel Rack, Paul Lorigan, Fiona Blackhall, Matthew Krebs, Louise Carter, Fiona Thistlethwaite, Donna Graham, Natalie Cook
Provision of study material or patients: Nadina Tinsley, Cong Zhou, Grace Tan, Samuel Rack, Paul Lorigan, Fiona Blackhall, Matthew Krebs, Louise Carter, Fiona Thistlethwaite, Donna Graham, Natalie Cook
Collection and/or assembly of data: Nadina Tinsley, Cong Zhou, Grace Tan, Samuel Rack, Paul Lorigan, Fiona Blackhall, Matthew Krebs, Louise Carter, Fiona Thistlethwaite, Donna Graham, Natalie Cook
Data analysis and interpretation: Nadina Tinsley, Cong Zhou, Grace Tan, Samuel Rack, Paul Lorigan, Fiona Blackhall, Matthew Krebs, Louise Carter, Fiona Thistlethwaite, Donna Graham, Natalie Cook
Manuscript writing: Nadina Tinsley, Cong Zhou, Grace Tan, Samuel Rack, Paul Lorigan, Fiona Blackhall, Matthew Krebs, Louise Carter, Fiona Thistlethwaite, Donna Graham, Natalie Cook
Final approval of manuscript: Nadina Tinsley, Cong Zhou, Grace Tan, Samuel Rack, Paul Lorigan, Fiona Blackhall, Matthew Krebs, Louise Carter, Fiona Thistlethwaite, Donna Graham, Natalie Cook
Disclosures
Paul Lorigan: Merck Sharp & Dohme, Bristol‐Myers Squibb, Novartis, Amgen, Pierre Fabre (C/A, RF, H); Fiona Blackhall: AstraZeneca, Novartis, Pfizer, Amgen, Bristol‐Myers Squibb (RF), Regeneron, Medivation, AbbVie, Takeda, Roche, Ibsen (SAB), CellMedica, Merck Sharp & Dohme (C/A), Boehringer Ingelheim (Other‐Speaker bureau); Louise Carter: Athenex, Sierra, Takeda (RF as PI on commercial clinical trials) Athenex, Bicycle Therapeutics (SAB); Fiona Thistlethwaite: Bristol‐Myers Squibb, Novartis, Medimmune, Incyte, Janssen, Cytomx, Pfizer, Synthon, GenMab (RF); Donna Graham: Merck Sharp & Dohme, Debiopharm, Taiho Oncology (RF); Natalie Cook: Tarveda Therapeutics, Neomed Therapeutics (SAB), Boehringer Ingelheim, Astra Zeneca, Roche, Taiho, RedX, Pfizer, Eisai, Novartis, Millenium (RF). The other authors indicated no financial relationships.
(C/A) Consulting/advisory relationship; (RF) Research funding; (E) Employment; (ET) Expert testimony; (H) Honoraria received; (OI) Ownership interests; (IP) Intellectual property rights/inventor/patent holder; (SAB) Scientific advisory board
Supporting information
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Figure
Supplemental Tables
Acknowledgments
This work was supported by Cancer Research U.K. via funding to the Cancer Research U.K. Manchester Centre [C5759/A25254]. Preparation of this manuscript was supported by The Systemic Therapy Research Group at The Christie NHS Foundation Trust, Manchester, U.K., in particular Dr Alison Backen, who assisted with manuscript preparation. This study was presented as a Poster Discussion at ASCO Annual Meeting 2018 [J Clin Oncol 36, 2018 (suppl; abstr 3010)].
Footnotes
For Further Reading: Hiroaki Akamatsu, Eriko Murakami, Jun Oyanagi et al. Immune‐Related Adverse Events by Immune Checkpoint Inhibitors Significantly Predict Durable Efficacy Even in Responders with Advanced Non‐Small Cell Lung Cancer. The Oncologist first published on November 19, 2019; doi:10.1634/theoncologist.2019‐0299
Implications for Practice: Although the predictive value of immune‐related adverse events (irAEs) induced by immune checkpoint inhibitors (ICIs) has been suggested by several studies, it has not been elucidated whether irAEs also play a significant role even in responders. This study showed that more than 60% of responders had irAEs. It demonstrated the strong correlation between irAEs and efficacy even in responders. Investigation into the significance of irAEs in responders will contribute to the establishment of optimal administration of ICI.
Contributor Information
Donna Graham, Email: donna.graham@christie.nhs.uk.
Natalie Cook, Email: natalie.cook@christie.nhs.uk.
References
- 1. Ventola CL. Cancer immunotherapy, part 1: Current strategies and agents. P T 2017;42:375–383. [PMC free article] [PubMed] [Google Scholar]
- 2. Blank CU, Haanen JB, Ribas A et al. Cancer immunology. The “cancer immunogram.” Science 2016;352:658–660. [DOI] [PubMed] [Google Scholar]
- 3. Le DT, Uram JN, Wang H et al. PD‐1 blockade in tumors with mismatch‐repair deficiency. N Engl J Med 2015;372:2509–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Yi M, Jiao D, Xu H et al. Biomarkers for predicting efficacy of PD‐1/PD‐L1 inhibitors. Mol Cancer 2018;17:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan TA, Yarchoan M, Jaffee E et al. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol 2019;30:44–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Viale G, Trapani D, Curigliano G. Mismatch repair deficiency as a predictive biomarker for immunotherapy efficacy. Biomed Res Int 2017;2017: 4719194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jenkins RW, Barbie DA, Flaherty KT. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer 2018;118:9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Arbour KC, Mezquita L, Long N et al. Impact of baseline steroids on efficacy of programmed cell death‐1 and programmed death‐ligand 1 blockade in patients with non–small‐cell lung cancer. J Clin Oncol 2018;36:2872–2878. [DOI] [PubMed] [Google Scholar]
- 9. Quigley EM. Gut bacteria in health and disease. Gastroenterol Hepatol (N Y) 2013;9:560–569. [PMC free article] [PubMed] [Google Scholar]
- 10. Modi SR, Collins JJ, Relman DA. Antibiotics and the gut microbiota. J Clin Invest 2014;124:4212–4218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Routy B, Le Chatelier E, Derosa L et al. Gut microbiome influences efficacy of PD‐1‐based immunotherapy against epithelial tumors. Science 2018;359:91–97. [DOI] [PubMed] [Google Scholar]
- 12. Vétizou M, Pitt JM, Daillère R et al. Anticancer immunotherapy by CTLA‐4 blockade relies on the gut microbiota. Science 2015;350:1079–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kuczma MP, Ding Z‐C, Li T et al. The impact of antibiotic usage on the efficacy of chemoimmunotherapy is contingent on the source of tumor‐reactive T cells. Oncotarget 2017;8:111931‐111942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xue J, Wang Z, Guo S et al. Intestinal microbiota may dynamically facilitate the anti‐PD‐L1 immunotherapy. Cancer Res 2017;77(13 Suppl):2008a. [Google Scholar]
- 15. Chaput N, Lepage P, Coutzac C et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Ann Oncol 2017;28:1368–1379. [DOI] [PubMed] [Google Scholar]
- 16. Ledford H. Gut microbes can shape responses to cancer immunotherapy. Nature 2017. Nov 3;358(6363):573. [DOI] [PubMed] [Google Scholar]
- 17. Gopalakrishnan V, Spencer CN, Nezi L et al. Gut microbiome modulates response to anti‐PD‐1 immunotherapy in melanoma patients. Science 2018;359:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Matson V, Fessler J, Bao R et al. The commensal microbiome is associated with anti‐PD‐1 efficacy in metastatic melanoma patients. 2018;359:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang Y, Wiesnoski DH, Helmink BA et al. Fecal microbiota transplantation for refractory immune checkpoint inhibitor‐associated colitis. Nat Med 201812;24:1804–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Eisenhauer EA, Therasse P, Bogaerts J et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228–247. [DOI] [PubMed] [Google Scholar]
- 21. Bohnsack O, Hoos A, Ludajic K. 1070P Adaptation of the immune related response criteria: irRECIST. Ann Oncol 2014. ;25(suppl 4):iv369. [Google Scholar]
- 22. Fehske H, Schneider R, Weiße A, eds. Computational many‐particle physics. Berlin, Heidelberg: Springer, 2008. [Google Scholar]
- 23. Carroll KJ. Analysis of progression‐free survival in oncology trials: Some common statistical issues. 2007;6:99–113. [DOI] [PubMed] [Google Scholar]
- 24. Gill R, Schumacher M. A simple test of the proportional hazards assumption. Biometrika 1987;74:289–300. [Google Scholar]
- 25. Therneau TM, Grambsch PM, Fleming TR. Martingale‐based residuals for survival models. Biometrika 1990;77:147–160. [Google Scholar]
- 26. Altman DG, McShane LM, Sauerbrei W et al. Reporting recommendations for tumor marker prognostic studies (REMARK): Explanation and elaboration. PLoS Med 2012;9:e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Galloway‐Peña JR, Jenq RR, Shelburne SA. Can consideration of the microbiome improve antimicrobial utilization and treatment outcomes in the oncology patient? Clin Cancer Res 2017;23:3263–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Derosa L, Routy B, Kroemer G et al. The intestinal microbiota determines the clinical efficacy of immune checkpoint blockers targeting PD‐1/PD‐L1. Oncoimmunology 2018;7:e1434468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Derosa L, Routy B, Mezquita L et al. Antibiotics prescription to decrease progression‐free survival (PFS) and overall survival (OS) in patients with advanced cancers treated with PD1/PDL1 immune checkpoint inhibitors. J Clin Oncol 2017;35(suppl):3015a. [Google Scholar]
- 30. Huemer F, Rinnerthaler G, Westphal T et al. Impact of antibiotic treatment on immune‐checkpoint blockade efficacy in advanced non‐squamous non‐small cell lung cancer. Oncotarget 2018;9(23):16512–16520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ahmed J, Kumar A, Parikh K et al. Use of broad‐spectrum antibiotics impacts outcome in patients treated with immune checkpoint inhibitors. Oncoimmunology 2018;7:e1507670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sen S, Carmagnani Pestana R, Hess K et al. Impact of antibiotic use on survival in patients with advanced cancers treated on immune checkpoint inhibitor phase I clinical trials. Ann Oncol 2018;29:2396–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pinato DJ, Howlett S, Ottaviani D et al. Antibiotic treatment prior to immune checkpoint inhibitor therapy as a tumor‐agnostic predictive correlate of response in routine clinical practice. J Clin Oncol 2019;37(suppl 8):147a. [Google Scholar]
- 34. Kaderbhai C, Richard C, Fumet JD et al. Antibiotic use does not appear to influence response to nivolumab. Anticancer Res 2017;37:3195–3200. [DOI] [PubMed] [Google Scholar]
- 35. Hakozaki T, Okuma Y, Omori M et al. Impact of prior antibiotic use on the efficacy of nivolumab for non‐small cell lung cancer. Oncol Lett 2019;17:2946–2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils, and pitfalls. J Clin Microbiol 2007;45:2761–2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Woo PCY, Lau SKP, Teng JLL et al. Then and now: Use of 16S rDNA gene sequencing for bacterial identification and discovery of novel bacteria in clinical microbiology laboratories. Clin Microbiol Infect 2008;14:908–934. [DOI] [PubMed] [Google Scholar]
- 38. Hodi FS, Hwu WJ, Kefford R et al. Evaluation of immune‐related response criteria and RECIST v1.1 in patients with advanced melanoma treated with pembrolizumab. J Clin Oncol 2016;34:1510–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
See http://www.TheOncologist.com for supplemental material available online.
Supplemental Figure
Supplemental Tables


